Abstract
Hsp104 is a hexameric protein chaperone that resolubilizes stress-damaged proteins from aggregates. Hsp104 promotes [PSI+] prion propagation by breaking prion aggregates, which propagate as amyloid fibers, into more numerous prion “seeds.” Inactivating Hsp104 cures cells of [PSI+] and other amyloid-like yeast prions. Overexpressing Hsp104 also eliminates [PSI+], presumably by completely resolubilizing prion aggregates. Inexplicably, however, excess Hsp104 does not cure the other prions. Here we identify missense mutations in Hsp104's amino-terminal domain (NTD), which is conserved among Hsp100 proteins but whose function is unknown, that improve [PSI+] propagation. Hsp104Δ147, engineered to lack the NTD, supported [PSI+] and functioned normally in thermotolerance and protein disaggregation. Hsp104Δ147 failed to cure [PSI+] when overexpressed, however, implying that excess Hsp104 does not eliminate [PSI+] by direct dissolution of prion aggregates. Curing of [PSI+] by overexpressing catalytically inactive Hsp104 (Hsp104KT), which interferes with endogenous Hsp104, did not require the NTD. We further found that Hsp104 mutants defective in threading peptides through the hexamer pore had reduced ability to support [PSI+] in proportion to protein resolubilization defects, suggesting that [PSI+] propagation depends on this threading and that Hsp104 “breaks” prion aggregates by extracting protein monomers from the amyloid fibers.
HSP104 is a member of the ClpB family of Hsp100 AAA+ ATPases that form ring-shaped hexamers with an axial pore (Schirmer et al. 1996; Lupas and Martin 2002; Lee et al. 2003). These proteins help cells recover from stress by resolubilizing proteins from aggregates through continuous extrusion of monomers (Parsell et al. 1994; Glover and Lindquist 1998; Weibezahn et al. 2004). This reaction requires the assistance of Hsp70, a protein chaperone that helps proteins adopt and maintain native conformation and traverse membranes and ribosomes (Murakami et al. 1988; Nelson et al. 1992). Hsp70 also protects cells from stress by binding partially unfolded proteins and preventing protein aggregation. In the homologous chaperone system of Escherichia coli, there is evidence that Hsp70 binds aggregates first, possibly increasing exposure of surfaces for subsequent ClpB interaction (Weibezahn et al. 2003; Zietkiewicz et al. 2004). Recent data show that peptides transit the pore of Hsp104 and ClpB during the disaggregation reaction (Lum et al. 2004; Weibezahn et al. 2004). Hsp70 also might act in the disaggregation process by helping extrude peptides from the exit site of the pore, perhaps in a manner similar to assisting peptides through membranes or ribosomes (Glover and Tkach 2001).
Hsp104 is required for propagation of the yeast prions [PSI+], [URE3], and [PIN+]/[RNQ1+] (Chernoff et al. 1995; Derkatch et al. 1997; Moriyama et al. 2000), which are self-replicating, amyloid-like aggregates of the proteins Sup35p, Ure2p, and Rnq1p, respectively (Wickner 1994; Derkatch et al. 1997; Sondheimer and Lindquist 2000). Amyloid is an insoluble, highly structured, fibrous aggregate that propagates by recruiting the soluble form of the protein and converting it into the amyloid form as it joins the growing polymer. For these prions to be maintained in a growing yeast population, prion aggregates must continually replicate as cells divide. Hsp104 is a primary factor in yeast prion replication, thought to act by breaking the aggregates into smaller pieces, thereby generating new self-replicating particles, or seeds (Paushkin et al. 1996; Ness et al. 2002; Kryndushkin et al. 2003). Specifically inactivating Hsp104 by adding millimolar amounts of guanidine to growth media causes cells to lose [PSI+] and the other amyloid prions of yeast (Ferreira et al. 2001; Jung and Masison 2001; Grimminger et al. 2004). Overexpression of Hsp104 also cures [PSI+]. The mechanism of curing by excess Hsp104 is unknown but has been presumed to be due to complete dissolution of the prion aggregates by Hsp104's protein disaggregation activity (Kushnirov and Ter-Avanesyan 1998; Paushkin et al. 1996; Wegrzyn et al. 2001). Unexpectedly, however, this curing is specific for [PSI+], as propagation of [URE3] and [PIN+] is unaffected by excess Hsp104 (Derkatch et al. 1997; Moriyama et al. 2000).
We earlier showed that [PSI+] prion aggregates are larger in cells expressing the Hsp70 mutant Ssa1-21p than in those expressing wild-type Ssa1p, suggesting that Ssa1p normally helps to break up prion aggregates (Song et al. 2005). The molecular mechanism by which the altered Hsp70 function impairs [PSI+] propagation is not yet known, but, as SSA1-21 cells are hypersensitive to guanidine curing of [PSI+] (Jung et al. 2000), Ssa1-21p might interfere with the Hsp104-mediated process of breaking [PSI+] prion aggregates. On this premise we isolated compensatory mutations in Hsp104 that overcome the impairment of [PSI+] by Ssa1-21p. Characterization of the mutations showed that they enhance [PSI+] propagation independently of Hsp70 genotype and that Hsp104's amino-terminal domain is dispensable for Hsp104 function but necessary for elimination of [PSI+] by overproduced Hsp104.
MATERIALS AND METHODS
Strains and growth conditions:
Our wild-type yeast strain is 779-6A (MATα, kar1-1, SUQ5, ade2-1, his3Δ202, leu2Δ1, trp1Δ63, ura3-52). Strain JC39K1 (Jung et al. 2002) is isogenic but has hsp104D184Y, which weakens [PSI+] propagation and makes [PSI+] cells incapable of growing without adenine at 30°. Strains with chromosomal alleles of mutant HSP104 genes were constructed by replacing the first half of the hsp104D184Y gene in strain JC39K1 by cotransformation of plasmid pRS315 (see below) and 1.5-kb BamHI–BglII DNA fragments of HSP104 alleles excised from wild-type and Hsp104 mutant versions of plasmid pJ309 (see below). Plasmid transformants that incorporated the cotransforming HSP104 DNA were selected on plates without adenine at 30°. Integration of desired alleles was verified by sequencing of PCR-amplified DNA from the HSP104 locus. Strain J104NC (Jones and Masison 2003) is 779-6A with HSP104 replaced by KanMX. Strain 932 (Jung et al. 2002) is SSA1-21, hsp104∷KanMX, and carries HSP104 on plasmid pJ312 (see below). Strains with different HSP70 genotypes were generated through crosses with isogenic mutant strains (Jones et al. 2004).
