Abstract
We have studied diversity in Arabidopsis lyrata of sequences orthologous to the ARK3 gene of A. thaliana. Our main goal was to test for recombination in the S-locus region. In A. thaliana, the single-copy ARK3 gene is closely linked to the non-functional copies of the self-incompatibility loci, and the ortholog in A. lyrata (a self-incompatible species) is in the homologous genome region and is known as Aly8. It is thus of interest to test whether Aly8 sequence diversity is elevated due to close linkage to the highly polymorphic incompatibility locus, as is theoretically predicted. However, Aly8 is not a single-copy gene, and the presence of paralogs could also lead to the appearance of elevated diversity. We established a typing approach based on different lengths of Aly8 PCR products and show that most A. lyrata haplotypes have a single copy, but some have two gene copies, both closely linked to the incompatibility locus, one being a pseudogene. We determined the phase of multiple haplotypes in families of plants from Icelandic and other populations. Different Aly8 sequence types are associated with different SRK alleles, while haplotypes with the same SRK sequences tend to have the same Aly8 sequence. There is evidence of some exchange of sequences between different Aly8 sequences, making it difficult to determine which ones are allelic or to estimate the diversity. However, the homogeneity of the Aly8 sequences of each S-haplotype suggests that recombination between the loci has been very infrequent over the evolutionary history of these populations. Overall, the results suggest that recombination rarely occurs in the interval between the S-loci and Aly8 and that linkage to the S-loci can probably account for the observed high Aly8 diversity.
PLANT self-incompatibility loci are subject to frequency-dependent selection with a fertility advantage to rare alleles, a form of balancing selection that acts to keep allele frequencies intermediate and prevents loss of alleles from populations (Wright 1939; Vekemans and Slatkin 1994; Schierup et al. 1998; Uyenoyama 2000). This long-term maintenance of alleles is predicted to lead to high diversity at sites genetically closely linked to the sites where selection acts (Nordborg et al. 1996; Charlesworth et al. 1997; Innan and Nordborg 2003; Schierup et al. 2000a; Wiuf et al. 2004). In SRK, which encodes the pistil receptor kinase, high diversity has indeed been found in the intracellular kinase domain, which is unlikely to be subject to balancing selection (Charlesworth et al. 2003a).
In Brassica, there are three closely linked S-locus genes, only two of which are involved in the incompatibility recognition response. SRK encodes the stigma receptor kinase, and SCR encodes the pollen ligand recognized by the SRK's extracellular S-domain to give the self-incompatibility (SI) phenotype (Nasrallah 2000). To maintain functional incompatibility, the SCR and SRK genes must be in linkage disequilibrium, with each haplotype carrying alleles at the two loci that are recognized as incompatible. Recombination between these S-genes will lead to nonfunctional (self-compatible) haplotypes. Low recombination may thus have evolved in the S-locus region (Casselman et al. 2000) to maintain incompatible combinations of SRK and SCR alleles during the long time periods during which incompatibility alleles exist.
If recombination is reduced in the whole region, diversity should be high in loci across the nearby genome region. Several nonincompatibility loci are present in the S-locus region, physically close to the genes involved in the SI response (Kusaba et al. 2001), and molecular diversity studies of such genes are thus of interest in testing these ideas and gaining an understanding of the combined effects of both balancing selection and recombination across this region. Here we study one such locus, Aly8, in the self-incompatible plant, A. lyrata, a distantly related species in the same plant family as Brassica, which is closely related to A. thaliana. Aly8 is the ortholog of the A. thaliana ARK3 gene, which is not involved in incompatibility (Kusaba et al. 2001). In two haplotypes physically mapped by Kusaba et al. (2001), Aly8 is located at differing distances from the S-genes (in haplotype a, it is ∼5 kb from SCR and 10 kb from SRK, and in haplotype b the distances are 50 and 30 kb, respectively). Other haplotypes may have gene arrangements and physical distances different from both of these, as large size differences are seen in Brassica (Suzuki et al. 2000).
Like SRK, Aly8 has an S-domain sequence and a kinase region, and it has been shown to have high diversity (Charlesworth et al. 2003b), but it is not involved in SI. Expression studies of ARK3 in A. thaliana and of the probable ortholog, SFR2, in Brassica oleracea (Kai et al. 2001) show responses to bacterial infection and wounding (Pastuglia et al. 2002) and expression in vegetative, not reproductive, tissues (Dwyer et al. 1994).
ARK3 is a single-copy gene in A. thaliana, at least in the Col-0 strain, and in both physically mapped A. lyrata haplotypes (Kusaba et al. 2001). However, when PCR amplification products from this locus were cloned and sequenced from individual A. lyrata plants, more than two sequences were sometimes found, suggesting the presence of multiple, paralogous loci (Charlesworth et al. 2003b), which may partly explain Aly8's high sequence diversity (Charlesworth et al. 2003b). To study diversity at this locus, and to test whether it is elevated because of linkage to the S-locus region, it is therefore necessary to establish how many loci are present, to assign sequences to individual loci and determine which are linked to the S-loci. Here, we show, using family data, that some, but not all, S-haplotypes have two copies, and that, in such haplotypes, both Aly8 copies are linked to the S-locus.
To test for close linkage over evolutionary time, which is expected if the S-locus region recombines unusually rarely, we established the phase of SRK-Aly8 haplotypes, using families made by crossing plants with known SRK alleles. Finding consistent associations between SRK and Aly8 alleles across multiple populations (as we observe) suggests linkage disequilibrium maintained for considerable times. This constitutes much stronger evidence for low recombination in the S-locus region than family data, where even a few genotyping errors can give incorrect estimates (e.g., Isidore et al. 2003). Population genetic data can stringently test for the occurrence of recombination and potentially restrict estimates of the recombination frequency to lower values than can be detected in family data, because even very infrequent recombination eliminates associations over multiple generations.
MATERIALS AND METHODS
Plant materials and notation:
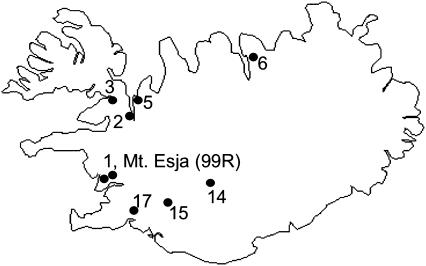
To investigate the genetics of the Aly8 sequences, and determine whether there are only two loci or more than two, and their linkage relations, we initially analyzed five full-sib families of Arabidopsis lyrata, originating from crosses between plants from a single population at Mt. Esja near Reykjavik, Iceland, with known SRK sequences; the parents of these crosses are designated as 00B or 99R, followed by individual identification numbers (see also Bechsgaard et al. 2004; Mable et al. 2004). The numbering system for the families and sequences that is used in the tables, based on the sequence lengths, is described below in Fragment length analysis. To test whether the associations between SRK and Aly8 are maintained in independent plants with the same S-alleles, we also studied segregation in five additional families—42, 43, 44, 47, and 49—whose parents originated from several different Icelandic populations (Bechsgaard 2002). The locations of the populations are shown in Figure 1.
Figure 1.
Locations of the Icelandic populations from which the plants were sampled.
Among the A. lyrata haplotypes tested, we included genotypes carrying SRK corresponding to two of the three A. thaliana SRK haplogroups (Shimizu et al. 2004). These were haplotypes carrying the A. lyrata S16 allele (Schierup et al. 2001) and A. lyrata S37 (similar to Hap A); amplification from the S36 haplotype (Hap C) showed that the Aly8 sequence was probably of the long type, but no useful sequence was obtained. In addition, BAC clones from the two physically mapped haplotypes, carrying the S13 and S20 alleles, originating from Indiana (Kusaba et al. 2001), were kindly provided by June Nasrallah (Cornell University). To assess Aly8 sequence variability within haplotypes with a given SRK, we included as many non-Icelandic individuals as possible with known SRK alleles. In what follows, we refer to the SRK sequences as S-alleles and denote them by subscript numbers, for brevity. S1 and S9 haplotypes were studied in plants from an A. lyrata mapping family derived from crossing plants from two populations, Mjallom from Sweden, and Karhumaki from Russia (Kuittinen et al. 2004), and an S1 haplotype was studied from an S1/S34 heterozygote individual from Plech, Germany, provided by M. H. Schierup (the Aly8 sequence from the S34 haplotype was not determined).
PCR and sequencing:
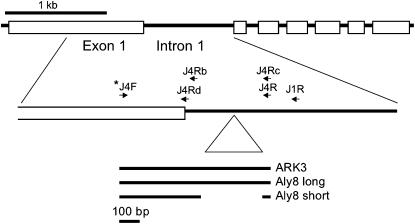
Previously published Aly8 S-domain sequences were compared with sequences from the paralogous loci Aly13 and Aly10 (Charlesworth et al. 2003b) and primers specific for different portions of Aly8 were designed; the regions studied include parts of the S-domain and parts of intron 1 (Figure 2). To find genetic markers for studying segregation, we first sequenced Aly8 from plants in families from one population, Mt. Esja. PCR reactions were run using genomic DNA with QIAGEN (Valencia, CA) Taq polymerase under standard conditions, using the primers J4F 5′-GGTCAGATGGGTGTGTGCGA-3′ and J4R 5′-CCGAACGCGTCAACATCTCAATCA-3′ or J4F and J1R 5′-GGCATTTGGGTGTACCTTAAACCA-3′. Other primers used are listed in supplemental Table 1 at http://www.genetics.org/supplemental/. The Aly8 allele linked to allele S6 (see below) did not amplify with the original primer pair (see results), but could be amplified and sequenced using the J1R reverse primer, allowing a primer specific for this allele to be designed (J4Rc 5′-CACATACGAACGCGTCAAGAAGT-3′). When possible, parental plant alleles were sequenced, but offspring were used if the parental DNA samples failed to amplify. GenBank accession numbers are given in the Figure 3 legend.
