Abstract
Sex differences occur in most species and affect a variety of biological traits including morphology, behavior, and life history. The nematode Caenorhabditis elegans exists as a population of self-fertile hermaphrodites with occasional males, which differ anatomically and behaviorally from hermaphrodites. Here we show that male C. elegans also differ from hermaphrodites in their susceptibility to a fungal pathogen, Cryptococcus neoformans. Wild-type males show greater resistance than hermaphrodite animals to killing by this pathogen and this resistance can be induced in hermaphrodite animals by inappropriate activation of the male sex-determination pathway. Resistance is molecularly determined, rather than resulting from behavioral changes or reproductive differences, and requires the activity of the stress-response transcription factor DAF-16. Finally, we demonstrate that resistance to C. neoformans correlates broadly with longevity within the Caenorhabditis genus. Our results hint at an overlap between the pathways controlling immunity and longevity and raise the possibility that differential regulation of these pathways may contribute to sex-dependent and species-dependent variation.
IN species that have more than one gender, sex differences occur in both physical (e.g., size, coloration) and behavioral (e.g., aggression, motility) traits. In addition, there are often differences between the sexes in terms of disease susceptibility (Barna et al. 1996; Komukai et al. 1999; Whitacre et al. 1999), many of which cannot be explained in terms of behavioral or anatomical differences. Such observations suggest that there may be intrinsic differences at the molecular level between sexes in terms of their ability to respond to disease.
The nematode Caenorhabditis elegans has been widely used as an experimental organism for >30 years (Brenner 1974; Riddle et al. 1997). C. elegans occurs as a population of self-fertilizing hermaphrodites that are diploid for the sex-determining X chromosome, together with a low percentage of males (which possess only a single X chromosome). Males are produced either as a result of nondisjunction of the X chromosomes during gametogenesis in the hermaphrodite (which occurs with a frequency of ∼0.2%) or from a male–hermaphrodite mating (which produces ∼50% male offspring) (Hope 1999).
C. elegans males are morphologically distinct from hermaphrodites: the body is thinner and lacks female germline and the tail is specialized to deliver sperm to the hermaphrodite during mating. There are also clear behavioral differences between the sexes. The male is more active and engages in mate searching and frequent “backing” behavior, which is never seen in hermaphrodites. In addition, the male life span is intrinsically longer than that of hermaphrodite animals, although in a normal population setting this increased longevity is dramatically reduced due to male-specific behavior (e.g., mating) (Gems and Riddle 2000).
The yeast Cryptococcus neoformans is an opportunistic pathogen of immunocompromised humans and other mammals (Casadevall and Perfect 1998). Along with numerous other human pathogens (Couillault and Ewbank 2002; Kurz and Ewbank 2003), it has been demonstrated that a C. neoformans infection can be modeled in C. elegans: following acquisition of the pathogen in the diet, animals become sick and die with a highly reproducible median survival time of 4–5 days (Mylonakis et al. 2002). This system has been used to probe the molecular basis of virulence in C. neoformans and has successfully identified novel genes that play an important role in virulence in this pathogen (Mylonakis et al. 2004). However, little is known about factors that determine host susceptibility to infection by C. neoformans. Here we demonstrate that male C. elegans are more resistant than hermaphrodites to C. neoformans-mediated killing. This phenotype is a direct result of gender specification and requires the stress-response transcription factor DAF-16. In addition, male-specific resistance is not common to all rhabditid nematodes and varies according to longevity, suggesting that there may be overlap between pathways that regulate both longevity and immunity.
MATERIALS AND METHODS
C. elegans culture and strains:
C. elegans were cultured using standard methods (Hope 1999). Strains used in this work were: AF16 (Caenorhabditis briggsae wild type), BA1 [fer-1(hc1)I], CB2590 [tra-1(e1099)/dpy-18(e1096)III], CB2823 [tra-1(e1488)III; eDp6(III;f)], CB4856 (C. elegans wild type, Hawaiian isolate), CB5161 (C. sp. wild type), CB5475 [her-1 (e1518) V; sdc-2(y15) X], CF1038 [daf-16(mu86) I], DF5078 (C. japonica wild type), EM464 [C. remanei wild type], JK987 [tra-2 (q276) II/mnC1; dpy-10(e128)II; unc-52(e444)II], KR314 (C. elegans wild type, Vancouver isolate), N2 (C. elegans wild type, Bristol isolate), NL4317 [rmIs126(Punc-54∷Q0∷YFP)V in N2 background], RC301 (C. elegans wild type, German isolate), SS104 [glp-4(bn2)I], VT847 (C. briggsae wild type, Hawaiian isolate).