Yeast was grown at 30° unless indicated otherwise. Complex and synthetic media were as described (Sherman 1994; Schwimmer and Masison 2002). To overexpress Hsp104, cells were grown in synthetic dextrose medium overnight and shifted to synthetic galactose medium, which contains both 2% galactose and 1% raffinose in place of glucose.
Plasmids:
Plasmids pRS316, pRS315, and pRS314 are single-copy URA3-, LEU2-, and TRP1-based vectors (Sikorski and Hieter 1989). Plasmid pJ309 is pRS315 with Hsp104 and 500 bp of 5′ and 3′ flanking DNA on a BamHI fragment. Plasmid pJ312 is pRS316 with the same BamHI fragment. Plasmid pDCM65, which contains galactose-inducible HSP104, was described previously (Schwimmer and Masison 2002). The ClaI–SacI fragment of pDCM65 was excised and ligated to pRS314 digested with the same enzymes to produce pGCH17. The BamHI–BglII fragment of pGCH17 was replaced by similar fragments of HSP104 alleles encoding Hsp104H25Y, Hsp104A37T, and Hsp104T160M to produce pGCH18, pGCH19, and pGCH20, respectively.
Plasmid pGCH35, used to overexpress the Hsp104 amino-terminal domain (NTD), was made by first cutting pDCM65 with EagI, blunting the ends using Klenow enzyme, and then cutting with ClaI. The resulting fragment containing the galactose promoter and HSP104 codons 1–188 was then inserted into pRS314 cut with ClaI and SmaI. Plasmids pDCM94 and pDCM95 are pRS314 with HSP104KTΔ147 and HSP104KTT160M, respectively. They were constructed by replacing the EagI–SacI fragments from plasmids pGCH23 and pGCH20 with that from pFL39-Gal-HSP104-KT (Wegrzyn et al. 2001). Plasmid pDCM58 is pRS315 with the ApaI–SmaI fragment containing luxAB from pGPDluxAB(HIS) (Parsell et al. 1994; Jung and Masison 2001).
HSP104 with NTD repeat deletion was constructed in pJ309 using the overlap extension PCR method for deletion mutagenesis as described (Lee et al. 2004). First, an NcoI site with an in-frame ATG at the initiator methionine codon of HSP104 was created by PCR mutagenesis. The primer at the 5′ end used to amplify the Δ147 allele contained an NcoI site to introduce an ATG codon at codon 147. Hsp104 codons 2–147 were deleted in the resulting plasmid (pGCH16). All PCR amplified clones were verified by DNA sequencing.
Mutagenesis and Hsp104 mutant selection:
A previously prepared library of hydroxylamine-mutagenized plasmid pJ309 (Jung et al. 2002) was used to transform SSA1-21 strain 932. Transformants were transferred to plates lacking leucine and adenine and containing 5-fluoro-orotic acid, which kills cells expressing URA3, and incubated at 30°. To grow under these conditions cells must have lost pJ312 and gained improved [PSI+] propagation, presumably due to a mutated HSP104 allele on pJ309 that overcomes the [PSI+]-impairing effects of SSA1-21. Retransformation was done to verify plasmid dependency of phenotype.
Thermotolerance assays:
Experiments were done as described (Jung and Masison 2001). Briefly, [psi−] cells in early log phase in YPAD at 30° were preincubated at 39° for 30 min to induce Hsp104 and the heat-shock response and then transferred to a 52° water bath. Aliquots of cells were transferred to ice immediately before exposure to 52° and periodically thereafter. Survival of cells in these samples was determined by titering on YPAD.
Luciferase reactivation assays:
Experiments using cells transformed by pDCM58 were done as described (Jung and Masison 2001). Briefly, log phase [psi−] cells expressing a thermolabile luciferase reporter protein were preconditioned at 39° for 30 min, exposed to 42° for 1 hr to cause luciferase to aggregate, and then allowed to recover at 25°. Cycloheximide was added to cultures 10 min before removing them from 42° to prevent luciferase synthesis during the recovery period.
RESULTS
Isolation of Hsp104 alleles that overcome impairment of [PSI+] by SSA1-21:
The Saccharomyces cerevisiae [PSI+] prion is a self-replicating aggregated form of the translation termination factor Sup35p. Aggregation of Sup35p in [PSI+] cells reduces termination efficiency, which causes readthrough of nonsense codons (Firoozan et al. 1991). Mutants with the ade2-1 nonsense allele lack an enzyme required for adenine biosynthesis and are unable to grow without exogenous adenine. When adenine is limiting, colonies appear red due to accumulation of a metabolite of the Ade2p substrate (Silver and Eaton 1969). In certain strains, suppression of ade2-1 by [PSI+] restores adenine prototrophy and white colony color, which provides a sensitive and straightforward way to monitor the presence or absence of [PSI+] (Cox 1965). The degree of suppression of ade2-1 caused by [PSI+] is related to the degree of Sup35p aggregation, which reflects efficiency of prion propagation. Thus, stronger [PSI+] propagation results in whiter colonies and faster growth without adenine and weaker-than-normal propagation is manifested as a pinker colony color and slower or temperature-sensitive growth without adenine (Jung et al. 2000).
Cells expressing Ssa1-21p, a mutant of the Hsp70 Ssa1p with the L483W substitution, are defective for [PSI+] propagation and frequently lose [PSI+] during mitosis. Because nonsense suppression caused by [PSI+] is reduced in SSA1-21 cells, at the optimal growth temperature of 30° they are pink rather than white. Without adenine, these cells grow very slowly at 25° and do not grow at 30°. SSA1-21 cells are also hypersensitive to curing of [PSI+] by millimolar amounts of guanidine in the growth medium, which inactivates Hsp104 (Jung et al. 2000). Reasoning that this hypersensitivity was due to altered cooperation between Ssa1-21p and Hsp104, we screened for compensatory mutations in Hsp104 that improved [PSI+] propagation in SSA1-21 cells, allowing them to grow without adenine at 30° (see materials and methods). None of the Hsp104 mutants described in this study affect ade2-1 suppression independently of [PSI+] (data not shown).
