Abstract
Identifying genes responsible for quantitative variation remains a major challenge. We previously identified a quantitative trait locus (QTL) affecting body size that segregated between two inbred strains of mice. By fine mapping, we have refined the location of this QTL to a genomic region containing only four protein-coding genes. One of these genes, PAPPA2, is a strong candidate because it codes for an enzyme that cleaves insulin-like growth-factor-binding protein 5 (IGFBP-5), an important stimulator of bone formation. Among littermates that segregate only for the four-gene region, we show that the QTL has a significant effect on the circulating levels of IGFBP-5 and IGFBP-3 (the latter subject to limited degradation by PAPPA2), but not on levels of IGFBP-2 and IGFBP-4, which are not cleaved by PAPPA2. There are 14 nonsynonymous SNPs among QTL alleles, which may affect the activity of the translated protein. The refinement of the target region to four genes and the finding that the QTL affects IGFBP-5 levels suggest that PAPPA2 may be involved with normal postnatal growth. Our mapping results also illustrate the potentially fractal nature of QTL: as we mapped our QTL with increasing resolution, what appeared to be a single QTL resolved into three closely linked QTL (previous work), and then one of these was further dissected into two in this study.
IN recent years, there have been enormous efforts to identify genes responsible for quantitative variation, and numerous studies have mapped quantitative trait loci (QTL) affecting traits of medical, ecological, and agricultural importance (Flint et al. 2005; Slate 2005). However, identifying the genes underlying QTL has proven extremely difficult, and despite an increasing number of successes (e.g., Liang et al. 2003; Klein et al. 2004; Yalcin et al. 2004; Oliver et al. 2005), there is a critical need for studies and approaches that implicate specific genes, rather than identify more QTL (Flint et al. 2005). For this reason, we have focused on fine mapping a single QTL, originally identified in a cross between DBA/2J (DBA) and C57BL/6J (C57) (Morris et al. 1999). This QTL has a general effect on body size, affecting the length of the tail and most bones studied, as well as having a weaker effect on mass (Christians et al. 2003). Previous work narrowed the location of the QTL to an ∼4-Mb region of chromosome 1 containing 33 genes and demonstrated the presence of linked QTL on either side of the central QTL (Christians and Keightley 2004). In this study, we used congenic progeny testing to further refine the location of the QTL to a region of only four genes. This work identified a strong candidate gene; therefore we examined its expression levels, searched for sequence polymorphisms in its coding region, and investigated downstream physiological pathways.
MATERIALS AND METHODS
Mice:
Previous work introgressed the C57 allele of the QTL into the DBA background and produced a number of congenic lines segregating for different segments of the chromosome 1 QTL region (Christians and Keightley 2004). Recombinants from these lines yielded the 10 new congenic lines used in this study (see Figure 1, Table 1). These 10 lines all segregate within the region between D1Mit265 and D1Mit219, the region containing the primary QTL (Christians and Keightley 2004), thereby eliminating effects from linked QTL outside this region. New recombinants were mated to a DBA mouse to yield offspring that were at backcross generation 7. Thereafter, lines were maintained by inter se mating of individuals heterozygous for the segregating segment. Tail length and body mass of 1680 progeny were measured at 10 weeks of age. All experiments were carried out in accordance with the United Kingdom Home Office regulations.
Figure 1.—
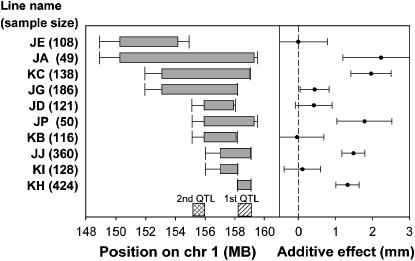
The 10 congenic lines used to map the QTL. (Left) Horizontal bars indicate the region for which each line segregates and error bars indicate uncertainty in the size of the segment; these regions are determined by the positions of microsatellite markers (supplemental Table 1 at http://www.genetics.org/supplemental/). The hatched box indicates the location of the primary QTL. The cross-hatched box indicates the between-marker interval that provides the highest support for the presence of a secondary QTL and is not a confidence interval for the location of the QTL. Physical positions are from Ensembl Release 34 (Ensembl 2006). (Right) The additive effect on 10-week tail length estimated within each family; error bars indicate the 95% confidence interval (i.e., 1.96 ± standard error).
TABLE 1.