The strain used as a standard wild type in this work is NL4317. This strain is derived from the canonical laboratory strain N2 and carries an integrated transgene (rmIs126) that expresses yellow fluorescent protein in the body-wall muscle (Morley et al. 2002). The transgene enables rapid location of the animals inside the thick lawn of C. neoformans using a fluorescence-equipped stereomicroscope, as well as being a useful marker for genetic crosses. Following integration, the strain was backcrossed nine times to N2 to produce NL4317. Behavior, longevity under normal laboratory conditions, and survival on C. neoformans JEC21 have been extensively analyzed and do not differ from that of C. elegans N2 (data not shown).
CB2590 [tra-1(e1099)/dpy-18(e1096)III], CB2823 [tra-1(e1488)III; eDp6(III;f)], and JK987 [tra-2 (q276) II/mnC1; dpy-10(e128)II; unc-52(e444)II] are all strains in which hermaphrodite animals are transformed into males. Maintenance of males is ensured by the presence of genetic “balancers” (Hope 1999).
CB5475 [her-1 (e1518) V; sdc-2(y15) X] was kindly provided by Jonathan Hodgkin. This strain is null for the her-1 gene, causing males to adopt an hermaphrodite fate, and is additionally null for sdc-2, the loss of which causes a failure in dosage compensation and consequent death of XX individuals. Thus all “hermaphrodite” CB5475 animals are transformed males, rather than true hermaphrodites.
Male stocks:
With the exception of the transformed strains described above, male animals were produced by picking spontaneously occurring males from culture plates and crossing to an hermaphrodite animal from the same strain. Once produced, males were maintained by regularly crossing to hermaphrodite animals of the same strain.
Survival assays:
C. neoformans strain JEC21 was grown overnight in YPG medium (1% yeast extract, 1% bacto peptone, 2% glucose), shaking at 25°. For survival assays, 5 μl of this overnight culture was inoculated onto each well of a 24-well plate filled with nematode growth medium (NGM; Hope 1999) supplemented with kanamycin (30 μg/ml) to prevent bacterial contamination. Yeast colonies were allowed to grow for 48–72 hr at 25° before animals at the fourth larval stage (L4) were singled onto each well. Plates were incubated at 25° and animals were examined at 24-hr intervals for 8 days and scored for survival using a Leica M7.5 stereomicroscope. Death was defined as a failure to move in response to mechanical stimulus.
Life-span analyses:
For the analysis of wild-type hermaphrodite life span, L4 animals were transferred onto NGM plates seeded with OP50 and then removed to fresh-seeded plates every 2–3 days during the fertile period. For analysis of male and female life span in gonochoristic species, L4 stage animals were singled onto 24-well plates and transferred less frequently (since these individuals do not produce progeny that would otherwise interfere with the analysis). This additionally avoids the problem of male–male interactions, which has previously been shown to reduce male life span (Gems and Riddle 2000).
Statistical analysis:
Each survival curve is based on data from 40 to 200 animals. Curves are based on the Kaplan–Meier method and were generated using Microsoft Excel. Statistical significance was tested by the log-rank method. The median was used to calculate average survival time, since survival times do not show a normal distribution. Regression analysis to assess the correlation between longevity and resistance to C. neoformans was carried out using the “regression” function in Microsoft Excel.
RESULTS
Male C. elegans live longer than hermaphrodites following exposure to C. neoformans:
Exposing wild-type C. elegans hermaphrodites to lawns of C. neoformans (see materials and methods) results in highly reproducible killing with a median survival time of 4–5 days (using the C. neoformans strain JEC21 at 25°) (Mylonakis et al. 2002). Given the well-documented differences in a variety of biological traits between male and hermaphrodite C. elegans (Gems and Riddle 2000; Emmons and Lipton 2003), we asked whether the kinetics of killing by C. neoformans differ between the two sexes. Surprisingly, C. elegans males were far more resistant to killing by C. neoformans JEC21 than hermaphrodites (P < 0.0001), with a median survival time of ∼9 days (Figure 1 and data not shown).