A diagram of Hsp104 with the locations of mutations identified in the screen is shown in Figure 1A. Hsp104 has a conserved amino-terminal domain of unknown function that contains two structurally similar direct repeats (R1 and R2) (Lo et al. 2001). The NTD is followed by two nucleotide-binding domains (NBD1 and NBD2) of which the first is thought to act mainly in Hsp104's enzymatic function and the second in its oligomerization (Schirmer et al. 1998). An intervening middle region is important for regulation of Hsp104 function (Schirmer et al. 2004), and the C-terminal domain has been shown to have a role in substrate discrimination (Schirmer et al. 1996; Cashikar et al. 2002). Half of the mutant Hsp104 proteins isolated have two or more alterations. Among these mutants, amino acid substitutions were found in all of the defined domains, each mutant had substitutions in two different domains, and none of the mutants had alterations in the same two domains. To avoid complications in interpreting results obtained with these mutants, we focused on the seven proteins with single mutations. All mutations in these proteins clustered in the NTD, which implies that the NTD is important for an aspect of Hsp104 function with regard to [PSI+] propagation.
Figure 1.
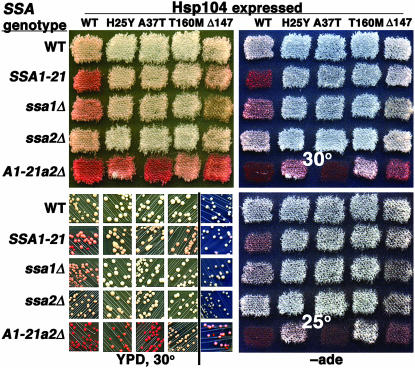
Hsp104 structure and location of mutations that improve [PSI+] propagation. (A) Hsp104 coding region with indicated domains: NTD, N-terminal domain with repeats R1 and R2 shown in red; NBD, nucleotide-binding domains (1 and 2); MR, middle region; CTD, C-terminal domain. Numbers indicate amino acid positions, and substitutions that improve [PSI+] are indicated. (Bottom) Mutants with multiple alterations, each color representing substitutions in a single protein, are shown. (Top) Mutants with single substitutions are shown. (B) Western analysis of Hsp104 abundance. Abundance of wild-type (wt) and three mutant Hsp104 proteins (top) were assessed in five strains of different Hsp70 genotype (bottom). (Top, Hsp104) The blot probed with Hsp104 antibody is shown. (Bottom, load) A region of the same blots subsequently stained with amido black as a loading and transfer control is shown. The wild-type sample on the far left was run with molecular weight markers so the band in the center should be used as the loading control. (C) Similar blot as in B, except strains are also hsp104Δ and express either Hsp104 or Hsp104Δ147, as indicated, from single-copy plasmids. The sample to the right of the vertical line is the parental hsp104Δ strain with empty plasmid. Asterisk indicates Hsp104 degradation product.
Hsp104 abundance is normal in cells expressing mutant Hsp104's:
To assess the possibility that Hsp104 mutations were affecting phenotypes by altering levels of Hsp104 expression, we compared abundance of Hsp104 in cells expressing the different alleles. Alleles encoding each of the NTD mutant Hsp104's were integrated into the chromosome in place of the endogenous HSP104. As shown in Figure 1B, the amount of the different Hsp104 proteins focused on in this study (see below) was similar overall. Any appreciable differences in signal intensity could be attributed to differences in amount of total protein on the blots (compare protein in Figure 1B, “load”). Thus, effects of the Hsp104 mutations did not appear to be due to differences in Hsp104 abundance but rather to effects on Hsp104 function.
Hsp104 mutations improve [PSI+] propagation in cells expressing different Hsp70's:
SSA1-21 cells that lack the functionally redundant Ssa2p are unable to support [PSI+] propagation under any condition (Jung et al. 2000). Thus, a more stringent test for ability of the Hsp104 mutants to suppress [PSI+]-inhibitory effects of Ssa1-21p is whether they can restore [PSI+] propagation in this strain. The ability of [PSI+] to propagate in SSA1-21 ssa2Δ cells with mutant HSP104 alleles integrated in place of wild-type HSP104 is shown in Figure 2. Two mutants (Hsp104P28L, Hsp104T160M) restored [PSI+] completely, showing normal white coloration and no mitotic loss of [PSI+]. Three mutants (Hsp104H25Y, Hsp104G58D, Hsp104E85K) restored [PSI+] propagation only weakly, which was seen as partial pigmentation and notable mitotic loss of [PSI+] (Figure 2 and data not shown). Although selected as improving [PSI+] propagation in SSA1-21 cells, two mutants (Hsp104A37T, Hsp104E41K) failed to restore [PSI+] propagation in SSA1-21 ssa2Δ cells. These [psi−] strains are always red and unable to grow without adenine.
Figure 2.
[PSI+] phenotypes of SSA1-21 strains with (SSA2, top) or without (ssa2Δ, bottom) Ssa2p, expressing wild-type (WT) and mutant (as indicated) Hsp104. Patches of cells grown on YPAD at 30° were replica plated onto medium lacking adenine and grown for 2 days at 30°. Relative strength of [PSI+] is reflected in extent of growth and in degree of pigmentation. As strength of [PSI+] increases, growth becomes denser and pigmentation decreases. SSA1-21 SSA2 cells expressing Hsp104T160M have a wild-type [PSI+] phenotype and SSA1-21 ssa2Δ cells expressing wild-type Hsp104 cannot support [PSI+] and represent a [psi−] phenotype.
These results showed that Hsp104 NTD mutants could be separated into different classes on the basis of the extent to which they restore [PSI+] propagation in SSA1-21 ssa2Δ cells. We selected Hsp104T160M, Hsp104H25Y, and Hsp104A37T as representatives of these classes for further study. We also evaluated effects of the Hsp104 mutations in wild-type cells and in cells lacking Ssa1p or Ssa2p to determine if Hsp104 function was affected differently by different Hsp70 isoforms. The various Hsp70 and Hsp104 proteins were expressed from normal chromosomal loci in these isogenic strains.
