Abstract
A common feature of interspecific animal and plant hybrids is the uniparental silencing of ribosomal RNA gene transcription, or nucleolar dominance. A leading explanation for the genetic basis of nucleolar dominance in animal hybrids is the enhancer-imbalance model. The model proposes that limiting transcription factors are titrated by a greater number of enhancer-bearing subrepeat elements in the intergenic spacer (IGS) of the dominant cluster of genes. The importance of subrepeats for nucleolar dominance has repeatedly been supported in competition assays between Xenopus laevis and X. borealis minigene constructs injected into oocytes. However, a more general test of the importance of IGS subrepeats for nuclear dominance in vivo has not been conducted. In this report, rRNA gene expression was examined in interpopulation hybrids of the marine copepod Tigriopus californicus. This species offers a rare opportunity to test the role of IGS subrepeats in nucleolar dominance because the internal subrepeat structure, found in the IGS of virtually all animal and plant species, is absent in T. californicus. Our results clearly establish that nucleolar dominance occurs in F1 and F2 interpopulation hybrids of this species. In the F2 generation, nucleolar dominance appears to break down in some hybrids in a fashion that is inconsistent with a transcription factor titration model. These results are significant because they indicate that nucleolar dominance can be established and maintained without enhancer-bearing repeat elements in the IGS. This challenges the generality of the enhancer-imbalance model for nucleolar dominance and suggests that dominance of rRNA transcription in animals may be determined by epigenetic factors as has been established in plants.
A common pattern of altered gene expression in interspecific hybrids is the uniparental transcriptional silencing of ribosomal RNA (rRNA) genes. In nonhybrid diploid genomes, both rRNA gene clusters on homologous chromosomes are expressed, but in interspecific hybrids, transcription of the array from one parent is frequently silenced (e.g., Blackler and Gecking 1972). This pattern of expression, called nucleolar dominance, is a hybrid phenomenon that has been documented in many interspecific plant and animal hybridizations (Reeder 1985; Pikaard 2000a,b; Grummt and Pikaard 2003). Despite the widespread occurrence of nucleolar dominance little is known about how the pattern of dominance is established or why it occurs.
The rRNA gene family in plants and animals consists of tandemly arrayed repeats of the 18S, 5.8S, and 28S structural genes (Long and Dawid 1980). Each repeat unit consists of all three structural genes and the cis-regulatory elements necessary for transcription initiation and termination. Regulatory features are located in the intergenic spacer (IGS) and typically include a promoter and arrays of internal subrepeats thought to act as enhancers of transcription (Busby and Reeder 1983; Labhart and Reeder 1984). The broad conservation of enhancer function of these subrepeats in Drosophila (Grimaldi and Di Nocera 1988), Xenopus (Pape et al. 1989), and mouse (Kuhn et al. 1990) and structural similarities across eukaryotes suggest broad conservation of function (Reeder 1990). Transcription of the ribosomal genes is accomplished by RNA polymerase I (Pol I). This polymerase and its associated transcription factors are dedicated solely to transcription of the rRNA genes and promoter recognition is specific to the promoter in the IGS (Grummt 2003). This dedication of Pol I transcription apparatus to a single family of genes may facilitate rapid coevolution of components of the Pol I transcription apparatus to the rRNA gene promoter (Grummt et al. 1982). Tests of this hypothesis have demonstrated that the RNA Pol I transcription apparatus is not interchangeable among distantly related species such as mouse and human (Grummt et al. 1982) and Drosophila virilis and D. melanogaster (Kohorn and Rae 1982). In most cases, it is unclear how this unusual feature of the rRNA transcription apparatus may contribute to nucleolar dominance (Miesfeld et al. 1984; Reeder and Roan 1984; Reeder 1985; Frieman et al. 1999).
Studies of interspecific hybrids between Xenopus laevis and X. borealis have suggested a mechanism for nucleolar dominance. In hybrids, X. laevis rRNA genes are transcriptionally dominant over X. borealis (Honjo and Reeder 1973). Transcriptional silencing of X. borealis genes has been hypothesized to result from the ability of the X. laevis cluster to more effectively sequester limiting transcription factors (Reeder and Roan 1984). Support for this model has come from transcription assays with artificial minigene constructs. When constructs bearing X. laevis regulatory elements are co-injected into oocytes with constructs containing X. borealis regulatory features, the X. laevis construct shows transcriptional dominance, thus mimicking the situation observed in interspecific hybrids. These assays have suggested that transcriptional dominance could be conferred to X. laevis genes as a result of fourfold more enhancers (Reeder and Roan 1984) or stronger spacer promoters (i.e., repeated promoter-like sequences necessary for full enhancer function in Xenopus; Caudy and Pikaard 2002) in the X. laevis IGS. Either of these mechanisms could deprive the X. borealis genes of transcription factors and result in transcriptional dominance of X. laevis arrays.
This enhancer-imbalance model is attractive because it explains all general features of nucleolar dominance and simultaneously explains how transcriptional dominance is established and maintained through subsequent cell divisions (Reeder 1985). However, this model has recently come under scrutiny because of evidence that enhancer imbalance may not contribute to nucleolar dominance in plant hybrids (Chen et al. 1998; Frieman et al. 1999). Studies of allotetraploid hybrids in Brassica and Arabidopsis have clearly established that modifications to chromatin structure are at least partially responsible for rRNA gene silencing. Nucleolar dominance in these species appears to be an epigenetic phenomenon that is inherited through chromatin modifications that are introduced by a self-reinforcing cycle of cytosine methylation and histone deacetylation (Chen and Pikaard 1997; Lawrence et al. 2004). Although it is not clear how the repressive chromatin state of the underdominant array is originally specified, modifications to chromatin structure account for how transcriptional silencing occurs and how the transcriptional status of genes can be transmitted through subsequent cell divisions.
