Abstract
We report a detailed analysis of maize chromosome structure with respect to seven histone H3 methylation states (dimethylation at lysine 4 and mono-, di-, and trimethylation at lysines 9 and 27). Three-dimensional light microscopy and the fine cytological resolution of maize pachytene chromosomes made it possible to compare the distribution of individual histone methylation events to each other and to DNA staining intensity. Major conclusions are that (1) H3K27me2 marks classical heterochromatin; (2) H3K4me2 is limited to areas between and around H3K27me2-marked chromomeres, clearly demarcating the euchromatic gene space; (3) H3K9me2 is restricted to the euchromatic gene space; (4) H3K27me3 occurs in a few (roughly seven) focused euchromatic domains; (5) centromeres and CENP-C are closely associated with H3K9me2 and H3K9me3; and (6) histone H4K20 di- and trimethylation are nearly or completely absent in maize. Each methylation state identifies different regions of the epigenome. We discuss the evolutionary lability of histone methylation profiles and draw a distinction between H3K9me2-mediated gene silencing and heterochromatin formation.
IN eukaryotes the fundamental unit of DNA packing is the nucleosome, a protein octamer/DNA complex composed of four core histones, H2A, H2B, H3, and H4, that are wrapped twice with ∼146 bp of DNA (Luger et al. 1997). The histone amino termini are targets for a series of post-translational modifications, including acetylation, phosphorylation, and methylation. These modifications regulate chromatin structure and gene expression (Strahl and Allis 2000; Jenuwein and Allis 2001; Turner 2002). Multiple modifications in various combinations are thought to form a “histone code” that extends the information capacity of the associated DNA (Strahl and Allis 2000). For example, histone H3 and H4 acetylation is consistently associated with transcriptionally active euchromatin, while methylation can be associated with either active or inactive chromatin depending on the residue. Methylation at H3K4, H3K36, and H3K79 is a hallmark for active transcription, whereas methylation at H3K9, H3K27, and H4K20 is correlated with transcriptionally inert heterochromatin (Fischle et al. 2003; Lachner et al. 2003; Margueron et al. 2005; Peters and Schubeler 2005). Lysine can be monomethylated (me1), dimethylated (me2), or trimethylated (me3), and each methylation state may have unique biological functions, increasing the potential complexity of the histone code (Dutnall 2003).
At the chromosomal level, transcriptionally inactive regions tend to be associated with heterochromatin (Brown 1966) and the brightest immunostaining for “off” marks such as methylated H3K9, H3K27, and H4K20 (Peters et al. 2003; Schotta et al. 2004). In animal interphase cells, H3K9me3, H3K27me1, and H4K20me3 mark the most deeply stained regions while less condensed heterochromatin contains H3K9me1, H3K9me2, and H3K27me3 (Peters et al. 2003; Plath et al. 2003; Rice et al. 2003; Silva et al. 2003; Schotta et al. 2004; Okamoto et al. 2004). Available data suggest that at least part of this description applies to plants as well. Several reports indicate that the mono- and dimethylated forms of H3K9 and H3K27 are enriched in heterochromatin (Jackson et al. 2004; Lindroth et al. 2004; Mathieu et al. 2005; Naumann et al. 2005), although there is variation among small- and large-genome plants (Houben et al. 2003). Unlike in animals, Arabidopsis H3K27me3 is associated with euchromatin and H3K9me3 is extremely rare (Lindroth et al. 2004; Mathieu et al. 2005; Naumann et al. 2005). There is also evidence that H4K20 methylation is present in Arabidopsis (Naumann et al. 2005; Ng et al. 2006). It is not clear, however, whether there is a direct relationship between heterochromatin and histone methylation in any species since quantitative comparisons are not yet available. Further, very little is known about histone methylation in large-genome plants (Houben et al. 2003), which make up the bulk of the angiosperms (Arumuganthan and Earle 1991).
Although most of the genome interacts with histone H3, centromeric DNA also interacts with centromeric histone H3 (CENH3) (Henikoff et al. 2001). CENH3 is an important variant of H3 that recruits inner kinetochore proteins such as CENP-C (centromere protein C) and mediates kinetochore formation (Van Hooser et al. 2001). Arrays of CENH3-containing nucleosomes are not continuous, but intervened by blocks of histone H3-containing nucleosomes (Blower et al. 2002; Sullivan and Karpen 2004; Chueh et al. 2005; Yan et al. 2005). Centromeres are also transcribed to produce long stable RNAs—an observation that sets them apart from other highly repetitive regions of the genome (Saffery et al. 2003; Nagaki et al. 2004; Topp et al. 2004; May et al. 2005). The available data suggest that the chromatin structure of centromeres is quite different from flanking heterochromatin, but the extent and importance of the differences are poorly understood.
Here we provide a quantitative whole-genome view of the distribution of mono-, di-, and trimethylation at H3K9 and H3K27 in maize, a model species with a large genome. The excellent cytological resolution of maize pachytene chromosomes, three-dimensional light microscopy, and the ability to quantify staining patterns relative to cytological features provide an unparalleled view of histone methylation. The data reveal that three marks (H3K9me1, H3K27me1, and H3K27me2) correlate with DAPI (DNA) staining, but that only H3K27me2 is specifically enriched in condensed areas. Contrary to expectations we observed that H3K9me2 is not abundant in heterochromatin, but is instead enriched between chromomeres along with H3K4me2. Other data demonstrate that centromeres contain H3K9me2 and H3K9me3, that H3K27me3 occurs at several brightly focused euchromatic domains, and that H4K20 methylation is rare or absent.
MATERIALS AND METHODS
Maize strains and antibodies:
The maize inbred line KYS (with four knobs, Dawe et al. 1992), the inbred line B73 (with five knobs, Kato et al. 2004), and a strain with five knobs that has been extensively backcrossed to W23 (Hiatt et al. 2002, referred to as W23 here) were grown in the Department of Plant Biology greenhouse of the University of Georgia.
Chicken anti-maize CENP-C (1:50 for immunofluorescence) was described in a previous study (Zhong et al. 2002). Other antibodies (Table 1) were purchased from Upstate (Lake Placid, NY) and Abcam (Cambridge, MA).
TABLE 1.