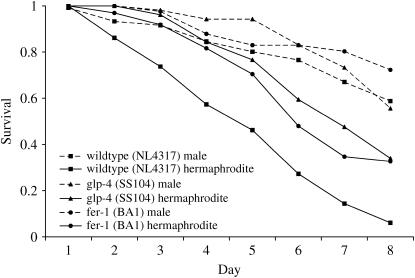
Figure 1.
Male and hermaphrodite animals at the L4 stage were singled onto wells preseeded with C. neoformans JEC21 and monitored for survival over 8 days. While hermaphrodite animals show almost 100% lethality over this period, male survival is significantly higher (P < 0.0001), with >50% of animals surviving >8 days. This increase in survival rate is not due simply to the absence of “matricidal killing” in males, since sterile hermaphrodite animals (BA1 and SS104) do not suffer from matricidal killing but remain more sensitive than males (P < 0.001) to Cryptococcus-mediated killing.
One obvious explanation for this effect could be that single males do not produce progeny. It has previously been shown that infection of hermaphrodite C. elegans with a variety of pathogens (Aballay et al. 2000; O'Quinn et al. 2001; Sifri et al. 2003), including C. neoformans (Mylonakis et al. 2002), results in a high incidence of “bagging”; rather than being laid, eggs are retained within the body and hatch internally, resulting in so-called “matricidal killing.” To control for this effect, we tested two C. elegans strains that show temperature-sensitive sterility. In agreement with previous work (Mylonakis et al. 2002), we found that sterility increased survival in hermaphrodite animals fed on C. neoformans JEC21 by ∼2 days (P < 0.001; median survival of 7 days) (Figure 1). However, this level of resistance was still significantly (P < 0.001) lower than that of wild-type C. elegans males or males derived from either of the sterile strains. This, together with the tra-1 results discussed below, suggests that male-dependent Cryptococcus resistance does not result simply from the absence of matricidal killing.
A male phenotype, but not genotype, is required to confer resistance to C. neoformans-mediated killing:
The molecular basis of sex determination in C. elegans has been well documented (Stothard and Pilgrim 2003) and centers around a repressor-based signal cascade. Briefly, in animals destined to become hermaphrodite (i.e., with two X chromosomes) the extracellular ligand HER-1 is not expressed and the transmembrane protein TRA-2 thus remains active, triggering a pathway that culminates in the activation of the transcription factor TRA-1 and the adoption of an hermaphrodite fate. Conversely, male animals (XO) transcribe the her-1 gene, resulting in inhibition of TRA-2 and, consequently, repression of TRA-1 (Hodgkin and Brenner 1977; Hodgkin 1980).
Loss of many of these signaling components can trigger sexually reversed phenotypes [e.g., animals that have an XO (genetically male) chromosome complement but are morphologically hermaphrodite]. We exploited several such mutants to test the importance of a male karyotype in specifying resistance to Cryptococcus-mediated killing.
her-1 animals lack expression of the HER-1 protein, even in males, causing the latter to adopt an hermaphrodite fate (Hodgkin 1980). We measured the survival of XO (genetically male) her-1 animals exposed to C. neoformans JEC21. Despite their male genotype, these animals were just as susceptible to killing by C. neoformans as wild-type hermaphrodites were (Figure 2), suggesting that the male phenotype, and not the male genotype, is required for resistance.
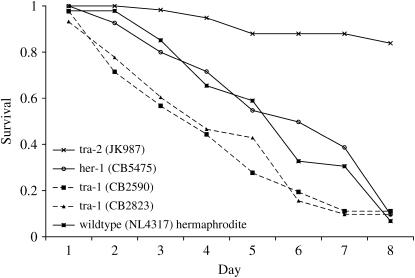
Figure 2.
Phenotypically male (JK987, CB2823, CB2590) and hermaphrodite (NL4317, CB5475) L4 animals were singled onto wells preseeded with C. neoformans JEC21 and monitored for survival over 8 days. Note that sexually transformed tra-2 animals are significantly more resistant than either wild-type or sexually transformed hermaphrodite animals (P < 0.0001) to Cryptococcus-mediated killing. Sexually transformed males carrying mutations in tra-1, however, are fully susceptible to Cryptococcus-mediated killing.