The [PSI+] phenotypes of the various strains are shown in Figure 3. The first column in each quadrant of Figure 3 shows cells expressing the different Hsp70 isoforms in combination with wild-type (WT) Hsp104. Those with wild-type SSA genotype (expressing both Ssa1p and Ssa2p) are faintly pink on both YPD and −ade plates when grown at 30°. The [PSI+] phenotype of ssa2Δ mutants (expressing only Ssa1p) was similar to wild type, but that of ssa1Δ cells (expressing only Ssa2p) was slightly weakened, which is seen as pinker color on YPD and −ade plates, and reduced growth without adenine at 30°. These data suggest that with regard to [PSI+] propagation wild-type Hsp104 functioned better with Ssa1p than with Ssa2p. SSA1-21 [PSI+] cells are even more red, spontaneously lose [PSI+] during mitosis (Figure 3, red [psi−] colonies in YPD streaks), and grow weakly at 25° and not at all at 30° on −ade. As noted above, [PSI+] cannot propagate in SSA1-21ssa2Δ cells (Figure 3, bottom left in each quadrant) so these cells give rise to uniformly red colonies on YPD and are unable to grow on −ade at any temperature.
Figure 3.
[PSI+] phenotypes of wild-type and SSA mutant strains expressing different Hsp104 proteins. Strains with Hsp70 genotype indicated on left express the Hsp104 protein indicated at top from the chromosomal HSP104 locus. Patches of cells were replica plated from an original YPD master plate onto YPD (top left) and −ade (right) plates. Cells streaked for colonies (bottom left quadrant) are from the same YPD master plate and strains are arranged in the same pattern. Cells indicated Δ147 are hsp104Δ strains expressing Hsp104Δ147 from a plasmid. Colonies of these cells (right of the line in bottom left quadrant) were grown on medium that lacks leucine to maintain selection for the plasmid, and contains limiting adenine, which allows pigmentation. YPD plates (left) were incubated for 2 days (top) or 3 days (bottom) at 30°. Cells on −ade plates (right) were grown for 2 days at 30° (top) or 25° (bottom). A1-21 a2Δ is abbreviation for SSA1-21 ssa2Δ.
Although the mutations were selected on the basis of improving [PSI+] propagation in SSA1-21 cells, all the Hsp104 mutants improved [PSI+] propagation in wild-type, ssa1Δ, and ssa2Δ cells (Figure 3), which is seen as whiter color on both YPD and −ade plates, and improved growth without adenine (Figure 3 and data not shown). Thus, the Hsp104 mutant proteins enhanced [PSI+] propagation regardless of the Hsp70 isoforms expressed, indicating that the mutations had a general effect that was not specific to an interaction with the Ssa1-21 protein.
Hsp104 NTD mutations variably affect Hsp104 function in Hsp70 mutant strains:
Hsp104 protects cells from lethal effects of exposure to elevated temperature by resolubilizing proteins that have aggregated (Parsell et al. 1994). This reaction is assisted by Hsp70 and its cochaperone Hsp40 (Glover and Lindquist 1998). Preconditioning yeast with a brief exposure to elevated temperature (37°–42°) induces expression of heat-shock proteins including Hsp104 and significantly improves survival after subsequent exposure to lethal heat (52°). This acquired thermotolerance is essentially absent in cells that lack Hsp104 (Parsell et al. 1994).
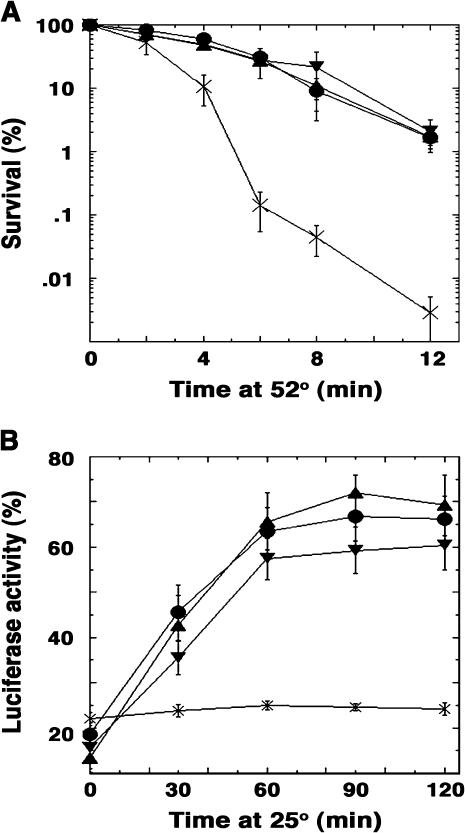
Figure 4 shows survival after exposure to 52° of preconditioned cells that express different Hsp70's with wild-type or mutant Hsp104. For cells expressing both Ssa1p and Ssa2p, the mutants Hsp104T160M, Hsp104H25Y, and Hsp104A37T all provided normal thermotolerance (Figure 4A), indicating that they functioned normally in otherwise wild-type cells. In cells expressing only Ssa1p (ssa2Δ), all mutant Hsp104's also functioned normally (data not shown). In contrast, the mutant Hsp104's were variably reduced in ability to provide thermotolerance in cells expressing only Ssa2p (Figure 4B) or only Ssa1-21p (Figure 4C). The greatest reduction in thermotolerance was observed when Hsp104A37T was combined with only Ssa1-21p (Figure 4C, triangles). This same combination was weakest with regard to [PSI+] propagation (see Figures 2 and 3). In SSA1-21 cells expressing Ssa2p, Hsp104T160M was notably reduced in ability to provide thermotolerance, but the wild-type and other mutant Hsp104 proteins functioned normally (Figure 4D). Thus, although the Hsp104 mutations improved [PSI+] propagation in the Hsp70 mutants, they each caused detectable impairment of Hsp104 thermotolerance function in some of these mutants.
Figure 4.
Thermotolerance and protein resolubilization mediated by mutant Hsp104's. (A–D) Thermotolerance. Cells grown in YPAD at 30° were shifted to 39° for 30 min and then shifted to 52°. The percentage of viable cells is plotted as a function of time exposed to 52°. Wild-type and hsp104Δ cells are shown as filled circles and X's, respectively. Mutant proteins Hsp104H25Y, Hsp104A37T, and Hsp104T160M are represented as squares, triangles, and diamonds, respectively. SSA genotype of strains assayed is indicated in the bottom left corner of A–H. (E–H) Protein resolubilization. Cells expressing a thermolabile form of bacterial luciferase were heat-shocked to cause luciferase aggregation. Resolubilization of the aggregates by Hsp104 was monitored as restoration of luciferase activity during a recovery period at 25°. The same symbols as in A–D are used to represent the different Hsp104 proteins.