List of tested antibodies, their localization, and dilutions used
| Antibody | Source/catalog no. | Host | Pachytene immunofluorescence (IF) staining | WB | Dilution for IF | Dilution for WB |
|---|---|---|---|---|---|---|
| H3K4me2 | Upstate/07-030 | Rabbit | Between chromomeres | + | 1:50 | 1:2,000 |
| H3K9me1 | Upstate/07-450 | Rabbit | Pericentromeres, chromomeres, and between chromomeres | + | 1:75 | 1:10,000 |
| H3K9me2 | Upstate/05-768 | Rabbit | Between chromomeres and in/around kinetochores (weak) | + | 1:50 | 1:5,000 |
| H3K9me2 | Upstate/07-212 | Rabbit | Between chromomeres and in/around kinetochores (strong) | + | 1:50 | 1:2,000 |
| H3K9me2 | Upstate/07-441 | Rabbit | Between chromomeres and in/around kinetochores (medium) | ND | 1:50 | ND |
| H3K9me3 | Abcam/ab8898 | Rabbit | Between chromomeres and in/around kinetochores | + | 1:50 | 1:2,000 |
| H3K27me1 | Upstate/07-448 | Rabbit | Pericentromeres, chromomeres, and continuously between chromomeres | + | 1:100 | 1:12,000 |
| H3K27me2 | Upstate/07-452 | Rabbit | Pericentromeres and chromomeres | + | 1:50 | 1:2,000 |
| H3K27me2 | Abcam/ab14222 | Rat | Pericentromeres and chromomeres | ND | 1:50 | ND |
| H3K27me3 | Upstate/07-449 | Rabbit | Several discrete domains | + | 1:50 | 1:1,000 |
| H3K27me3 | Abcam/ab6002 | Mouse | Several discrete domains (only stained in W23) | ND | 1:25 | ND |
| Acetyl H4 | Upstate/06-598 | Rabbit | ND | + | 1:50 (mouse) | 1:2,500 |
| H4K20me1 | Upstate/07-440 | Rabbit | No visible signals | + | 1:25–1:200 | 1:2,000 |
| H4K20me2 | Upstate/07-367 | Rabbit | No visible signals | − | 1:25–1:200 | 1:2,000 |
| H4K20me3 | Upstate/07-463 | Rabbit | No visible signals | − | 1:25–1:200 | 1:2,000 |
| H4K20me3 | Abcam/ab9053 | Rabbit | No visible signals | ND | 1:25–1:200 | ND |
ND, no data available; WB, Western blot.
Cytological preparation and indirect immunofluorescence:
Anthers from the inbred lines KYS, W23, and B73 were fixed and processed as described previously (Dawe et al. 1994), except that 2% paraformaldehyde and 0.1% Triton X-100 were added during fixation. All data described here were confirmed in at least two inbreds (KYS and either W23 or B73). Cells were adhered to polylysine-coated coverslips by spinning at 100 × g. The coverslips were then incubated overnight with primary antibodies, rinsed in PBS three times, and blocked with 10% goat serum for 2 hr. Secondary antibodies (FITC-conjugated goat anti-rabbit, rhodamine-conjugated donkey anti-chicken, or rhodamine conjugated donkey anti-rat, Jackson ImmunoResearch, West Grove, PA) were diluted 1:100 and applied for 2 hr. These preparations contain both meiotic cells and anther wall cells known as tapetal cells; the tapetal cells were used as material for the analysis of interphase (Figure 5). Cultured mouse neocortical neuron cells were obtained from the laboratory of Thomas F. Murray (University of Georgia) and processed for immunocytochemistry as previously reported (Dravid et al. 2005). Mouse cells were blocked with 5% BSA in PBS for 30 min, primary antibodies were applied for 1 hr at 1:200 dilution in PBS, and secondary antibodies were applied for 30 min. All cells were stained with 0.01 mg/ml DAPI (4,6-diamidino-2-phenylindole) for 10 min, mounted in Mowiol mounting medium (Harlow and Lane 1988), and sealed with nail polish.
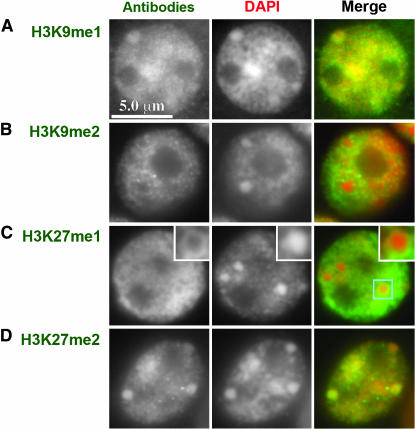
Figure 5.—
Histone H3K9 and H3K27 mono- and dimethylation in interphase cells. (A) H3K9me1 stains knobs weakly. (B) H3K9me2 does not stain knobs. (C) H3K27me1 stains the outer surface of knobs (inset is a 2× magnification of the knob). (D) H3K27me2 stains knobs brightly.
Protein blot analysis:
Maize protein was extracted from young ears ∼10 cm in length (Zhang et al. 2005) and from cultured mouse neocortical neuron cells (Dravid et al. 2005). Protein extraction and protein blot analysis were carried out as previously described (Zhang et al. 2005). A single nitrocellulose membrane was cut into four pieces and incubated with different primary antibodies. After detection, the membrane was stripped and reused for other antibodies.
Microscopy and image processing:
The images shown in Figures 1, 2, 3C, 6B, and 7A were captured and analyzed using a DeltaVision 3D light microscopy system and associated software (Applied Precision, Issaquah, WA). The remaining data were captured and processed using a Zeiss Axio Imager and SlideBook 4.0 software (Intelligent Imaging Innovations, Denver). Whole nucleus three-dimensional data sets were processed by mathematical deconvolution. Single optical sections or partial projections from several contiguous sections were chosen for presentation. For Figure 6A, three sections from different parts of the nucleus were combined (and projected) to show a complete view of the cell. The steps of chromosome straightening were described previously (Dawe et al. 1994). Each of the histograms in Figure 2 represents the average staining intensity data from two straightened chromosomes 9. Statistical smoothing by the Lowess method (Cleveland 1979, using GraphPad Software, San Diego) was applied to the resulting histograms.
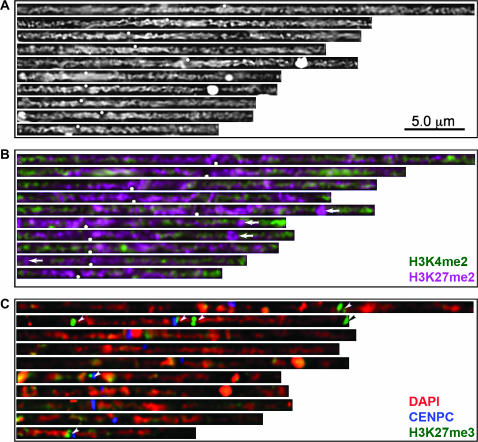
Figure 1.—
Complete KYS karyotypes showing DAPI, H3K4me2, H3K27me2, and H3K27me3. (A) The KYS karyotype as it appears after staining with DAPI. (B) The KYS karyotype after staining with anti-H3K4me2 (green) and anti-H3K27me2 (magenta). (C) The KYS karyotype showing DAPI (red), anti-H3K27me3 (green), and CENP-C (blue). The white dots in A and B show the position of centromeres, arrows point to the positions of knobs, and the arrowheads point to H3K27me3-rich domains.
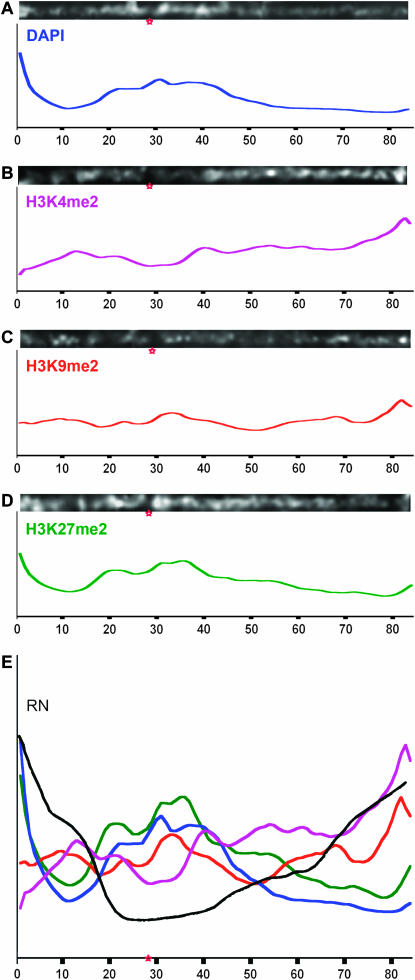
Figure 2.—
Staining patterns on chromosome 9. (A–D) Lowess-smoothed curves showing the general trends for several staining patterns. (A) DAPI staining, showing the distribution of heterochromatin. (B) H3K4me2 staining, showing a distribution that is skewed up toward chromosome ends and negatively correlated with heterochromatin. (C) H3K9me2 staining, showing a flat distribution that trends upward similar to H3K4me2. (D) H3K27me2 staining, showing a distribution that closely follows heterochromatin. (E) A combination plot of A–D and the recombination nodules map for chromosome 9 in black (adapted from Anderson et al. 2003). Red stars show the positions of centromeres.