The importance of chromatin configuration raises questions about the relationship between the nucleolar dominance-like pattern observed in assays with artificial minigene constructs in Xenopus and the phenomenon of nucleolar dominance. It may be that the enhancer-imbalance mechanism at work in transcription assays does not contribute to nucleolar dominance in the chromosomal context of a hybrid nucleus (Caudy and Pikaard 2002). If so, this may indicate that enhancer-bearing subrepeat elements, which are thought to be important in nucleolar dominance in Xenopus, may not be necessary for either the establishment or the maintenance of nucleolar dominance.
An unusual feature of the rRNA gene array in the marine copepod Tigriopus californicus provides a means for assessing the role of subrepeat elements in nucleolar dominance. In a recent survey of genetic variation in the rRNA gene cluster, we found substantial interpopulation divergence in the most conserved section of the IGS and the external transcribed spacer (ETS) (Burton et al. 2005). Uncorrected interpopulation divergences in this region were found to exceed 10% among all populations and exceeded 35% between northern populations and a partially reproductively isolated population in Baja California, Mexico (Ganz and Burton 1995). In addition to extensive IGS divergences, the region upstream of the transcription initiation site does not contain subrepeats (Burton et al. 2005). In other species, these subrepeats are known to contain enhancers of transcription and are otherwise a characteristic feature of animal IGS sequences (Reeder 1990). Because these repeats are hypothesized to contribute to nuclear dominance (Reeder and Roan 1984), studies of ribosomal gene expression in T. californicus hybrids may provide new insight into the mechanism(s) responsible for uniparental silencing of rRNA genes.
Here we report patterns of rRNA gene expression in interpopulation hybrids of T. californicus. We assess whether transcription of rRNA is biased in F1 and F2 hybrids and demonstrate a clear pattern of nucleolar dominance in the F1 generation. In F2 hybrids, nucleolar dominance appears to partially break down, with some hybrids showing complete dominance and others partial dominance or codominance in rRNA gene expression. Given the unusual structure of the T. californicus IGS, these results imply that repetitive elements in the IGS are not required for uniparental gene silencing of rRNA transcription.
MATERIALS AND METHODS
Life history:
T. californicus is an obligately sexual harpacticoid copepod species that inhabits intertidal rocky shelves along the Pacific Coast of North America. Limited gene flow occurs among populations of T. californicus and populations are frequently divergent at mitochondrial and nuclear gene loci (e.g., Burton and Feldman 1981; Burton and Lee 1994; Edmands 2001). In laboratory crosses, F1 interpopulation hybrids are vigorous, while F2 hybrids suffer from outbreeding depression (Burton 1990; Edmands 1999; Edmands et al. 2005). The mechanism of sex determination is unknown in T. californicus, although it appears to be influenced by both genetic and environmental factors (Voordouw and Anholt 2002). There is no evidence of sex chromosomes in this species (Ar-rushdi 1963).
Experimental crosses:
Samples were collected from Abalone Cove, Los Angeles County (AB), San Diego (SD), and Santa Cruz (SC), California, and maintained in laboratory cultures at 20° with a 12:12 light-dark cycle. Collecting locales are the same as those in previous studies (e.g., Burton and Feldman 1981; Burton and Lee 1994; Ganz and Burton 1995). Interpopulation hybrids were obtained from crosses between AB × SD and SD × SC populations and their reciprocals. Pair crosses were established following the method of Burton and Feldman (1981) by isolating virgin females and mating them in mass culture with males from an alternate population. F1 hybrid males were either isolated for DNA and RNA extraction or allowed to mate with F1 females for production of F2 intercross hybrids. Second generation hybrids were subsequently obtained by transferring fertilized F1 females to new cultures for production of F2 progeny. All experiments were conducted on F1 and F2 adult males.
SNP detection:
Detection of population-specific rRNA genes and transcripts in interpopulation hybrids is limited in T. californicus by low levels of among-population divergence in the 18S and 28S structural genes (Burton et al. 2005). Among the focal populations, a single nucleotide polymorphism (SNP) at position 189 of the 18S is fixed for cytosine in SD and thymine in AB and SC populations (see GenBank accessions: AY588143.1, AY588134.1, and AY588140.1; Burton et al. 2005). The AB and SC populations are indistinguishable in the 18S and 28S rRNA genes. This restricted our analysis to SD × AB and SD × SC interpopulation crosses.