In agreement with this, tra-2 (q276) animals, in which absence of the TRA-2 protein causes hermaphrodites to adopt a male fate, were resistant to Cryptococcus-mediated killing (Figure 2) (P < 0.0001 vs. wild-type hermaphrodites). Thus animals that are genetically hermaphrodite but morphologically male nonetheless show a “male” pattern of resistance to C. neoformans. In contrast, however, both tra-1 (e1488) and tra-1 (e1099) animals show an hermaphrodite-like response to Cryptococcus (i.e., sensitivity). Although tra-1(e1488) animals are only partially transformed into males (they retain an hermaphrodite gonad and intestine and are self-fertile), tra-1(e1099) animals are not self-fertile and show very few hermaphrodite characteristics. Thus it appears that male specification at the level of the TRA-1 transcription factor is insufficient to trigger Cryptococcus resistance, whereas intervention at an earlier stage of the sex-determination pathway (tra-2) recapitulates both the male phenotype and Cryptococcus resistance.
Male-specific resistance is dependent on the DAF-16 transcription factor:
Solitary male C. elegans are ∼20% longer lived than hermaphrodite animals and this longevity is largely, but not entirely, attributable to increased activity of the stress-response transcription factor DAF-16 (Gems and Riddle 2000). We asked whether DAF-16 activity was also responsible for the observed male-dependent resistance to Cryptococcus-mediated killing. Hermaphrodite daf-16 mutant animals exposed to C. neoformans JEC21 died at a rate indistinguishable from wild-type hermaphrodites (Figure 3). However, daf-16 males were dramatically more sensitive than their wild-type counterparts to C. neoformans-mediated killing (Figure 3, P < 0.0001) although, interestingly, not as sensitive as hermaphrodite animals (P < 0.02, daf-16 males vs. wild-type hermaphrodites). Thus, as with life span, DAF-16 activity is largely, but not solely, responsible for increased resistance to C. neoformans.
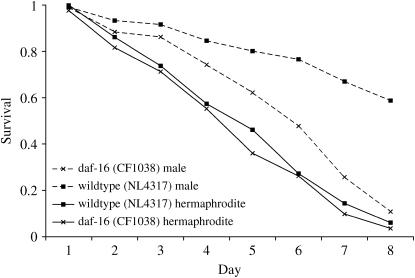
Figure 3.
Male and hermaphrodite L4 animals were exposed to JEC21 and monitored for survival over 8 days. Wild-type and daf-16-deficient hermaphrodite animals show identical susceptibility to Cryptococcus-mediated killing. daf-16 males, however, are far more susceptible than wild-type males (P < 0.0001) to the pathogen, but remain significantly more resistant to killing than either wild-type or daf-16 hermaphrodites (P < 0.01). Thus DAF-16 is largely, but not exclusively, required for male-dependent resistance to Cryptococcus-mediated killing.
Different patterns of resistance to Cryptococcus-mediated killing within the Caenorhabditis genus:
We asked whether the phenomenon of male-specific resistance to Cryptococcus-mediated killing was widespread among related nematode species. There are up to 21 species currently known within the genus Caenorhabditis, of which 4 are further classified into the so-called “elegans group” of which C. elegans is a member (Cho et al. 2004; Kiontke et al. 2004). The genus is genetically highly diverse, such that even the elegans subgroup shows more variation than the entire mammalian lineage (Kiontke et al. 2004). Interestingly, recent data suggest that hermaphroditism has evolved multiple times within the Caenorhabditis genus and that the molecular mechanism to generate hermaphroditism differs between C. elegans and the related hermaphroditic species C. briggsae (Kiontke et al. 2004; Nayak et al. 2005). Given this diversity, we wondered whether male-dependent resistance to Cryptococcus-mediated killing was widespread within the clade.
First, we analyzed the survival of hermaphrodite or female animals (depending on species) from representative strains within the genus following exposure to C. neoformans JEC21. Surprisingly, we found considerable variation in survival depending on species, with females of two species (C. remanei and C. sp. CB5161) proving highly resistant to killing by the pathogen (Figure 4A; median survival time ∼13 days for both species; P < 0.0001 vs. N2 hermaphrodites). This resistance is not a general feature of female (in contrast to hermaphrodite) animals, since C. japonica females were equally susceptible to killing as C. elegans hermaphrodites (Figure 4A). Although hermaphrodite/female resistance varies considerably within the genus, there is little if any variation within the C. elegans species, since hermaphrodites of several, geographically widespread C. elegans isolates (including the polymorphic Hawaiian strain CB4856) show identical susceptibility to C. neoformans (Figure 4A and data not shown).