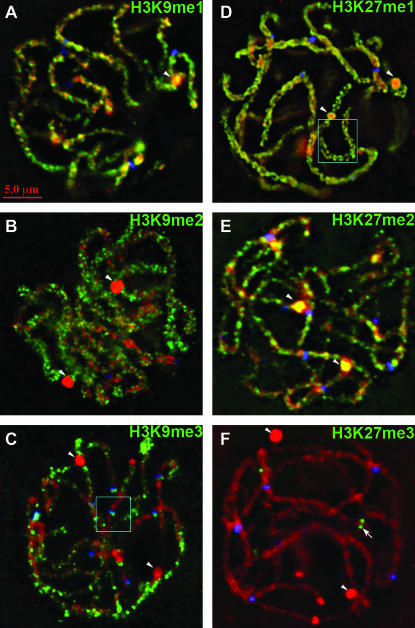
Figure 3.—
Whole-nucleus distribution of histone H3K9 and K27 mono-, di-, and trimethylation at pachytene. Methylated histone H3's are shown in green, DAPI (DNA) in red, and CENP-C (kinetochores) in blue. (A) H3K9me1 stains heterochromatin in a spotty pattern. (B) H3K9me2 stains euchromatin (i.e., regions that do not stain well with DAPI). (C) H3K9me3 stains euchromatin weakly. A 2× magnification of centromere staining with this antiserum (boxed region) is shown in Figure 7C. (D) H3K27me1 stains chromosomes uniformly. The boxed region is also shown as a 2× magnification in Figure 4A. (E) H3K27me2 stains heterochromatin. (F) H3K27me3 staining is enriched in several focused domains (one is shown in the center, arrow). Arrowheads indicate knobs.
Figure 6.—
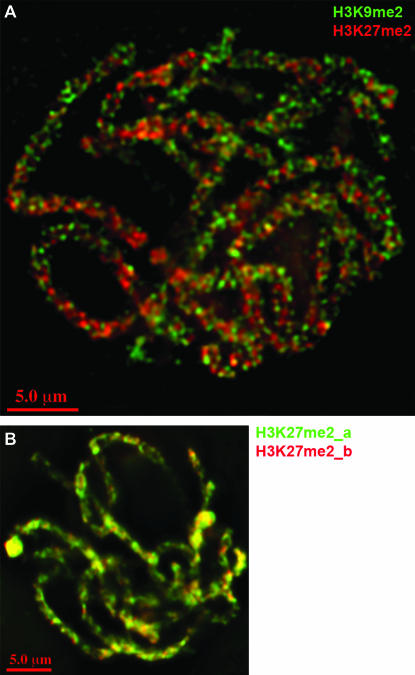
H3K9me2 and H3K27me2 staining do not overlap. (A) Colabeling with anti-H3K9me2 and anti-H3K27me2 reveals no significant overlap. (B) Colabeling control showing that two different antibodies against H3K27me2 (Upstate 07-452 from rabbit and Abcam 14222 from rat, labeled H3K27me2_a and H3K27me2_b) when applied together label the same regions of pachytene chromosomes. Minor nonconcordance can be attributed to chromatic aberration.
Figure 7.—
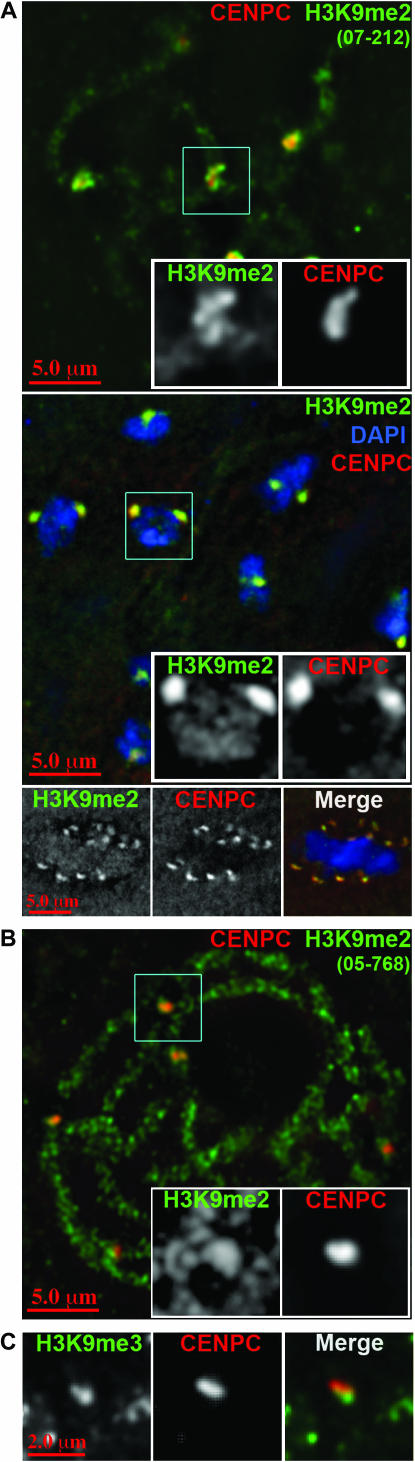
K9-methylated H3 is intermingled in primary constrictions but does not perfectly overlap with CENP-C. (A) Anti-H3K9me2 (07-212) stains kinetochores very brightly at all stages of meiosis. Pachytene (top), diakinesis (middle), and metaphase II (bottom) are shown, where blue represents DAPI. Insets show boxed regions at higher magnification. At pachytene H3K9me2 and CENP-C show only partial colocalization. (B) Anti-H3K9me2 (05-768) stains chromosome arms brightly but kinetochores weakly. (C) Kinetochore staining with anti-H3K9me3 antisera.
RESULTS
H3K4me2 identifies euchromatin—the between-chromomere space in distal halves of chromosome arms:
The major features of the maize karyotype can be observed on chromosomes stained with DAPI. As shown in Figure 1, the 10 chromosomes vary in length and arm ratio. Pericentromeric heterochromatin is evident as deeply stained regions flanking centromeres, and the distal portions of chromosome arms contain numerous small regions of heterochromatin known as chromomeres. In addition, maize and most other grasses contain dense heterochromatic domains called knobs (see Dawe and Hiatt 2004). In KYS there are four knobs—on 5L, 6L, 7L, and 9S (Figure 1). As in other species the majority of maize genes are known to lie in the distal halves of chromosome arms (Anderson et al. 2004).
