Abstract
An understanding of the determinants of trait variation and the selective forces acting on it in natural populations would give insights into the process of evolution. The combination of long-term studies of individuals living in the wild and better genomic resources for nonmodel organisms makes achieving this goal feasible. This article reports the development of a complete linkage map in a pedigree of free-living Soay sheep on St. Kilda and its application to mapping the loci responsible for three morphological polymorphisms for which the maintenance of variation demands explanation. The map was derived from 251 microsatellite and four allozyme markers and covers 3350 cM (∼90% of the sheep genome) at ∼15-cM intervals. Marker order was consistent with the published sheep map with the exception of one region on chromosome 1 and one on chromosome 12. Coat color maps to chromosome 2 where a strong candidate gene, tyrosinase-related protein 1 (TYRP1), has also been mapped. Coat pattern maps to chromosome 13, close to the candidate locus Agouti. Horn type maps to chromosome 10, a location similar to that previously identified in domestic sheep. These findings represent an advance in the dissection of the genetic diversity in the wild and provide the foundation for QTL analyses in the study population.
AN area of fundamental research in evolutionary genetics concerns the closely related issues of understanding the determinants of trait variation in natural populations and understanding how genetic variation for traits is maintained in the face of natural selection. The first of these problems is often summarized as the “genetic architecture” question: in general we would like to know whether genes of large effect commonly segregate in natural populations or whether the infinitesimal model, i.e., that most traits are controlled by many genes of small effect, is appropriate—or, perhaps more likely, some configuration in between (Barton and Keightley 2002; Brem and Kruglyak 2005). Similarly, we would like to know to what extent genetic interactions such as dominance, pleiotropy, and epistasis contribute to the evolutionary dynamics of a population. The second problem was long ago identified by Fisher (1958): How is it that genetic variation for traits persists when selection is so often directional? The answer to this question must lie not only in the genetic architecture question, but also in the modes of selection and their temporal and spatial stability.
In principle, the arrival of abundant molecular markers, genetic maps, and whole-genome sequences allows us to address both genetic architecture and selection in much greater depth than ever before, since the role of variation at individual loci can be assessed. Mapping trait loci is a starting point for providing information on the genetic architecture of a trait in terms of the number of genes involved, relative effect, and mode of expression (Erickson et al. 2004; Slate 2005). In turn, this allows study of the relationship between phenotype and genotype and inference of the selective forces acting on the critical locus. Furthermore, by mapping genes, it is possible to test for the presence of gene-by-gene (epistatic) and gene-by-environment interactions, which are thought to contribute to phenotypic variation in natural and controlled settings (Carlborg and Haley 2004; Erickson 2005). In addition, the discovery of the map location of genes that influence phenotypic variation means that patterns of linkage disequilibrium (LD) and haplotype structure can be examined, which may provide insights about population history and selection. Unfortunately, some of the characteristics that make experimental populations so practical for linkage mapping also restrict the degree to which findings can be extrapolated to natural populations. Usually, geneticists generate segregating populations derived from one or a few pair of parents, which are often inbred and selected for the extreme phenotypes. In addition, the population is raised in a uniform and controlled environment (e.g., a greenhouse) where nongenetic sources of phenotypic variation are minimized. On the one hand, this strategy maximizes the power of analysis; i.e., it increases the probability of finding a statistical association between marker genotype and phenotypic trait, but on the other hand, as the aim of genetic research becomes the understanding of how selection shapes genomes, the findings in experimental crosses are of limited applicability (Roff and Simons 1997; Conner 2002). In the wild, individuals are exposed to environmental and genetic forces (e.g., genotype-by-environment interaction, pleiotropy, epistasis, maternal effects), some of which are unwittingly or deliberately diminished in experimental settings and may conceal important effects in the wild (Kroymann and Mitchell-Olds 2005; Wilson et al. 2005a,b). Although these forces are particularly difficult to detect in the wild, their possible absence from an experimental design may lead to biased conclusions.
A refined understanding of the process of evolution can be expected if the precise loci underlying trait variation can be identified and their behavior studied in free-living populations. Hence, a recent development is the application of genomic analyses to studies of free-living populations. Techniques for generating large numbers of genetic markers (e.g., AFLPs and microsatellites) and the availability of markers from related model species means that genetic maps and quantitative trait locus (QTL) searches in organisms originally studied in the wild are becoming more common (Erickson et al. 2004; Slate 2005). To date, most of these studies have involved wild plants or animals brought into and bred in the laboratory. Although in some cases the experimental design makes use of pedigrees generated from several lines (Zhang et al. 2005) and investigates fitness-related traits (Laurie et al. 2004), such studies do not directly address the action of natural selection as the study organisms are the product of breeding programs. Other projects have been designed to answer specific ecological or evolutionary questions and to this end have employed individuals drawn from the wild and crossed under controlled conditions (Hawthorne and Via 2001; Lexer et al. 2003). The artificial development of the mapping populations, however, may generate genetic variation that may not occur in the wild (Erickson et al. 2004; Slate 2005). Given the existence of several studies of individually monitored, pedigreed individuals living in the wild, an obvious extension of these studies is to generate genetic maps and attempt to map genes underlying trait variation in nature. To date, however, we know of only two such studies pursuing this line (excluding studies of humans, where cultural factors make extension of findings to animal populations difficult). In red deer (Cervus elaphus) living on the island of Rum, Slate et al. (2002) obtained a partial map (∼62% genome coverage) using microsatellite marker genotypes and then searched for QTL for a phenotypic trait, birth weight, finding three candidate regions for further investigation. Second, Hansson et al. (2005) have recently generated a preliminary genetic map (∼25% genome coverage) for the great reed warbler (Acrocephalus arundinaceus) population at Lake Kvismaren, Sweden, again using microsatellites.