Figure 2.
Diagram of the Aly8 locus, showing the whole gene, the region sequenced, and the locations of the primers used for amplification and detection of alleles, including the primer labeled with the fluorescent dye (indicated by *). The forward primer is located in the first exon and all reverse primers are in the first intron. The triangle indicates the large indel distinguishing the “L” alleles. The part of Aly8 previously sequenced (Schierup et al. 2001) is located just upstream of the region studied here.
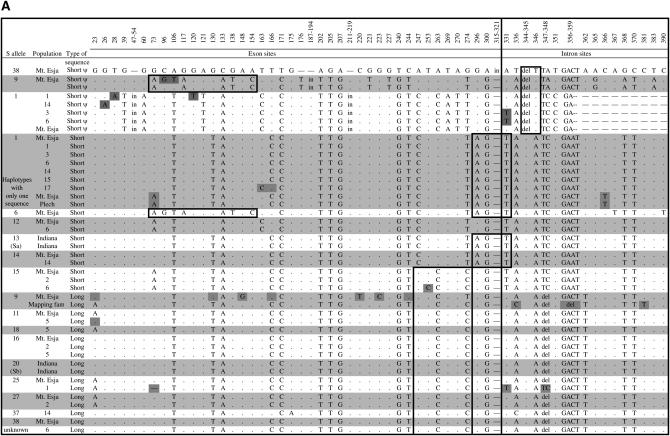
Figure 3.
Polymorphisms in the region of the Aly8 gene sequenced. The positions of all SNPs and indel variants are shown (positions are numbered according to an alignment with the A. thaliana ARK3 sequence). The different S-haplotypes are indicated by alternating white and gray stripes. Variants among sequences from the same S-haplotypes are indicated in dark gray. The top sequences are the short sequences, and those that are evidently pseudogenes are indicated, while the long sequences are given below. Regions possibly involved in gene conversion events are boxed. Note that the single-nucleotide deletion at position 73 in the coding region of one Aly8 allele from an S25 haplotype was found in the only clone from this individual that yielded an S25 sequence, and so this variant is not certain. GenBank accession nos. are DQ394305–DQ394346.
We tested offspring selected to maximize the chances of finding and identifying all Aly8 alleles, on the assumption (based on preliminary results of M. H. Schierup) that at least one of the Aly8 loci should be linked to SRK (this is confirmed by our results, see below). Amplified fragments from several plants were cloned using the Invitrogen (San Diego) TOPO TA cloning kit for sequencing. Bacterial colonies were picked and used for an additional round of PCR with the same primers, followed by automated sequencing on an ABI 3730 capillary sequencer (Applied Biosystems, Foster City, CA). Fragments were sequenced in both forward and reverse directions, and the resulting sequences were examined using the Sequencher software (version 4.2.2; GeneCodes, Ann Arbor, MI) with alignment and base-calling adjustments after manual comparison of the chromatograms. Multiple colonies were sequenced from each individual to check for PCR errors and PCR recombination. The sequences from a set of 18 Mt. Esja plants (see Table 2 and Figure 3 for details) have many single-nucleotide differences and a strikingly high indel-to-polymorphism ratio.
TABLE 2.
Summary of the S-haplotypes found among the parents of the families from all seven Icelandic populations (Mt. Esja and six other populations), and the Aly8 sequences found, using the numbering system for different sequences explained in materialsandmethods
|
Aly8 sequences detected
|
||||
|---|---|---|---|---|
| SRK allele | Long | Short | Populations | Families |
| S1 | None | 477 and 570395 | Mt. Esja | 12, 14, BM02-H5A, H5B |
| 1 | 44 | |||
| 6 | 43 and 47 | |||
| 14 | 42 | |||
| Russia or Sweden | Mapping family | |||
| S1 | None | 570395 | 15 | 49 |
| S6 | None | 539 | Mt. Esja | 23, BM02-H5A, H5B |
| S9 | L415 | 575 | Mt. Esja | 12, 23, BM02-H2A, H2B, H4B |
| L419 | 575 | Russia or Sweden | Mapping family | |
| S11 | L419f | None | 5 | 47 |
| S12 | None | 572 | Mt. Esja | BM02-H2A, H2B, H4B |
| 6 | 47 and 49 | |||
| S14 | None | 562 | Mt. Esja | 12 |
| 14 | 42 | |||
| S15 | None | 570 | 2 | 44 |
| 6 | 43 | |||
| S16 | L419d | None | Mt. Esja | BM02-H2A, H2B, H4B |
| 5 | 42 | |||
| 2 | 43 | |||
| S18 | L419g | None | 5 | 42, 47 |
| S22 | L419e | None | Mt. Esja | BM02-H2A, H2B, H4B |
| 15 | 49 | |||
| S25 | L419a | None | Mt. Esja | 14, BM02-H5A, H5B |
| 1 | 44 | |||
| S27 | L419c | None | Mt. Esja | 23 |
| 2 | 43, 44 | |||
| S38 | L419b | 570399 | Mt. Esja | 12, 14 |
| Sunknown | L419i | None | 6 | 49 |
Segregation data for the Mt. Esja families are in Table 1 (detailed data from the other families are not shown).
Fragment length analysis:
Length differences were used to genotype individuals in the families (see below). To score presence of sequence variants in the progeny plants from our families, PCR was run using the forward primer J4F labeled with a fluorescent dye (6-Fam, Applied Biosystems), together with the reverse primer J4R. The PCR products were separated and the alleles detected in an ABI 3730 capillary sequencer (Applied Biosystems) and compared with a size standard ladder (ABI GeneScan LIZ500). The resulting output was analyzed with GeneMapper 3.0 (Applied Biosystems). Aly8 sequences were denoted by their lengths (e.g., 477 denotes an estimated size of 477 bp). However, the amplified products are rather large for this kind of analysis, and sequencing showed that the true lengths of alleles 560–575 are typically ∼10 bp shorter than the values these names suggest.
The initial results showed that some sequences contain a large insertion relative to others. However, fragments approaching an expected size of 800 bp could not be detected in the fragment length analysis or, if detected, could not be sized accurately. A second, internal, primer, J4Rb 5′-GGTGGGAAGACGACAAATCATTC-3′ (Figure 2), was therefore designed to amplify a reliably scorable portion of the longer alleles. In what follows, an “L” is added to the lengths of sequences that were amplified with this reverse primer.
In three families (43, 47, and 49), the region used in the typing included no length polymorphism between the alleles of one parent. The Aly8 sequences in the first two families were resolved by sequencing, while in family 49 the S12- and S1-linked Aly8 sequences are distinguishable after digestion with MfeI (which cut the S12-linked Aly8 sequence, but not that linked to S1). In addition, two haplotypes, carrying S-alleles S38 and S1 (see below), both amplified a sequence 570 bp long. These were distinguished by an additional PCR amplification with labeled J4F primer together with a new primer, J4Rd 5′-GCCCATTATCTATCAACCGTTA-3′, which gave a band of length 399 bp for haplotype S38 and 395 bp for the S1 haplotype; these alleles were thus labeled 570399 and 570395, respectively. This primer combination also amplified some of the other alleles, helping to confirm presence or absence of these sequences.
To determine the number of loci, it was important to detect all Aly8 sequences present. We therefore made repeated PCR amplifications, using multiple primers, decreased annealing temperatures, and a range of template concentrations. All discrepancies were rechecked with new PCR reactions. In some cases (see below), allele-specific primers were designed to detect Aly8 sequences.
Sequences from other individuals with known S genotypes:
When length typing did not differentiate alleles, we inferred phases from the alleles' sequences whenever informative variants allowed. Since our initially established haplotypes allowed prediction of Aly8 sequences, we assumed that a band length consistent with the expected one, given the haplotype's SRK sequence, represents the same Aly8 sequence and then checked by sequencing. For instance, S15 haplotypes from the Mt. Esja population yield a band of length 570, but we cannot always distinguish this from the 570 band of the S1 haplotype (in the Mt. Esja population, these haplotypes are distinguished by the absence of the 477 bp band, indicating a second Aly8 copy, in S15, but this copy is also absent from some S1 haplotypes, see below); in a family from a different population segregating for S15, we assumed that this band represents the same Aly8 sequence as the S15-associated allele, not the S1 one.
As just described, Aly8 bands of the same size were sometimes obtained from haplotypes with different SRK alleles (particularly with primer J4Rb, specific to the long alleles). Multiple alleles of the same length were sequenced to test whether these alleles differ within or between different S-haplotypes, and sequence differences were often found between S-haplotypes (see results). These different Aly8 sequences were given a superscript, as above, or an additional letter (a, b, c, etc.) to identify them uniquely.
Sequence analysis:
Sequence differences between clones from the same S-haplotype of a single individual were treated as errors during amplification and cloning, and a consensus sequence for the Aly8 from the haplotype was deduced on the basis of all clones of the haplotype from the plant. For some individuals, however, only one clone of a given allele was sequenced, so that some of our variants may not be true differences. Since our aim is to test for associations between particular SRK and Aly8 sequences, any such errors will be conservative for our conclusions (i.e., will overestimate variability within a given S-haplotype and obscure associations).