On the basis of prior data we would expect H3K4me2 staining to reflect the number or activity of genes present and H3K27me2 staining to be most pronounced in pericentromeric regions and knobs. Analysis of computationally straightened chromosomes confirmed these expectations, showing that H3K4me2-stained regions are located in the distal halves of chromosome arms and that H3K27me2 is enriched in heterochromatin (Figures 1B, 2B, 2D, and 3E).
For quantitative interpretations we focused on chromosome 9, which has been thoroughly studied at the cytological level. Gene and recombination frequencies have been carefully documented for this chromosome (Anderson et al. 2003, 2004) and a cytogenetic map of chromosome 9 was recently constructed using high-resolution single-copy gene FISH (Wang et al. 2006). Chromosome 9 from cells stained with DAPI and H3K4me2 were straightened, intensity histograms extracted from the linear axes, and statistical smoothing used to reveal general staining trends. The data show that DAPI and H3K4me2 staining complement each other, with H3K4me2 being distributed toward chromosome ends away from pericentromeric heterochromatin (Figure 2, A and B). Further conforming to expectations, the data show that H3K4me2 staining is roughly correlated with the gene/recombination map of Anderson and colleagues (Figure 2E; Anderson et al. 2003).
High-magnification views revealed that H3K4me2 staining is limited to the between-chromomere spaces where DAPI staining is weak or absent (Figure 4E). When combined with the trend analysis showing a general correlation with genes (Figure 2E), these data strongly suggest that the between-chromomere space represents euchromatin in maize.
Figure 4.—
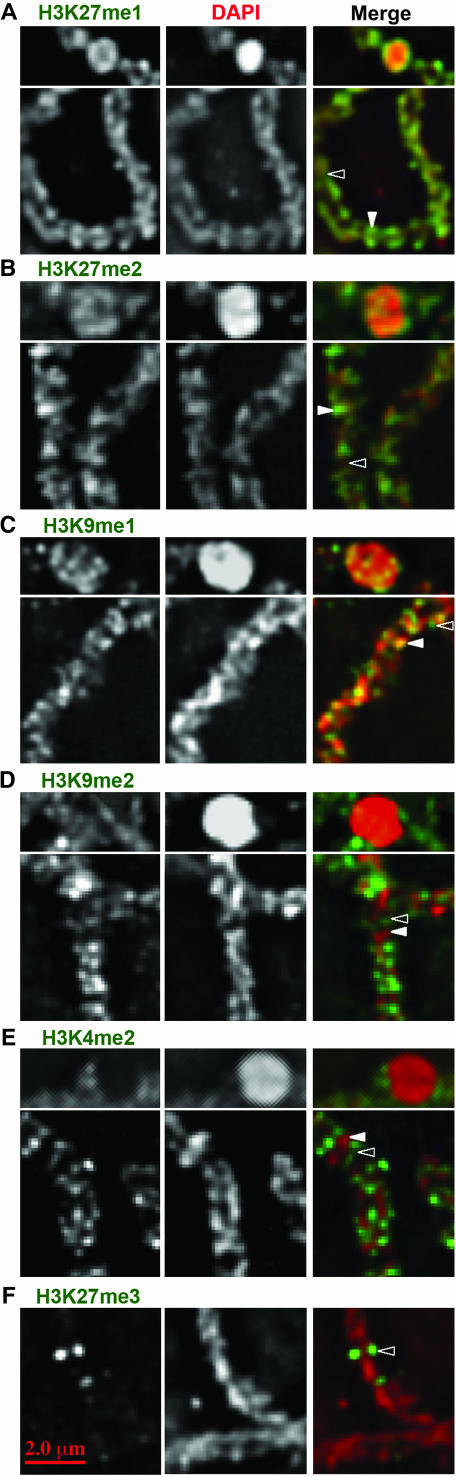
Histone methylation staining relative to chromomeres. (A) H3K27me1 uniformly stains DAPI-rich chromomeres and between-chromomere regions, but only stains the exposed surfaces of knobs. (B) H3K27me2 is enriched in chromomeres. Staining occurs unevenly throughout knobs. (C) H3K9me1 is enriched in chromomeres as well as in occasional between-chromomere regions. H3K9me1 staining is pronounced in the middle of knobs. (D) H3K9me2 stains between-chromomere spaces and rarely within knobs. (E) H3K4me2 only stains between and around chromomeres. (F) H3K27me3-rich domains do not overlap with chromomeres. A—E, top, show the knob staining for each methylation state. Solid arrowheads point to chromomeres, while open arrowheads point to between-chromomere spaces.
H3K9me2 is associated with euchromatin:
Localization with three different anti-H3K9me2 antisera on maize pachytene chromosomes failed to support the general consensus that H3K9me2 is a heterochromatic marker in plants (Houben et al. 2003; Jackson et al. 2004; Lindroth et al. 2004; Mathieu et al. 2005; Naumann et al. 2005). Analysis of >50 pachytene nuclei revealed consistently bright signal along chromosome arms and little staining in knobs and pericentromeres (Figure 3B). Similarly, no significant staining of knobs was observed in mitotic interphase cells (Figure 5B). H3K9me2 is abundant at the end of chromosome 9L (Figure 2C) and between chromomeres (Figure 4D), mirroring the staining patterns for H3K4me2. It is important to note, however, that H3K9me2 staining did not match H3K4me2 precisely. For instance, H3K9me2 showed light staining in knobs, whereas H3K4me2 did not (Figures 2C, 3B, 4D).
H3K9me1, H3K27me1, and H3K27me2 stain heterochromatin, but only H3K27me2 is enriched there:
Pachytene staining patterns for the mono-, di-, and trimethylated forms of H3K9 and H3K27 are shown in Figure 3. Anti-H3K27me1 and DAPI colocalized almost perfectly, both within and between chromomeres (Figures 3D and 4A). The only exception was in knobs where staining was limited to exposed surfaces (at both pachytene and mitotic interphase; Figures 4A and 5). Similarly, although H3K9me1 was abundant in pericentromeric heterochromatin and knobs (Figures 3A and 5), there was consistent evidence of localized accumulation between chromomeres (Figure 4C). These data suggest that H3K27me1 and H3K9me1 do not fall neatly into either the heterochromatin or euchromatin categories since they are represented within both domains.
In contrast, chromosomes labeled for H3K27me2 revealed a pattern that closely matches expectations for a marker of classical heterochromatin. In whole pachytene nuclei (Figure 3E) and straightened chromosomes (Figures 1B and 2D), staining was not uniform but enriched in pericentromeres and knobs. H3K27me2 was also the only modification that strongly stained knobs in interphase (Figure 5). Fine-scale analyses of chromomeres provided the expected colocalization of H3K27me2 with chromomeres, but unlike H3K27me1, it was not uniformly distributed between chromomeres. With few exceptions the between-chromomere space was either devoid of staining or stained very weakly (Figure 4B).
As an independent test of both the heterochromatic distribution of H3K27me2 and the euchromatic distribution of H3K9me2, we doubly labeled cells for both markers (Figure 6A). The data confirm that there is very little overlap between H3K27me2 and H3K9me2 in maize: chromosomes appeared as a collage of red and green, with virtually none of the yellow color that indicates colocalization. We can rule out the possibility that the results are confounded by antibody competition of some form (i.e., that the presence of one antibody excludes the binding of a second in the same vicinity). Control localizations using two different primary antibodies to the same epitope (H3K27me2) showed nearly perfect overlap (Figure 6B).