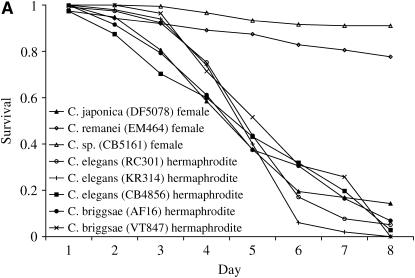
Figure 4.
(A) L4-stage hermaphrodite or female animals were singled onto C. neoformans JEC21 and monitored for survival. Female C. remanei and C. sp CB5161 animals were significantly more resistant to C. neoformans-mediated killing than wild-type C. elegans hermaphrodites (P < 0.0001 for both) or C. japonica females (P < 0.0001 for both). In contrast, females/hermaphrodites of the other species tested showed remarkably similar levels of susceptibility (P > 0.05 for each pairwise comparison). (B) L4-stage male and hermaphrodite/female animals were singled onto C. neoformans JEC21 and monitored for survival. Hermaphrodite/female animals from each of the four species tested showed similar levels of susceptibility to killing (P > 0.05), while both C. elegans (CB4856) males and C. japonica (DF5078) males were significantly more resistant to killing than hermaphrodites/females of the same species (P < 0.0001 and P < 0.001, respectively). Interestingly, however, male C. briggsae were either equally (AF16) or more (VT847) susceptible to killing by Cryptococcus than were hermaphrodite animals. Thus male-dependent resistance is not a ubiquitous feature of the Caenorhabditis genus.
We then asked whether the males of “susceptible” Caenorhabditis species (those in which hermaphrodites/females show significant death rates within 8 days) always showed enhanced resistance to Cryptococcus-mediated killing relative to hermaphrodites–females of the same species. C. elegans males from all strains tested always showed enhanced resistance to Cryptococcus-mediated killing (Figure 4B). Similarly, males of the closely related gonochoristic (male/female) species C. japonica were also more resistant to killing relative to C. japonica females. In sharp contrast, however, C. briggsae males were either equally (AF16) or more (VT847) susceptible than C. briggsae hermaphrodites to Cryptococcus-mediated killing (Figure 4B). Male-dependent resistance to Cryptococcus is thus not a conserved feature of all rhabditid nematodes but rather varies according to longevity.
Finally, we also considered the possibility that males may show altered resistance to Cryptococcus-mediated killing even in species in which females are already highly resistant. However, extended survival analysis of C. remanei (EM464) and C. sp. (CB5161) on C. neoformans JEC21 (data not shown) showed no significant difference in median survival between the sexes (P > 0.02). Note, however, that this survival time is markedly shorter than the normal median survival for either species when fed a standard diet of Escherichia coli OP50. Thus even resistant species are not “immune” to the effects of C. neoformans, but rather are simply able to delay the rate at which they succumb to Cryptococcus-mediated killing.
A correlation between Cryptococcus resistance and longevity:
We were struck by the general similarity between our data on varying resistance to Cryptococcus in different nematode species and previously published data on the longevity of many of these species (McCulloch and Gems 2003). To further explore this potential link, we added to previously published longevity data (Hsin and Kenyon 1999; Gems and Riddle 2000; Patel et al. 2002; McCulloch and Gems 2003) and then compared life span on OP50 with Cryptococcus resistance across all strains (Table 1). Regression analysis shows a statistically significant correlation (F-value = 0.0022, n = 15) between the two variables, with long-lived species tending to show increased resistance to Cryptococcus-mediated killing. In addition, the pattern of longevity between the two sexes within a species is also recapitulated in their Cryptococcus resistance: in most nematodes, males are both longer lived and more resistant to Cryptococcus, whereas in both C. briggsae strains tested, this pattern is reversed (Table 1). We note, however, that there are marked exceptions to this trend. For example, C. remanei (EM464) males and females are equally resistant to Cryptococcus and yet male longevity is twice that of females in this species, indicating that there are likely to be many additional factors that impact upon both immunity and longevity.