Population-specific copies of rDNA and the corresponding rRNA transcripts were distinguished on the basis of the T189C SNP with a fluorescence resonance energy transfer (FRET) hybridization probe assay. The method discriminates between SNP variants on the basis of the loss of fluorescence associated with melting of hybridization probes in a post-PCR melting curve assay (see below). Total RNA and genomic DNA were simultaneously isolated from single adult copepods. Individual animals (∼30 μg wet weight) were submersed live in 40 μl of TRI reagent (Sigma-Aldrich, St. Louis) with ∼150 mg of zirconia/silica beads (Biospec, Bartlesville, OK) in 500 μl RNase/DNase-free microtubes. Cells were disrupted by bead-beating each sample for 20 sec in a FastPrep 120 bead beater (Qbiogene, Carlsbad, CA) with a rotor speed of 4.5 m/sec. RNA and DNA were subsequently extracted following the TRI reagent extraction protocol. RNA was DNase treated and first-strand cDNA synthesis was performed with Superscript II (Invitrogen, Carlsbad, CA) with an 18S rRNA gene-specific primer, 5′-GTGCGATCAGCAAAAAGT-3′. A 200-bp fragment containing the T189C polymorphism was PCR amplified with the primer used for cDNA synthesis and a forward primer, 5′-AGGTGAAGCTGCGAAC-3′. The cycling profile consisted of 95° for 30 sec, 50° for 30 sec, and 72° for 30 sec for 40 cycles in a conventional thermal cycler. Identical conditions were used to amplify genomic DNA. PCR products were visualized with ethidium bromide-stained agarose gels and quantified with Hoechst dye on a fluorescence plate reader. PCR controls for contamination of RNA with genomic DNA were conducted for all samples. In this control, DNase-treated RNA was added directly to a PCR reaction. Samples were considered free of contaminating DNA if PCR amplification failed. In some cases, contaminating genomic DNA was detected and a second DNase treatment was conducted. These samples were checked a second time for DNA contamination prior to cDNA synthesis.
The donor probe (5′-GCGGTAATTCTGGAGCTAATACATGCTACAA-3′) for the melting curve assay was end-labeled with a 3′ fluorescein. The acceptor probe (5′-CCCTTCACGTTGTGTGAGGG-3′) was positioned 2 bp downstream of the 3′ end of the donor probe and was end-labeled with a 5′ LC Red 640 and a 3′ phosphate group (Synthegen, Houston). A cytosine located 5 bp from the 5′ end of the acceptor probe produces a C:A heteroduplex when hybridized with SD-derived DNA and a C:G base pair with SC- and AB-derived DNA.
Melting curves were generated in a LightCycler (Roche, Mannheim, Germany) with equal concentrations of unpurified PCR-amplified DNA, 3.25 mm MgCl2, and 300 nm of each probe in glass capillaries (Roche Molecular Biochemicals, Indianapolis). Each assay consisted of an initial denaturation step of 0 sec at 95° followed by annealing at 45° for 15 sec and ramping of the reaction to 80° at rates between 0.2°/sec and 0.5°/sec. Fluorescence was detected continuously and the instantaneous rate of fluorescence change was determined by calculating the negative derivative of the melting curve. Plots of the negative derivative of fluorescence vs. temperature yielded one or two distinct peaks depending on whether one or both SNP variants were present in the sample. These peaks correspond to the temperatures at which there is a maximum instantaneous rate of fluorescence loss due to melting of the acceptor probe from homo- and heteroduplexes.
In our assay, there was a tight linear correlation between the relative frequency of SNP variants and relative melting peak heights (Figure 1). This relationship was used to quantify the relative frequencies of each SNP in a DNA or cDNA pool [see Lyon et al. (2001) for a related method]. Standard curves were generated by combining population-specific PCR-amplified products in ratios of 5:1, 2:1, 1:1, 1:2, and 1:5 (i.e., relative frequencies of 5/6, 2/3, 1/2, 1/3, and 1/6) followed by melting curve analysis in parallel with unknown samples. In a few samples, the ratio of SNP variants inferred from a hybrid fell outside of the range of the standard curve. Ratios from these samples were calculated by extrapolating from the standard curve. This was done to indicate that both SNP variants were detected in such samples and to distinguish them from samples in which we found no evidence of both SNP variants. When only one SNP variant was detected, the frequency of the undetected SNP was inferred to be less than 1/6 and assigned a value of zero.
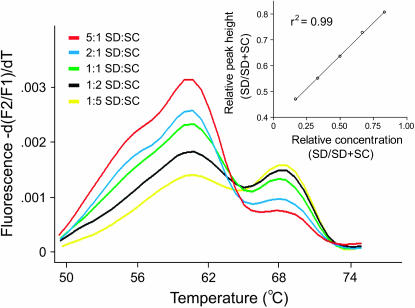
Figure 1.—
Melting peak profile derived from melting curve assays with the specified ratios of SC and SD PCR-amplified genomic DNA. The inset is a standard curve derived from these peaks.
The validity of the SNP quantification method was independently verified with a restriction assay. The 18S rRNA gene was amplified with a second pair of primers (F: 5′-AGTCATATGCTTGTCTCAAAG-3′, R: 5′-GGATGAGTCCGGTATCG-3′) that flanked the T189C SNP. PCR products were digested with DraIII and population-specific fragments were quantified by densitometry with a ChemiImager 5500 CCD imager with AlphaEase FC software (Alpha Innotech).