A possible explanation for the differences in thermotolerance between ssa1Δ and ssa2Δ strains might be that deleting SSA1 reduced overall Hsp70 levels more than deleting SSA2. However, in previous studies we did not detect differences in Hsp70 abundance between our ssa1Δ and ssa2Δ strains (Jung et al. 2000; Jones et al. 2004). Together, our data suggest that for thermotolerance, as with [PSI+] propagation, Hsp104 might function better with Ssa1p than with Ssa2p.
As a measure of the degree to which the NTD mutations affect ability of Hsp104 to resolubilize thermally denatured protein aggregates in vivo, we heat-shocked cells expressing a thermolabile form of bacterial luciferase and then monitored recovery of luciferase activity. Reactivation of such aggregated luciferase requires Hsp104 (Parsell et al. 1994). Luciferase reactivation in all cells expressing Ssa2p was similar to that of wild type, regardless of the Hsp104 present (Figure 4, E and F, and data not shown). The Hsp104 mutants varied only modestly in ability to reactivate luciferase in ssa1Δ and SSA1-21 cells (Figure 4, E and F, respectively). Thus, the >10-fold reduction in thermotolerance seen for Hsp104A37T in ssa1Δ cells (Figure 4B) was not reflected in a similar impairment of resolubilization of a model substrate (Figure 4E). Possibly the mutations have a more significant effect on resolubilization of substrates that are important for cell survival than for a substrate not normally expressed in yeast.
Cells lacking Ssa2p and expressing wild-type Ssa1p and Hsp104 have normal luciferase reactivation (Figure 4G) (Song et al. 2005), but in the same strain expressing the mutant Hsp104 proteins efficiency of resolubilization was significantly less efficient (Figure 4G). Similarly, in SSA1-21 ssa2Δ cells, which express only Ssa1-21p, all Hsp104 mutants showed reduced resolubilization activity (Figure 4H). In these cells the degree to which the Hsp104 mutants were reduced in ability to reactivate luciferase did not correlate with their reduced ability to confer thermotolerance. Thus, the luciferase reactivation assay also revealed subtle differences in cooperation of Hsp70's with Hsp104. Together our data indicate that although this assay is useful for quantifying protein resolubilization in vivo, it does not always accurately reflect Hsp104 function in cytoprotection.
Hsp104 NTD is dispensable for [PSI+] propagation, thermotolerance, and luciferase reactivation:
To test the importance of the NTD for Hsp104 functions we engineered a protein lacking the R1 and R2 repeats, designated Hsp104Δ147. [PSI+] hsp104Δ strains expressing wild-type Hsp104 from a plasmid were first transformed with a plasmid encoding Hsp104Δ147. Transformants retaining both plasmids remained [PSI+] (data not shown). Those that subsequently lost the plasmid encoding wild-type Hsp104 and retained the plasmid encoding Hsp104Δ147 were then isolated. The expression level of Hsp104Δ147 in these strains was compared to that of wild-type Hsp104 in the isogenic strains. For the SSA wild-type strains, Hsp104Δ147 appeared somewhat elevated compared with wild-type Hsp104, but in all the Hsp70 mutant strains the abundance of Hsp104 and Hsp104Δ147 was similar (Figure 1C). As observed with the proteins having single mutations, Hsp104Δ147 had a general effect in enhancing nonsense suppression caused by [PSI+] (Figure 3, far right column in each quadrant). Hsp104Δ147 also restored ability of [PSI+] to propagate in SSA1-21 ssa2Δ cells, albeit weakly. Thus, deletion of the entire NTD improved [PSI+] propagation but did not have a greater effect than the single missense mutations.
The ability of Hsp104Δ147 to function in prion propagation led us to expect that it also retained cytoprotective functions. Indeed, the function of Hsp104Δ147 in thermotolerance and luciferase reactivation experiments, assayed as described above, was very nearly normal. For both wild-type SSA (Figure 5, A and B, triangles) and SSA1-21 (Figure 5, A and B, inverted triangles) strains, deleting the NTD had little effect in either assay. Thus, the NTD was dispensable for Hsp104 function in protecting cells from exposure to lethal heat and resolubilizing thermally denatured proteins from aggregates.
Figure 5.
Thermotolerance and protein resolubilization mediated by Hsp104Δ147. (A) Thermotolerance was assayed as described in Figure 4, A–D. (B) Protein resolubilization was assayed as described in Figure 4, E–H. In A and B HSP104 wild type and hsp104Δ cells are indicated as filled circles and X's, respectively. SSA wild type and SSA1-21 cells (both hsp104Δ) expressing Hsp104Δ147 from a plasmid are indicated by triangles and inverted triangles, respectively.
Hsp104 extrudes proteins from aggregates by threading monomers from the aggregates through the central channel of the Hsp104 hexamer (Lum et al. 2004). Although Hsp104 has been suggested to aid prion propagation by shearing or cleaving prion fibers (Borchsenius et al. 2001; Kryndushkin et al. 2003; Inoue et al. 2004; Shorter and Lindquist 2004), our results show a correlation in ability of Hsp104 to support [PSI+] and to function in protein resolubilization. To test if substrate threading is required for prion propagation, we compared [PSI+] propagation in cells expressing Hsp104 mutants specifically defective in this function. Tyrosine residue 662 is critical for the threading process, and mutants in which this residue has been changed to alanine (Hsp104Y662A) or lysine (Hsp104Y662K) are incapable of resolubilizing aggregated luciferase in vitro or providing stress protection (Lum et al. 2004). Mutants with more conservative substitutions of tryptophan (Hsp104Y662W) or phenylalanine (Hsp104Y662F) retain partial function in these assays, with Hsp104Y662F functioning somewhat better than Hsp104Y662W (Lum et al. 2004).
We used a plasmid shuffle similar to that used with Hsp104Δ147 to express these Y662 mutants in hsp104Δ cells. We found that [PSI+] was not supported by Hsp104Y662A or Hsp104Y662K, but it was supported weakly by Hsp104Y662W and more strongly, but not to normal levels, by Hsp104Y662F (Figure 6). We also assessed [PSI+] propagation in cells expressing Hsp104E645K, which alters nucleotide-induced movement of Y662 and has detectable but very weak disaggregation function (Lum et al. 2004). Cells expressing Hsp104E645K did not support [PSI+] propagation. The differences in [PSI+] phenotype in cells expressing these threading mutants were not due to differences in expression levels of the proteins (Figure 6, top). Therefore, the ability of these mutants to support [PSI+] correlated directly with their ability to thread peptides though their axial pores.