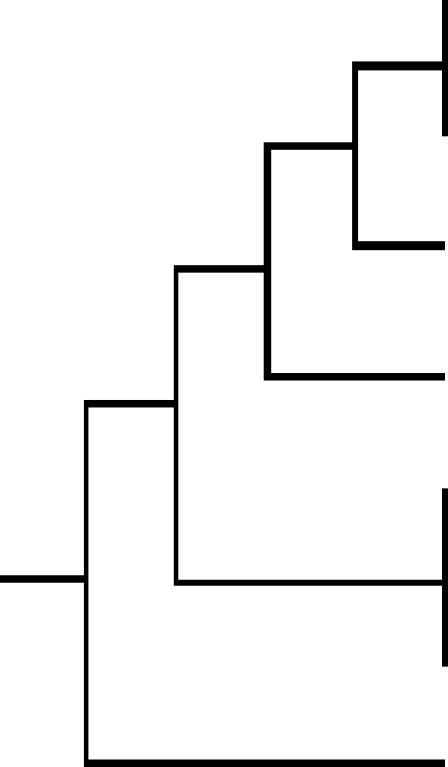
TABLE 1.
Comparison of longevity and Cryptococcus resistance in different nematode strains
| Median longevity on OP50
|
Survival on C. neoformans
|
||||
|---|---|---|---|---|---|
| Species | H/Fa | Mb | H/Fa | Mb | |
 |
C. briggsae AF16 | 19 | 17 | 5 | 4 |
| C. briggsae VT847 | 17 | 10 | 5 | 2 | |
| C. remanei EM464 | 36 | 61 | 13 | 13 | |
| C. sp CB5161 | 27 | 27 | 13 | 13 | |
| C. elegans N2 | 18 | 20 | 4 | 9 | |
| C. elegans CB4856 | 23 | 25 | 5 | 10 | |
| C. elegans RC301 | 21 | 27 | 5 | ND | |
| C. japonica DF5078 | 20 | 22 | 4 | 8 | |
Species tested are positioned according to recent phylogenetic analysis (Cho et al. 2004; Kiontke et al. 2004). Note that branch length is not indicative of genetic distance. Longevity is expressed as median survival time in days when fed on live E. coli OP50 at 20°. Longevity data are derived from the following references: Hsin and Kenyon (1999); Gems and Riddle (2000); Patel et al. (2002); and McCulloch and Gems (2003), with the exception of strains DF5078 and CB5161, for which life-span analyses were carried out as part of this work. Survival on Cryptococcus is expressed as median survival time in days after transfer onto C. neoformans JEC21 (at L4 stage) at 25°. Regression analysis shows a significant (F = 0.0022) correlation between median longevity on OP50 and survival on C. neoformans.
Hermaphrodites/females.
Males.
DISCUSSION
Pathogen resistance is an attribute of profound importance in the life of an organism and, as such, is likely to be under strong evolutionary selection. The force and direction of this selection will depend on numerous factors, including exposure and virulence of the pathogen and life history traits of the host.
In C. elegans and several related nematode species, solitary male animals are longer lived than hermaphrodites/females (McCulloch and Gems 2003) [note that, when kept in groups, male life span is reduced significantly by homosexual interactions between males (Gems and Riddle 2000)]. The exception to this rule is C. briggsae, in which males are shorter lived (McCulloch and Gems 2003). Interestingly, we find a similar pattern with respect to Cryptococcus resistance; C. briggsae males are more susceptible than C. briggsae hermaphrodites to Cryptococcus-mediated killing, while in all other species tested either the males (C. elegans, C. japonica) or both sexes (C. sp CB5161, C. remanei) are resistant to this pathogen. Indeed, on the basis of our own data and previously published analyses of longevity (Hsin and Kenyon 1999; Gems and Riddle 2000; Patel et al. 2002; McCulloch and Gems 2003), it appears that, in general, animals with increased longevity also show increased resistance to Cryptococcus.
Why might this be? The most likely explanation is that many of the molecular determinants of longevity also determine pathogen resistance. Recent microarray analyses of long-lived C. elegans strains indicate that these strains do indeed upregulate numerous antimicrobial molecules (Murphy et al. 2003; McElwee et al. 2004; Gems and McElwee 2005), suggesting that the expression of immunity factors is partially under the control of pathways that also contribute to longevity.
It is tempting to speculate that evolution has favored this coregulation of longevity and immunity. For example, animals whose highest probability of reproduction occurs at later ages are, by definition, under selective pressure to have an increased life span (to maximize the chance of successful reproduction) (Williams 1957; McCulloch and Gems 2003). Since life-threatening infections would compromise this longevity, it is reasonable to suppose that longer-lived animals are under increased pressure to evolve some form of immunity to maintain that longevity advantage. Under this scenario, C. elegans males (which must find and mate with an hermaphrodite to reproduce) must resist lethal pathogen infections for longer than hermaphrodites (which are self-fertile at ∼3 days of age and become sterile once they exhaust their sperm supply 4 days later).