In this article, we describe the construction of a relatively much more complete genetic map for a free-living population, the Soay sheep (Ovis aries) living on St. Kilda, taking advantage of existing genomic resources available for domestic sheep. This population is the subject of a long-term, individual-based multidisciplinary study, which includes the collection of extensive phenotypic, ecological, and genetic information (Clutton-Brock and Pemberton 2004). Soay sheep are highly variable in appearance, with two independent polymorphisms of coat pigmentation (coat color and coat pattern) and polymorphic horns [normal, deformed (“scurred”), or polled horns]. Selection acting on two of these polymorphisms, coat color and horn type, has been previously documented (Milner et al. 2004). We demonstrate the utility of the genetic map by mapping the genes underlying these three polymorphic traits, setting the scene for better understanding of selection on these traits and for future QTL searches in the study population.
MATERIALS AND METHODS
Mapping population:
The Soay sheep on the islands of Soay and Hirta (St. Kilda archipelago, northwestern Scotland, UK, 57°49′ N, 08°34′ W) are feral populations of a breed regarded as the most primitive in Europe (Campbell 1974; Doney et al. 1974); today, the sheep population of Hirta varies between 600 and 2000 individuals. Since 1985 regular expeditions have been sent to St. Kilda to monitor the population dynamics and to record the entire history of individuals living in Village Bay, Hirta (Clutton-Brock et al. 2004a). No predators are present on St. Kilda.
The mapping population analyzed in this study was selected from a larger Soay sheep data set comprising >3300 individuals with phenotypic records. In this population, maternity is determined by observation, and paternity is inferred through molecular analysis (Overall et al. 2005). The mating system is polygynous and promiscuous, such that very few full-sibs occur in the population. To trade off between power of linkage mapping and amount of genotyping work, we selected and genotyped half-sibships with 12 or more well-phenotyped individuals and their common parent, plus half-sibships with at least 10 animals that were related to previously selected animals. In addition, we included in the mapping pedigree file, but did not genotype, the noncommon parents and the ancestors of the half-sib families. Although not genotyped and in some cases phenotypically less well characterized, these additional individuals link different sibships, which otherwise would appear as unrelated. This strategy increases the number of informative meioses as missing genotypes and marker phases, in some cases, can be inferred from the knowledge that different individuals share the same ancestors. In total, the mapping pedigree numbers 882 animals with 571 paternal links and 663 maternal links, of which 588 animals were genotyped (supplemental Figure S1 at http://www.genetics.org/supplemental/).
Polymorphic traits:
In Soay sheep the color of the pelage is determined by two independently segregating polymorphisms, one affecting the color of the coat (hereafter referred to as coat color, locus Coat color, Figure 1A), and the other determining the contrast in color between belly and coat (hereafter referred to as coat pattern, locus Coat pattern, Figure 1B). Coat color can be classified into two distinct phenotypes, dark and light, which occur in a ratio of ∼3:1. Segregation analyses in mainland Soays (Doney et al. 1974) and in resolved pedigrees on St. Kilda (Coltman and Pemberton 2004) suggest that a single biallelic locus, in which dark is completely dominant to light, determines the two classes (see Table 1). With respect to coat pattern, Soay sheep with the “wild-type” morph have a paler belly and rump than the rest of the coat while the “self” morph is characterized by a uniform and more intense coat color. The wild and self morphs occur in a ratio of ∼20:1. This variation is also determined by a single biallelic locus with wild type completely dominant to self (Coltman and Pemberton 2004). Wild-type sheep have hairs in which the dark color is alternated by pale bands, a pattern commonly found in wild mammals and usually due to the Agouti locus (Bennett and Lamoreux 2003). Conversely, in self sheep the hairs have no banding pattern (Clutton-Brock et al. 2004b).
Figure 1.—
Soay sheep showing the three traits subjected to linkage mapping. (A) Coat color polymorphism: dark (left) and light (right) lambs. (B) Coat pattern polymorphism: self (left) and wild-type (right) lambs; note lack of contrast in color between belly and rest of the body and the intensified coat color in the self individual. (C) Horn-type morphs in adult males: normal (left) and extreme scurred (right).
TABLE 1.
Phenotypic distributions and underlying genotypes of the study traits in the genotyped members of the Soay mapping pedigree (maximum n = 588)
| Trait (n) | Phenotype | Genotype | Frequency |
|---|---|---|---|
| Coat color (560) | Dark | Dark/− | 0.74 |
| Light | Light/Light | 0.26 | |
| Coat pattern (560) | Wild | Wild/− | 0.94 |
| Self | Self/Self | 0.06 | |
| Horn type—females (286) | Normal | Ho+/Ho+ | 0.38 |
| Ho+/HoL | |||
| Scurred | HoL/HoL | 0.24 | |
| Ho+/HoP | |||
| Polled | HoP/HoP | 0.38 | |
| HoP/HoL | |||
| Horn type—males (270) | Normal | Ho+/− | 0.90 |
| HoL/− | |||
| Scurred | HoP/HoP | 0.10 |
With respect to horn type (locus Horn type), Soay sheep are polymorphic for horns in both sexes. Females are classified into three horn types: normal (33%), scurred (vestigial and deformed, 28%), and polled (hornless, 39%), whereas in males only the normal (87%) and scurred (13%) phenotypes occur (Figure 1C, Table 1). Although the inheritance of the horn phenotype is not completely understood, pedigree data on St. Kilda are consistent with a single locus with three alleles (normal horned, sex-limited horned, and polled) showing sex-specific expression and dominance (Coltman and Pemberton 2004), a model originally proposed for Merino sheep (Dolling 1961).