Microsatellite markers delimiting the regions segregating within each of the congenic lines
| Line name | Outer proximal marker/physical position | Inner proximal marker/physical position | Inner distal marker/physical position | Outer distal marker/physical position |
|---|---|---|---|---|
| JE | D1Mit265 | D1Mit200 | D1Mit42 | D1Icp156 |
| 148.77 | 150.24 | 154.18 | 154.99 | |
| JA | D1Mit265 | D1Mit200 | D1Icp11 | D1Mit219 |
| 148.77 | 150.24 | 159.32 | 159.52 | |
| KC | D1Mit513 | D1Mit396 | D1Icp8 | D1Icp9 |
| 151.82 | 153.08 | 159.03 | 159.06 | |
| JG | D1Mit513 | D1Mit396 | D1Icp7 | D1Icp1 |
| 151.82 | 153.08 | 158.18 | 158.19 | |
| JD | D1Icp157 | D1Mit451 | D1Mit290 | D1Icp5 |
| 155.04 | 155.92 | 157.92 | 158.09 | |
| JP | D1Icp157 | D1Mit451 | D1Icp11 | D1Mit219 |
| 155.04 | 155.92 | 159.32 | 159.52 | |
| KB | D1Icp157 | D1Mit451 | D1Icp6 | D1Icp7 |
| 155.04 | 155.92 | 158.11 | 158.18 | |
| JJ | D1Mit451 | D1Mit202 | D1Icp9 | D1Icp10 |
| 155.92 | 157.00 | 159.06 | 159.12 | |
| KI | D1Mit451 | D1Mit202 | D1Icp1 | D1Icp4 |
| 155.92 | 157.00 | 158.19 | 158.22 | |
| KH | D1Icp7 | D1Icp1 | D1Icp9 | D1Icp10 |
| 158.18 | 158.19 | 159.06 | 159.12 |
“Outer proximal” markers are those closest to the segregating region that do not segregate within the line, whereas “inner proximal” markers do segregate within the line. Positions are in megabases and are from Ensembl Release 38 (Ensembl 2006). Primer sequences are listed in supplemental Table 1 at http://www.genetics.org/supplemental/.
Microsatellite genotyping:
Extraction of genomic DNA from ear clips and microsatellite genotyping were performed by standard methods. Mice from each line were typed for the two markers that delimited the segment segregating in their line (Table 1); primer sequences for these markers are listed in supplemental Table 1 at http://www.genetics.org/supplemental/.
QTL analyses:
Preliminary analyses were performed to assess whether each line segregated for a QTL. Analyzing each line separately, a general linear model was used to determine whether there was an additive effect of genotype (in the region for which the line segregated) on phenotype; parental pair, parity (nested within parental pair), and sex were also included in the model as fixed effects. A line segregating for an ∼0.93-Mb region was found to segregate for a QTL (see Figure 1). Because this line unambiguously demonstrated the presence of at least one QTL in this small region, we fitted a first QTL in this region when searching for a second QTL in other between-marker intervals (rather than performing a two-dimensional scan of all possible pairs of locations), analyzing all lines together. Specifically, we fitted a second QTL in various between-marker intervals to find the interval that yielded the highest support for a second QTL. For each interval, a general linear model was performed while fitting additive and dominance effects for the first QTL and for a second QTL, as well as effects of line, parental pair (nested within line), parity (nested within parental pair and line), and sex. No interval was found to have statistical support for a dominance effect for a second QTL (using a significance threshold of α = 0.05, unadjusted for multiple tests), and so the scan was repeated, fitting only an additive effect for the second QTL. Support for a second QTL was assessed using the F statistic for this additive effect (F2nd QTL).
To assess the significance of the support for the second QTL, we used simulations to determine the significance threshold for F2nd QTL. This was achieved by discarding the real phenotypes from the data set and simulating new ones on the basis of the additive and dominance effects estimated from line KH, the line segregating for the 0.93-Mb region. Families segregating for this region were modeled to segregate for this QTL whereas families not segregating for this region were modeled to show no differences among genotypes. For each simulated data set, a search for a second QTL was performed as described above, and the region with the highest support for a second QTL and its F2nd QTL was determined. A total of 10,000 simulations were performed to determine the distribution of F2nd QTL under the null hypothesis that there was only one QTL, and the F2nd QTL from the real data was compared with this distribution.
Sequencing:
Products for sequencing were amplified from adult kidney RNA using a one-step RT–PCR kit (QIAGEN, Chatsworth, CA) and extracted from agarose gels. Sequencing was carried out in forward and reverse directions using DYEnamic ET terminator cycle sequencing kits (Amersham, Buckinghamshire, UK) on an ABI Prism 3730 DNA analyzer. Sequencing primer sequences are shown in supplemental Table 2 at http://www.genetics.org/supplemental/.
