Abstract
Studying gene expression in polyploids is complicated by genomewide gene duplication and the problem of distinguishing transcript pools derived from each of the two homeologous genomes such as the A- and D-genomes of allotetraploid Gossypium. Short oligonucleotide probes designed to specifically target several hundred homeologous gene pairs of Gossypium were printed on custom NimbleGen microarrays. These results demonstrate that relative expression levels of homeologous genes may be measured by microarrays and that deviation from equal expression levels of homeologous loci may be common in the allotetraploid nucleus of Gossypium.
WHOLE-genome duplication, or polyploidy, has been a prominent force in angiosperm evolution (Grant 1981; Leitch and Bennett 1997). Recently formed allopolyploids, such as cotton, retain duplicated copies of most genes on homeologous chromosomes. These homeologous loci typically have sufficiently high sequence identity that their transcripts cross-hybridize on standard microarray platforms, thereby obscuring the genomic origin of expressed genes. Because of this technical limitation, the contribution of each homeolog from each constituent genome of a polyploid to the transcriptome has remained largely unexplored. Recent work indicates, however, that these contributions need not be equal and, in fact, that altered gene expression in allopolyploids is common (Kashkush et al. 2002; Adams et al. 2003; Osborn et al. 2003; Adams and Wendel 2005; Wang et al. 2006).
Domesticated cotton (Gossypium hirsutum) is an allotetraploid derived from two diploid genomes, “A” and “D.” Accumulated evidence indicates a relatively recent origin of the allopolyploid lineage, probably in the past 1–2 million years, from diploid parents similar to modern A- (G. arboreum or G. herbaceum) and D- (G. raimondii) genome species (Wendel and Cronn 2003). Most genes of A- and D-genome diploid Gossypium species are 98–99% similar in exon sequence, as are their homeologous counterparts in the allotetraploids (Senchina et al. 2003). Because of this high sequence identity, ESTs from diploid and allopolyploid species may be combined during contig assembly (Udall et al. 2006).
In this Note, we describe a novel bioinformatic and molecular methodology for simultaneously monitoring transcript accumulation for thousands of pairs of homeologous genes. The methodology involves custom short-oligonucleotide microarrays based on A- and D-genome-specific single nucleotide polymorphism (SNPs) or small insertion/deletions (indels), identified following assembly of ESTs of three different Gossypium species (Figure 1; Udall et al. 2006). Through comparisons of the progenitor diploid genomes, ortholog- and homeolog-specific polymorphisms were identified by scanning the 24,363 assembled contigs for polymorphisms between the A- and D-genome ESTs (Figure 1; supplemental Table S1 at http://www.genetics.org/supplemental/). A total of 2277 SNPs and 98 small indels from 701 genes were identified and probe pairs targeting these polymorphisms were included on a custom DNA microarray (supplemental Figure S1 at http://www.genetics.org/supplemental/; Nuwaysir et al. 2002; NimbleGen Systems).
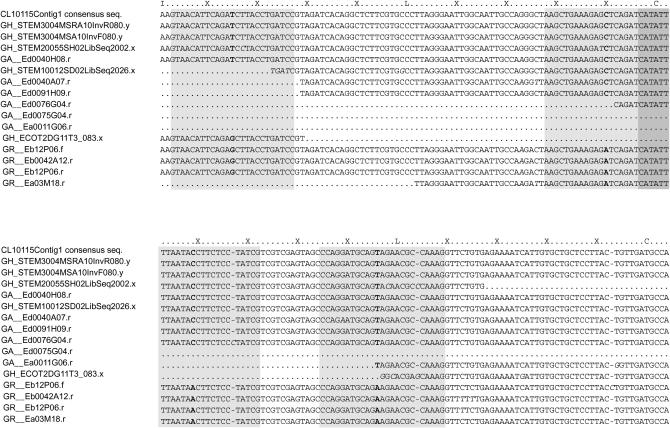
Figure 1.—
SNPs were identified between A- and D-genome ESTs, leading to assignment of genomic origin for ESTs from allopolyploid G. hirsutum. A portion (positions 811–1095) of the alignment for contig CL10115Contig1 is shown and a two-letter prefix of each EST name indicates its respective Gossypium species [GA, G. arboreum (A-genome diploid); GH, G. hirsutum (AD-genome); GR, G. raimondii (D-genome diploid)]. Sites of species-specific or homeolog-specific polymorphisms are in boldface type and allelic and/or sequencing errors are in italic type. Shaded boxes represent 25-mer probes designed to target A- or D-genomes where genome specificity is conferred by the central SNP. The darkly shaded portion represents overlapping probe sequences of two independently targeted SNPs. Contig CL10115Contig1 was created in an EST assembly: a preliminary assembly of ∼150,000 ESTs collected from 30 different cDNA libraries from three different Gossypium species was constructed using PAVE (Program for Assembling and Viewing ESTs; http://agcol.arizona.edu/; Udall et al. 2006). Most cDNA libraries were derived from G. hirsutum and composed 38% of the total number of ESTs in the assembly. The remaining ESTs were derived from three deeply sampled cDNA libraries generated from the two diploids composing 24 and 38% of the total number of ESTs, respectively. For homeolog identification, contigs were scanned using a custom perl script facilitated by BioPerl modules (Stajich et al. 2002) to identify SNPs and small indels characteristic of the A- and D-genomes of Gossypium. Internally, a consensus sequence was created for both A- (including A and AT sequences) and D-genomes (including D and DT sequences), and then target polymorphisms were found by comparing these two sequences. Probes were designed to target those polymorphisms by placing the distinguishing SNP or first base pair of the small indel centrally in a 25-mer oligonucleotide (Forman et al. 1997).