DNA extraction and microsatellite genotyping:
Commercial kits were used to isolate DNA from blood samples (GFX genomic blood purification kit, Amersham Biosciences) or ear punches (GenomicPrep cells and tissue DNA isolation kit, Amersham Biosciences) following the manufacturer's instructions. When the amount of starting material was too small or degraded to allow the use of these methods, the DNA was extracted using Chelex resin beads (Chelex 100 Resin, Bio-Rad Laboratories, Hercules, CA). About 1–5 mg of blood or tissue was incubated at 56° overnight in 300 μl of a 5% Chelex and 0.1 μg/μl proteinase K solution followed by 5 min at 95°. Before PCR amplification, the DNA solution extracted with either method was diluted 1:4 with ddH2O and 2 μl was air dried in 96-well PCR plates.
To construct a map with markers evenly distributed throughout the genome, the Australian Sheep Gene Mapping website (http://rubens.its.unimelb.edu.au/∼jillm/jill.htm) was taken as a reference to select microsatellite markers on the basis of their location and information content. PCR amplifications were performed in 5 μl volume, and MgCl2 concentration was adjusted between 1.5 and 4.0 mm to achieve optimal quality of the reaction. Two touchdown PCR programs were initially tested for each marker on a panel of eight sheep: one in which the annealing temperature was progressively decreased during the first 10 of 29 cycles from 60° to 50°, and the other in which the decrease was from 65° to 55°. Fluorescent primers (6FAM, VIC, PET, or NED fluorescence) were synthesized by Applied Biosystems (Foster City, CA). Fragment lengths were analyzed on an ABI3730 DNA Analyzer and genotypes were determined using GeneMapper v3.0 (Applied Biosystems).
To estimate the genotyping error rate, 84–258 randomly chosen individuals were regenotyped at 10 loci with average polymorphism. Genotyping error rate was also assessed on the basis of mother–offspring mismatches using CERVUS 2.0 (Marshall et al. 1998).
Linkage map and genome scan:
Parent–offspring genotype inconsistencies arising from incorrect paternity assignment (32 incorrect links found) or typing errors were detected through the program PedCheck (O'Connell and Weeks 1998) and either resolved by rechecking the parentage records and genotypes or scored as untyped. Some cases of paternity mis-assignment were expected since in the original data set paternity was assigned with only 80% confidence (Overall et al. 2005).
Linkage mapping was performed using CRI-MAP v2.4 (Green et al. 1990) to determine the marker order, intermarker intervals, two-point LOD scores, and number of informative meioses. The complexity of the pedigree and the number of markers employed made a systematic testing of all the possible map combinations impractical. In most cases, the size of the pedigree did not allow the analysis of more than seven or eight markers at a time; therefore, sets of overlapping markers were tested sequentially until a whole chromosome was mapped. Markers expected to belong to the same chromosome were first input into CRI-MAP following the order reported in domestic sheep (Australian Sheep Gene Mapping website at http://rubens.its.unimelb.edu.au/∼jillm/jill.htm). The log 10 likelihood of the initial marker order was then compared with that of alternative orders (flips2 or flips3 function) to detect more likely combinations (i.e., higher log 10 likelihood). An increase in log 10 likelihood of three or more was considered evidence of a significantly more probable map (Morton 1955). In cases of inconsistency between Soay and domestic sheep, the most probable Soay order was retained after having ruled out possible human or technical mistakes. Markers mapping to unexpected locations or supported by a weak LOD score (<1.8) were also tested for linkage (two-point function) against all the other markers in the database to detect whether better positions could be found.
Coat color and Coat pattern loci were mapped using CRI-MAP assuming each trait was encoded by a single locus with two alleles showing complete dominance: the Dark allele dominant over Light and the Wild dominant over Self (Coltman and Pemberton 2004; Table 1). A test for linkage between the target locus and any of the mapped markers was performed by means of the two-point function of CRI-MAP. The best position of a candidate locus was searched for by means of the flips and fixed functions of CRI-MAP to test alternative map positions. Consistent with the criteria used in the map construction, an increase in the log 10 likelihood of the map of three or more was taken as evidence of a significantly more likely position. In the case of Coat pattern, the low frequency of Self morphs resulted in a low number of informative meioses. To confirm or reject a suggestive linkage, more sheep in families segregating for Coat pattern were genotyped at markers in the relevant region (see results).
The Horn type locus was first investigated using CRI-MAP under a simplified model to scan the whole genome, and then the LINKAGE package (Terwilliger and Ott 1994) was employed to perform parametric multipoint analysis in target region(s) identified by the preliminary scan. In CRI-MAP, Horn type was coded as a single biallelic locus where the Normal (Ho+) allele was dominant and the Polled (HoP) allele was recessive in males (Ho+ allele conferring normal horns when heterozygote or homozygote and HoP allele resulting in scurred horns when homozygote), whereas in females Ho+ and HoP alleles were expressed codominantly (normal, scurred, or polled horns given by Ho+/Ho+, Ho+/HoP, and HoP/HoP, respectively). This model was simplified in that the presumptive Sex-limited allele (HoL) was not explicitly considered (see Table 1); modeling three alleles in CRI-MAP would have resulted in too many missing genotypes since this program does not allow a trait (or a marker) phenotype to be coded by more than one genotype. Although this simplification reduces the power of analysis, it does not bias the results.