K9-methylated H3 is intermingled with CENP-C in primary constrictions:
In several species histone H3 is interspersed with CENH3 in alternating blocks, such that an extended array of CENH3-containing nucleosomes is followed by an array of histone H3-containing blocks and so on (Blower et al. 2002; Sullivan and Karpen 2004; Chueh et al. 2005; Yan et al. 2005). Although H3K4me2 has been documented in both Drosophila and rice centromeres (Sullivan and Karpen 2004; Yan et al. 2005), we found no evidence by immunolocalization that H3K4me2 is present in maize centromeres. By our assays the major H3 modifications in centromeres are H3K9me2 and H3K9me3 (Figures 3C and 7). Both modifications are localized in the vicinity of CENP-C, but surprisingly, we detected very little colocalization (Figure 7). Most of the K9-methylated histone H3 was detected just outside of the main concentrations of CENP-C (Figure 7). These data suggest that the specialized histone H3 arrays around (and perhaps within) the kinetochore may not directly facilitate kinetochore assembly.
We observed distinct variability among H3K9me2 antisera with respect to centromere staining patterns. One anti-H3K9me2 antiserum rarely stained kinetochores (Upstate 05-768, Figures 3B and 7B), one stained kinetochores more consistently (Upstate 07-441), and one was almost entirely limited to kinetochores (Upstate 07-212, Figure 7A). Different labeling affinities could be caused by different chromatin accessibility/condensation features or by other post-translational modifications that interfere with (or promote) the binding of the antibodies.
H3K9me3 and H3K27me3 are specialized euchromatic marks:
Although the trimethylated form of lysine 27 marks inactive chromatin in animals (Plath et al. 2003; Cao and Zhang 2004; Okamoto et al. 2004), in Arabidopsis it is localized to euchromatin (Lindroth et al. 2004; Mathieu et al. 2005). In maize, two anti-H3K27me3 antisera stained chromosomes weakly except for several very bright focused domains. These H3K27me3-rich domains did not overlap DAPI-rich chromomeres (Figure 4F) and mapped cytologically to chromosomes 1, 2, 6, and 10 in the KYS inbred (Figures 1C and 3F). Areas of rich H3K27me3 staining were found flanking centromeres, in mid-chromosome arm locations, and near a telomere. Likewise in the W23 inbred (where chromosomes are not easily identified) there were at least three spots near centromeres and one at the end of a chromosome.
In Arabidopsis the trimethylated state of lysine 9 (H3K9me3) has been reported to be absent (Jackson et al. 2004) or extremely rare (Johnson et al. 2004). However, we found clear evidence of H3K9me3 in pachytene cells (Figure 3C), root tip cells (not shown), and Western blots (Figure 8B). The staining patterns resembled what we observed with H3K9me2 antisera but with much weaker signal. H3K9me3 was undetectable in pericentromeric heterochromatin and knobs and enriched in the distal portions of chromosome arms (Figure 3C).
Figure 8.—
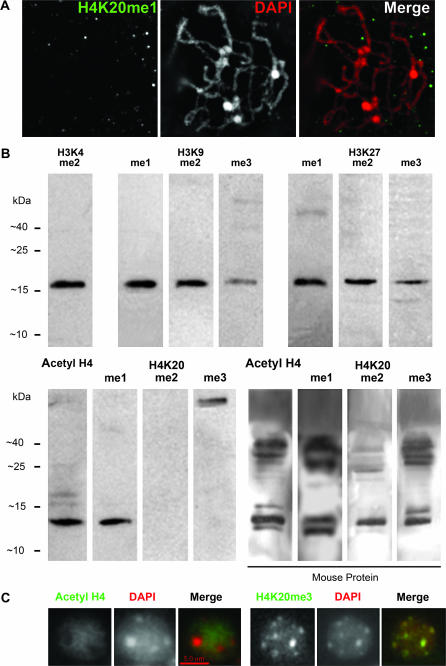
Methylation at H4K20 is rare in maize. (A) H4K20me1 was not detectable by immunolocalization in maize. (B) Western blots showing that each of seven histone H3 methylation states is detectable in maize protein extracts. Di- and trimethylated H4K20 were not detectable in maize extracts but were observed in mouse extracts. (C) Controls showing that in mouse cells anti-acetyl H4 antibodies stained euchromatin (left) and anti-H4K20me3 antibodies stained heterochromatin (right).
H4K20 di- and trimethylation is undetectable in maize:
Methylation of H4K20 is one of the most important markers of inactive chromatin in animals (Sims et al. 2003). However, we failed to detect any specific immunolocalization using antibodies to the mono-, di-, or trimethylated forms of H4K20 (Figure 8A). It is unlikely that the absence of staining is a consequence of improperly handled or inactive antisera. When the same antibodies to H4K20me3 were incubated with mouse neuronal cells (Figure 8C), strong punctate staining coinciding with DAPI-rich pericentromeres was observed, consistent with prior results from the same species (Sarg et al. 2004).
For the di- and trimethylated forms of H4K20, the immunolocalization data were confirmed by Western analysis of maize protein samples. Bands of the expected size were observed for nine of the H3 antibodies used in this study and for an antibody to acetylated H4 (Figure 8B). However, among the three anti-H4K20 antibodies, only those to anti-H4K20me1 revealed an expected 14-kDa band (Figure 8B). H4K20me2 and H4K20me3 were consistently undetectable in maize, although in mouse extracts the appropriate bands were clearly visible (Figure 8B). The very poor representation of H4K20 suggests that it is not a major repressive mark in maize.
The primary H4K20 methylases in animals are SET8/PR-Set7 and Suv-20 (Schotta et al. 2004; Couture et al. 2005; Xiao et al. 2005). We failed to identify any significant sequence homology to SET8/PR-Set7 or Suv-20 in either the complete Arabidopsis or near-complete rice genomes. However, in Arabidopsis, SUVH2 can catalyze monomethylation at H4K20 (Naumann et al. 2005) and this may be responsible for the monomethylation we observed on Western blots.
DISCUSSION
The term heterochromatin was first used by E. Heitz in 1928 to describe segments of chromosomes that stain deeply with DNA stains and do not fully decondense in interphase (Zacharias 1995). Using Drosophila as a model, Heitz went on to conclude that “. . . the density of genes in a chromosome is related to the longitudinal differentiation in euchromatin and heterochromatin. Euchromatic pieces are rich, whereas heterochromatic ones are at least poor in genes” (Heitz 1934, as translated by Zacharias 1995). Maintaining this integrated view of genetics and chromosome structure has become increasingly difficult as cyto-based genetics has transitioned to DNA-based genomics. However, recent results showing that heterochromatin is marked by specific histone methylation events have the potential to bridge the divide between genomics and chromosome structure. In principle, assays for histone methylation at lysines 9 and 27 can provide high-resolution cytological markers and add new clarity to the relationship between chromosome and gene.
Although both classical heterochromatin and histone modification are generally assayed by their staining intensities, heterochromatin and histone modification have not been quantified and carefully compared. As a result it is not clear how well histone methylation marks heterochromatic regions, which methylation states provide the best markers, or whether the modifications are conserved among organisms. Here we address these questions in maize, an important model for large-genome cereal grains and one of a handful of species with well-developed genetics and cytogenetics. Transposon-rich intergenic spaces in maize can extend for hundreds of kilobases (Chan et al. 2006), and in this context heterochromatin and euchromatin have clear foundations in differential condensation. Our data show that heterochromatin contains a relatively simple set of histone modifications that is distinct from the more complex mixture of on and off histone modifications that make up the gene-rich euchromatin space.