Expression profiling of PAPPA2:
The tissue distribution pattern of expression of the pregnancy-associated plasma protein-A2 (PAPPA2) gene was surveyed using mouse Rapid-Scan gene expression panels (OriGene Technologies; MSCB101), which included cDNA from brain, heart, kidney, spleen, thymus, liver, stomach, small intestine, muscle, lung, testis, skin, adrenal gland, pancreas, uterus, prostate gland, breast (virgin, pregnant, lactating, and involuting), and embryo (e8.5, e9.5, e12.5, and e19). The kits were used in accordance with the manufacturer's instructions using primer pairs Cr21 and Cr13 (supplemental Table 2 at http://www.genetics.org/supplemental/). Earlier versions of the Ensembl database (Ensembl 2006) annotated two transcripts for this gene and so one primer pair was used to target each transcript; the most recent version of the database (v38) lists a single transcript that incorporates both previously annotated transcripts. One plate was amplified with Cr21 for 35 cycles and another was amplified with Cr13 for 40 cycles. The results for the two primer pairs were very similar.
Real-time quantitative PCR:
Forty-eight 6-week-old mice from line KH (Figure 1) were collected for the quantification of PAPPA2 and IGFBP-5 mRNA levels. Individuals were collected in pairs, where each pair comprised littermates of the same sex but different genotypes. In all pairs, one of the individuals had the homozygous DBA genotype; comparison of heterozygotes with the C57 homozygotes would be less powerful because the C57 allele of the QTL is partially dominant (Christians and Keightley 2004). Whole kidneys were collected into RNAlater solution (QIAGEN) and stored at −20° until required. Total RNA was isolated using Qiashredder homogenizers (QIAGEN) and RNAEasy extraction kits (QIAGEN) according to the manufacturer's instructions.
The relative expression levels of genes were determined by real-time quantitative RT–PCR with the ABI PRISM 7000 sequence detection system using TaqMan one-step RT–PCR Master Mix reagent kits (Applied Biosystems, Foster City, CA). Each reaction contained 250 ng of RNA template for PAPPA2 and 100 ng of RNA template for IGFBP-5 and β-actin. Less template was used for IGFBP-5 and β-actin because preliminary experiments indicated that the transcripts for these two genes were much more abundant than those for PAPPA2. Two primer/probe sets were used for PAPPA2 to target each of the two transcripts previously described in the Ensembl database (Ensembl 2006); primer and probe sequences are provided in supplemental Table 3 at http://www.genetics.org/supplemental/. Each sample was amplified for 40 cycles and the cycle at which the signal rose above a fixed threshold (Ct) was determined. Samples were measured in triplicate with intra- and interassay coefficients of variation for Ct values of 0.4–1.0% and 0.5–1.0%, respectively. The Ct values for PAPPA2 and IGFBP-5 were normalized to those of β-actin using the method of Pfaffl (2001): i.e., the numbers reported are relative to a standard RNA pool made up of many samples; e.g., a value of 1.5 indicates a sample has 50% more RNA than the standard pool, correcting for β-actin.
Measurement of circulating IGFBPs:
Thirty-six 6-week-old mice were blood sampled by tail tipping using the paired design described above for the quantification of mRNA levels. Blood was centrifuged to obtain serum, “Complete” protease inhibitor cocktail (Roche) was added, and samples were frozen at −20° until analysis. Serum concentrations of IGFBP-5 were quantified by using a commercial ELISA specific for mouse IGFBP-5 (R&D Systems, Wiesbaden-Nordenstadt, Germany) according to the manufacturer's instructions. Serum levels of IGFBP-2, -3, and -4 were determined by Western ligand blot according to the method of Hossenlopp et al. (1986), with modifications (Hoeflich et al. 2004). In brief, samples were diluted 1:2 with sample buffer [62.5 mm Tris–HCl (pH 6.8), 2% (w/v) sodium dodecyl sulfate (SDS), 10% (w/v) sucrose], boiled (5 min), and electrophoresed on a 5% stacking/12% separating SDS–polyacrylamide gel using the Mini Protean II system (Bio-Rad, Munich). Separated proteins were transferred to PVDF membrane (Millipore, Eschborn, Germany). The blots were blocked with 1% fish gelatin and incubated with [125I]IGF-II (106 cpm/blot). Binding proteins were visualized and quantified using the Phosphor-Imager and ImageQuant software (Amersham Biosciences, Freiburg, Germany).