RESULTS
SNP detection:
Detection of biased transcription in interpopulation hybrids required development of a genetic marker to distinguish between population-specific copies of 18S rDNA and the corresponding rRNA transcripts. This was accomplished with a FRET hybridization probe assay that discriminates between the population-specific T189C SNP. The method is a post-PCR melting curve assay that capitalizes on melting temperature differences between homo- and heteroduplexes formed between an “acceptor” probe and the two SNP variants. The acceptor probe hybridizes to the SNP and flanking sites, while a “donor” probe anneals to the template immediately 5′ of the acceptor. When both probes are hybridized to the template, excitation of a 3′ fluorescein on the donor probe initiates a resonance energy transfer to a 5′ fluorophore on the adjacent acceptor. This results in a fluorescent emission. During the melting curve assay, the acceptor probe denatures from the homo- and heteroduplexes formed between the two SNP variants at different temperatures because of differences in their thermal stabilities. This can be monitored as a loss of fluorescence associated with the cessation of FRET between the donor and acceptor probes. Plots of the negative derivative of fluorescence vs. temperature yield one or two distinct melting peaks depending on whether one or both SNP variants are present in the sample. This method is commonly used for SNP genotyping (Bernard et al. 1998) and has also been used to quantify the relative frequency of SNP variants in a PCR product pool (Lyon et al. 2001).
The FRET hybridization probe assay readily distinguished between the two SNP variants in PCR-amplified DNA and cDNA pools. In melting curve assays, the melting temperatures of the acceptor probe from the targets were 60–62° for the SD-derived SNP variant and ∼67–69° for the variant specific to the SC and AB populations. In Figure 1, the difference in melting temperature can be seen in a plot of the negative derivative of the melting curve (“melting peak profile”), where the peak at ∼60° corresponds to the SNP variant from SD and the peak at ∼68° corresponds to the SNP variant from SC. This facilitated easy discrimination of population-specific rRNA transcripts and rRNA genes from individual hybrid copepods.
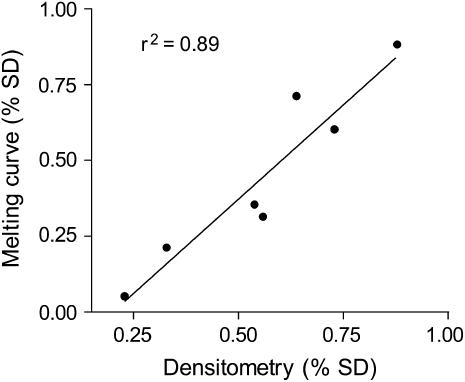
In our assay, the relative melting peak height from each polymorphism was tightly correlated (r2 = 0.99) with the relative frequency of the two SNP variants in a sample (Figure 1). This relationship allowed us to quantify the relative abundance of population-specific transcripts and multicopy rRNA genes from cDNA and genomic DNA derived from a single hybrid. The relationship between the ratio of melting peak height and the ratio of each SNP variant was linear over a range of values between 5:1 and 1:5 (Figure 1). However, inferences outside this range were frequently impossible because more skewed ratios sometimes failed to yield two distinct melting peaks. As a result, the presence of an SNP at low frequency (<1/6) may not have been distinguishable from its absence in a pool of PCR products. The linear relationship established in the melting curve analysis was independently verified with a restriction enzyme-based densitometry assay (Figure 2). A plot of the relative frequencies of the T189C SNP variants shows a strong correlation (r2 = 0.89) between estimates from the two techniques.
Figure 2.—
The relationship between SNP frequencies estimated with densitometry and the FRET melting curve assay.
F1 hybrids:
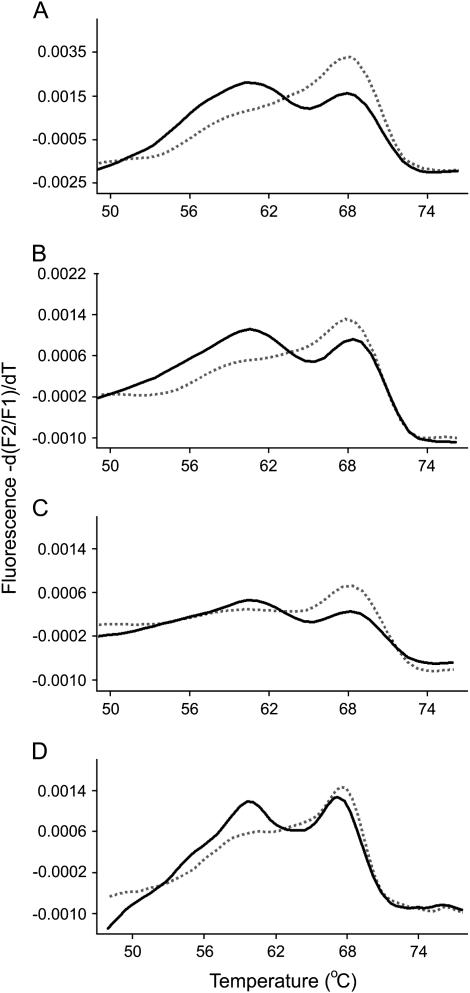
Figure 3 presents melting peak profiles from typical F1 hybrids from the SD × SC and AB × SC crosses and their reciprocals. In each plot, one profile represents the melting curve obtained from rDNA amplified from genomic DNA isolated from a single hybrid. The other represents the cDNA profile derived from rRNA extracted from the same animal. rRNA gene copies from both populations were evident in genomic DNA of F1 hybrids as expected. In the majority of F1 samples, transcripts representing only one of the two parental rRNAs were detected in the cDNA (Figure 3). This pattern of transcriptional dominance was the same for all F1 hybrids with AB genes dominant over SD in the SD × AB cross and SC genes dominant over SD in the SD × SC cross. These results establish a clear pattern of nucleolar dominance in the F1 generation and show that the dominance relationship is independent of the population origin of the maternal parent.