H3K27me2 marks classical heterochromatin:
Mass spectrometry of Arabidopsis H3K9 and H3K27 revealed that the monomethylated forms were predominant, the dimethyl forms were less abundant, and the trimethyl forms were present at levels several-fold lower (Johnson et al. 2004). H3K27me1 was found on >60% of all canonical histone H3 in inflorescence tissue (Johnson et al. 2004). Our observations using multiple antisera (Table 1) in several inbred lines generally confirm these conclusions. The intensity of signals from anti-H3K27me1 antibodies was as bright or brighter than DAPI staining, while the H3K27me2 signals were less intense, and H3K27me3 was sparsely distributed and difficult to detect on Western blots (Figure 8B). A similar trend was observed with antibodies against the H3K9 mono-, di-, and trimethylated epitopes.
One of the goals of our study was to determine which, if any, of the six methylation states at H3K9 and H3K27 accurately mark heterochromatin. Although three appear to mark heterochromatin in some capacity (Figure 9: H3K9me1, H3K27me1, and H3K27me2) our analysis suggests that H3K27me2 is the only modification that marks heterochromatin specifically. At pachytene, H3K27me2 is enriched in pericentromeres (Figures 1B, 2D, and 3E), chromomeres (Figure 4B), and knobs (Figure 3B). It is also the only modification we tested that thoroughly stains knobs at interphase (Figure 5) and the only marker that shows a clear reduction in staining between chromomeres at pachytene (Figure 4B). At present H3K9me1 and H3K27me1 are difficult to interpret since little is known about them from chromatin immunoprecipitation (ChIP) studies and they appear to stain cytologically condensed regions as well as noncondensed regions.
Figure 9.—
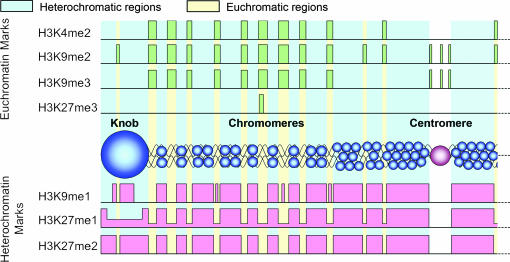
Summary of histone methylation patterns in maize. H3K4me2, H3K9me2, and H3K9me3 are distributed between chromomeres in the presumed euchromatin, with H3K9me2 also showing limited staining in knobs. H3K27me3 is enriched in specific euchromatic domains—one is shown here as an example—although in reality few chromosome arms contain such domains. H3K9me1 is observed in heterochromatic regions as well as in a subset of the between-chromomere spaces. H3K27me1 is abundant in heterochromatin but is also uniformly present between chromomeres. H3K27me2 is the only marker that is found specifically in heterochromatin.
A cytological definition of euchromatin in maize:
As a highly conserved marker of transcribed or “poised” genes (Schneider et al. 2004; Alvarez-Venegas and Avramova 2005), H3K4me2 provides an excellent molecular marker of the active gene space. Our data show that H3K4me2 staining complements the staining for H3K27me2, in terms of trends both along the linear axis of chromosome 9 (Figure 2) and among the chromomeres that dot distal regions of chromosome arms (Figures 4E and 6A). H3K4me2-stained chromatin lies not only between but also around H3K27me2-stained chromomeres, with some of the brightest H3K4me2 labeling occurring over areas with the weakest DAPI staining (Figure 4E). These data appear to confirm the interpretation first made by McClintock (1944) that the between-chromomere space is where the majority of genes are located in maize.
The distribution of H3K9 di- and trimethylation within euchromatin:
Prior data from Arabidopsis (Houben et al. 2003; Jackson et al. 2004; Lindroth et al. 2004; Mathieu et al. 2005; Naumann et al. 2005) indicate that H3K9me2 is a heterochromatin mark. Houben et al. (2003) showed that in faba bean cells and other large-genome plants, H3K9me2 was also distributed toward the ends of chromosomes. The authors suggested that in large-genome plants with many more retroelements the heterochromatin is more widely distributed along chromosomes. We show that the unexpected distribution of H3K9me2 in maize is not because heterochromatin is broadly distributed but because H3K9me2 is a euchromatic mark.
We make this conclusion on the basis of three forms of data. General observations and trend analyses indicate that H3K9me2 is preferentially distributed toward chromosome ends where it is correlated with H3K4me2 and estimated gene frequency (Figure 2). Also like H3K4me2, H3K9me2 staining is most intense in the between-chromomere gene space where DAPI staining is weakest (Figure 4D). Finally and perhaps most convincingly, when H3K9me2 and the bona fide heterochromatin mark H3K27me2 are labeled simultaneously there is very little overlap (Figure 6A). Staining patterns were essentially identical with either of two independently generated anti-H3K9me2 antisera (Figures 3B, 6A, and 7B). The fact that H3K9me2 can be observed at low frequency in pericentromeres and knobs (Figures 4D and 9) is not contrary to the interpretation that it is a euchromatic mark. Small genic (or at least low-copy) sequences are distributed at low frequencies in pericentromeric heterochromatin and knobs, as suggested by the presence of recombination proteins (visualized as recombination nodules at pachytene; Anderson et al. 2003; summarized in Figure 2E). The idea that H3K9me2 functions in euchromatin is also supported by several Arabidopsis studies: mutations of kryptonite [a Su(var)3-9 family histone methyltransferase] and dnmt1 (DNA methyltransferase 1) show significant reductions in H3K9me2 but no changes in the structure of pericentromeric heterochromatin (Jasencakova et al. 2003; Tariq et al. 2003; Jackson et al. 2004; Johnson et al. 2004).
The presence of both H3K4me2 and H3K9me2 in euchromatin can be interpreted as supporting the generally held views that H3K4me2 marks active or poised genes (Santos-Rosa et al. 2002; Schneider et al. 2004), while H3K9me2 marks those that are temporally or spatially inactive (Jenuwein and Allis 2001). The data further suggest that in maize most genes are at least poised for transcription and that the on and off marks are frequently in close proximity. This interpretation is supported by recent ChIP data that suggest that active genes may have substantial amounts of H3K9me2 and, vice versa, that inactive genes often contain H3K4me2 (Santos-Rosa et al. 2002; Bastow et al. 2004; Alvarez-Venegas and Avramova 2005). Alvarez-Venegas and Avramova (2005) showed that H3K4me2 and H3K9me2 coexisted at nearly equal levels in several Arabidopsis genes. This was a consistent result for inactive or moderately expressed sequences; only when gene expression was very high was there evidence that H3K4me2 dominated over H3K9me2.
H3K27me3 is limited to a small number of bright, focused euchromatic regions:
In animals H3K27me3 is a marker of inactive chromatin (e.g., Plath et al., 2003; Cao and Zhang 2004; Okamoto et al. 2004) while in Arabidopsis it marks euchromatin (Mathieu et al. 2005), and in maize H3K27me3 accumulates in a small number of highly focused domains (Figure 3F). The H3K27me3-rich domains lie between chromomeres (Figure 4F) and map to disparate regions, including adjacent to centromeres, mid-chromosome arm, and close to a telomere (Figure 1C). It is not clear what these regions of the genome have in common, and comparisons to the genetic map provide no obvious clues. What sequences underlie the H3K27me3-rich spots remain a mystery, although we assume they are either clusters of specific repeats or clusters of coregulated genes as in Drosophila (Ringrose et al. 2004).