RESULTS AND DISCUSSION
Fine mapping of QTL:
We developed 10 congenic lines, which segregate only within the region containing the primary QTL identified previously (Christians and Keightley 2004), thereby eliminating effects from linked QTL outside this region (Figure 1). One of these lines (KH) segregates for a 0.93-Mb region (D1Icp7–D1Icp10; supplemental Table 1 at http://www.genetics.org/supplemental/), and the additive effect on 10-week tail length is highly significant in this line (F1,349 = 69.2; P < 0.0001; Figure 1), demonstrating unambiguously that at least part of the effect of the QTL is due to this small region. In two other lines that do not segregate for this region (JD and JG, Figure 1), there appears to be a smaller additive effect. Although this smaller effect is not significant in either line when a correction for multiple tests is applied, these results suggested that there may be a second QTL in the region.
To test for the presence of a second QTL, we analyzed all 10 lines together and fitted a primary QTL in the 0.93-Mb region while also fitting a second QTL in various between-marker intervals to find the interval that yielded the highest support for a second QTL. This interval is that between D1Icp157 and D1Mit451 (Figure 1; support for second QTL: F1,1401 = 9.2; P = 0.0024, uncorrected for multiple tests). To determine whether this second QTL was significant when taking multiple tests into account, we simulated a single QTL (using the additive and dominance effects estimated in line KH) and repeated our scan for a second QTL. In 10,000 simulations, the support for a second QTL was equal to or greater than that in the real data only 112 times; therefore, the null hypothesis that there is a single QTL in the region can be rejected (P = 0.0112). In the model with both QTL, the additive and dominance effects of the primary QTL on 10-week tail length are 1.3 ± 0.1 mm (F1,1401 = 152.2; P < 0.0001) and 0.6 ± 0.1 mm (F1,1401 = 25.2; P < 0.0001), respectively, with C57 homozygotes larger than DBA homozygotes and the phenotype of heterozygotes closer to that of C57 homozygotes. The additive effect of the secondary QTL is 0.4 ± 0.1 mm (values are least squares means ± standard errors). The sum of the additive effects of the two QTL is very close to the additive effect of the primary QTL estimated in two previous populations while controlling for the effects of linked QTL (1.6–2.0 mm) (Christians and Keightley 2004). Therefore, the secondary QTL in this study is not a previously identified linked QTL, but rather the primary QTL identified previously is actually composed of at least two separate QTL. The estimated locations and effect sizes of the two QTL are consistent with the estimated additive effects from all of the lines except line KB. Line KB would be expected to segregate for the same effect as line JD, given that the proximal location of recombination is the same in these two lines, and yet the estimated additive effect in line KB is close to zero (Figure 1). However, the confidence interval from line KB includes the estimated effect of line JD, and some variation in estimated effect size due to sampling error is expected, given that effects have been estimated in 10 different lines.
Analyzing 10-week body mass with the two-QTL model used for tail length estimates the additive and dominance effects of the primary QTL to be 0.27 ± 0.08 g (F1,1508 = 10.7; P = 0.001) and 0.26 ± 0.10 g (F1,1508 = 6.7; P = 0.01), respectively, and the additive effect of the secondary QTL to be 0.20 ± 0.10 g, although the latter is marginally nonsignificant (F1,1508 = 3.8; P = 0.051). As with tail length, the sum of these estimates agrees very well with previous estimates (additive: 0.42 g; dominance: 0.28 g) (Christians and Keightley 2004).