Figure 3.—
Melting peak profiles derived from PCR-amplified cDNA and genomic DNA from typical F1 hybrids. In each plot, the solid line represents genomic DNA and the dashed line represents cDNA. (A) SD♀ × AB♂ F1 male. (B) AB♀ × SD♂ F1 male. (C) SD♀ × SC♂ F1 male. (D) SC♀ × SD♂ F1 male.
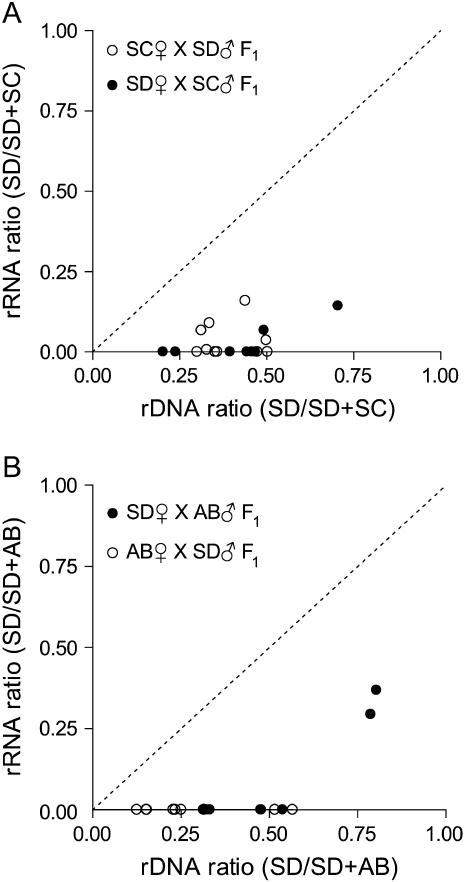
The results from 39 F1 samples are illustrated in Figure 4, A and B. In each plot, ratios of population-specific copies of rRNA (y-axis) or rDNA (x-axis) are expressed as SD/(SD + SC) for the SD × SC cross (Figure 4A) or SD/(SD + AB) for the SD × AB cross (Figure 4B). Each point represents the ratio of population-specific rRNA transcripts and the rDNA ratio estimated from the genome of a single F1 hybrid. The codominant expectation in these plots is indicated by a dotted line. Points above or below the line suggest dominance in rRNA gene expression. The results in Figure 4 indicate a strong bias in F1 hybrids favoring rRNA transcription of AB or SC over SD rRNA genes. In a few F1 hybrids, both population-specific transcripts were detected (Figure 4), but transcription in all cases was strongly biased against SD genes.
Figure 4.—
Plot of rRNA and rDNA ratios for F1 interpopulation hybrids. (A) SD × SC. (B) AB × SD. The dashed line represents the expectation for hybrids with codominant rRNA expression.
F2 hybrids:
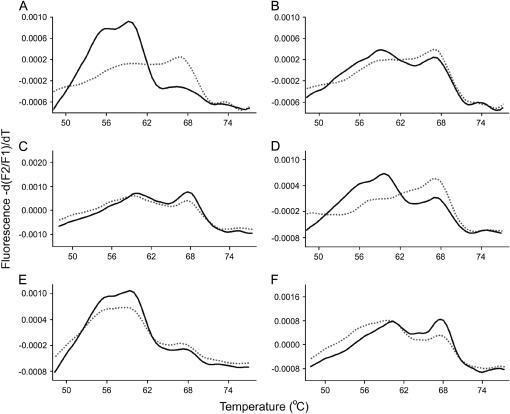
Melting peak profiles from F2 hybrids showed a range of expression phenotypes in the SC♀ × SD♂ cross (Figures 5 and 6A). In contrast to F1 hybrids, in some cases F2 hybrids showed partial dominance or codominance in rRNA expression. This is illustrated in melting peak profiles in which SNP variants were detected at approximately equal proportions in both genomic DNA and cDNA (e.g., Figure 5, C, E, and F). Hybrids with partial dominance or codominant phenotypes are located on or near the dotted line in Figure 6A and had a range of SD rDNA gene dosages. This is evident in Figure 6A, which shows that hybrids with relative complements of SD rDNA of <1/6 to >5/6 showed evidence of partial dominance or codominance in rRNA transcription.
Figure 5.—
Melting peak profiles derived from PCR-amplified cDNA and genomic DNA from F2 hybrids from the SC♀ × SD♂ cross. In each plot the solid line represents genomic DNA and the dashed line represents cDNA. A–F demonstrate the range of profiles observed in this cross.
Figure 6.—
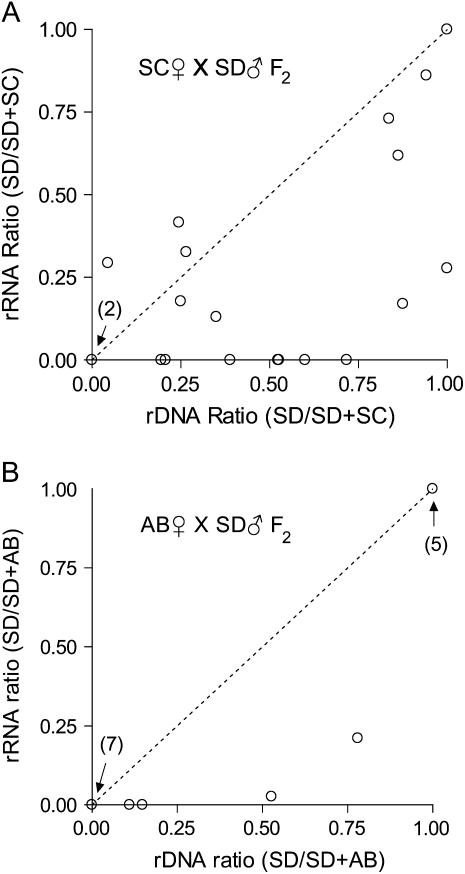
Plot of rRNA and rDNA ratios for F2 interpopulation hybrids. (A) SC♀ × SD♂ F2 adult males. (B) AB♀ × SD♂ F2 adult males. The dashed line represents the expectation for hybrids with codominant rRNA expression.