Diversity estimates were calculated using the software DNAsp (Rozas and Rozas 1999), which was also used to test for selection (see below) and to detect gene conversion tracts. We also tested for gene conversion using GeneConv (http://www.math.wustl.edu/∼sawyer/geneconv/) (Sawyer 1989). The alignment was also used to construct neighbor-joining (NJ) trees using MEGA software (version 3, Kumar et al. 2004). When the sequences of the A. thaliana paralog of ARK3, or the Brassica ortholog, SFR2 (accession no. X98520), were included, only the exon sequences aligned with the Aly8 sequences.
RESULTS
Sequences and polymorphisms of Aly8:
Table 1 lists the Mt. Esja families initially studied and their Aly8 sequences, using the notation described in materials and methods for Aly8 allele lengths and superscripts or additional letters to distinguish between alleles with the same length but different sequences. The number of Aly8 sequences detected in individual plants ranged from one, in some individuals homozygous for the S6 allele, to at most four sequences in some individuals in family 12 (see below). Thus, no more than two copies of the locus appear to exist. All attempts to detect more alleles, including additional PCR and cloning using the alternative reverse primers, were unsuccessful.
TABLE 1.
Segregation results in five full-sib families from the Mt. Esja (Iceland) population
| Parent 1 sequences
|
||||||
|---|---|---|---|---|---|---|
| 477 | 570395 | 570399 | L419b | No. of offspring | S-allele | Conclusion |
| Family 12 (parents 00B-17/3 × 00B-17/5) | ||||||
| + | + | − | − | 28 | S1 | Parental |
| − | − | + | + | 27 | S38 | Parental |
| Parent 2 sequences
| ||||||
| 562 | 575 | L415 | No. of offspring | S-allele | Conclusion | |
| Family 12 (parents 00B-17/3 × 00B-17/5) | ||||||
| + | − | − | 19 | S14 | Parental | |
| − | + | + | 34 | S9 | Parental | |
| + | − | + | 1 | S9 | Possible recombinant? | |
| Parent 1 sequences
| ||||||
| 477 | 570395 | 570399 | L419b | No. of offspring | S-allele | Conclusion |
| Family 14 (parents 00B-17/3 × 00B-22/1) | ||||||
| + | + | − | − | 14 | S1 | Parental |
| − | − | + | + | 18 | S38 | Parental |
| + | + | + | + | 1 | S38 | Possible triploid |
| Parent 2 sequences
| ||||||
| 477
|
570395
|
L419a
|
No. of offspring
|
S-allele
|
Conclusion
|
|
| Family 14 (parents 00B-17/3 × 00B-22/1) | ||||||
| + | + | − | 14 | S1 | Parental | |
| − | − | + | 18 | S25 | Parental | |
| + | + | + | 1 | S25 | Possible triploid | |
| Parent 1 sequences
| ||||||
| 539
|
L419c
|
No. of offspring
|
S-allele
|
Conclusion
|
||
| Family 23 (parents 00B-27/3 × 00B-29/3) | ||||||
| + | − | 17 | S6a | Parental | ||
| − | + | 10 | S27 | Parental | ||
| Parent 2 sequences
| |||||
|---|---|---|---|---|---|
| 539 | 575 | L415 | No. of offspring | S-allele | Conclusion |
| Family 23 (parents 00B-27/3 × 00B-29/3) | |||||
| + | − | − | 16 | S6 | Parental |
| − | + | + | 11 | S9 | Parental |
| Parent 1 sequences
| |||||
| 572
|
L419d
|
No. of offspring
|
S-allele
|
Conclusion
|
|
| Family BM02-H2A, -H2B, and -H4B (parent 99R 14/1 × 99R 35/5) | |||||
| + | − | 9 | S12 | Parental | |
| − | + | 9 | S16 | Parental | |
| Parent 2 sequences
| |||||
| L419e | 575 | L415 | No. of offspring | S-allele | Conclusion |
| Family BM02-H2A, -H2B, and -H4B (parent 99R 14/1 × 99R 35/5) | |||||
| + | − | − | 9 | S22 | Parental |
| − | + | + | 9 | S9 | Parental |
| Parent 1 sequences
| |||||
| 477 | 570 | L419a | No. of offspring | S-allele | Conclusion |
| Family BM02-H5A and H5B (parents 99R 38/1 × parent 99R 19/2) | |||||
| + | + | − | 17 | S1 | Parental |
| − | − | + | 16 | S25 | Parental |
| + | + | + | 1 | S1 | Possible recombinant |
| Parent 2 sequences
| |||||
| 477 | 570 | 539 | No. of offspring | S-allele | Conclusion |
| Family BM02-H5A and H5B (parents 99R 38/1 × parent 99R 19/2) | |||||
| + | + | − | 11 | S1 | Parental |
| − | − | + | 23 | S6 | Parental |
For each family, parents 1 and 2 are named in the column heading, in that order, and each row shows the SRK alleles of each parent and the Aly8 sequences we infer to be in each of the S-haplotypes present in the family.
The primer combination J4F and J4R, used for PCR for the other plants, did not yield a product in the individuals homozygous for the S6 allele, but amplification using the J1R reverse primer with J4F yielded a product, which was sequenced; a primer specific to this sequence was used to genotype 27 individuals (2 failed to amplify with all primers).
In three of the haplotypes with two Aly8 copies, one sequence is a pseudogene. S9 and S38 haplotypes have long plus short (pseudogene) sequences, while in the S1 haplotypes, both sequences were short. Each of the three pseudogene sequences has a unique mutation making it nonfunctional, although all three share a deletion of 2 bp near the start of intron 1 (see Figure 3 below). The 477 (S1) and 570399 (S38) sequences have indels causing frameshift mutations, while the 575 (S9) sequence has an in-frame insertion that introduces a stop codon. Indels in exons were present only in these pseudogene sequences.
We estimated divergence values from ARK3 separately for the short (nonpseudogene) and long Aly8's, since they may be different loci (see below). For synonymous sites, these are 0.190 and 0.170, respectively (0.191 for all short sequences), somewhat higher than the average of 0.154 based on a set of 34 genes (Wright et al. 2003); net divergence values, after correcting for the high Aly8 diversity (Nei 1987), are very similar to this mean. Nonsynonymous site divergence values (Ka) for short (nonpseudogene) and long Aly8's are 0.047 and 0.045, and both sets have very similar Ka/Ks values, close to 20%. Thus the Aly8 sequences that are not evident pseudogenes show evidence of selective constraint and are probably functional, or were until recently.
Testing linkage to AlSRK:
To test Aly8 sequences for linkage to the SRK gene in A. lyrata, and to test for the presence of multiple gene copies, we studied segregation of Aly8 variants in five families from the Mt. Esja population (Table 1) and in further families from six other Icelandic populations. Using a combination of allele-specific PCR, fragment length analysis, and, where necessary, sequencing (mostly to distinguish between sequences of length 419), we identified, as far as was possible, the sequences present in each individual. Cosegregation with SRK, indicating linkage, was found, and we inferred the phase of the alleles in the haplotypes. In some families, haplotypes were not unambiguously evident, and some of our conclusions (e.g., for the Mt. Esja family 23, see Table 1) are based on consistency with haplotypes in other families; i.e., we assumed linkage disequilibrium between SRK and Aly8. This is supported by finding identical haplotypes in multiple families, including ones derived from different populations, only one of which is close to Mt. Esja (Figure 1, Table 1). The detailed data are not shown, for populations other than Mt. Esja, but the overall results are summarized in Table 2.
In total, 328 of 333 haplotypes with confirmed SRK sequences were genotyped for Aly8 in the Mt. Esja families, yielding at most three possibly recombinant haplotypes, all in plants from which DNA was not available to check the SRK sequences; for all other possible recombinant plants, rechecking ruled out recombination. Given that S-locus genotyping is difficult, due to the presence of paralogous S-domain genes, and checking is always desirable, we infer linkage between the S-locus and the Aly8 locus (or loci, in some haplotypes), as has also been shown in A. halleri (X. Vekemans and V. Castric, personal communication). Below, we briefly highlight some important points and describe the few discrepancies.
Both parents of family 12 were heterozygous for two different SRK alleles (Table 1), and all offspring had three or four Aly8 sequences. Three of the four inferred parental haplotypes carried two Aly8 copies. Of 110 progeny-inferred haplotypes in this family, there are two apparent discrepancies from the parental phases of SRK and Aly8 alleles (all others proved, on retyping, to be S-locus typing errors). The discrepant individuals apparently inherited Aly8 sequences of length 562 with S9, instead of the expected S14. However, not enough DNA was available to confirm their SRK genotypes, which may, therefore, have been mistyped. Thus these plants do not provide strong evidence for recombination between SRK and Aly8.
In family 14, one individual yielded all three Aly8 sequences present in the family and also apparently carries all three parental S-alleles. This is the one confirmed exceptional plant, but it does not suggest recombination. It could be a triploid, or the sample may be contaminated (it cannot be tested further, as the DNA is exhausted).