Diploid leaf complementary RNA (cRNA) was used to empirically identify probe pairs that would distinguish between the AT and DT homeologs (where AT and DT refer to the two genomes in the allopolyploid). For example, the A-genome-specific probes hybridized better to the A-genome cRNA than to the D-genome cRNA (Figure 2A; supplemental Figure S2 at http://www.genetics.org/supplemental/). Many A-genome-specific probes also hybridized equally well to the D-genome cRNA, but this was not entirely unexpected, as our probe pairs were developed in silico without prior testing, and some probes had weak support for the existence of the putative SNP (e.g., few ESTs from the diploids; supplemental Figure S3 at http://www.genetics.org/supplemental/). Thus, to identify diagnostic probes, we conducted a mixed linear model analysis for each probe pair to find probe pairs for which the A-genome cRNA gave significantly higher signal than the D-genome cRNA for the A-genome probe, while the D-genome cRNA gave significantly higher signal than the A-genome cRNA for the D-genome probe. Significance was determined using P-values conservatively adjusted to control the false discovery rate (FDR; Benjamini and Hochberg 1995). A total of 1210 probes (461 genes) were found be diagnostic [adjusted (adj.) P < 0.05] with respect to AT and DT transcript levels; therefore, probes that hybridized significantly better to their targeted cRNA than to the alternative cRNA were considered diagnostic (Figure 2, Table 1).
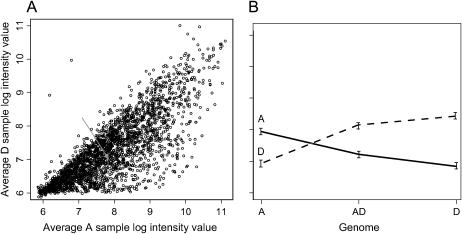
Figure 2.—
Intensities of diagnostic probes on the custom Nimblegen microarray. (A) Many probes designed to target the A-genome had a genome-specific bias when hybridized with D-genome cRNA. While each microarray had both A- and D-genome-specific probes, only the results of A-genome-specific probes are illustrated in A. Each log-transformed, median-adjusted dot in the scatter plot represents the average signal intensities of four replicate microarray hybridizations for the A-genome cRNA and D-genome cRNA on the x- and y-axis, respectively. The shaded arrow points to a dot representing the A and D hybridization values of the A-genome probe (solid line) in B. (B) An example of a reciprocally diagnostic pair of probes (CL10115Contig1 at position 895; see Figure 1 and supplemental Table S3 at http://www.genetics.org/supplemental/), showing significantly different expression levels when hybridized with labeled A- and D-genome cRNA from diploid leaves. Each probe pair (micorarrays) was also hybridized with cRNA from allotetraploid (AD) leaves. Thus once diagnostic probe pairs were identified, putative expression levels of AT and DT loci were tested for equal bias (null hypothesis; Table 1). For plant material, leaf tissue samples were collected from two plants of G. arboreum (5265), G. hirsutum (Acala Maxxa), and G. raimondii (GN33). The G. hirsutum and G. raimondii leaf samples were collected from mature plants grown under supplemental lighting (16-hr day) in the Pohl Conservatory in Bessey Hall at Iowa State University. The G. arboreum leaf samples were collected from plants grown inside a growth chamber with 16-hr days incandescent and fluorescent lights at 25°. RNA was extracted from each sample using a hot-borate method (Wilkins and Smart 1996). For microarray hybridization, six cRNA samples were prepared according to standard protocols of the NimbleGen hybridization service (NimbleGen Systems, Madison, WI) using a modified Eberwine procedure (Eberwine et al. 1992). RNA was first checked on an Agilent Bioanalyzer, followed by first- and second-strand cDNA synthesis with the inclusion of a T7-RNA polymerase promoter. cRNA was produced from the double-stranded cDNA product using Ambion (Austin, TX) MegaScript via in vitro transcription with biotinylated cytidine triphosphate and biotinylated UTP. The cRNA was fragmented and split into two samples for independent hybridization on 12 NimbleGen microarrays, providing a technical hybridization replication. Once hybridized to the arrays, the bound cRNA was stained with Cy-3-strepavidin (Amersham Biosciences, Piscataway, NJ). Slides were scanned with a GenePix Scanner (Molecular Devices, Sunnyvale, CA) and spot intensity data were extracted using NimbleGen proprietary software. For microarray data analysis, raw spot intensity data from NimbleGen were imported in the R statistical package (R Development Core Team 2005). The data were log transformed and median normalized. Subsequent box plots were used to visualize potential hybridization inconsistencies and scatter plots were used to visualize consistencies between technical and biological replications (supplemental Figure S2 at http://www.genetics.org/supplemental/). The data were analyzed as a split-plot design where the “plots” were the plants of type A, D, or AD and the “split plots” were the probes of type A or D. A mixed linear analysis was conducted separately for each pair of probes using PROC MIXED in SAS (Cary, NC). Each mixed linear model included fixed effects for plant types, probe types, and their interaction, along with random effects for biological replicates, technical replicates, and interaction between probe type and biological replication to allow for proper treatment of technical replication in the split-plot analyses. Significance values were adjusted for a false-discovery rate of 5 and 1% (Benjamini and Hochberg 1995).
TABLE 1.
Diagnostic oligonucleotide probes for diploid Gossypium and expression bias in their derived allopolyploid
| Both probes are significantly different (diagnostic probes) | No. of duplicated genes where the two homeologs exhibited unequal expression | |||
|---|---|---|---|---|
| Level of FDR | Adj. P < 0.05 | Adj. P < 0.01 | Adj. P < 0.05 | Adj. P < 0.01 |
| Probe pairs (n = 2375) | 1210 | 964 | 716 | 471 |
| A > D = 391a | A > D = 263a | |||
| D > A = 325a | D > A = 208a | |||
| Genes (n = 701) | 461 | 393 | 276b | 234b |
| A > D = 150 | A > D = 131 | |||
| D > A = 126 | D > A = 103 | |||
The adjusted P-value (FDR) was used to determine significant differences among probe intensities (Benjamini and Hochberg 1995). On the basis of the expectation of equal expression, there was a significant difference in the number of genes with an A-genome bias compared to those with a D-genome bias. A relatively small difference in total gene number was observed when probes were considered diagnostic at the 0.05 or 0.01 level.
χ2 significant at the 0.011 < adj. P < 0.014 level on the basis of an expectation of an equal number of probes.
The number of genes exhibiting homeolog bias includes genes targeted by a single diagnostic probe pair, genes where all probe pairs agreed in the direction of transcriptional bias, and 14 or 10 genes (adj. P < 0.05 and adj. P < 0.01, respectively) where four or more probe pairs had a consistent bias.
When the microarray probe sets were challenged with cRNA from the G. hirsutum allotetraploid, which contains both AT- and DT-genomes, many diagnostic probes were found to have unequal expression levels (Table 1). Within the subset of 1210 diagnostic probe pairs, our null hypothesis for each gene was equal expression of the AT and DT homeologs in the allotetraploid transcript pool. The null hypothesis was rejected for 716 probe pairs, indicating unequal AT and DT expression levels (adj. P < 0.05) of many genes. Two hundred and seventy six of the 461 genes containing diagnostic probes had significantly different AT and DT expression levels. Ninety-nine of these loci were biased in a consistent direction when a gene was targeted by multiple probes while 77 other loci with multiple probes had ambiguous results (supplemental Figure S1 at http://www.genetics.org/supplemental/). This percentage (199 of 461; 43%) of biased expression in a polyploid genome is higher than that previously reported on much smaller scales (Adams et al. 2003; Mochida et al. 2003). Among the sampled genes reported here, the types of genes that had biased expression appeared to be random (supplemental Table S2 at http://www.genetics.org/supplemental/), much like transcription biases in wheat (Mochida et al. 2003). The data in Table 1 are suggestive, however, of a consistent preference for transcription of A-genome homeologs although χ2-tests indicated only the differences at the probe level to be significant.