The pattern of rRNA expression inferred from 7 SC♀ × SD♂ F2 hybrids showed complete dominance similar to the F1 generation. In these samples, population-specific transcripts were detected from only one source population (e.g., 5D). In all cases where complete dominance was inferred, the dominance relationship was the same as that observed in the F1 with SC rRNA genes dominant over SD. Finally, 2 hybrids in the SC♀ × SD♂ cross were inferred to have rDNA from only one population consistent with recovery of parental rDNA genotypes. In 1 SC♀ × SD♂ hybrid we were unable to detect the presence of SC ribosomal genes, but we detected both SD and SC SNP variants in the cDNA. If SC genes were present at low frequency (e.g., as a result of segregation of two or more unlinked arrays, see discussion) then they may not have been detected in our assay. Results from the AB♀ × SD♂ cross are summarized in Figure 6B. Only 4 of 16 F2 hybrids were inferred to have rRNA genes from both source populations, but all 4 hybrids showed nucleolar dominance with the same dominance relationship observed in the F1 generation.
DISCUSSION
The enhancer-imbalance model proposes that enhancer-bearing subrepeat elements in the IGS are responsible for nucleolar dominance. In Xenopus, dominance of the X. laevis cluster has been attributed to fourfold more enhancers in the X. laevis IGS and/or higher activity of the X. laevis spacer promoter (Reeder and Roan 1984; Caudy and Pikaard 2002). Either of these mechanisms could limit the availability of transcription factors to the underdominant cluster and result in silencing of X. borealis genes. This enhancer-imbalance model is attractive because it accounts for the absence of maternal effects on the pattern of dominance (Honjo and Reeder 1973), why nucleolar dominance is observed in hybrid nuclei (Blackler and Gecking 1972), and also provides a mechanism for the establishment of dominance early in development (Reeder and Roan 1984; Reeder 1985). However, much of what is known about nucleolar dominance in Xenopus has come from competition assays between minigene constructs (Reeder and Roan 1984). When constructs with X. laevis regulatory elements are co-injected into oocytes with constructs containing X. borealis regulatory sequences, the pattern of transcription mimics the dominance relationship observed in the hybrids (i.e., minigenes containing the X. laevis spacer are dominant to constructs with the X. borealis spacer). While these assays have clearly established that transcriptional dominance could be determined by competition for limiting transcription factors, definitive evidence for the action of enhancer imbalance in a chromosomal context has not been established in hybrids of Xenopus or any other organism (Caudy and Pikaard 2002).
The unusual structure of T. californicus IGS provided an opportunity to evaluate whether enhancer-bearing subrepeats are essential for nucleolar dominance in vivo. We found that F1 hybrids from two interpopulation crosses show uniparental silencing of rRNA transcription despite the absence of internal subrepeats in the IGS. Consistent with other studies of nucleolar dominance, the pattern of dominance was highly repeatable among hybrid individuals from both crosses and was independent of the direction of the cross. This finding is significant because it suggests that nucleolar dominance in other species may be established and maintained in the absence of IGS subrepeats and challenges the generality of the enhancer-imbalance model. This highlights additional questions about whether the mechanism that produces nucleolar dominance-like expression of Xenopus minigene constructs is responsible for nucleolar dominance in the chromosomal context of a hybrid nucleus (Caudy and Pikaard 2002).
The pattern of partial dominance or codominance observed in F2 hybrids from the SC♀ × SD♂ cross is also inconsistent with predictions of the enhancer-imbalance model. A simple transcription factor titration model predicts that transcriptional dominance may be relaxed when there is a low dosage of the dominant array (e.g., Chen et al. 1998). In this situation, transcription factors may not be limiting and transcription of the underdominant cluster could occur. In T. californicus, there are at least two unlinked clusters of rRNA genes (Burton et al. 2005). Therefore, F2 intercross hybrids in the SC♀ × SD♂ cross can differ in dosage of the dominant and underdominant genes due to segregation in the F1. If transcription factor titration contributes to nucleolar dominance in T. californicus, hybrids with a low dosage of the dominant SC cluster may be expected to codominantly express their rRNA genes, but hybrids with high dosage are not. This was not observed. Rather the relaxation of dominance appears to be independent of SC rRNA gene dosage and genotypes with high and low relative copy numbers of rRNA genes codominantly express their ribosomal RNA genes. This suggests that some feature other than dosage of the dominant array of genes and the corresponding availability of transcription factors is responsible for nucleolar dominance in T. californicus. A similar observation in Arabidopsis suecica backcross hybrids has been interpreted as evidence against a transcription factor titration model (Chen et al. 1998).