Computational constraints due to the size of the pedigree and the number of inbreeding loops prevented the processing of the entire pedigree by parametric multipoint linkage analysis. To circumvent this problem, the mapping panel was split into 39 unlinked families. For the parametric multipoint analysis in LINKAGE (Terwilliger and Ott 1994), each sheep was assigned to one of five liability classes on the basis of their horn phenotype and sex: three classes for females (normal if Ho+/Ho+ or Ho+/HoL, scurred if HoL/HoL or Ho+/HoP, and polled if HoL/HoP or HoP/HoP; Table 1) and two classes for males (scurred horns if HoP/HoP, normal horns otherwise; males do not express the polled condition; Table 1). Finally, a sixth (fictitious) class was assigned to animals without phenotypic information; the underlying genotypes were assumed to have complete penetrance.
Horn allele frequencies were taken from Coltman and Pemberton (2004) as 0.441, 0.170, and 0.389 for Ho+, HoL, and HoP, respectively. Marker allele frequencies were estimated from the whole pedigree by 100,000 Markov chain Monte Carlo iterations implemented in Loki (Heath 1997); this procedure is based on a stochastic process and as such does not provide an exact result, but allows the handling of very large and complex pedigrees.
The LOD threshold of 3.3 to declare evidence of linkage corresponds to the value usually applied to human pedigrees (Lander and Kruglyak 1995). This decision was taken on the basis that the sizes of the sheep and human linkage maps are comparable.
RESULTS
Soay sheep linkage map:
The Soay sheep linkage map was developed with 247 microsatellite and four allozyme markers, giving a total of ∼124,000 genotypes, which generated a map with ∼15-cM intermarker spacing across 3350 cM, equivalent to ∼3080 cM on the International Mapping Flock (IMF) map and corresponding to ∼90% of the sheep genome. Figure 2 compares the Soay sheep linkage map with the domestic sheep map (Maddox et al. 2001); the appendix lists the mapped markers and their characteristics. The mean number of alleles per locus was 4.6 with a mean polymorphism information content (PIC) of 0.52, which are lower values than those recorded on the Australian Sheep Gene Mapping website (http://rubens.its.unimelb.edu.au/∼jillm/jill.htm) for the same markers typed in the IMF (10 alleles and PIC = 0.75); this is perhaps not surprising since the latter figures are for a pedigree derived from several sheep breeds. On average, each marker was typed in 510 sheep (86% of the 588 sheep selected for genotyping) and generated 310 informative meioses. Genomewide, the mean LOD score for linkage between two adjacent markers was 14.6. Twenty-two marker intervals were linked with a LOD score of <2, but their marker positions were retained since they were in agreement with the domestic sheep map (Maddox et al. 2001). Marker order was checked by means of the flips function of CRI-MAP and was consistent between the Soay and domestic sheep map in all but two cases: one on chromosome 1, where there is evidence for varying gene order in different sheep breeds (McRae and Beraldi 2006), and the other on chromosome 12, which we have not investigated further. Of the 1290 duplicated genotypes, 2.4% showed inconsistency with the first screening. The error rate based on mother–offspring mismatching was 1.49% as estimated by CERVUS (Marshall et al. 1998).
Figure 2.—
Soay sheep map compared with the domestic (IMF) sheep map v4.3 (Australian Sheep Gene Mapping at http://rubens.its.unimelb.edu.au/∼jillm/jill.htm). In each pair, the Soay chromosome is on the left side; dotted lines connect markers shared by both maps. The ruler at the top left corner represents a centimorgan scale.
Linkage mapping of Coat color, Coat pattern, and Horn type loci:
The phenotypic distributions of Coat color, Coat pattern, and Horn type in the mapping panel are reported in Table 1; these proportions do not differ significantly from the entire Soay sheep data set (χ2 test P > 0.1).
Coat color:
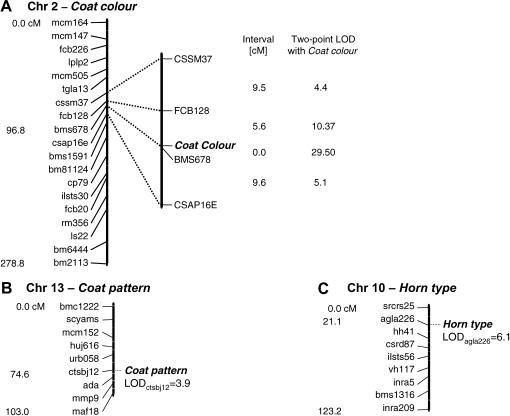
The highest LOD score for linkage was found with BMS678 (two-point LOD = 29.5 at 0 cM), a microsatellite located on chromosome 2 (Figure 3). Other markers on chromosome 2 were significantly linked to the target locus, namely FCB128 (LOD = 10.4), CSAP16E (LOD = 5.1), and CSSM37 (LOD = 4.4), whereas none of the other markers in the Soay sheep map produced a significant result for linkage (LOD <2). Figure 4A shows in detail the best position for the Coat color locus in the map of chromosome 2; any other map order results in a significant decrease (>3) in the log 10 likelihood of the map.
Figure 3.—
Two-point LOD score profiles for linkage between study traits (on the right side of the graph) and markers (data points) in the Soay sheep map. The x-axis represents the cumulative map distance of the genome (morgan) with chromosome boundaries marked on top of the graph and by dotted vertical lines. y-Axes report the LOD scale. The dotted lines denote the theoretical genomewide significance threshold (LOD = 3.3). The asterisk in the middle panel shows the LOD score after having typed additional animals at marker CTSBJ12 (see text).
Figure 4.—
Target regions identified by the genome scan for the three study traits. (A) Chromosome 2 full map and detailed map of the region carrying the Coat color locus. (B) Suggestive region for the Coat pattern locus on chromosome 13. (C) The Horn type location detected on chromosome 10 in the vicinity of AGLA226.