Figure 6.
Effects of Hsp104 substrate threading mutations on [PSI+] propagation. Strains are hsp104Δ and express Hsp104, with amino acid substitutions as indicated at top, from LEU2-based plasmids. WT, plasmid has wild type HSP104; Δ104, cells carry empty plasmid. (Top) Western analysis as described in Figure 1B is shown. (Bottom) [PSI+] phenotype is shown. Cells were grown on medium lacking leucine (−leu) to select for the plasmids and then replica plated onto −leu plates with limiting adenine (low ade). The latter plates (shown) were incubated at 30° for 2 days. Colonies (bottom) were grown on similar plates for 2 days at 30° and 1 day at 25°.
Curing of [PSI+] by overexpression of Hsp104 requires the NTD:
Transient overexpression of Hsp104 under conditions in which it is not normally induced cures cells of [PSI+] very efficiently. This process is thought to be caused by complete dissolution of Sup35p polymers by the excess Hsp104 disaggregating activity. To test if our Hsp104 mutants were able to cure [PSI+] when in excess, they were transiently overexpressed in cells expressing a chromosomal allele encoding the same Hsp104. The frequency of appearance of [psi−] cells after such treatment is shown in Table 1. All of the point mutations reduced the ability of Hsp104 to cure [PSI+], and in cells expressing wild-type Ssa1p, Hsp104T160M was incapable of curing. For SSA1-21 cells, in which [PSI+] is already somewhat unstable, all Hsp104 mutants further reduced [PSI+] stability when overexpressed, but only Hsp104A37T cured as well as wild type. Thus, point mutations in Hsp104's NTD interfered with Hsp104's ability to eliminate [PSI+] when overexpressed.
TABLE 1.
Curing of [PSI+] by excess Hsp104
| [psi−] cells (% ± SD)
|
||
|---|---|---|
| Hsp104 protein overexpressed | WT | SSA1-21 |
| None | <0.1 | <0.1 |
| Hsp104 (WT) | 38 ± 3 | 52 ± 5 |
| Hsp104H25Y | 6 ± 1 | 17 ± 2 |
| Hsp104A37T | 12 ± 1 | 52 ± 3 |
| Hsp104T160M | <0.1 | 12 ± 3 |
| Hsp104Δ147a | <0.1 | 5 ± 2 |
| NTD onlyb | <0.1 | <0.1 |
| Hsp104KTb | 24 ± 2 | ND |
| Hsp104KTT160Mb | 20 ± 1 | ND |
| Hsp104KTΔ147b | 19 ± 2 | ND |
[PSI+] cells with chromosomal HSP104 alleles and carrying a plasmid with the same galactose-inducible HSP104 were grown on galactose 2 days and then spread for colonies onto 1/2YPD. Entirely red colonies were scored as arising from [psi−] cells. Roughly 1500 colonies were counted in at least three separate experiments for each strain. WT, wild type; ND, not determined.
In hsp104Δ cells expressing Hsp104Δ147.
In HSP104 wild-type cells.
In similar experiments, overproduced Hsp104Δ147 was unable to cure [PSI+] from hsp104Δ cells expressing Hsp104Δ147 from a plasmid or from cells expressing endogenous wild-type Hsp104 (Table 1 and data not shown). These results show that an intact NTD is required for excess Hsp104 to eliminate [PSI+]. Although Hsp104Δ147 has normal function in both thermotolerance and luciferase resolubilization (Figure 5), increasing this resolubilization activity by overexpressing the protein was not sufficient for curing [PSI+], which implies that excess Hsp104 does not cure [PSI+] by completely disaggregating Sup35p polymers.
To test if the NTD alone mediated curing of [PSI+], Hsp104 residues 1–188 were overproduced in [PSI+] cells expressing endogenous wild-type Hsp104. This polypeptide was expressed abundantly under inducing conditions (data not shown). Transient overexpression of the NTD in both wild-type and SSA1-21 cells had no effect on [PSI+] stability (Table 1), showing that excess NTD alone could not cure [PSI+]. Apparently, a functional Hsp104 with a covalently attached NTD is necessary for [PSI+] curing.
A catalytically inactive form of Hsp104 that has point mutations in both ATPase domains (Hsp104KT, Parsell et al. 1991) can eliminate [PSI+] when overexpressed (Chernoff et al. 1995) by dominantly interfering with endogenous Hsp104 function (Ferreira et al. 2001). We tested if this different type of curing required NTD function by overexpressing variants of Hsp104KT lacking the NTD (Hsp104KTΔ147) or containing the T160M substitution (Hsp104KTT160M) in a wild-type [PSI+] strain. As we show above (Table 1), both of these alterations in Hsp104 abolished curing of [PSI+]. When overexpressed, both Hsp104KTΔ147 and Hsp104KTT160M cured [PSI+] nearly as efficiently as Hsp104KT (Table 1), indicating that curing of [PSI+] by dominantly interfering with Hsp104 function does not require the NTD.
DISCUSSION
We identified mutations in Hsp104 that improve propagation of the yeast [PSI+] prion. All mutant proteins with single mutations have alterations in the NTD. This clustering of [PSI+]-enhancing mutations in the NTD and our finding that Hsp104 lacking the NTD also improved [PSI+] indicate that the NTD is involved in Hsp104 function in a way that affects [PSI+] propagation. Our finding that curing of [PSI+] by excess Hsp104 required the NTD supports this conclusion. Nevertheless, Hsp104Δ147 supported [PSI+] propagation and functioned normally in thermotolerance and protein disaggregation. Therefore, the NTD was not required for functional interaction of Hsp104 with substrates, including the prion form of Sup35p, and Hsp104Δ147's protein disaggregation activity was not sufficient for curing [PSI+] when in excess.