Studies of interecotype hybrids of A. thaliana also show partially dominant or codominant transcription of rRNA in advanced generation hybrids. Lewis et al. (2004) reported that nucleolar dominance in F2 hybrids between the Cvi and Ler ecotypes ranged from partial dominance to complete dominance, similar to the results reported here. Examination of F8 hybrids in the same study also found a range of dominance phenotypes. However, complete dominance was restricted to genotypes homozygous for the Cvi chromosome 4 nucleolar organizer (NOR) and the Ler chromosome 8 NOR. Interestingly, the pattern of rRNA expression was found to segregate in the F8 generation as not all hybrids with the Cvi NOR4/Ler NOR8 genotype showed nucleolar dominance. This observation was partially attributable to an unlinked QTL (Lewis et al. 2004). The presence of hybrids with partial dominance/codominance and others with complete dominance in the SC♀ × SD♂ cross is also suggestive of a modifier locus segregating in this cross.
Is the enhancer-imbalance model consistent with observations from other hybrid organisms? In animals there is only limited data for comparison with Xenopus. Hybrids between D. melanogaster and D. simulans show some similarities to the situation observed in Xenopus. For example, while D. simulans genes are silenced in female F1 hybrids (Durica and Krider 1977), hybrid females with large deletions in the dominant D. melanogaster rRNA gene cluster express both D. melanogaster and D. simulans genes (Goodrich-Young and Krider 1989). This is consistent with a prediction of the enhancer-imbalance model—when transcription factors are not limiting, the underdominant genes may be transcribed. However, demonstration of chromosomal position effects on the pattern of dominance (Durica and Krider 1978) suggests that enhancer imbalance by itself is insufficient to explain the situation in Drosophila (Reeder 1985; Pikaard 2000a). In plants, tests of the enhancer-imbalance hypothesis have suggested that nucleolar dominance is unlikely to be attributable to competitive differences in the ability of rRNA gene clusters to sequester transcription factors (Chen et al. 1998; Frieman et al. 1999; Pikaard 2000a,b). Instead, there is direct evidence that epigenetic modification, including DNA methylation and histone deacetylation, contributes to rRNA silencing (Chen and Pikaard 1997; Lawrence et al. 2004). Such epigenetic changes are thought to collectively modify the chromatin state of the underdominant array making it refractory to transcription.
These observations from plants suggest a general model for how nucleolar dominance may be enforced in plants and animals (for review see Pikaard 2000a,b; Grummt and Pikaard 2003). While there is no direct support for an epigenetic mechanism in nucleolar dominance in animals, the results reported here and inconsistencies with the enhancer-imbalance model in other animals could point to a role for epigenetics. Future work on nucleolar dominance in animals should focus on whether epigenetic mechanisms are at work and how the dominance relationship is originally established. Additional insight into these questions may be gained from studies of T. californicus and other animal hybrids that initially should evaluate the methylation status of silenced rRNA genes.
Acknowledgments
We thank S. Harrison, C. Willett, and L. Chao for helpful discussions. We thank two anonymous reviewers whose comments greatly improved the manuscript. We also thank H. Huynh for assistance with laboratory crosses. This work was supported by National Science Foundation grants DEB9815424 and DEB0236363.
References
- Ar-rushdi, A. H., 1963. The cytology of achiasmatic meiosis in the female Tigriopus (Copepoda). Chromosoma 13: 526. [Google Scholar]
- Blackler, A. W., and C. A. Gecking, 1972. Transmission of sex cells of one species through the body of a second species in the genus Xenopus. II. Interspecific matings. Dev. Biol. 27: 385–394. [DOI] [PubMed] [Google Scholar]
- Bernard, P. S., M. J. Lay and C. T. Wittwer, 1998. Integrated amplification and detection of the C677T point mutation in the methylenetetrahydrofolate reductase gene by fluorescence resonance energy transfer and probe melting curves. Anal. Biochem 255: 101–107. [DOI] [PubMed] [Google Scholar]
- Burton, R. S, 1990. Hybrid breakdown in developmental time in the copepod Tigriopus californicus. Evolution 44: 1814–1822. [DOI] [PubMed] [Google Scholar]
- Burton, R. S., and M. W. Feldman, 1981. Population genetics of Tigriopus californicus. 2. Differentiation among neighboring populations. Evolution 35: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Burton, R. S., and B.-N. Lee, 1994. Nuclear and mitochondrial gene geneologies and allozyme polymorphism across a major phylogeographic break in the copepod Tigriopus californicus. Proc. Natl. Acad. Sci. USA 91: 5197–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, R. S., E. C. Metz, J. M. Flowers and C. S. Willett, 2005. Unusual structure of ribosomal DNA in the copepod Tigriopus californicus: intergenic spacer sequences lack internal subrepeats. Gene 344: 105–113. [DOI] [PubMed] [Google Scholar]
- Busby, S. J., and R. H. Reeder, 1983. Spacer sequences regulate transcription of ribosomal gene plasmids injected into Xenopus embryos. Cell 34: 989–996. [DOI] [PubMed] [Google Scholar]
- Caudy, A. A., and C. S. Pikaard, 2002. Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes. J. Biol. Chem. 277: 31577–31584. [DOI] [PubMed] [Google Scholar]