The fourth family group, BM02-H2A, -H2B and -H4B, confirms the phase of the S9 haplotype already inferred and establishes three new S-haplotypes: 572-S12, L419d-S16, and L419e-S22, while the fifth family, BM02-H5A (and H5B, with the same parents), includes three previously established ones (Table 1). Five progeny in this family had previously been tentatively scored as S1 homozygotes, but with some uncertainty (B. Mable, personal communication); our Aly8 sequences confirm this S-locus genotype. Two progeny (not shown in Table 1) do not fully conform to the expected patterns, but neither one strongly indicates recombination. One individual had the 477, 570395, and 539 Aly8 sequences, suggesting the SRK genotype S1S6, but only S1 is confirmed. The other has the 477 and 570395 sequences (suggesting that it carries S1) plus the L419a sequence, which, given the haplotypes inferred in this family, predicts the presence of the S25 allele, but this was not detected. This plant could be mistyped at the S-locus or possibly a recombinant.
The results, taken together, clearly support the conclusion of at most two Aly8 loci. Most S-haplotypes have only one Aly8 copy, but the three haplotypes carrying S1,S9, and S38 have two different closely linked Aly8 sequences, one of which is nonfunctional.
Linkage disequilibrium in the Mt. Esja population and conservation of haplotypes in additional populations:
The homogeneity in lengths that is seen in the Mt. Esja families just described, with the Aly8 sequence types from plants with the same SRK alleles being similar, is consistent with linkage disequilibrium between these two loci, but the sample size is small, due to the laborious linkage testing involved in inferring haplotypes. We therefore examined 20 additional individuals from the same population. Plants with known S-alleles were tested for Aly8 sequences with distinctive lengths known from the family data to be present in haplotypes with those particular S-alleles; finding the same associations of Aly8 and SRK sequences would support the hypothesis of linkage disequilibrium. For this test, we used two primer pairs, J4F and J4R, which amplify most Aly8 alleles, plus J4Rc, which amplified the sequence found in S6 haplotypes. Unlike the family analyses, the sequences were not verified by sequencing; we simply recorded presence or absence of bands of the expected lengths, and thus we do not record the long sequences seen with this set of primers, because they cannot be distinguished without sequencing. In two cases the 477 band found in the initial S1 plants was absent in a plant carrying S1. This appears to be genuine heterogeneity among S1 haplotypes, as other instances with only the 570395 band were also found in other populations (see Figure 3 below). Apart from this, 16 of the plants had the expected Aly8 bands (Table 3). The absence of an expected sequence in two plants suggests recombination in the ancestry of these haplotypes (S14 and S6). Failure to see unexpected bands in the majority of the 120 tests supports linkage disequilibrium, since each plant was tested with both primer pairs. The two instances when unexpected bands were seen do not necessarily indicate recombination, as contamination is possible. Alternatively, the 477 band in plant 99R-28/3, in which S1 has not been detected, may simply be a case of a missed S-allele, and the 539-bp band in 99R-19/1 (which has S1 and S25, but not the S6 allele), may be another instance of a plant with three SRK sequences, like those occasionally found (see above).
TABLE 3.
Aly8 sequences present in individuals with known S-alleles from the Mt. Esja population
| Individual | S-alleles detected | 477 (S1) | 539 (S6) | 562 (S14) | 572 (S12) | 575 (S9) | L415 (S9) |
|---|---|---|---|---|---|---|---|
| 99R-5/4 | S1 | +a | —b | —b | —b | —b | —b |
| 99R-9/5 | S1 S14 | +a | —b | —ac | —b | —b | —b |
| 99R-10/2 | S1 Sx | +a | —b | —b | —b | —b | —b |
| 99R-11/1 | S1 | +a | —b | —b | —b | —b | —b |
| 99R-11/3 | S6 | —b | +a | —b | —b | —b | —b |
| 99R-13/3 | S1 Sx | +a | —b | —b | —b | —b | —b |
| 99R-15/2 | S1? S25 | +a | —b | —b | —b | —b | —b |
| 99R-16/1 | S1 S6 | —a | —ac | —b | —b | —b | —b |
| 99R-18/1 | S1 Sx | +a | —b | —b | —b | —b | —b |
| 99R-19/1 | S1 S25? | +a | +bc | —b | —b | —b | —b |
| 99R-20/2 | S1 S15 | +a | —b | —b | —b | —b | —b |
| 99R-25/1 | S1 S9 | +a | —b | —b | —b | +a | +a |
| 99R-28/1 | S1S25 | +a | —b | —b | —b | —b | —b |
| 99R-28/3 | S12 | +bcd | —b | —b | +a | —b | —b |
| 99R-32/1 | S1 S25 | +a | —b | —b | —b | —b | —b |
| 99R-32/2 | S1 S25 | —a | —b | —b | —b | —b | —b |
| 99R-36/3 | S1 S22 | +a | —b | —b | —b | —b | —b |
| 99R-37/4 | S1 S9 | +a | —b | —b | —b | +a | +a |
| 99R-43/1 | S1 S25 | +a | —b | —b | —b | —b | —b |
| 99R-44/4 | S12 | —b | —b | —b | +a | —b | —b |
Each column shows results for a given sequence length and the S-allele with which the sequence was associated in the families in Tables 2 and 3. “+” denotes cases when a sequence was detected in the individual, while “—” symbols indicate that the sequence in the column was not found.
An Aly8 sequence was expected in the plant represented by the row.
Sequences that were not expected to be present.
All cases that deviate from the expected pattern, except for two plants with S1 but with no 477 band (see text).
99R-28/3 has the 477 band, but no S1 allele has been detected.
Since the individuals described so far all originated from the same population as the parental plants of the families, close relationships may account for some of the linkage disequilibrium (Wakeley and Lessard 2003). To test whether the same associations are found in other populations, we determined phases of haplotypes from five additional families, involving parents from different Icelandic populations, most at considerable distances from Mt. Esja (Figure 1). Only one plant did not behave as expected, of 194 progeny chromosomes. One haplotype classified as S1 had the L419a sequence, normally found with S25; this individual may be triploid, like the plant in family 14 (see above). The results confirm the presence of seven haplotypes with associations of Aly8 and SRK identical to those in Mt. Esja (S1, S12, S14, S16, S22, S25, and S27), plus four new haplotypes, S15 (in two families, derived from different populations), and three with L419 sequences: L419f-S11, L419g-S18 (in two different population 5 parents), and a haplotype with an unknown S-allele, denoted L419i-Sunknown (Tables 2 and 3).
Rigorous tests for linkage disequilibrium between Aly8 and the S-locus require testing whether different S-haplotypes always, or generally, carry different Aly8 alleles, and, even more importantly, whether different examples of the same S-haplotype generally carry the same Aly8 sequence. We therefore cloned and sequenced Aly8 from as many S-haplotypes from our families as possible, including the same S-haplotype from different populations, when available (supplemental Table 1 at http://www.genetics.org/supplemental/). Sequencing allowed us to increase our sample size by inferring sequences from additional plants whose SRK sequences were known, but from which progeny were not available, using reasoning similar to that in the previous section. If a plant sampled from nature carries a known SRK sequence, and we have determined the phase of haplotypes carrying this allele in one or more families, we can predict the Aly8 sequence that this haplotype should carry if the associations observed in the families are maintained in unrelated plants. If sequencing reveals the presence in the plant of an Aly8 sequence identical to the expected one, we can infer that this plant carries the previously observed SRK-Aly8 haplotype. In total, we sequenced Aly8 from 16 S-haplotypes.
One indication of linkage disequilibrium is that, if SRK and Aly8 alleles are indeed associated over long evolutionary times, haplotypes with different SRK alleles should always have different Aly8 alleles. The two haplotypes from the United States studied by Kusaba et al. (2001) were therefore added to the S-haplotypes tested. The physically longer Sb is equivalent to allele S20 of Schierup et al. (2001) and has a long Aly8 sequence of 824 bp, while the Sa (equivalent to allele S13 of Schierup et al. 2001) has a short Aly8 sequence (557 bp). No genomic DNA was available from the plant from which the BAC clones were made, so only the clones containing the S-locus were studied, but independent plants with the S13 and S20 alleles were studied (98E17/10 and BM04-A1/7 provided by Barbara Mable or already available in the lab), originating from the same U.S. population (Charlesworth et al. 2003b). These yielded the same band lengths and sequences as those from the BAC clones, indicating that no second Aly8 allele is present in the genome, outside the cloned region. Thus, only the three S-haplotypes already mentioned carry two Aly8 sequences. The S13 and S20 haplotypes were found to carry different Aly8 alleles, each differing at multiple sites from other sequences, and they are again homogeneous in independent individuals. The S37 haplotype, corresponding to the A. thaliana haplogroup A (Shimizu et al. 2004), also differed at several (five) sites from the most similar other Aly8 sequence.
Nine S-haplotypes appeared in families from at least two populations (1, 9, 12, 14, 15, 16, 22, 25, and 27), including S9 (Kuittinen et al. 2004). Three more were found in multiple families from the same population (S6 and S38 from Mt. Esja and S18 from population 5); multiple copies of these were not sequenced from the same population, as finding the same associations in different populations is much better evidence of linkage disequilibrium (LD), but the Aly8 lengths were the same in each of the pairs with the same SRK and different between haplotypes with different SRK sequences. In addition, we sequenced S11 from two different Icelandic populations (5 and 1, with the phase established only from the former). Despite repeated efforts with the available reverse primers, no full-length S22 sequence was obtained. Thus, in total, 9 S-haplotypes (Table 2 and Figure 3) could be tested for homogeneous associations between populations, the strongest test for linkage disequilibrium, and 11 could be tested for the presence of different Aly8 sequences in haplotypes with different SRK alleles. Ones with the same SRK allele generally had very similar or identical Aly8 sequences, even when they originated from different populations. Most haplotypes contained only one or two sequence variants, including indels (Figure 3).