Coat pattern:
The highest linkage score (LOD = 2.1) was detected on chromosome 13 (Figure 3). This LOD score fell short of genomewide significance, but this is likely to be a consequence of the low frequency of the self morph (6%), which meant that the Coat pattern locus was segregating in only a few families and there were few informative meioses for mapping (N = 32). To confirm or reject this suggestive linkage, another 78 animals composing 15 families segregating for coat pattern were genotyped for the two microsatellite markers encompassing the LOD score peak (CTSBJ12 and MMP9), and the association between marker CTSBJ12 and the Coat pattern locus rose to LOD = 3.9 with no recombinants between these loci (Figures 3 and 4B).
Horn type:
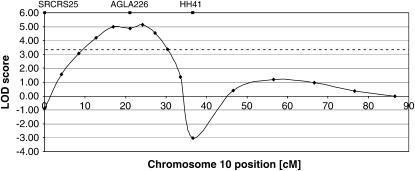
Consistent with Montgomery et al. (1996), CRI-MAP detected linkage between Horn type and AGLA226 on chromosome 10 (LOD = 6.1, Figure 3), but no other marker on chromosome 10 or elsewhere in the genome showed any significant linkage. Once the best location for Horn type was established on the chromosome 10 map (by use of the fixed function), CRI-MAP positioned Horn type distal to SRCRS25, the most telomeric marker on chromosome 10 (21.1 cM away from AGLA226). However, the likelihood of Horn type at this position was not significantly greater than in the interval between AGLA226 and SRCRS25 (log 10 likelihood: −150.16 vs. −151.44), although significantly better than in the interval AGLA226–HH41 (log 10 likelihood: −153.17). Therefore, at this stage we concluded that Horn type is located on chromosome 10 distal to or in the vicinity of AGLA226 (Figure 4C), but an accurate map position could not be assigned.
As described in materials and methods, the CRI-MAP model of Horn type is simplified and does not account for the Horn type and marker allele frequencies. Therefore, the analysis was improved by performing multipoint parametric mapping to derive a more accurate estimate of the Horn type locus position. The Horn type locus was tested for linkage against AGLA226 and its two flanking markers. The LOD profile found by the multipoint analysis (Figure 5) suggests that the 1-LOD support interval for the presence of the target locus spans ∼16 cM.
Figure 5.—
Parametric four-point mapping of the Horn type locus. AGLA226 on chromosome 10, the marker showing the strongest two-point linkage in CRI-MAP, and two adjacent markers (SRCRS25 and HH41) were simultaneously tested against Horn type. The location of the three markers is shown at the top. The Horn type position was tested every 5 cM (data point). The dashed line denotes the theoretical genomewide significance threshold (LOD = 3.3).
DISCUSSION
As a step toward the comprehension of the genetic dynamics of wild populations, this article reports the development of a genetic map in a free-living population, the Soay sheep on St. Kilda, and its use in a genome scan to map the loci responsible for three morphological traits. To the best of our knowledge, this is one of the first accomplishments of gene mapping in a free-living population. The Soay sheep on St. Kilda present interesting features from an evolutionary and genetic point of view: their number is naturally regulated by a combination of food availability, parasite burden, and winter weather (Coulson et al. 2001; Clutton-Brock et al. 2004a; Wilson et al. 2004), factors that, together, cause substantial fluctuations in population size (Coulson et al. 2001; Clutton-Brock et al. 2004a).
Development of the Soay sheep linkage map:
The map presented here has been developed with the primary purpose of localizing genes of evolutionary interest. The map position of a locus can integrate extant models to describe the population dynamics of Soay sheep. Especially when the phenotype conveys little information about the underlying genotype, as is the case for many quantitative characters, the monitoring of the target trait is improved and complemented by the genotype inferred through linked markers.
Patterns of allelic association in terms of linkage disequilibrium and population structure provide insights into history and selection of a population (Abecasis et al. 2005). To this end, a linkage map is a starting point to enrich regions of interest with markers to assess the extension of the association and to compare the latter with theoretical expectations (McRae et al. 2005). Genomic tools such as comparative mapping will facilitate the discovery of additional markers and candidate genes in target regions.
Mapping of Coat color, Coat pattern, and Horn type:
The attempt to map the locus responsible for coat color variation successfully yielded a region on chromosome 2 (Figure 3) defined by a window of ∼15 cM (Figure 4A) in which the Coat color locus cosegregates with BMS678. Independently, J. Gratten, D. Beraldi, B. Lowder, A. McRae, P. Visscher, J. Pemberton and J. Slate (unpublished results), following a candidate gene method, have tested for association with different genes known to affect coat color in mammals and have identified the responsible gene (TYRP1) and its causal mutation.
The interest in coat color in Soay sheep stems from the differential survival between dark and light animals although no predators are present on St. Kilda and no obvious environmental conditions should favor one color over the other. It has previously been found that dark coats are positively selected during some high-mortality winters, but this is inconsistent and in other winters selection favors light-colored sheep or neither morph (Moorcroft et al. 1996; Milner et al. 2004). Dark animals are significantly heavier than light ones, providing a possible mechanism for their better survival (Clutton-Brock et al. 1997). There is no difference in female fecundity between dark and light sheep (Clutton-Brock et al. 1997). At present, there is no explanation for why the light-color morph is maintained in the population; clearly, being able to distinguish the three genotypes may shed light on this puzzle. Hypotheses and future work to explain the difference in survival will take advantage of the map position and molecular characterization of the Coat color gene. A comparison between LD in the FCB128–CSAP16E interval and background LD in the Soay sheep genome should also provide information about the origin and evolutionary consequences of coat color variation.