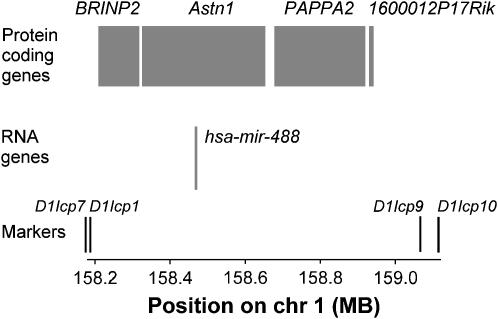
The results from line KH unambiguously refine the region containing the primary QTL to a 0.93-Mb region (D1Icp7–D1Icp10) that harbors only four protein-coding genes and one noncoding microRNA according to the Ensembl database (Ensembl 2006) (Figure 2). Functional or expression studies have been performed for only three of the genes within the target region. Astrotactin 1 (Astn1) is involved in the development of the brain, and knocking out Astn1 does not appear to affect body size in mice (Adams et al. 2002). Bone morphogenetic protein/retinoic acid-inducible neural-specific protein (BRINP2) appears to be expressed only in the nervous system, as determined by Northern blotting of embryonic and adult rat tissues and in situ hybridization of whole mouse embryo (Kawano et al. 2004). Thus, Astn1 and BRINP2 do not stand out as strong candidates. In contrast, PAPPA2 is homologous to the human PAPPA2 gene (Ensembl 2006), the protein product of which cleaves insulin-like growth-factor-binding protein 5 in vitro (IGFBP-5) (Overgaard et al. 2001). IGFBP-5 is known to be an important stimulator of bone formation (Govoni et al. 2005); therefore PAPPA2 is a strong candidate gene for a QTL that affects skeletal growth. Further evidence that this gene family is involved in growth comes from targeted disruption of a homolog of PAPPA2, PAPPA. PAPPA knockout mice are 60% the size of wild-type littermates at birth and remain smaller through postnatal development (Conover et al. 2004).
Figure 2.—
Detailed view of the target region for the primary QTL, including protein coding and RNA genes annotated in the Ensembl database (Ensembl 2006). Synonyms and/or Ensembl gene IDs for the protein coding genes are as follows: BRINP2, 6430517E21Rik, ENSMUSG00000004031; Astn1, ENSMUSG00000026587; PAPPA2, Creg1, ENSMUSG00000026732; 1600012P17Rik, ENSMUSG00000047661. 1600012P17Rik was identified from placenta cDNA. Other genes are described in the text.
Sequence variation at PAPPA2:
Fourteen nonsynonymous and 15 synonymous SNPs were found between DBA and C57 (supplemental Table 4 at http://www.genetics.org/supplemental/). Because little is known about the functional domains of the protein, it is not possible at present to predict the consequences of the nonsynonymous SNPs on the proteolytic activity of the PAPPA2 protein. However, it is plausible that one or more of these SNPs could affect its ability to cleave IGFBP-5, given that single amino-acid substitutions can drastically affect the proteolytic activity of PAPPA (Boldt et al. 2004).
Expression of PAPPA2:
To examine where PAPPA2 is expressed, we used a 24-tissue gene expression panel. Expression is strongest in stomach, skin, and embryo, with expression also observed in kidney, brain, heart, lung, testis, pancreas, and prostate gland. We chose to quantify PAPPA2 expression in kidney because this organ is one of the major sources of IGFBP-5 (the putative target of PAPPA2) (Schneider et al. 2000) and because expression levels of another gene in this tissue have been associated with whole-organism skeletal phenotype (Klein et al. 2004). Furthermore, as in the mouse, PAPPA2 is also expressed at intermediate levels in kidney in humans (Page et al. 2001), suggesting conservation of function.
We examined expression in 6-week-old mice because the effect of the QTL is strong but still increasing at this age (Christians and Keightley 2004) and because the physiological differences responsible for the QTL may have already exerted their effect and may no longer be apparent in older mice. Among 6-week-old littermates segregating only for the four-gene region, the expression of β-actin does not differ significantly among genotypes (F2,22 = 0.7, P = 0.51; Table 2), indicating that this gene is suitable for use as a reference or “housekeeping” gene. Using expression levels normalized with those of β-actin, we found no effect of the QTL on the expression of PAPPA2 using either of the primer/probe sets (probe a: F2,22 = 2.4, P = 0.12; probe b: F2,22 = 1.2, P = 0.33; Table 2). Furthermore, we found no effect of the QTL on the expression of IGFBP-5 (F2,22 = 0.3, P = 0.77; Table 2), the putative target of PAPPA2. However, there is a significant difference in tail lengths among genotypes in this group of mice, as expected (F2,21 = 5.3, P = 0.01; Table 2).
TABLE 2.