Histone H3 in centromeres is di- and trimethylated at K9:
Centromeres can be defined by their interactions with the specialized histone variant CENH3 and associated proteins such as CENP-C (Dawe et al. 1999; Henikoff et al. 2001; Zhong et al. 2002). Prior data suggest that CENH3-containing nucleosomes are closely associated with intervening arrays of nucleosomes containing histone H3 (Blower et al. 2002; Chueh et al. 2005; Yan et al. 2005). The centromere-embedded H3 may be specially modified to facilitate centromere/kinetochore function (Sullivan and Karpen 2004) or may function as an extension of pericentromeric heterochromatin (Yan et al. 2005), perhaps mediating chromatid cohesion (Bernard et al. 2001). Our data from maize show that the H3 modifications in centromeres are distinctly different from those in flanking pericentromeric heterochromatin. We observed clear centromere labeling with antisera to H3K9me2 and H3K9me3, but found no evidence of H3K9me1, H3K27me1, and H3K27me2, which are enriched in pericentromeres. Although resolution is limited at the submicrometer level of the centromere, our data further suggest that histone H3 may be only partially associated with the CENP-C-marked kinetochore, at least at pachytene (Figure 7).
Differences in histone modifications between centromeres and pericentromeres are likely to reflect the transcriptional activity of the two domains (reviewed by Jiang et al. 2003). While pericentromeres are heterochromatic by our assays, maize centromeres are known to be transcribed (Topp et al. 2004) and, as shown here, contain euchromatic histone modifications (H3K9me2 and H3K9me3). Available data from other species are consistent with this view. H3K4me2 is abundant in Drosophila centromeres (Sullivan and Karpen 2004) where it is presumed to mark a transcriptionally poised state. Rice and Arabidopsis centromeric DNAs are transcribed and associated with both H3K9me2 and H3K4me2 (May et al. 2005; Yan et al. 2005). The relative abundance of centromeric H3K4me2 and H3K9me2 appears to vary among species, but it is not yet clear if these are biological differences or simply differences in the methods used (i.e., ChIP vs. immunolocalization). An initial study in rice detected only H3K9me2 within centromere cores (Nagaki et al. 2004), but H3K4me2 was also detected when more sensitive methods were used (Yan et al. 2005).
Evolutionary lability of histone methylation patterns:
The proposition that histone modification patterns might provide a universal code for interpreting eukaryotic gene activity (Strahl and Allis 2000; Jenuwein and Allis 2001) has in many ways been borne out in recent years, at least with respect to the acetylation events that activate gene expression (e.g., Ng et al. 2006). However, it was already apparent at the outset of our study that plants and animals differed with respect to the patterns of histone methylation in heterochromatin (Houben et al. 2003; Jackson et al. 2004; Lindroth et al. 2004; Mathieu et al. 2005; Naumann et al. 2005). Because we focused our efforts on a large-genome plant and used pachytene chromosomes as our subject material, we are now able to confirm and extend these comparisons to animals as well as within plants. The data suggest that the genomewide localization of histone methylation patterns is much more variable than previously recognized.
A tabular comparison of the pericentromeric staining patterns of maize, Arabidopsis, and human cells is shown in Table 2. Strikingly, of the three known histone methylation states in human pericentromeric heterochromatin, only one, H3K27me1, is also found in the corresponding regions of Arabidopsis and maize chromosomes (Jackson et al. 2004; Lindroth et al. 2004; Naumann et al. 2005). There is also an unexpected degree of variability within the angiosperms: two of the five marks present in Arabidopsis heterochromatin are rare or absent in the corresponding regions of maize. H3K9me2 is readily detectable in Arabidopsis heterochromatin (Houben et al. 2003; Jackson et al. 2004; Lindroth et al. 2004; Mathieu et al. 2005; Naumann et al. 2005) but is limited to euchromatin in maize. We also observed a near-complete absence of H4K20 methylation in maize, which is apparently prevalent in Arabidopsis (Naumann et al. 2005). H4K20me2 and H4K20me3 were undetectable by immunostaining or Western blot in our experiments. H4K20me1 could be detected on Western blots (Figure 8B), but either it is insufficiently abundant or the epitope is insufficiently exposed to be detected immunocytochemically.
TABLE 2.
Comparison of pericentromeric staining in maize, Arabidopsis, and human cells
| Methylation state | Maize | Arabidopsis | Human |
|---|---|---|---|
| H3K9me1a,b | + | + | |
| H3K9me2a,b,c | + | ||
| H3K9me3a,b | + | ||
| H3K27me1a,b,c,d | + | + | + |
| H3K27me2a,b,c,d | + | + | |
| H3K27me3a,b,c,d | |||
| H4K20me1b,e | ?f | + | |
| H4K20me2b,e | |||
| H4K20me3b,e | + |
Detectable by Western analysis but not by immunofluorescence.
These data demonstrate that over evolutionary time there have been important shifts in the distribution of the mono-, di-, and trimethylated forms of histone H3 lysines 9 and 27. As yet there is no evidence that these shifts affect the basic function of H3K9 and H3K27 in gene silencing. However, the results do suggest that the presumed epigenetic code has the capacity to evolve along with changes in genome architecture. In maize, where retroelements dominate as the most abundant form of repeat (SanMiguel and Benntzen 1998; Chan et al. 2006), histone marks that are specialized for retroelement inactivation (e.g., H3K27me2) may have played a larger role in differentiating chromosomes. The distribution of marks that function primarily in gene silencing (e.g., H3K9me2) may have a more striking euchromatic distribution in maize only because there is a sharper differentiation of chromosome structure in this species. In the larger context our data suggest that genetic inactivity is not always manifested as heterochromatin and that the repeat structure of an organism is likely to have a major impact on the distribution and prevalence of histone methylation.
Acknowledgments
We thank Thomas F. Murray and Zhengyu Cao for supplying materials and expertise during the analysis of mouse neuronal cells. This work was supported by a grant from the National Science Foundation (0421619).