A set of five genes was selected to verify the microarray results by single-strand conformational polymorphism (SSCP) analysis and by randomly sequencing cloned colonies (supplemental Table S2 at http://www.genetics.org/supplemental/). Primers were designed to amplify one or more targeted polymorphisms within contigs containing both A- and D-genome ESTs. Verification results for all of the genes agree with the microarray-based results in the direction of expression bias. CL15638Contig1 had a nonsignificant homeolog bias on the microarray, but was later found to have a bias via SSCP and sequencing (supplemental Table S2 at http://www.genetics.org/supplemental/). Four additional loci with ambiguous microarray results were further investigated for their expression bias (supplemental Table S3 at http://www.genetics.org/supplemental/). For two of the four, our verification results agreed with one of the two probes targeting these homeologous loci, suggesting that no expression bias existed. Another locus had several diagnostic probe sets in two different verification amplicons and significant biases were consistently supported by verification. For a fourth ambiguous locus, the correct direction of homeolog bias was determined by verification. Within these ambiguous results, perhaps cross-hybridization of probes to other family members could explain the inconsistent microarray results among the putatively diagnostic probe pairs. In summary, our microarray results suggest that homeologous expression level biases may be widespread in the allotetraploid nucleus; however, our investigation of ambiguous microarray results suggests that more probes per gene would be useful in future experiments.
We note that leaves, the only organ used in this study, consist of many different cell types including trichomes, epidermis, xylem, phloem, etc. Thus, homeologous transcript levels within a leaf RNA extract represent an average expression level of all these different cell types. In this light, perhaps it is not surprising that the largest biases between homeologous loci were found in differentiated tissues with fewer types of cells, such as petals (Adams et al. 2003). Because the methodology described here permits monitoring of homeolog-specific patterns of gene expression, custom microarrays may prove to be one of the tools necessary for the biotechnological improvement of cotton fiber. These and comparable arrays may also yield insights into fundamental processes of regulatory networks and transcriptional controls in cotton as well as other polyploid plants.
Acknowledgments
We gratefully acknowledge the National Science Foundation Plant Genome Program and the U. S. Department of Agriculture–National Research Initiative for financial support.
References
- Adams, K. L., and J. F. Wendel, 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Adams, K. L., R. Cronn, R. Percifield and J. F. Wendel, 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57: 289–300. [Google Scholar]
- Eberwine, J., H. Yeh, K. Miyashiro, Y. Cao, S. Nair et al., 1992. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA 89: 3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman, J. E., I. D. Wilson, D. Stern, R. P. Rava and M. O. Trulson, 1997. Thermodynamics of duplex formation and mismatch discrimination on photolithographically synthesized oligonucleotide arrays in molecular modeling of nucleic acids, pp. 206–228 in Molecular Modeling of Nucleic Acids, edited by N. B. Leontis and J. Santa Lucia, Jr. ACS Publications, Oxford University Press, Oxford.
- Grant, V., 1981. Plant Speciation. Columbia University Press, New York.
- Kashkush, K., M. Feldman and A. A. Levy, 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch, I. J., and M. D. Bennett, 1997. Polyploidy in angiosperms. Trends Plant Sci. 2: 470–476. [Google Scholar]
- Mochida, K., Y. Yamazaki and Y. Ogihara, 2003. Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol. Gen. Genet. 270: 371–377. [DOI] [PubMed] [Google Scholar]
- Nuwaysir, E. F., W. Huang, T. J. Albert, J. Singh, K. Nuwaysir et al., 2002. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 12: 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn, T. C., J. Chris Pires, J. A. Birchler, D. L. Auger, Z. Jeffery Chen et al., 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2005. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
- Senchina, D. S., I. Alvarez, R. C. Cronn, B. Liu, J. Rong et al., 2003. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20: 633–643. [DOI] [PubMed] [Google Scholar]
- Stajich, J. E., D. Block, K. Boulez, S. E. Brenner, S. A. Chervitz et al., 2002. The Bioperl toolkit: perl modules for the life sciences. Genome Res. 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall, J. A., J. M. Swanson, K. Haller, R. A. Rapp, M. E. Sparks et al., 2006. A global assembly of cotton ESTs. Genome Res. 16: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., L. Tian, H.-S. Lee, N. E. Wei, H. Jiang et al., 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., and R. C. Cronn, 2003. Polyploidy and the evolutionary history of cotton. Adv. Agron. 78: 139–186. [Google Scholar]
- Wilkins, T. A., and L. B. Smart, 1996. Isolation of RNA from plant tissue, pp. 21–41 in A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis, edited by P. A. Krieg. Wiley-Liss, New York.