What is the molecular basis of varying resistance to Cryptococcus-mediated killing? The mechanism(s) by which Cryptococcus kills its nematode host and the nature of the host response to infection by this pathogen are currently unknown (Mylonakis et al. 2002). However, the upregulation of a number of antimicrobial factors appears critical for host survival (R. C. May and M. C. W. van den Berg, unpublished results), and many of these factors appear to be constitutively upregulated in long-lived strains (Murphy et al. 2003; McElwee et al. 2004). On the basis of other C. elegans pathogen models (Mahajan-Miklos et al. 1999; Tan et al. 1999a,b; Jansen et al. 2002; Moy et al. 2004), Cryptococcus may also produce toxins while inside the nematode host. Interestingly, long-lived C. elegans strains also show dramatic upregulation of broad-spectrum detoxification mechanisms (Gems and McElwee 2005). It remains to be seen whether these, too, play a role in conferring resistance to Cryptococcus-mediated killing.
As with male longevity, the transcription factor DAF-16 appears central to male-dependent Cryptococcus resistance. DAF-16 acts downstream of, and is indirectly repressed by, the insulin-like growth factor DAF-2. daf-2 animals are remarkably long lived and, in addition, highly resistant to a number of pathogens, including C. neoformans (R. C. May, unpublished data) and the bacteria Enterococcus faecalis, Staphylococcus aureus, and Pseudomonas aeruginosa (Garsin et al. 2003). Interestingly, C. elegans males also show enhanced resistance to P. aeruginosa relative to hermaphrodites (Tan et al. 1999a). It is thus tempting to speculate that DAF-16 may be responsible for the resistance of male C. elegans to bacterial pathogens as well as to C. neoformans, further supporting the hypothesis that immunity and aging are intrinsically linked at the molecular level (Kurz and Tan 2004). Given that male C. elegans are long lived (Gems and Riddle 2000) and pathogen resistant (this work) and that both effects are, in large part, DAF-16 dependent, it appears likely that male C. elegans either constitutively activate DAF-16 or indirectly enhance its effects. We hypothesize that increased activity of DAF-16, its ortholog(s), or its downstream effectors may also be responsible for the enhanced life span and Cryptococcus resistance seen in C. japonica males and in both sexes of C. remanei and C. sp. CB5161.
In conclusion, we show that resistance to a lethal fungal pathogen, C. neoformans, is highly variable among related nematode species and, importantly, between different sexes of the same species. This resistance is largely dependent on DAF-16 and is specified at an early stage of the sex-determination pathway. These findings support the hypothesis that longevity and immunity are intrinsically linked at the molecular level and suggest that altered regulation of longevity pathways may underlie numerous biological differences that occur within and among species.
Acknowledgments
We are very grateful to David Gems, Teun Boekhout, and Marjan Bovers for helpful discussions and suggestions and to Ronald Plasterk and members of his group for constructive criticism and advice during the initial phase of this project. Many strains used in this work were provided by the Caenorhabditis Genetics Center. Strain CB5475 was kindly provided by Jonathan Hodgkin. This work was supported by funds from the Human Frontier Science Program, The Wijk–Stam–Caspers funds, and the Rowbotham Bequest.
References
- Aballay, A., P. Yorgey and F. M. Ausubel, 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10: 1539–1542. [DOI] [PubMed] [Google Scholar]
- Barna, M., T. Komatsu, Z. Bi and C. S. Reiss, 1996. Sex differences in susceptibility to viral infection of the central nervous system. J. Neuroimmunol. 67: 31–39. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall, A., and J. R. Perfect, 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, DC.