References
- Alvarez-Venegas, R., and Z. Avramova, 2005. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 33: 5199–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., G. G. Doyle, B. Brigham, J. Carter, K. D. Hooker et al., 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., N. Salameh, H. W. Bass, L. C. Harper, W. Z. Cande et al., 2004. Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics 166: 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganthan, K., and E. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218. [Google Scholar]
- Bastow, R., J. S. Mylne, C. Lister, Z. Lippman, R. A. Martienssen et al., 2004. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat et al., 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542. [DOI] [PubMed] [Google Scholar]
- Blower, M., B. Sullivan and G. Karpen, 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. W., 1966. Heterochromatin. Science 151: 417–425. [DOI] [PubMed] [Google Scholar]
- Cao, R., and Y. Zhang, 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14: 155–164. [DOI] [PubMed] [Google Scholar]
- Chan, A. P., G. Pertea, F. Cheung, D. Lee, L. Zheng et al., 2006. The TIGR maize database. Nucleic Acids Res. 34: D771–D776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh, A. C., L. H. Wong, N. Wong and K. H. Choo, 2005. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14: 85–93. [DOI] [PubMed] [Google Scholar]
- Cleveland, W. S., 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74: 829–836. [Google Scholar]
- Couture, J. F., E. Collazo, J. S. Brunzelle and R. C. Trievel, 2005. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 19: 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R. K., and E. N. Hiatt, 2004. Plant neocentromeres: fast, focused, and driven. Chromosome Res. 12: 655–669. [DOI] [PubMed] [Google Scholar]
- Dawe, R. K., D. A. Agard, J. W. Sedat and W. Z. Cande, 1992. Pachytene DAPI map. Maize Genet. Coop. News Lett. 66: 23–25. [Google Scholar]
- Dawe, R. K., J. W. Sedat, D. A. Agard and W. Z. Cande, 1994. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76: 901–912. [DOI] [PubMed] [Google Scholar]
- Dawe, R. K., L. Reed, H.-G. Yu, M. G. Muszynski and E. N. Hiatt, 1999. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11: 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid, S. M., D. G. Baden and T. F. Murray, 2005. Brevetoxin augments NMDA receptor signaling in murine neocortical neurons. Brain Res. 1031: 30–38. [DOI] [PubMed] [Google Scholar]
- Dutnall, R. N., 2003. Cracking the histone code: one, two, three methyls, you're out! Mol. Cell 12: 3–4. [DOI] [PubMed] [Google Scholar]
- Fischle, W., Y. Wang and C. D. Allis, 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15: 172–183. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane, 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Heitz, E. 1934. Die somatische Heteropyknose bei Drosophila melanogaster und ihre genetische Bedeutung (Cytologische Untersuchungen an Dipteren, III. Z. Zellforsch. Mikrosk. Anat. 20: 237–287. [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hiatt, E. N., E. K. Kentner and R. K. Dawe, 2002. Independently-regulated neocentromere activity of two classes of satellite sequences in maize. Plant Cell 14: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, A., D. Demidov, D. Gernand, A. Meister, C. R. Leach et al., 2003. Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 33: 967–973. [DOI] [PubMed] [Google Scholar]
- Jackson, J. P., L. Johnson, Z. Jasencakova, X. Zhang, L. PerezBurgos et al., 2004. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112: 308–315. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., W. J. Soppe, A. Meister, D. Gernand, B. M. Turner et al., 2003. Histone modifications in Arabidopsis- high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 33: 471–480. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and C. D. Allis, 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jiang, J., J. A. Birchler, W. A. Parrott and R. K. Dawe, 2003. A molecular view of plant centromeres. Trends Plant Sci. 8: 570–575. [DOI] [PubMed] [Google Scholar]
- Johnson, L., S. Mollah, B. A. Garcia, T. L. Muratore, J. Shabanowitz et al., 2004. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 32: 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., J. C. Lamb and J. A. Birchler, 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner, M., R. J. O'Sullivan and T. Jenuwein, 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116: 2117–2124. [DOI] [PubMed] [Google Scholar]
- Lindroth, A. M., D. Shultis, Z. Jasencakova, J. Fuchs, L. Johnson et al., 2004. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23: 4286–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent and T. J. Richmond, 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Margueron, R., P. Trojer and D. Reinberg, 2005. The key to development: Interpreting the histone code? Curr. Opin. Genet. Dev. 15: 163–176. [DOI] [PubMed] [Google Scholar]
- Mathieu, O., A. V. Probst and J. Paszkowski, 2005. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24: 2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, B. P., Z. B. Lippman, Y. Fang, D. L. Spector and R. A. Martienssen, 2005. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 1: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1944. The relation of homozygous deficiencies to mutations and allelic series in maize. Genetics 29: 478–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Z. Cheng, S. Ouyang, P. B. Talbert, M. Kim et al., 2004. Sequencing of a rice centromere reveals active genes. Nature Genet. 36: 138–145. [DOI] [PubMed] [Google Scholar]
- Naumann, K., A. Fischer, I. Hofmann, V. Krauss, S. Phalke et al., 2005. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24: 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D. W., M. B. Chandrasekharan and T. C. Hall, 2006. Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell 18: 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, I., A. P. Otte, C. D. Allis, D. Reinberg and E. Heard, 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649. [DOI] [PubMed] [Google Scholar]
- Peters, A. H., and D. Schubeler, 2005. Methylation of histones: playing memory with DNA. Curr. Opin. Cell Biol. 17: 230–238. [DOI] [PubMed] [Google Scholar]
- Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck et al., 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12: 1577–1589. [DOI] [PubMed] [Google Scholar]
- Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer et al., 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. [DOI] [PubMed] [Google Scholar]
- Rice, J. C., S. D. Briggs, B. Ueberheide, C. M. Barber, J. Shabanowitz et al., 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12: 1591–1598. [DOI] [PubMed] [Google Scholar]
- Ringrose, L., H. Ehret and R. Paro, 2004. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16: 641–653. [DOI] [PubMed] [Google Scholar]
- Saffery, R., H. Sumer, S. Hassan, L. H. Wong, J. M. Craig et al., 2003. Transcription within a functional human centromere. Mol. Cell 12: 509–516. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., and J. L. Benntzen, 1998. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82: 37–44. [Google Scholar]
- Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein et al., 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411. [DOI] [PubMed] [Google Scholar]
- Sarg, B., W. Helliger, H. Talasz, E. Koutzamani and H. H. Lindner, 2004. Histone H4 hyperacetylation precludes histone H4 lysine 20 trimethylation. J. Biol. Chem. 279: 53458–53464. [DOI] [PubMed] [Google Scholar]
- Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson et al., 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6: 73–77. [DOI] [PubMed] [Google Scholar]
- Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta et al., 2004. A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev. 18: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova et al., 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4: 481–495. [DOI] [PubMed] [Google Scholar]
- Sims, III, R. J., K. Nishioka and D. Reinberg, 2003. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19: 629–639. [DOI] [PubMed] [Google Scholar]
- Strahl, B. D., and C. D. Allis, 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Sullivan, B. A., and G. H. Karpen, 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11: 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, M., H. Saze, A. V. Probst, J. Lichota, Y. Habu et al., 2003. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100: 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp, C. N., C. X. Zhong and R. K. Dawe, 2004. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101: 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B. M., 2002. Cellular memory and the histone code. Cell 111: 285–291. [DOI] [PubMed] [Google Scholar]
- Van Hooser, A. A., I.I. Ouspenski, H. C. Gregson, D. A. Starr, T. J. Yen et al., 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114: 3529–3542. [DOI] [PubMed] [Google Scholar]
- Wang, C. R., L. Harper and W. Z. Cande, 2006. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18: 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, B., C. Jing, G. Kelly, P. A. Walker, F. W. Muskett et al., 2005. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 19: 1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H., W. Jin, K. Nagaki, S. Tian, S. Ouyang et al., 2005. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 18: 3227–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, H., 1995. Emil Heitz (1892–1965): chloroplasts, heterochromatin, and polytene chromosomes. Genetics 141: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., X. Li, J. B. Marshall, C. X. Zhong and R. K. Dawe, 2005. Phosphoserines on maize CENTROMERIC HISTONE H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell 17: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C. X., J. B. Marshall, C. Topp, R. Mroczek, A. Kato et al., 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14: 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]