There are two major exceptions to this uniformity of sequences sharing the same SRK allele. The S9 haplotype in the mapping family, like the Mt. Esja S9 haplotype, has two copies of the Aly8 gene. The short (pseudogene) sequences are almost identical (two differences), but the lengths and sequences of the long copies differ. The Mt. Esja S9 haplotype Aly8 allele differs from the sequence from the mapping family allele by 24 SNP variants, many more than within any other haplotype; the Mt. Esja sequence is also 4 bp shorter than the other L sequences (it has a length of 415, see Figure 4). The S1-linked Aly8 sequences also vary. In the Mt. Esja families and the A. lyrata mapping family (from distant populations), all haplotypes with the SRK S1 allele carried 477 (pseudogene) and 570395 Aly8 sequence types (Table 2). However, the S1 haplotype in a family from a different Icelandic population, 15, had no 477 sequence, and other Mt. Esja plants also carry S1 without this sequence (Table 3 and Figure 3). The S1-linked sequences of the 570-bp band also differ, mainly because of differences between S1 haplotypes with two Aly8 copies vs. only one (Figure 3), but there were also 6 variants among the four lacking it; 4 of these variants are shared with Aly8 sequences from other S-haplotypes, suggesting exchange events (see also below). The S1 haplotypes with the 477 sequence (from five different populations) include no variants and may therefore be derived from the haplotype lacking it.
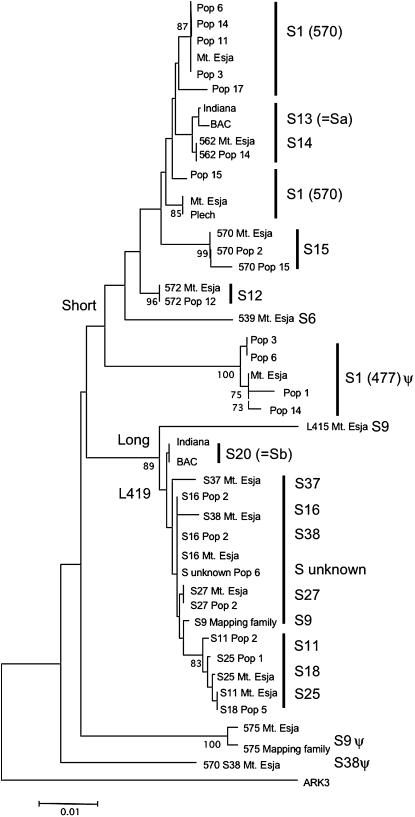
Figure 4.
NJ tree including all sequenced Aly8 alleles, with the sequence of the A. thaliana ortholog, ARK3 from the Col-0 strain of A. thaliana (GenBank accession no. AL161555) as an outgroup. The populations from which the sequences were derived are indicated, and the Aly8 sequence lengths and the SRK allele numbers are shown. The S1, S9, and S38 haplotypes all have two copies of Aly8 (Bechsgaard 2002). The analysis used all single-nucleotide variants in the alignment to estimate Kimura two-parameter distances, using the pairwise deletion option. Bootstrap values (based on 1000 bootstraps) are shown for nodes with values >66%. The BAC clones of alleles S13 and S20 were kindly provided by June Nasrallah (see materials and methods), and the other plants carrying these two SRK alleles were from plants grown from a natural population from Indiana (see materials and methods).
Homogeneity within haplotypes with a given SRK is thus not complete. Clusters of Aly8 sequences according to the haplotype's SRK allele are nevertheless clearly seen in the NJ tree (Figure 4). There is often good bootstrap support, even among the short sequences, which are only ∼500 bp long, but less often among the long sequences, which differ less. In either case, resolution is quite surprising. The numbers of differences between Aly8 sequences from the same S-haplotype are overestimates. Some variants within S-haplotypes are confirmed by sequencing multiple clones, but, for some haplotypes only one or a few clones were sequenced, for example, L419a-S25, and the variation detected may be due to PCR error (as was frequently observed when several clones from the same individual were sequenced). Note, however, that most such variants are also seen in other samples.
Tests for recombination or gene conversion:
If the SRK and Aly8 loci recombine, even occasionally, the same Aly8 sequence should occur in haplotypes with entirely different SRK alleles. We found only one clear instance of this. Among the set of S-haplotypes with similar long Aly8 alleles S11, S18, and S25 (Figures 3 and 4), the sequence from the S18 haplotype of population 5 was identical to the S11 haplotype sequence from Mt. Esja; the population 5 S11 Aly8 was sequenced from a single clone, and the single difference may be a PCR error. The S11 and S18 Aly8 sequences may differ elsewhere in the gene, but the similarity between them in the portion studied suggests recombination between these two haplotypes, whose SRK sequences are not closely similar and belong to different dominance classes (Mable et al. 2004). S22 may be another case, but is not shown in Figure 4, because only part of the S22-linked Aly8 was sequenced (with the J4Rb primer); this was identical to the Aly8 sequence from the S18 and S25 haplotypes, but it must differ in the primer site, at least, since it does not amplify with the J4R primer.
Small-scale exchanges may also have occurred between haplotypes. We tested for gene conversion between haplotypes with different S-alleles, using the test of Betran et al. (1997) implemented in DNAsp. Even between the long and short sequence types, the test detects several conversion events (boxed in Figure 3; the tests require at least three individuals in each sequence set, so few individual haplotype pairs could be tested). GeneConv detects a single event spanning a region of 218 bp, from the first to the last of the regions detected above (P = 0.0324, after correction for multiple tests).
Assigning Aly8 sequences to loci and estimating diversity:
The S-locus is subject to frequency-dependent selection, leading to high polymorphism. Given that Aly8 loci are close to the S-locus, probably with a very low recombination rate, we expect its diversity to be high, as explained in the Introduction. One goal of our work was to estimate diversity of the Aly8 genes, to test this prediction. However, to estimate diversity we need to establish how many loci are present and which sequences are allelic and which paralogous (i.e., to assign alleles to their respective loci) or else quantify the rate of genetic exchange of sequences between the two loci (Innan 2002).
It is unclear whether the long and short Aly8 sequence types represent different loci or whether the nonpseudogene short and long sequences are allelic. The A. thaliana ARK3 sequence of the Col-0 strain shares most of the large insertion (∼270 bp in the first intron) with the long Aly8 alleles, suggesting that the long state is ancestral and the short state derived within A. lyrata. Three smaller indels, plus a single fixed nucleotide difference, also distinguish the long sequences from all the short ones, and these are not found in ARK3 (in the region containing the third indel, the ARK3 sequence has a larger deletion with respect to the Aly8 sequences). We cannot obtain any information about the ancestry of the long and short sequences from the putatively orthologous B. oleracea SFR2 sequence, because only exon sequence could be aligned, precluding analysis of the indels, which are in the first intron.
To try to determine whether the nonpseudogene short and long Aly8 sequences are allelic, we estimated FST between the two sequence types. The values are high (0.67 for all sites and 0.55 for silent sites or slightly less if the duplicate, pseudogene sequences are included). However, these values are not based on random samples from the populations, as our sample includes at least two of many S-haplotypes; FST values estimated from just one of each nonpseudogene S-haplotype are much lower (<0.125 for all site types), but the NJ gene tree nevertheless clearly clusters the long sequences (Figure 4). In contrast, sequences from haplotypes with two copies, S38 (L419b and 570399), S9 (L415 and 575), and S1 (570395 and 477) do not suggest a distinct clade of haplotypes with these sequences, although they share the small deletion noted above, near the start on intron 1.
The long/short separation does not, however, necessarily imply that these sequences represent different loci. As in other cases with paralogous genes where there is diversity (Bosch et al. 2004), it is difficult to draw firm conclusions about which Aly8 sequences are allelic. The synonymous site divergence between the two functional sequence types in our sample is 5.4% (average number of single-nucleotide differences 17.9); excluding the pseudogene sequences, only 5 are fixed differences, plus at most three indels (Figure 3). However, the lack of fixed differences is largely due to high polymorphism within each set of sequences. Synonymous site diversity (πs) estimated from all the nonpseudogene sequences in our sample is 3.9% (3.7% within the short set of sequences and 1.4% for just the more uniform long ones); a similar, although smaller, difference is seen for all silent sites. The distinctiveness of the two types is thus overestimated by considering fixed differences, because, with our fairly small numbers of sequences, some polymorphisms will be missed. We tentatively conclude that they probably represent highly diverged alleles (with the pseudogene sequences being generated by occasional duplications in diverse S-haplotypes). Unequal crossing over generating haplotypes with different copy numbers after an initial duplication seems to be excluded, because the duplicates are each very distinctive in sequence (see Figure 4).