With respect to Coat pattern, the high frequency of the wild morph (94% of the sheep scored, Table 1) severely reduced the number of informative meioses (32) so that strong linkage to any marker was unlikely to be found. The power of linkage mapping is proportional to the fraction of parents heterozygous at both the target locus and the linked markers. This combination generates the necessary marker-trait association in the progeny (Lynch and Walsh 1998). It follows that if the target locus has a highly skewed allelic distribution, few heterozygous individuals are generated and more meioses need to be scored (the information content, estimated as PIC, reaches the highest value when all the alleles have the same frequency). Accordingly, the highest LOD score for Coat pattern reached only 2.1 on chromosome 13 (Figure 3) after an initial scan. However, the extension of the sample size confirmed this suggestive linkage. Interestingly, chromosome 13 harbors the Agouti locus, a candidate for Coat pattern (Parsons et al. 1999). Agouti encodes for an antagonist of the melanocortin receptor, causing a switch from eumelanin to pheomelanin production in the pigment-producing cells, which results in the characteristic banding pattern observed in Soay sheep hairs and other mammals (Bennett and Lamoreux 2003). To date, we have not detected selection acting on the Coat pattern locus.
Multipoint parametric linkage analysis was not performed for Coat color and Coat pattern because, in contrast to Horn type, the CRI-MAP model for Coat color and Coat pattern was already consistent with the most likely model, so that little or no improvement would have been gained by multipoint parametric analysis.
The mapping of Horn type returned a telomeric region on chromosome 10 previously detected by Montgomery et al. (1996; Figures 4C and 5). This work opens the way for multiple strategies to fine map and isolate the Horn type gene. These include exploitation of bioinformatic tools to enrich the target region with SNPs and other microsatellites and identification of positional candidates by comparison with the annotated genome assemblies of cattle and other species. Like coat color, horn phenotype is under selection in Soay sheep and other wild populations. In ruminants, horns are typically used in intrasexual conflict, particularly among males where they reach much greater size. Previous analyses of Soay sheep have suggested that normal-horned males and scurred females have the highest annual breeding success (Clutton-Brock et al. 1997; Stevenson et al. 2004), but that in winters characterized by high mortality, the scurred phenotype is generally favored in both sexes (Moorcroft et al. 1996). Exactly how these forces maintain variation in the population is the subject of current research and would clearly be helped by being able to distinguish individuals by genotype rather than by phenotype. Therefore, the Horn type region is an attractive target for molecular evolution studies.
Future directions:
The traits analyzed here are characterized by relatively simple inheritance patterns which, to some extent, may limit their applicability to the understanding of the process of evolution. However, this project opens the way to the more challenging task of detecting QTL affecting a variety of morphological and physiological traits. The Soay sheep has been the subject of a number of studies aimed at estimating quantitative genetic parameters for traits like birth weight and body size (Coltman et al. 1999; Milner et al. 2000, 2004). It has been found that the additive genetic variance of these traits is low but not null, despite the pressure of selection acting on them (Milner et al. 2000). As these previous studies have been conducted under the infinitesimal model framework, the dissection of these traits through QTL mapping to determine eventual Mendelian factors would represent a major breakthrough toward the comprehension of the evolutionary processes in the wild.