These results imply that excess Hsp104 does not cure [PSI+] by directly dissolving prion aggregates. However, excess NTD alone also failed to cure [PSI+] so Hsp104 disaggregation function might still be necessary for such curing. This possibility cannot be tested directly as overproduction of catalytically inactive Hsp104 cures [PSI+] by a different mechanism, i.e., dominant interference of endogenous Hsp104 function. We find this type of curing does not require the NTD. An alternative could be that the NTD directs Hsp104 function to subcellular locations required for eliminating [PSI+] or that excess Hsp104 disrupts an unknown process required for [PSI+] propagation. The mechanism by which excess Hsp104 causes [PSI+] to be lost remains to be uncovered. The identification of this mechanism could also reveal why excess Hsp104 does not affect propagation of other yeast prions.
Substitutions of pore residues known to be important for threading of substrates through the axial channel of Hsp104 hexamers impaired thermotolerance and [PSI+] propagation proportionately, indicating that Hsp104's ability to promote prion propagation requires this threading activity. These results suggest that amyloid polymers are not simply severed by a mechanical action of Hsp104, but are destabilized and then break following the extrusion of Sup35p monomers from the fibers. If so, then such a process would be consistent with Hsp104 acting at the sides of fibers.
Our finding that Hsp104Δ147 supported [PSI+] propagation was not entirely unexpected as homologs from several prokaryotic species that lack the NTD are nearly normal in protecting cells from stress. These truncated proteins function well in protein disaggregation and the role of the NTD in ClpB function is still speculative (Clarke and Eriksson 2000; Beinker et al. 2002; Mogk et al. 2003; Chow et al. 2005). The NTD of ClpB can affect interactions with aggregated model substrates (Beinker et al. 2002; Tek and Zolkiewski 2002; Mogk et al. 2003; Barnett et al. 2005), but this activity clearly is not essential for ClpB to recognize and act on protein aggregates. The NTD of other orthologs such as ClpA, which unfolds proteins while transferring them to a protease complex, is important for mediating interactions with adapter proteins that regulate its functions and affect substrate specificity (Dougan et al. 2002a,b). Although such adapter proteins for ClpB homologs are unknown, it is possible that the NTD of Hsp104 interacts with factors that influence its activities or substrate specificity.
The observation that single missense mutations in the NTD improved [PSI+] in a manner similar to deletion of the entire repeat region points to these residues as being critical for the function of the intact domain with regard to [PSI+] propagation. This disruption of function by these mutations could mean these residues are important for interactions with specific substrates or other components of the disaggregation machinery, such as Hsp70 or Hsp40. Properties of a specific Hsp40 (Sis1p) have been suggested to enhance interaction of the yeast [PIN+]/[RNQ+] prion determinant Rnq1p with Hsp70 (Lopez et al. 2003), and Hsp40 has been shown to physically interact with Hsp104 (Glover and Lindquist 1998). Whether this interaction requires Hsp104's NTD is unknown. Since the NTD is located near the entrance to the axial pore of the Hsp104 hexamer, interactions of Hsp70 or Hsp40 with the NTD would necessarily be near the site of initial interaction of Hsp104 with substrate. Recent in vitro evidence suggests that Hsp70 interacts with aggregates and alters them in ways that facilitate subsequent interaction with ClpB (Weibezahn et al. 2004; Zietkiewicz et al. 2004, 2006). Our data might reflect alterations in interactions between the chaperones on the face of the Hsp104 hexamer where substrate enters the pore or reduced dependency of Hsp104 on prior Hsp70 action on substrate.
Our screen for Hsp104 mutations that improve the weakened [PSI+] phenotype of SSA1-21 cells identified mutations that enhanced [PSI+] propagation regardless of the Hsp70 background, indicating that the effects of the mutations are not specific to an interaction between Hsp104 and Ssa1-21p. Nevertheless, we saw modest differences in Hsp104 function when the mutants were combined with different Hsp70 isoforms. For example, in the luciferase refolding assay, Hsp104H25Y and Hsp104A37T functioned less well than wild-type Hsp104 when combined only with Ssa1p but functioned normally when combined only with Ssa1-21p. Additionally, the strain expressing Hsp104A37T had reduced thermotolerance when combined only with Ssa2p but the other mutant Hsp104's functioned well in all assays when Ssa2p was present. Together, our data are consistent with previously observed ability to separate Hsp104 functions in stress protection and prion propagation (Jung et al. 2002) and suggest that Hsp104 might function differently with specific Hsp70 partners under different conditions. Such differences might be mediated by variations in relative abundance of the Hsp70's or in interactions of Hsp70 with cochaperones that are present under the different conditions.
Acknowledgments
We thank Seyung Chung and Gary Jones for assistance in preliminary transformations and mutant screening, sequence analysis of mutants, and preparation of Figure 1A. We thank Yury Chernoff (Georgia Institute of Technology) for plasmids and John Glover (University of Toronto) for plasmids and Hsp104 antibody. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes, Digestive, and Kidney diseases.