Consistent with the long and nonpseudogene short sequences being alleles, the evidence from diversity supports the conclusion above that both sequence sets are subject to selective constraint. πa/πs values are low (0.169 for long and 0.103 for short sequences or 0.125 for both sets of sequences combined). These are similar to the Ka/Ks value for divergence from ARK3 (see above) or between long and short sequences (0.117 using nonpseudogenes and 0.173 for all short sequences). These values suggest selective constraint, not balancing selection that could account for the high diversity. Tajima's D-values (not shown) were negative, but did not differ significantly from zero for the long or the short sequence sets or the combined set, so there is also no evidence for a selective sweep that could have been caused by recent spread of a duplicate copy. Fu and Li's tests (using ARK3 as an outgroup) were also nonsignificant. Haplotype tests were also nonsignificant and are discussed below. The pseudogene sequences had higher polymorphism, with πa- and πs-values of 0.064 and 0.027, and an elevated πa/πs of 0.42 (for all site types, π was 0.051 for the pseudogene sequences vs. 0.024 for the others), suggesting that they arose long ago, consistent with their wide distribution in the tree (Figure 4), and have accumulated sequence differences in addition to those causing loss of function (see above).
DISCUSSION
Tight linkage of Aly8 gene copies:
Absence of recombination between the pollen and pistil expressed S-genes is a prerequisite for keeping the incompatible S-allele pairs together in functional haplotypes (e.g., Casselman et al. 2000). Given the considerable rearrangement in the S-locus region (Kusaba et al. 2001), it is plausible that recombination is repressed across the whole region, but no independent evidence has so far been obtained. No recombination was detected between several genes close to SRK in B. oleracea families (Casselman et al. 2000). However, the four completely linked genes were located within only ∼40 kb, which, with the estimated recombination rate in A. lyrata of between ∼400 and 600 kb/cM (Wright et al. 2003) or even with the higher rate in A. thaliana (∼200 kb/cM for noncentromere regions, see Copenhaver et al. 1998), would require a family larger than the 250 plants studied to accurately estimate crossing over. A population genetic approach may be more likely to detect recombination, especially in the adjoining genome regions (Takebayashi et al. 2004).
To use such an approach, however, we must understand the genetic situation of Aly8 sequences. Previous work on Aly8 (Charlesworth et al. 2003b) did not establish the number of copies or whether more than one is linked to SRK, although there was some evidence of linkage disequilibrium between Aly8 and SRK. We now conclude that Aly8 is closely linked to the S-locus and, furthermore, that some haplotypes exist with two copies and there is no unlinked copy in the genome. The situation for Aly8 is thus similar to that sometimes found for S-loci; duplicates of both SCR and the S-linked SLG loci have been found in some S-haplotypes (Cabrillac et al. 1999; Kusaba et al. 2001). Interestingly, the three instances of haplotypes with two copies seem to have arisen independently. Whether the long and short sequences are allelic or not, we can use the Aly8 sequence types to test for long-term associations with SRK alleles.
Linkage disequilibrium between Aly8 and SRK:
Our association results suggest that recombination between Aly8 and the S-locus is rare over evolutionary time (as well as in the single generation tested in our families). Previous studies of the SRK gene sequences of a set of different S-haplotypes (either typed by pollination tests or inferred from the S-domain sequences to represent different S-alleles) showed extreme homogeneity of the kinase domains within each S-haplotype, whereas different alleles differ greatly. This suggests that little or no recombination occurs within the S-locus, across a physical distance of up to 3 kb (Charlesworth et al. 2003a). This was confirmed by the finding that linkage disequilibrium does not decrease with increasing distances between sites in the sequence and also that silent site diversity is as high at sites in the kinase domain as in the S-domain, which is the presumed location of the sites under balancing selection (Charlesworth et al. 2003a).
The results presented here for the more distant Aly8 gene, showing very similar Aly8 sequences in independent haplotypes with the same SRK, across several different A. lyrata populations, are similar to those for the SRK locus itself, but across a distance of at least several kilobases from SRK (Kusaba et al. 2001). Only occasional variants were found within haplotypes. Evidently the same S-haplotypes are maintained in different Icelandic populations and also in the few populations from other geographic regions from which we have samples. The fact that new Aly8 sequences are found, whenever we apply our typing method to A. lyrata plants with S-alleles different from ones already sequenced, also strongly suggests fairly long-term linkage disequilibrium between Aly8 and the S-locus. Thus Aly8 and the S-locus are not only linked, but also in linkage disequilibrium. Note that using SRK sequences in lieu of knowing that the haplotypes carry functionally different S-alleles is conservative. If SRK sequences differ among alleles of the same S-allele type, associations will be obscured.
It will be interesting to study the geographic scale of SRK-Aly8 associations further, to better estimate the timescale over which they are maintained. It is also not yet possible to compare our results with LD elsewhere in the A. lyrata genome, because no good data yet exist on genetic and physical maps for this species. Results from five nuclear loci within the Mt. Esja population alone suggested LD higher than expected with recombination rates per base pair as high as in A. thaliana (Wright et al. 2003). This could be due to low recombination in A. lyrata, but, until additional populations have been studied, we cannot exclude other plausible causes of LD, such as a bottleneck in the population's history. Data on recombination rates on four A. lyrata chromosomes suggest a recombination rate per megabase for noncentromere regions similar to that in A. thaliana or slightly lower (Hansson et al. 2006).
We did not attempt to analyze linkage disequilibrium explicitly or to quantify it, for several reasons. First, the duplication prevents our determining which Aly8 sequences are allelic. The distances between the Aly8 loci are also unknown. It is not reasonable to estimate LD between sites that may not be in alleles (they might fail to recombine simply because they are at different positions in the chromosome, yet the region around them might recombine, and it is recombination in the region, not within the Aly8 gene, that we wish to test). Second, the length of sequence is short, after excluding sites that are absent from the short sequences, plus gaps in the alignment between these very different sequences; this means that we have low ability to detect a decline in LD with distance between sites and could not conclude that recombination is absent, merely because no decline is detected.
Third, even just within SRK, we previously found low LD within either the S- or kinase domains, despite very clear evidence (reviewed above) for distinctive differences maintained by different S-alleles throughout the SRK sequences, over long evolutionary times (Charlesworth et al. 2003a). LD may be obscured by gene conversion between different S-alleles or because divergence times between alleles are very long, so that identical mutations may have occurred in different haplotypes; in either case, variants are expected to be shared between haplotypes, and this is indeed seen in SRK sequences (between 12 alleles analyzed, over a length of 782 bp of the highly polymorphic S-domain, 222 of the 496 variable sites are shared variants found in 2 or more S-alleles) and in Aly8 (between 16 S-haplotypes analyzed, of length 525 bp, excluding indels, 27 of the 50 variable sites were shared between different haplotypes).
A final, more subtle, reason for not quantifying linkage disequilibrium is that we have at present sequenced only a few instances of each S-haplotype. To establish Aly8 sequences of haplotypes and to have material for testing for associations, we aimed to get as many different haplotypes as possible, as well as to obtain sequence Aly8 from multiple haplotypes with the same SRK sequence. Thus our present sample is not a random population sample. The same is true for many sequence data sets from the S-loci themselves, where the emphasis has generally been on sequencing as many different functional allelic types as possible. Diversity is thus overestimated, although there may also be a possibility of underestimation due to failure of PCR primers to amplify some particularly divergent sequences. In our sample of Aly8 alleles, the number of haplotypes is consistent with neutral expectations assuming no recombination, given the number of SNPs in our nonpseudogene sequences (using coalescent simulations implemented in DNAsp), but is not significantly lower than expected with even a small amount of recombination (R per gene = 1); thus this test does not exclude a low recombination rate. However, if we include two copies of each S-haplotype (i.e., assuming that our observed low diversity within S-haplotypes would hold for haplotypes for which we currently have only one sequence), the data exclude even a low recombination rate. Clearly, therefore, more S-haplotypes must be investigated in the future.
A different type of LD test is to compare estimates of nucleotide diversity within and between S-haplotypes, defined by their SRK sequences. A very small recombination rate will lead to diversity within allelic classes being very similar to the diversity in the full sample (Charlesworth et al. 1997; Innan and Tajima 1999), just as rare migration in an island model of migration between demes leads to within-deme diversity being almost the same as total diversity (reviewed in Charlesworth et al. 2003). For the 11 S-haplotypes for which we have multiple Aly8 sequences, the mean within-haplotype diversity (πA of Charlesworth et al. 1997) is 0.0036 for all site types, including the two very different S9 haplotypes, while the diversity for our full Aly8 sequence sample (excluding the pseudogene sequences), πT, is six times higher, 0.022. We estimated the analog of FST between all pairs of such haplotypes, and the value is 0.965 for all site types. FST is 0.865 for all short nonpseudogene sequences and 0.798 for just the long ones, excluding S9 haplotypes, or 0.346 including these sequences (in all cases, the haplotypes are highly significantly differentiated using the Ks test of Hudson et al. 1992). The high isolation between different S-haplotypes is evidence that LD is strong, and the FST value estimates a quantity σd2 (Charlesworth et al. 1997; Innan and Tajima 1999) that is closely similar to the LD measure r2 (McVean 2002).
Do exchanges occur between different S-haplotypes?
Among all the sequences from different haplotypes, we found only one instance of the same Aly8 sequence associated with different S-haplotypes (an Aly8 sequence found in an S18 haplotype is identical with one of the very similar sequences found in an S11 haplotype). More such cases would be expected if the SRK and Aly8 loci recombine, even occasionally. However, there is some evidence for smaller-scale exchanges (see Figure 3). Taken together, our results suggest that linkages of particular Aly8 sequences and SRK alleles are maintained over long enough periods of time for migration to occur between different populations. Because (as mentioned above) associations will not persist unless recombination is very infrequent, this suggests that low recombination may extend to a wide region surrounding the S-locus. The S-region is, however, known to vary in both length and gene order between the only two haplotypes yet studied in detail (our S13 and S20, equivalent to Sa and Sb of Kusaba et al. 2001). It will be important in the future to determine physical distances in the S-locus region and also to further develop the necessary theory to predict the extent of the region in which LD is expected and the expected magnitude of FST between haplotypes at different recombination distances.