APPENDIX.
Details of the markers included in the Soay sheep genetic map
| IMF mapl
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosomea | Marker | Two-point LODb | Interc (cM) | Posd (cM) | Ne | InfMeif | No. allg | H(O)h | H(E)i | PICj | Est err ratek | Posd | No. allg | PICj |
| 1 | BMS2833 | 40.43 | 11 | 0 | 533 | 496 | 5 | 0.762 | 0.743 | 0.7 | 0.0053 | 14.2 | 9 | 0.7 |
| 1 | EPCDV22 | 10.17 | 32 | 11 | 543 | 463 | 6 | 0.773 | 0.743 | 0.707 | 0 | 26.4 | 7 | NA |
| 1 | BM3020 | 24.99 | 13.6 | 43 | 481 | 440 | 6 | 0.778 | 0.785 | 0.75 | 0.0052 | 59.5 | 13 | 0.74 |
| 1 | CP93 | 16.03 | 15.7 | 56.6 | 545 | 343 | 4 | 0.618 | 0.593 | 0.55 | 0 | 73.6 | 6 | 0.58 |
| 1 | BM6465 | 42.6 | 8.2 | 72.3 | 522 | 405 | 6 | 0.707 | 0.737 | 0.697 | 0.0107 | 87.7 | 8 | NA |
| 1 | CSAP36E | 10.8 | 13.8 | 80.5 | 525 | 484 | 5 | 0.73 | 0.739 | 0.692 | 0.0058 | 93.5 | 10 | 0.82 |
| 1 | INRA3 | 20.68 | 4.3 | 94.3 | 517 | 216 | 2 | 0.453 | 0.455 | 0.352 | 0 | 102.8 | 5 | 0.46 |
| 1 | TGLA263 | 30.03 | 10 | 98.6 | 402 | 377 | 7 | 0.816 | 0.787 | 0.755 | 0 | NA | NA | NA |
| 1 | MCM58 | 20.1 | 13.8 | 108.6 | 550 | 464 | 7 | 0.78 | 0.746 | 0.702 | 0 | 114 | 14 | 0.88 |
| 1 | AE57 | 3.48 | 18.9 | 122.4 | 502 | 290 | 4 | 0.552 | 0.534 | 0.474 | 0.0545 | 120.7 | 9 | 0.76 |
| 1 | BMS482 | 12.81 | 13.6 | 141.3 | 528 | 281 | 4 | 0.424 | 0.457 | 0.429 | 0.0707 | 132.3 | 11 | NA |
| 1 | INRA6 | 12.53 | 8.7 | 154.9 | 500 | 267 | 3 | 0.582 | 0.563 | 0.466 | 0 | 142.7 | 8 | 0.52 |
| 1 | BM6438 | 9.71 | 12.9 | 163.6 | 526 | 302 | 4 | 0.551 | 0.535 | 0.454 | 0 | 150.6 | 6 | 0.77 |
| 1 | BMS574 | 4 | 28.6 | 176.5 | 434 | 238 | 4 | 0.562 | 0.581 | 0.518 | 0.0164 | 157.5 | 12 | 0.75 |
| 1 | MCMA8 | 5.03 | 23.5 | 205.1 | 544 | 215 | 3 | 0.406 | 0.435 | 0.394 | 0 | 181.3 | 11 | 0.65 |
| 1 | CSSM04 | 17.51 | 2.9 | 228.6 | 528 | 236 | 3 | 0.492 | 0.452 | 0.359 | 0 | 200.9 | 8 | NA |
| 1 | BMS4000 | 3.47 | 23.6 | 231.5 | 425 | 202 | 5 | 0.595 | 0.569 | 0.496 | 0 | 204.1 | 15 | 0.82 |
| 1 | BMS527 | 7.6 | 24.2 | 255.1 | 526 | 292 | 5 | 0.555 | 0.535 | 0.491 | 0.0117 | 214.9 | 12 | 0.82 |
| 1 | BM7145 | 89.4 | 0.8 | 279.3 | 503 | 394 | 4 | 0.718 | 0.721 | 0.669 | 0 | 234.6 | 6 | 0.66 |
| 1 | MCM137 | 69.12 | 2.1 | 280.1 | 558 | 523 | 7 | 0.763 | 0.752 | 0.715 | 0 | 233.4 | 15 | 0.85 |
| 1 | BM6506 | 58.31 | 1.4 | 282.2 | 506 | 333 | 5 | 0.686 | 0.676 | 0.63 | 0.0075 | 235.6 | 6 | NA |
| 1 | BMS4008 | 35.13 | 6.8 | 283.6 | 522 | 411 | 7 | 0.787 | 0.765 | 0.728 | 0 | 236.7 | 10 | 0.75 |
| 1 | BM8246 | 10.2 | 15.6 | 290.4 | 525 | 350 | 5 | 0.617 | 0.599 | 0.558 | 0.009 | 242.2 | 9 | NA |
| 1 | MCM130 | 0 | 40.7 | 306 | 553 | 363 | 4 | 0.571 | 0.541 | 0.435 | 0.0114 | 256.5 | 16 | 0.73 |
| 1 | BM864 | 0 | 44.3 | 346.7 | 520 | 143 | 3 | 0.287 | 0.284 | 0.256 | 0 | 264.5 | 11 | 0.73 |
| 1 | BM1824 | 40.75 | 3.1 | 391 | 501 | 322 | 5 | 0.653 | 0.671 | 0.621 | 0 | 294.1 | 5 | 0.68 |
| 1 | TRF | 0 | 36.8 | 394.1 | 539 | 437 | 7 | 0.785 | 0.764 | 0.728 | 0.0135 | NA | NA | NA |
| 1 | MCM357 | 0 | 430.9 | 517 | 176 | 2 | 0.422 | 0.417 | 0.33 | 0.0631 | 332.6 | 11 | 0.85 | |
| 2 | MCM164 | 3.74 | 19.2 | 0 | 546 | 97 | 3 | 0.192 | 0.181 | 0.169 | 0 | 17.6 | 13 | 0.78 |
| 2 | MCM147 | 8.58 | 6.8 | 19.2 | 533 | 414 | 7 | 0.717 | 0.722 | 0.68 | 0.0114 | 39.8 | 12 | 0.83 |
| 2 | FCB226 | 3.49 | 14.7 | 26 | 529 | 127 | 4 | 0.386 | 0.377 | 0.348 | 0 | 44.6 | 12 | 0.79 |