- Chen, J. Z., and C. S. Pikaard, 1997. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11: 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J., L. Comai and C. S. Pikaard, 1998. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. USA 95: 14891–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durica, D. S., and H. M. Krider, 1977. Studies on the ribosomal RNA cistrons in interspecific Drosophila hybrids. I. Nucleolar dominance. Dev. Biol. 59: 62–74. [DOI] [PubMed] [Google Scholar]
- Durica, D. S., and H. M. Krider, 1978. Studies on ribosomal RNA cistrons in inter-specific Drosophila hybrids. 2. Heterochromatic regions mediating nucleolar dominance. Genetics 89: 37–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands, S., 1999. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution 53: 1757–1768. [DOI] [PubMed] [Google Scholar]
- Edmands, S., 2001. Phylogeography of the intertidal copepod Tigriopus californicus reveals substantially reduced population differentiation at northern latitudes. Mol. Ecol. 10: 1743–1750. [DOI] [PubMed] [Google Scholar]
- Edmands, S., H. V. Feaman, J. S. Harrison and C. C. Timmerman, 2005. Genetic consequences of many gernations of hybridization between divergent copepod populations. J. Hered. 96: 114–123. [DOI] [PubMed] [Google Scholar]
- Frieman, M., Z. J. Chen, J. Saez-Vasquez, L. A. Shen and C. S. Pikaard, 1999. RNA polymerase I transcription in a Brassica interspecific hybrid and its progenitors: tests of transcription factor involvement in nucleolar dominance. Genetics 152: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz, H. H., and R. S. Burton, 1995. Genetic differentiation and reproductive incompatibility among Baja California populations of the copepod Tigriopus californicus. Mar. Biol. 123: 821–827. [Google Scholar]
- Goodrich-Young, C., and H. M. Krider, 1989. Nucleolar dominance and replicative dominance in Drosophila interspecific hybrids. Genetics 123: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi, G., and P. P. Di Nocera, 1988. Multiple repeat units in Drosophila melanogaster ribosomal DNA spacer stimulate rRNA precursor transcription. Proc. Natl. Acad. Sci. USA 85: 5502–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt, I., 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17: 1691–1702. [DOI] [PubMed] [Google Scholar]
- Grummt, I., and C. S. Pikaard, 2003. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4: 641–649. [DOI] [PubMed] [Google Scholar]
- Grummt, I., E. Roth and M. R. Paul, 1982. rRNA transcription in vitro is species-specific. Nature 296: 173–174. [DOI] [PubMed] [Google Scholar]
- Honjo, T., and R. H. Reeder, 1973. Preferential transcription of Xenopus laevis ribosomal RNA in interspecies hybrids between Xenopus laevis and Xenopus mulleri. J. Mol. Biol. 80: 217–228. [DOI] [PubMed] [Google Scholar]
- Kohorn, B. D., and P. M. M. Rae, 1982. Accurate transcription of truncated ribosomal DNA templates in a Drosophila cell-free system. Proc. Natl. Acad. Sci. USA 79: 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A., U. Deppert and I. Grummt, 1990. A 140-base pair repetitive sequence element in the mouse ribosomal RNA gene spacer enhances transcription by RNA polymerase I in a cell-free system. Proc. Natl. Acad. Sci. USA 87: 7527–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart, P., and R. H. Reeder, 1984. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell 37: 285–289. [DOI] [PubMed] [Google Scholar]
- Lawrence, R. J., K. Earley, O. Pontes, M. Silva, Z. J. Chen et al., 2004. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Lewis, M. S., J. M. Cheverud and C. S. Pikaard, 2004. Evidence for nucleolus organizer regions as the units of regulation in nucleolar dominance in Arabidopsis thaliana interecotype hybrids. Genetics 167: 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, E. O., and I. B. Dawid, 1980. Repeated genes in eukaryotes. Ann. Rev. Biochem. 49: 727–764. [DOI] [PubMed] [Google Scholar]
- Lyon, E., A. Millson, M. C. Lowery, R. Woods and C. T. Wittwer, 2001. Quantification of HER2/neu gene amplification by competitive PCR using fluorescent melting curve analysis. Clin. Chem. 47: 844–851. [PubMed] [Google Scholar]
- Miesfeld, R., B. Sollner-Webb, C. Croce and N. Arnheim, 1984. The absence of a human-specific ribosomal DNA transcription factor leads to nucleolar dominance in mouse > human hybrid cells. Mol. Cell. Biol. 4: 1306–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, L. K., J. J. Windle, E. B. Mougey and B. Sollnerwebb, 1989. The Xenopus ribosomal DNA 60 base pair and 81 base pair repeats are position-dependent enhancers that function at the establishment of the preinitiation complex – analysis in vivo and in an enhancer-responsive in vitro system. Mol. Cell. Biol. 9: 5093–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard, C. S., 2000. a The epigenetics of nucleolar dominance. Trends Genet. 16: 495–500. [DOI] [PubMed] [Google Scholar]
- Pikaard, C. S., 2000. b Nucleolar dominance: uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant. Mol. Biol. 43: 163–177. [DOI] [PubMed] [Google Scholar]
- Reeder, R. H., 1985. Mechanisms of nucleolar dominance in animals and plants. J. Cell Biol. 101: 2013–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder, R. H., 1990. rRNA synthesis in the nucleolus. Trends Genet. 6: 390–394. [DOI] [PubMed] [Google Scholar]
- Reeder, R. H., and J. G. Roan, 1984. The mechanism of nucleolar dominance in Xenopus hybrids. Cell 38: 39–44. [DOI] [PubMed] [Google Scholar]
- Voordouw, M. J., and B. R. Anholt, 2002. Heritability of sex tendency in a harpacticoid copepod, Tigriopus californicus. Evolution 56: 1754–1763. [DOI] [PubMed] [Google Scholar]