Linkage across the S-locus region is, however, clearly not complete on an evolutionary timescale, since silent site diversity in the kinase domain of SRK is much higher than at Aly8 or the other flanking loci studied (Kamau and Charlesworth 2005), including the B80 gene (another gene in the region flanking the S-loci; see Kusaba et al. 2001). Recombination is also suggested by the results for the S-locus region in A. thaliana. Although this species is self-compatible, the orthologous region is still present (Kusaba et al. 2001). High diversity was found for the homologs of both Aly8 (ARK3 in A. thaliana) and B80 (a U-box gene also located in the region flanking the S-loci; see Kusaba et al. 2001). These genes appear to mark the boundaries of a region affected by a selective sweep at the pollen S-locus, SCR1, which may have been the site of the mutation causing loss of incompatibility in this species (Shimizu et al. 2004). On this interpretation, crossing over must be frequent enough to allow SCR1 to recombine with ARK3 and the U-box gene, so that allelic diversity at these loci remained very high.
Occasional recombination or other exchange events may also account for why the Aly8 sequence is not completely uniform within S-haplotypes (Figure 3). Even if crossing over is infrequent near the S-locus, gene conversion might still occur, as in low-recombination regions of Drosophila melanogaster (Langley et al. 2000; Jensen et al. 2002), and could homogenize parts of the alleles' sequences. However, the large fixed insertion in intron 1 may limit the frequency of gene conversion, allowing the long and short sequences to diverge. Our observation (see above) that the apparently derived short Aly8 sequences (even just those that are not pseudogenes) have at least as high diversity as the ancestral long ones supports the possibility that exchanges occur, and explicit tests for gene conversion are consistent with this (see Figure 3).
However, the same S-haplotype is rarely found associated with different Aly8 sequences. The S1 haplotype, where some variants are seen, corresponds to a very recessive S-allele (Mable et al. 2004). Recessive S-alleles are expected to persist in populations for longer than more dominant ones (Schierup et al. 1998; Uyenoyama 2000), and so there will have been more time during which mutation and recombination events can have occurred in their ancestry, compared with other alleles. Our result that the S1 haplotype is less homogeneous in its Aly8 sequences than other haplotypes is therefore consistent with this theoretical expectation, but recombination is not necessary to generate this variability. However, the S9 haplotypes, which differ much more in their Aly8 sequences, are in a higher S-allele dominance class (Mable et al. 2004).
Another expectation under our interpretation of long-term LD between the Aly8 and SRK loci is that Aly8 sequence clades should be congruent with those of SRK. Four different groups of SRK sequences have been distinguished (Mable et al. 2004), corresponding to SRK alleles with different dominance levels. However, we find no strictly congruent clustering of Aly8 sequences with the four SRK groups. There is also no correspondence between SRK sequence types and the number of Aly8 copies and therefore no suggestion of different physical arrangements of the S-loci and Aly8, although this should be studied explicitly in the future. Overall, these results suggest that Aly8 is sufficiently distant from SRK and SCR that exchanges have occurred over the long evolutionary times during which the S-alleles have been maintained.
Aly8 diversity:
Our results show that the high diversity of Aly8 sequences is largely, but not entirely, due to the presence of the two different sequence types. It seems clear that there are at most two loci, probably due to a tandem duplication in some haplotypes. We cannot definitively exclude the possibility that all haplotypes have two copies and that the second copy does not amplify with the primers we used; this, however, seems unlikely, because three different primer combinations all failed to amplify any new sequence types in plants that seem to have just one gene copy. If many haplotypes carry copies undiscovered due to variation in our primer sites, we should also score some haplotypes with no copies amplifying, which never happened. Moreover, Kusaba et al. (2001) found only single Aly8 copies in the two haplotypes they studied, and we confirmed this for the genomes of independent plants with these SRK alleles.
The uncertainty about the allelic status of the Aly8 genes does not affect our inferences about associations with SRK alleles, but it does impede our ability to estimate diversity. Unequal crossing over between tandem duplicates might generate haplotypes with different copy numbers, and exchanges of sequence may occur. Many such cases are known, including A. thaliana disease resistance gene clusters (Baumgarten et al. 2003) and the human Rhesus blood group D and E genes (Innan 2003), and there is evidence that exchanges between duplicates are common in yeast (Gao and Innan 2004). If allelic and nonallelic sequences can be reliably distinguished, as is often the case, diversity can be estimated. If, however, a distinction is not possible, rates of exchange can sometimes be estimated, if they are not high, and used to obtain diversity estimates (Innan 2002).
In the case of Aly8, exchanges may be frequent, relative to the age of the S-haplotypes, and the haplotypes do not all carry two copies, so that the exchange parameters cannot be estimated. If the short and long nonpseudogene sequences are allelic, nucleotide diversity can be estimated. As explained above, we have not yet studied random samples of S-haplotypes from natural populations, since our main goal was to study associations with SRK alleles. Given that there are many S-alleles in A. lyrata populations (Charlesworth et al. 2003b; Mable et al. 2003), such samples will consist largely of different alleles. Diversity estimated from the nonpseudogene Aly8 sequences of one of each S-haplotype is 5% for synonymous sites (4% for silent sites); a somewhat lower value is obtained if we use our sample with multiple instances of each S-haplotype (πs = 3.9%, πsilent = 3.7%). If the sequences are two distinct loci, not alleles, this overestimates the diversity at each locus. A crude underestimate, assuming free “migration” of sequences between two loci, i.e., taking the effective size for each locus to be at most twice that for other loci in the species, would halve our diversity estimates. If one sequence type is much more abundant than the other, this overcorrects the effect on diversity; however, our data include roughly equal numbers of the two sequence types. Even this Aly8 diversity estimate above is about twice as high as that for reference loci in this species (Wright 2003; Ramos-Onsins et al. 2004), suggesting that diversity is elevated due to linkage to the S-loci (consistent with our evidence for long-term associations). All diversity values are higher if the pseudogene copies are included as alleles (see results).
Overall, Aly8 thus shows some signs of elevated diversity compared with results from other loci in this species, which average ∼1% for silent sites (Wright et al. 2003). It is also consistent with a high Aly8 diversity that there are several indels among the different Aly8 sequences in the exon part of the sequence, but no fixed indel differences between A. thaliana and the Aly8 consensus sequence. Aly8 diversity is not, however, extremely high and is much lower than that for the A. lyrata SRK locus (Charlesworth et al. 2003b). This adds support to the evidence from the apparent gene conversion events in our sequences that some form of genetic exchange between haplotypes occurs in the region. This conclusion is similar to that for other genes in the region (Kamau and Charlesworth 2005).
If the high Aly8 diversity is caused by its linkage to the S-loci, we would expect ARK3 in A. thaliana, a plant lacking SI responses, to be much less polymorphic. Diversity of ARK3 has recently been studied, and its synonymous site diversity was estimated to be 0.0316 (Shimizu et al. 2004). This is much higher than the average for A. thaliana loci, for which mean intron or fourfold degenerate site specieswide diversity is estimated to be ∼0.008, with a large range (Nordborg et al. 2005), and, surprisingly, it is almost as high as that for the A. lyrata Aly8 loci or even slightly higher if our correction for duplicate gene copies is appropriate. However, the regions of the ARK3 sequence (5′ noncoding plus part of exon 1) studied do not overlap that studied in A. lyrata, so the comparison is only rough. Another large set of ARK3 sequences does partially overlap (C. Tang, personal communication), and these are also polymorphic. This is probably explained by recent loss of incompatibility in A. thaliana (Shimizu et al. 2004). It will be interesting in the future to compare the complete region in the two species, to understand whether A. thaliana has high diversity throughout the region or whether diversity is highly heterogeneous and is high only in the 5′ region. There is a large disease resistance gene cluster near the S-loci on A. thaliana chromosome 4 (Baumgarten et al. 2003), and it is possible that this may include highly polymorphic loci that affect diversity at genes in this region in A. thaliana.
Conclusions:
If linkage disequilibrium between Aly8 and SRK genes is indeed maintained for long enough to be consistent in different A. lyrata populations, it may be possible to use Aly8 typing to estimate frequencies of S-haplotypes, rather than using the S-locus sequences themselves, which are so polymorphic that obtaining reliable PCR amplification from all alleles present in populations, and getting sequences, is very difficult, and errors are frequent. Some SRK alleles do not amplify with currently available SRK primers, yielding alleles classified as “unknown”; with flanking loci, these can potentially be distinguished from one another, allowing haplotypes to be classified without obtaining SRK or SCR sequences, which currently requires screening genomic libraries for clones containing the S-loci (Kusaba et al. 2001). Aly8 typing can also help to confirm hypotheses from PCR–RFLP results, currently used to suggest which allele-specific primers are most promising for attempting to determine S-allele sequences present in individual plants (Schierup et al. 2001; Bechsgaard 2002; Mable et al. 2004). This approach should be helpful for testing the predictions of theoretical models about allele frequencies in natural populations (e.g., Schierup 1998; Uyenoyama 2000; Muirhead 2001), which requires ascertaining the haplotypes of large numbers of plants. For example, SRK alleles show slight population structure and low FST (Charlesworth et al. 2003b), as predicted for a locus under balancing selection (Schierup et al. 2000b), and our results suggest that this is also true for Aly8. Current data cannot yet test this rigorously, but it should be tested in the future.