Effects of QTL genotype on phenotype, gene expression, and circulating IGFBP levels in 6-week-old mice segregating only for a four-gene region
| Genotype
|
|||
|---|---|---|---|
| DBA homozygote | Heterozygote | C57 homozygote | |
| Phenotypes | 23 | 14 | 9 |
| Tail length (mm) | 72.8 ± 0.4 | 74.3 ± 0.6 | 75.2 ± 0.8 |
| Body mass (g) | 17.6 ± 0.3 | 18.2 ± 0.5 | 19.1 ± 0.7 |
| Expression levelsa | 24 | 14 | 10 |
| PAPPA2 (probe a) | 1.18 ± 0.09 | 1.05 ± 0.14 | 0.78 ± 0.18 |
| PAPPA2 (probe b) | 1.48 ± 0.08 | 1.55 ± 0.13 | 1.21 ± 0.16 |
| IGFBP-5 | 1.07 ± 0.03 | 1.03 ± 0.05 | 1.05 ± 0.06 |
| β-actin | 17.81 ± 0.06 | 17.86 ± 0.10 | 17.66 ± 0.12 |
| Circulating protein levels | 18 | 5 | 13 |
| IGFBP-2b | 15.2 ± 0.8 | 14.0 ± 2.0 | 14.0 ± 1.1 |
| IGFBP-3b | 13.0 ± 0.9 | 11.8 ± 2.1 | 8.7 ± 1.1 |
| IGFBP-4b | 5.7 ± 0.3 | 6.5 ± 0.7 | 5.5 ± 0.4 |
| IGFBP-5 (ng/ml) | 22.6 ± 1.6 | 30.2 ± 3.9 | 15.9 ± 2.1 |
Numbers in the same row as each category name indicate sample sizes. Traits for which there are significant differences between genotypes are underlined. For all traits, values are least squares means ± standard errors from a general linear model including genotype and pair.
Normalized values for PAPPA2 and IGFBP-5 are expressed relative to a standard RNA pool made up of many samples; e.g., a value of 1.5 indicates levels 50% higher than those of the standard pool. Ct values are provided for β-actin.
Circulating protein levels for IGFBP-2, IGFBP-3, and IGFBP-4 were measured using IGF-binding activity and are expressed in arbitrary units.
Circulating IGFBP-5:
To investigate whether the QTL affected physiological pathways downstream from PAPPA2, we measured the circulating concentrations of IGFBP-5 in 6-week-old mice from litters segregating only for the four-gene region (line KH). There is significant variation in IGFBP-5 levels among genotypes (F2,16 = 5.0, P = 0.02; Table 2), with heterozygotes having the highest levels, C57 homozygotes having the lowest levels, and DBA homozygotes being intermediate (Table 2). The significant effect of the QTL on IGFBP-5 levels is not due entirely to the heterozygotes; the difference between the two homozygous genotypes is significant (F1,12 = 5.5, P = 0.04).
The effect of the QTL on PAPPA2's target suggests that PAPPA2 is the causal gene underlying the QTL, or at least that there is functional variation in PAPPA2 between QTL alleles. At this stage it is difficult to interpret the relationship between IGFBP-5 levels and phenotype, although the overdominance observed in IGFBP-5 levels suggests nonlinear relationships among PAPPA2 proteolytic activity, IGFBP-5 levels, and phenotype. This is not surprising, given that IGFBP-5 may have IGF-dependent effects when intact and IGF-independent effects when intact or cleaved (Govoni et al. 2005). IGFBP-5 could not be detected in serum by Western immunoblotting (data not shown) and therefore we quantified IGFBP-5 levels using ELISA, which did not distinguish between the intact protein and fragments generated by proteolysis. However, even if we could have made this distinction, the relative potency of IGFBP-5 fragments compared with the intact protein is not known (Govoni et al. 2005).
Other IGFBPs:
In littermates segregating only for the four-gene region, we also found a significant effect of genotype on circulating IGFBP-3 levels (F2,16 = 4.5, P = 0.03), with lower levels in C57 homozygotes than in DBA homozygotes and intermediate levels in heterozygotes (Table 2). Although IGFBP-5 appears to be the primary target of PAPPA2, limited degradation of IGFBP-3 by this proteinase has been observed (Overgaard et al. 2001). In contrast, we found no effect of genotype on the circulating levels of IGFBP-2 (F2,16 = 0.6, P = 0.58; Table 2) or IGFBP-4 (F2,16 = 0.8, P = 0.48; Table 2), two binding proteins that do not appear to be cleaved by PAPPA2 (Overgaard et al. 2001).
Conclusions:
Our results illustrate the potentially fractal nature of QTL; i.e., as we mapped our QTL with increasing resolution, what initially appeared to be a single QTL resolved into three QTL (Christians and Keightley 2004), and then one of these was further dissected into two (this study). We have refined the location of the primary QTL to a region containing only four genes and shown that variation in this four-gene region also affects circulating levels of IGFBP-5, the target of PAPPA2. Although we found no evidence of differences in the regulation of PAPPA2 between QTL alleles, the nonsynonymous SNPs that we identified may affect the activity of the translated protein.