References
- Barnett, M. E., M. Nagy, S. Kedzierska and M. Zolkiewski, 2005. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 280: 34940–34945. [DOI] [PubMed] [Google Scholar]
- Beinker, P., S. Schlee, Y. Groemping, R. Seidel and J. Reinstein, 2002. The N terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J. Biol. Chem. 277: 47160–47166. [DOI] [PubMed] [Google Scholar]
- Borchsenius, A. S., R. D. Wegrzyn, G. P. Newnam, S. G. Inge-Vechtomov and Y. O. Chernoff, 2001. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J. 20: 6683–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar, A. G., E. C. Schirmer, D. A. Hattendorf, J. R. Glover, M. S. Ramakrishnan et al., 2002. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9: 751–760. [DOI] [PubMed] [Google Scholar]
- Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov and S. W. Liebman, 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor. [psi+] Science 268: 880–884. [DOI] [PubMed] [Google Scholar]
- Chow, I. T., M. E. Barnett, M. Zolkiewski and F. Baneyx, 2005. The N-terminal domain of Escherichia coli ClpB enhances chaperone function. FEBS Lett. 579: 4242–4248. [DOI] [PubMed] [Google Scholar]
- Clarke, A. K., and M. J. Eriksson, 2000. The truncated form of the bacterial heat shock protein ClpB/HSP100 contributes to development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 182: 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, B. S., 1965. “Ψ” a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20: 505–521. [Google Scholar]
- Derkatch, I. L., M. E. Bradley, P. Zhou, Y. O. Chernoff and S. W. Liebman, 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan, D. A., A. Mogk, K. Zeth, K. Turgay and B. Bukau, 2002. a AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529: 6–10. [DOI] [PubMed] [Google Scholar]
- Dougan, D. A., B. G. Reid, A. L. Horwich and B. Bukau, 2002. b ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9: 673–683. [DOI] [PubMed] [Google Scholar]
- Ferreira, P. C., F. Ness, S. R. Edwards, B. S. Cox and M. F. Tuite, 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40: 1357–1369. [DOI] [PubMed] [Google Scholar]
- Firoozan, M., C. M. Grant, J. A. Duarte and M. F. Tuite, 1991. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast 7: 173–183. [DOI] [PubMed] [Google Scholar]
- Glover, J. R., and S. Lindquist, 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. [DOI] [PubMed] [Google Scholar]
- Glover, J. R., and J. M. Tkach, 2001. Crowbars and ratchets: hsp100 chaperones as tools in reversing protein aggregation. Biochem. Cell Biol. 79: 557–568. [PubMed] [Google Scholar]
- Grimminger, V., K. Richter, A. Imhof, J. Buchner and S. Walter, 2004. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J. Biol. Chem. 279: 7378–7383. [DOI] [PubMed] [Google Scholar]
- Inoue, Y., H. Taguchi, A. Kishimoto and M. Yoshida, 2004. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 279: 52319–52323. [DOI] [PubMed] [Google Scholar]
- Jones, G. W., and D. C. Masison, 2003. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. W., Y. Song, S. Chung and D. C. Masison, 2004. Propagation of yeast [PSI+] is prion impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 24: 3928–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., and D. C. Masison, 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43: 7–10. [DOI] [PubMed] [Google Scholar]
- Jung, G., G. Jones, R. D. Wegrzyn and D. C. Masison, 2000. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., G. Jones and D. C. Masison, 2002. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. USA 99: 9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin, D. S., I. M. Alexandrov, M. D. Ter-Avanesyan and V. V. Kushnirov, 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278: 49636–49643. [DOI] [PubMed] [Google Scholar]
- Kushnirov, V. V., and M. D. Ter-Avanesyan, 1998. Structure and replication of yeast prions. Cell 94: 13–16. [DOI] [PubMed] [Google Scholar]
- Lee, J., H. J. Lee, M. K. Shin and W. S. Ryu, 2004. Versatile PCR-mediated insertion or deletion mutagenesis. Biotechniques 36: 398–400. [DOI] [PubMed] [Google Scholar]
- Lee, S., M. E. Sowa, Y. H. Watanabe, P. B. Sigler, W. Chiu et al., 2003. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115: 229–240. [DOI] [PubMed] [Google Scholar]
- Lo, J. H., T. A. Baker and R. T. Sauer, 2001. Characterization of the N-terminal repeat domain of Escherichia coli ClpA-A class I Clp/HSP100 ATPase. Protein Sci. 10: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, N., R. Aron and E. A. Craig, 2003. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol. Biol. Cell 14: 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum, R., J. M. Tkach, E. Vierling and J. R. Glover, 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279: 29139–29146. [DOI] [PubMed] [Google Scholar]
- Lupas, A. N., and J. Martin, 2002. AAA proteins. Curr. Opin. Struct. Biol. 12: 746–753. [DOI] [PubMed] [Google Scholar]
- Mogk, A., C. Schlieker, C. Strub, W. Rist, J. Weibezahn et al., 2003. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 278: 17615–17624. [DOI] [PubMed] [Google Scholar]
- Moriyama, H., H. K. Edskes and R. B. Wickner, 2000. [URE3] Prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20: 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, H., D. Pain and G. Blobel, 1988. 70-kD heat shock-related protein is one of at least two distinct cytosolic factors stimulating protein import into mitochondria. J. Cell. Biol. 107: 2051–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne and E. A. Craig, 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71: 97–105. [DOI] [PubMed] [Google Scholar]
- Ness, F., P. Ferreira, B. S. Cox and M. F. Tuite, 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22: 5593–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell, D. A., Y. Sanchez, J. D. Stitzel and S. Lindquist, 1991. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature 353: 270–273. [DOI] [PubMed] [Google Scholar]
- Parsell, D. A., A. S. Kowal, M. A. Singer and S. Lindquist, 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478. [DOI] [PubMed] [Google Scholar]
- Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov and M. D. Ter-Avanesyan, 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15: 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Schirmer, E. C., J. R. Glover, M. A. Singer and S. Lindquist, 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21: 289–296. [PubMed] [Google Scholar]
- Schirmer, E. C., C. Queitsch, A. S. Kowal, D. A. Parsell and S. Lindquist, 1998. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J. Biol. Chem. 273: 15546–15552. [DOI] [PubMed] [Google Scholar]
- Schirmer, E. C., O. R. Homann, A. S. Kowal and S. Lindquist, 2004. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol. Biol. Cell 15: 2061–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer, C., and D. C. Masison, 2002. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22: 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., 1994. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Shorter, J., and S. Lindquist, 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304: 1793–1797. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, J. M., and N. R. Eaton, 1969. Functional blocks of the ad-1 and ad-2 mutants of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 34: 301–305. [DOI] [PubMed] [Google Scholar]
- Sondheimer, N., and S. Lindquist, 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5: 163–172. [DOI] [PubMed] [Google Scholar]
- Song, Y., Y. X. Wu, G. Jung, Y. Tutar, E. Eisenberg et al., 2005. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 4: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tek, V., and M. Zolkiewski, 2002. Stability and interactions of the amino-terminal domain of ClpB from Escherichia coli. Protein Sci. 11: 1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn, R. D., K. Bapat, G. P. Newnam, A. D. Zink and Y. O. Chernoff, 2001. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 21: 4656–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn, J., C. Schlieker, B. Bukau and A. Mogk, 2003. Characterization of a Trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 278: 32608–32617. [DOI] [PubMed] [Google Scholar]
- Weibezahn, J., P. Tessarz, C. Schlieker, R. Zahn, Z. Maglica et al., 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., 1994. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science 264: 566–569. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz, S., J. Krzewska and K. Liberek, 2004. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J. Biol. Chem. 279: 44376–44383. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz, S., A. Lewandowska, P. Stocki and K. Liberek, 2006. HSP70 chaperone machine remodels protein aggregates at the initial step of HSP70-HSP100 dependent disaggregation. J. Biol. Chem. 281: 7022–7029. [DOI] [PubMed] [Google Scholar]