- Cho, S., S. W. Jin, A. Cohen and R. E. Ellis, 2004. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 14: 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault, C., and J. J. Ewbank, 2002. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 70: 4705–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons, S. W., and J. Lipton, 2003. Genetic basis of male sexual behavior. J. Neurobiol. 54: 93–110. [DOI] [PubMed] [Google Scholar]
- Garsin, D. A., J. M. Villanueva, J. Begun, D. H. Kim, C. D. Sifri et al., 2003. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300: 1921. [DOI] [PubMed] [Google Scholar]
- Gems, D., and J. J. McElwee, 2005. Broad spectrum detoxification: The major longevity assurance process regulated by insulin/IGF-1 signaling? Mech. Ageing Dev. 126: 381–387. [DOI] [PubMed] [Google Scholar]
- Gems, D., and D. L. Riddle, 2000. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics 154: 1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1980. More sex-determination mutants of Caenorhabditis elegans. Genetics 96: 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J. A., and S. Brenner, 1977. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics 86: 275–287. [PMC free article] [PubMed] [Google Scholar]
- Hope, I. A., 1999. C. elegans: A Practical Approach. Oxford University Press, Oxford.
- Hsin, H., and C. Kenyon, 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362–366. [DOI] [PubMed] [Google Scholar]
- Jansen, W. T., M. Bolm, R. Balling, G. S. Chhatwal and R. Schnabel, 2002. Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect. Immun. 70: 5202–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101: 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komukai, Y., H. Amao, N. Goto, Y. Kusajima, T. Sawada et al., 1999. Sex differences in susceptibility of ICR mice to oral infection with Corynebacterium kutscheri. Exp. Anim. 48: 37–42. [DOI] [PubMed] [Google Scholar]
- Kurz, C. L., and J. J. Ewbank, 2003. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat. Rev. Genet. 4: 380–390. [DOI] [PubMed] [Google Scholar]
- Kurz, C. L., and M. W. Tan, 2004. Regulation of aging and innate immunity in C. elegans. Aging Cell 3: 185–193. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos, S., M. W. Tan, L. G. Rahme and F. M. Ausubel, 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96: 47–56. [DOI] [PubMed] [Google Scholar]
- McCulloch, D., and D. Gems, 2003. Evolution of male longevity bias in nematodes. Aging Cell 2: 165–173. [DOI] [PubMed] [Google Scholar]
- McElwee, J. J., E. Schuster, E. Blanc, J. H. Thomas and D. Gems, 2004. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279: 44533–44543. [DOI] [PubMed] [Google Scholar]
- Morley, J. F., H. R. Brignull, J. J. Weyers and R. I. Morimoto, 2002. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99: 10417–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy, T. I., E. Mylonakis, S. B. Calderwood and F. M. Ausubel, 2004. Cytotoxicity of hydrogen peroxide produced by Enterococcus faecium. Infect. Immun. 72: 4512–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C. T., S. A. McCarroll, C. I. Bargmann, A. Fraser, R. S. Kamath et al., 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- Mylonakis, E., F. M. Ausubel, J. R. Perfect, J. Heitman and S. B. Calderwood, 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99: 15675–15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis, E., A. Idnurm, R. Moreno, J. El Khoury, J. B. Rottman et al., 2004. Cryptococcus neoformans Kin1 protein kinase homologue, identified through a Caenorhabditis elegans screen, promotes virulence in mammals. Mol. Microbiol. 54: 407–419. [DOI] [PubMed] [Google Scholar]
- Nayak, S., J. Goree and T. Schedl, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Quinn, A. L., E. M. Wiegand and J. A. Jeddeloh, 2001. Burkholderia pseudomallei kills the nematode Caenorhabditis elegans using an endotoxin-mediated paralysis. Cell. Microbiol. 3: 381–393. [DOI] [PubMed] [Google Scholar]
- Patel, M. N., C. G. Knight, C. Karageorgi and A. M. Leroi, 2002. Evolution of germ-line signals that regulate growth and aging in nematodes. Proc. Natl. Acad. Sci. USA 99: 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess, 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Sifri, C. D., J. Begun, F. M. Ausubel and S. B. Calderwood, 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71: 2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard, P., and D. Pilgrim, 2003. Sex-determination gene and pathway evolution in nematodes. BioEssays 25: 221–231. [DOI] [PubMed] [Google Scholar]
- Tan, M. W., S. Mahajan-Miklos and F. M. Ausubel, 1999. a Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins and F. M. Ausubel, 1999. b Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96: 2408–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre, C. C., S. C. Reingold and P. A. O'Looney, 1999. A gender gap in autoimmunity. Science 283: 1277–1278. [DOI] [PubMed] [Google Scholar]
- Williams, G. C., 1957. Pleiotropy, natural selection and the evolution of senescence. Evolution 11: 398–411. [Google Scholar]