Numerous studies have mapped QTL affecting skeletal morphology in mice to better understand the genes that predispose humans to low bone mineral density (BMD) and higher risk of osteoporotic fracture (Klein et al. 2001; Shultz et al. 2003; Bouxsein et al. 2004). The QTL that is the focus of this study affects the length of a number of bones (Christians et al. 2003) and would be expected to have pleiotropic effects on other aspects of skeletal morphology (e.g., BMD) if PAPPA2 is the causative gene. IGFBP-5 is known to play a role in bone physiology (Schneider et al. 2002; Govoni et al. 2005) and transgenic overexpression of IGFBP-5 affects BMD (Devlin et al. 2002; Salih et al. 2005). Furthermore, Mohan et al. (2003) identified a QTL affecting both IGFBP-5 and BMD in exactly the same location as the QTL in this study.
This study suggests that PAPPA2 may be involved in normal postnatal growth. This QTL may therefore serve as an important model for understanding the physiological roles of IGFBP-5 and PAPPA2 in growth and maintenance, complementing transgenic models (Wolf et al. 2005). While the functions of IGFBP-5 and its ligands, the IGFs, have been well-studied, the regulation of the IGFBPs by proteolysis is less well understood (Bunn and Fowlkes 2003; Govoni et al. 2005), and the in vivo significance of PAPPA2 is unknown.
Acknowledgments
We thank Andrew Bell, Andrew Wargo, and Andrew Read for assistance with real-time quantitative PCR, Olga Fettscher for assistance with the Western ligand blots, Claus Oxvig, Greg Gibson, and two anonymous reviewers for comments on the manuscript, Fiona Oliver for technical help, and the Biotechnology and Biological Sciences Research Council for funding.
References
- Adams, N. C., T. Tomoda, M. Cooper, G. Dietz and M. E. Hatten, 2002. Mice that lack astrotactin have slowed neuronal migration. Development 129: 965–972. [DOI] [PubMed] [Google Scholar]
- Boldt, H. B., K. Kjaer-Sorensen, M. T. Overgaard, K. Weyer, C. B. Poulsen et al., 2004. The Lin12-Notch repeats of pregnancy-associated plasma protein-A bind calcium and determine its proteolytic specificity. J. Biol. Chem. 279: 38525–38531. [DOI] [PubMed] [Google Scholar]
- Bouxsein, M. L., T. Uchiyama, C. J. Rosen, K. L. Shultz, L. R. Donahue et al., 2004. Mapping quantitative trait loci for vertebral trabecular bone volume fraction and microarchitecture in mice. J. Bone Miner. Res. 19: 587–599. [DOI] [PubMed] [Google Scholar]
- Bunn, R. C., and J. L. Fowlkes, 2003. Insulin-like growth factor binding protein proteolysis. Trends Endocrinol. Metab. 14: 176–181. [DOI] [PubMed] [Google Scholar]
- Christians, J. K., and P. D. Keightley, 2004. Fine mapping of a murine growth locus to a 1.4-cM region and resolution of linked QTL. Mamm. Genome 15: 482–491. [DOI] [PubMed] [Google Scholar]
- Christians, J. K., V. Bingham, F. Oliver, T. T. Heath and P. D. Keightley, 2003. Characterization of a QTL affecting skeletal size in mice. Mamm. Genome 14: 175–183. [DOI] [PubMed] [Google Scholar]
- Conover, C. A., L. K. Bale, M. T. Overgaard, E. W. Johnstone, U. H. Laursen et al., 2004. Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development 131: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Devlin, R. D., Z. Du, V. Buccilli, V. Jorgetti and E. Canalis, 2002. Transgenic mice overexpressing insulin-like growth factor binding protein-5 display transiently decreased osteoblastic function and osteopenia. Endocrinology 143: 3955–3962. [DOI] [PubMed] [Google Scholar]
- Ensembl, 2006. Ensembl Mouse Genome Server Database, v38 (http://www.ensembl.org/Mus_musculus/).