This approach may not be limited to the species studied here or to the sporophytic incompatibility system of Brassicaceae. Genes closely linked to the S-loci have now been identified in many self-incompatible plants. Diversity studies of S-alleles and flanking loci genes will be a first kind of valuable evidence; if there is linkage disequilibrium, these loci should have higher diversity than other loci in the genome (Charlesworth et al. 1997; McVean 2002), because both the linkage disequilibrium and the diversity in these situations reflect divergence between alleles associated over long evolutionary times with functionally different S-alleles. More data from natural population studies of diversity in S-locus regions of other plant genomes are thus needed.
Acknowledgments
We thank B. K. Mable (University of Glasgow) for DNA samples from families and for help with SRK typing, June Nasrallah (Cornell University) for BAC clones of Sa and Sb SRK haplotypes, Xavier Vekemans and Vincent Castric (University of Lille) for information about linkage of the A. halleri Aly8 and SRK orthologs, Chunlao Tang (University of Southern California) for ARK3 sequence data, and the Natural Environment Research Council of the United Kingdom for funding.
References
- Baumgarten, A., S. Cannon, R. Spangler and G. May, 2003. Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard, J., 2002. Population genetic dynamics of homomorphic self-incompatibility systems: evidence of different selection pressures on the different self-incompatibility alleles above that of frequency-dependent selection. Masters Thesis, University of Aarhus, Aarhus, Denmark.
- Bechsgaard, J., T. Bataillon and M. H. Schierup, 2004. Uneven segregation of sporophytic self-incompatibility alleles in Arabidopsis lyrata. J. Evol. Biol. 17: 554–561. [DOI] [PubMed] [Google Scholar]
- Betran, E., J. Rozas, A. Navarro and A. Barbadilla, 1997. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, E., M. E. Hurles, A. Navarro and M. A. Jobling, 2004. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 14: 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrillac, D., V. Delorme, J. Garin, V. Ruffio-Chable, J.-L. Giranton et al., 1999. The S15 self-incompatibility haplotype in Brassica oleracea includes three S gene family members expressed in stigmas. Plant Cell 11: 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselman, A. L., J. Vrebalov, J. A. Conner, A. Singhal, J. Giovanni et al., 2000. Determining the physical limits of the Brassica S-locus by recombinational analysis. Plant Cell 12: 23–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., M. Nordborg and D. Charlesworth, 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided inbreeding and outcrossing populations. Genet. Res. 70: 155–174. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., D. Charlesworth and N. H. Barton, 2003. The effects of genetic and geographic structure on neutral variation. Annu. Rev. Ecol. Evol. Syst. 34: 99–125. [Google Scholar]
- Charlesworth, D., C. Bartolomé, M. H. Schierup and B. K. Mable, 2003. a Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol. Biol. Evol. 20: 1741–1753. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., B. K. Mable, M. H. Schierup, C. Bartolomé and P. Awadalla, 2003. b Diversity and linkage of genes in the self-incompatibility gene family in Arabidopsis lyrata. Genetics 164: 1519–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G. P., W. E. Browne and D. Preuss, 1998. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer, K. G., M. K. Kandasamy, D. I. Mahosky, J. Axxiai, B. I. Kudish et al., 1994. A superfamily of S locus-related sequences in Arabidopsis: diverse structures and expression patterns. Plant Cell 6: 1829–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L.-z., and H. Innan, 2004. Very low gene duplication rate in the yeast genome. Science 306: 1367–1370. [DOI] [PubMed] [Google Scholar]
- Hansson, B., A. Kawabe, S. Preuss, H. Kuittinen and D. Charlesworth, 2006. Comparative gene mapping in Arabidopsis lyrata chromosomes 1 and 2 and the corresponding A. thaliana chromosome 1: recombination rates, rearrangements and centromere location. Genet. Res. 87: 75–85. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151. [DOI] [PubMed] [Google Scholar]
- Innan, H., 2002. A method for estimating the mutation, gene conversion and recombination parameters in small multigene families. Genetics 161: 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., 2003. A two-locus gene conversion model with selection and its application to the human RHCE and RHD genes. Proc. Natl. Acad. Sci. USA 100: 8793–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., and M. Nordborg, 2003. The extent of linkage disequilibrium and haplotype sharing around a polymorphic site. Genetics 165: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., and F. Tajima, 1999. The effect of selection on the amounts of nucleotide variation within and between allelic classes. Genet. Res. 73: 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidore, E., H. van Os, S. Andrzejewski, J. Bakker, I. Barrena et al., 2003. Toward a marker-dense meiotic map of the potato genome: lessons from linkage group 1. Genetics 165: 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, M. A., B. Charlesworth and M. Kreitman, 2002. Patterns of genetic variation at a chromosome 4 locus of Drosophila melanogaster and D. simulans. Genetics 160: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, N., G. Suzuki, M. Watanabe, A. Isogai and K. Hinata, 2001. Sequence comparisons among dispersed members of the Brassica S multigene family in an S-9 genome. Mol. Genet. Genomics 265: 526–534. [DOI] [PubMed] [Google Scholar]
- Kamau, E., and D. Charlesworth, 2005. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr. Biol. 15: 1773–1778. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., A. A. D.. Haan, C. Vogl, S. Oikarinen, J. Leppälä et al., 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and Arabidopsis thaliana. Genetics 168: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Kusaba, M., K. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative, A. thaliana. Plant Cell 13: 627–643. [PMC free article] [PubMed] [Google Scholar]
- Langley, C. H., B. P. Lazzaro, W. Phillips, E. Heikkinen and J. M. Braverman, 2000. Linkage disequilibria and the site frequency spectra in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics 156: 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable, B. K., M. H. Schierup and D. Charlesworth, 2003. Estimating the number of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90: 422–431. [DOI] [PubMed] [Google Scholar]
- Mable, B. K., J. Beland and C. D. Berardo, 2004. Inheritance and dominance of self-incompatibility alleles in polyploid Arabidopsis lyrata. Heredity 93: 476–486. [DOI] [PubMed] [Google Scholar]
- McVean, G. A. T., 2002. A genealogical interpretation of linkage disequilibrium. Genetics 162: 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muirhead, C. A., 2001. Consequences of population structure on genes under balancing selection. Evolution 55: 1532–1541. [DOI] [PubMed] [Google Scholar]
- Nasrallah, J. B., 2000. Cell-cell signaling in the self-incompatibility response. Curr. Opin. Plant Biol. 3: 368–373. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nordborg, M., B. Charlesworth and D. Charlesworth, 1996. Increased levels of polymorphism surrounding selectively maintained sites in highly selfing species. Proc. R. Soc. Lond. Ser. B. 163: 1033–1039. [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, Y. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia, M., R. Swarup, A. Rocher, P. Saindrenan, D. Roby et al., 2002. Comparison of the expression patterns of two small gene families of S gene family receptor kinase genes during the defence response in Brassica oleracea and Arabidopsis thaliana. Gene 282: 215–225. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., B. E. Stranger, T. Mitchell-Olds and M. Aguadé, 2004. Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics 166: 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3.0: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Sawyer, S. A., 1989. Statistical test for determining gene conversion. Mol. Biol. Evol. 6: 526–538. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., 1998. The number of self-incompatibility alleles in a finite, subdivided population. Genetics 149: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and F. B. Christiansen, 1998. Allelic genealogies in sporophytic self-incompatibility systems in plants. Genetics 150: 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and D. Charlesworth, 2000. a The effect of hitch-hiking on genes linked to a balanced polymorphism in a subdivided population. Genet. Res. 76: 63–73. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and D. Charlesworth, 2000. b The effect of subdivision on variation at multi-allelic loci under balancing selection. Genet. Res. 76: 51–62. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., B. K. Mable, P. Awadalla and D. Charlesworth, 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K. K., J. M. Cork, A. L. Caicedo, C. A. Mays, R. C. Moore et al., 2004. Darwinian selection on a selfing locus Science 306: 2081–2084. [DOI] [PubMed]
- Suzuki, G., M. Watanabe and T. Nishio, 2000. Physical distances between S-locus genes in various S haplotypes of Brassica rapa and B. oleracea. Theor. Appl. Genet. 101: 80–85. [Google Scholar]
- Takebayashi, N., E. Newbigin and M. K. Uyenoyama, 2004. Maximum-likelihood estimation of rates of recombination within mating-type regions. Genetics 167: 2097–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama, M. K., 2000. Evolutionary dynamics of self-incompatibility. Genetics 156: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans, X., and M. Slatkin, 1994. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics 137: 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley, J., and S. Lessard, 2003. Theory of the effects of population structure and sampling on patterns of linkage disequilibrium applied to genomic data from humans. Genetics 164: 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf, C., K. Zhao, H. Innan and M. Nordborg, 2004. The probability and chromosomal extent of trans-specific polymorphism. Genetics 168: 2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1939. The distribution of self-sterility alleles in populations. Genetics 24: 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., 2003. Effects of recombination rate and mating system on genome evolution and diversity in Arabidopsis lyrata. Ph.D. Thesis, Institute of Cell, Animal and Population Biology, University of Edinburgh, Edinburgh.
- Wright, S. I., B. Lauga and D. Charlesworth, 2003. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 12: 1247–1263. [DOI] [PubMed] [Google Scholar]