- Flint, J., W. Valdar, S. Shifman and R. Mott, 2005. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 6: 271–286. [DOI] [PubMed] [Google Scholar]
- Govoni, K. E., D. J. Baylink and S. Mohan, 2005. The multi-functional role of insulin-like growth factor binding proteins in bone. Pediatr. Nephrol. 20: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich, A., L. Bunger, S. Nedbal, U. Renne, M. W. Elmlinger et al., 2004. Growth selection in mice reveals conserved and redundant expression patterns of the insulin-like growth factor system. Gen. Comp. Endocrinol. 136: 248–259. [DOI] [PubMed] [Google Scholar]
- Hossenlopp, P., D. Seurin, B. Segoviaquinson, S. Hardouin and M. Binoux, 1986. Analysis of serum insulin-like growth-factor binding proteins using Western blotting: use of the method for titration of the binding proteins and competitive-binding studies. Anal. Biochem. 154: 138–143. [DOI] [PubMed] [Google Scholar]
- Kawano, H., T. Nakatani, T. Mori, S. Ueno, M. Fukaya et al., 2004. Identification and characterization of novel developmentally regulated neural-specific proteins, BRINP family. Mol. Brain Res. 125: 60–75. [DOI] [PubMed] [Google Scholar]
- Klein, R. F., A. S. Carlos, K. A. Vartanian, V. K. Chambers, R. J. Turner et al., 2001. Confirmation and fine mapping of chromosomal regions influencing peak bone mass in mice. J. Bone Miner. Res. 16: 1953–1961. [DOI] [PubMed] [Google Scholar]
- Klein, R. F., J. Allard, Z. Avnur, T. Nikolcheva, D. Rotstein et al., 2004. Regulation of bone mass in mice by the lipoxygenase gene Alox15. Science 303: 229–232. [DOI] [PubMed] [Google Scholar]
- Liang, T. B., J. Spence, L. X. Liu, W. N. Strother, H. W. Chang et al., 2003. α-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc. Natl. Acad. Sci. USA 100: 4690–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan, S., G. Masinde, X. M. Li and D. J. Baylink, 2003. Mapping quantitative trait loci that influence serum insulin-like growth factor binding protein-5 levels in F2 mice (MRL/MpJ × SJL/J). Endocrinology 144: 3491–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, K. H., A. Ishikawa and P. D. Keightley, 1999. Quantitative trait loci for growth traits in C57BL/6J × DBA/2J mice. Mamm. Genome 10: 225–228. [DOI] [PubMed] [Google Scholar]
- Oliver, F., J. K. Christians, X. J. Liu, S. Rhind, V. Verma et al., 2005. Regulatory variation at glypican-3 underlies a major growth QTL in mice. PLoS Biol. 3: 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard, M. T., H. B. Boldt, L. S. Laursen, L. Sottrup-Jensen, C. A. Conover et al., 2001. Pregnancy-associated plasma protein-A2 (PAPP-A2), a novel insulin-like growth factor-binding protein-5 proteinase. J. Biol. Chem. 276: 21849–21853. [DOI] [PubMed] [Google Scholar]
- Page, N. M., D. J. Butlin, K. Lomthaisong and P. J. Lowry, 2001. The characterization of pregnancy associated plasma protein-E and the identification of an alternative splice variant. Placenta 22: 681–687. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih, D. A. M., S. Mohan, Y. Kasukawa, G. Tripathi, F. A. Lovett et al., 2005. Insulin-like growth factor-binding protein-5 induces a gender-related decrease in bone mineral density in transgenic mice. Endocrinology 146: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, M. R., H. Lahm, M. Y. Wu, A. Hoeflich and E. Wolf, 2000. Transgenic mouse models for studying the functions of insulin-like growth factor-binding proteins. FASEB J. 14: 629–640. [DOI] [PubMed] [Google Scholar]
- Schneider, M. R., E. Wolf, A. Hoeflich and H. Lahm, 2002. IGF-binding protein-5: flexible player in the IGF system and effector on its own. J. Endocrinol. 172: 423–440. [DOI] [PubMed] [Google Scholar]
- Shultz, K. L., L. R. Donahue, M. L. Bouxsein, D. J. Baylink, C. J. Rosen et al., 2003. Congenic strains of mice for verification and genetic decomposition of quantitative trait loci for femoral bone mineral density. J. Bone Miner. Res. 18: 175–185. [DOI] [PubMed] [Google Scholar]
- Slate, J., 2005. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 14: 363–379. [DOI] [PubMed] [Google Scholar]
- Wolf, E., M. R. Schneider, R. Zhou, T. M. Fisch, N. Herbach et al., 2005. Functional consequences of IGFBP excess: lessons from transgenic mice. Pediatr. Nephrol. 20: 269–278. [DOI] [PubMed] [Google Scholar]
- Yalcin, B., S. A. G. Willis-Owen, J. Fullerton, A. Meesaq, R. M. Deacon et al., 2004. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat. Genet. 36: 1197–1202. [DOI] [PubMed] [Google Scholar]