| 2 | LPLP2 | 4.35 | 13 | 40.7 | 546 | 441 | 6 | 0.749 | 0.739 | 0.696 | 0.0051 | 65.5 | 14 | 0.84 |
| 2 | MCM505 | 3.15 | 10.2 | 53.7 | 551 | 80 | 5 | 0.181 | 0.182 | 0.176 | 0 | 71.4 | 8 | 0.7 |
| 2 | TGLA13 | 8.47 | 17.8 | 63.9 | 397 | 277 | 4 | 0.718 | 0.737 | 0.687 | 0.0165 | 77.8 | 6 | NA |
| 2 | CSSM37 | 30.66 | 9.5 | 81.7 | 545 | 374 | 5 | 0.556 | 0.566 | 0.537 | 0 | 91.1 | 12 | 0.54 |
| 2 | FCB128 | 48.92 | 5.6 | 91.2 | 544 | 439 | 4 | 0.662 | 0.645 | 0.597 | 0.0241 | 99.4 | 8 | 0.72 |
| 2 | BMS678 | 16.38 | 9.6 | 96.8 | 549 | 426 | 5 | 0.619 | 0.614 | 0.542 | 0 | 106.1 | 14 | 0.83 |
| 2 | CSAP16E | 6.24 | 9.8 | 106.4 | 523 | 146 | 2 | 0.283 | 0.285 | 0.244 | 0.0468 | 112.4 | 6 | 0.43 |
| 2 | BMS1591 | 10.86 | 17.1 | 116.2 | 484 | 264 | 5 | 0.556 | 0.521 | 0.481 | 0.0131 | 127 | 16 | 0.85 |
| 2 | BM81124 | 9.89 | 21.8 | 133.3 | 526 | 410 | 4 | 0.679 | 0.638 | 0.566 | 0 | 148.2 | 11 | NA |
| 2 | CP79 | 4.61 | 19.8 | 155.1 | 473 | 306 | 6 | 0.668 | 0.667 | 0.611 | 0.0248 | 161 | 8 | NA |
| 2 | ILSTS30 | 8.73 | 5.4 | 174.9 | 550 | 120 | 2 | 0.213 | 0.21 | 0.188 | 0 | 182.4 | 10 | 0.7 |
| 2 | FCB20 | 7.47 | 20.6 | 180.3 | 400 | 236 | 7 | 0.61 | 0.635 | 0.6 | 0.075 | 194 | 12 | 0.8 |
| 2 | RM356 | 13.78 | 15.6 | 200.9 | 496 | 360 | 6 | 0.752 | 0.748 | 0.705 | 0.0244 | 211.1 | 16 | NA |
| 2 | LS22 | 3.41 | 36.2 | 216.5 | 546 | 349 | 5 | 0.668 | 0.638 | 0.57 | 0 | 225.8 | 8 | 0.78 |
| 2 | BM6444 | 7.72 | 26.1 | 252.7 | 521 | 425 | 6 | 0.758 | 0.755 | 0.713 | 0 | 260.2 | 13 | 0.88 |
| 2 | BM2113 | 278.8 | 495 | 356 | 4 | 0.614 | 0.595 | 0.514 | 0.0218 | 291.9 | 8 | 0.67 | ||
| 3 | BMS1350 | 4.76 | 25 | 0 | 438 | 187 | 5 | 0.393 | 0.403 | 0.377 | 0 | 0 | 12 | NA |
| 3 | ILSTS28 | 15.04 | 9.9 | 25 | 553 | 356 | 5 | 0.633 | 0.607 | 0.56 | 0 | 32 | 14 | 0.82 |
| 3 | BM746 | 5.17 | 21.9 | 34.9 | 537 | 287 | 3 | 0.529 | 0.537 | 0.43 | 0.0243 | 45.4 | 9 | 0.52 |
| 3 | FCB129 | 3.49 | 22.7 | 56.8 | 528 | 347 | 4 | 0.595 | 0.625 | 0.577 | 0.0084 | 70.2 | 10 | 0.71 |
| 3 | RM150 | 2.3 | 24.8 | 79.5 | 536 | 160 | 5 | 0.326 | 0.34 | 0.316 | 0 | 85.9 | 6 | NA |
| 3 | INRA131 | 12.56 | 16.5 | 104.3 | 523 | 327 | 3 | 0.639 | 0.657 | 0.582 | 0.0278 | 111.4 | 7 | NA |
| 3 | RM96 | 3.81 | 23.4 | 120.8 | 521 | 344 | 4 | 0.601 | 0.55 | 0.494 | 0.0129 | 126.7 | 6 | 0.56 |
| 3 | BM2818 | 2.89 | 24.7 | 144.2 | 496 | 214 | 3 | 0.528 | 0.531 | 0.45 | 0.0726 | 134.9 | 5 | NA |
| 3 | BMS2131 | 6.94 | 19.8 | 168.9 | 543 | 327 | 3 | 0.652 | 0.634 | 0.559 | 0.0753 | 150.4 | 5 | NA |
| 3 | ILSTS42 | 15.04 | 9.9 | 188.7 | 530 | 365 | 7 | 0.675 | 0.654 | 0.612 | 0.007 | 159.1 | 10 | NA |
| 3 | BP1 | 4.84 | 12.6 | 198.6 | 521 | 257 | 2 | 0.53 | 0.5 | 0.375 | 0 | 166 | 4 | 0.38 |
| 3 | AGLA293 | 10.34 | 2.7 | 211.2 | 557 | 241 | 2 | 0.434 | 0.433 | 0.339 | 0.0525 | 184.7 | 4 | 0.36 |
| 3 | FCB5 | 0.61 | 28.6 | 213.9 | 547 | 163 | 2 | 0.333 | 0.313 | 0.264 | 0 | 186.3 | 3 | 0.42 |
| 3 | BL4 | 36.8 | 1 | 242.5 | 569 | 301 | 3 | 0.559 | 0.581 | 0.488 | 0.0095 | 202.5 | 8 | NA |
| 3 | FNG | 12.43 | 7.4 | 243.5 | 563 | 238 | 2 | 0.448 | 0.465 | 0.357 | 0 | NA | NA | NA |
| 3 | VH34 | 0 | 30.3 | 250.9 | 572 | 329 | 5 | 0.579 | 0.576 | 0.532 | 0 | 210.7 | 9 | 0.68 |
| 3 | RM29 | 0.75 | 11.7 | 281.2 | 177 | 69 | 5 | 0.644 | 0.592 | 0.53 | 0.0347 | 238.6 | 8 | 0.67 |
| 3 | BMS1248 | 5.83 | 17.9 | 292.9 | 530 | 204 | 2 | 0.413 | 0.421 | 0.332 | 0.0588 | 251.1 | 11 | 0.67 |
| 3 | CSAP39E | 2.41 | 17.4 | 310.8 | 517 | 236 | 3 | 0.596 | 0.545 | 0.482 | 0.0761 | 268.7 | 11 | 0.75 |
| 3 | CSSME76 | 328.2 | 542 | 106 | 3 | 0.231 | 0.236 | 0.22 | 0 | 290.4 | 9 | 0.68 | ||
| 4 | CSRD100 | 9.49 | 19.5 | 0 | 549 | 207 | 3 | 0.364 | 0.364 | 0.309 | 0 | 9.2 | 8 | 0.71 |
| 4 | MCM218 | 38.21 | 12 | 19.5 | 529 | 437 | 5 | 0.773 | 0.748 | 0.705 | 0.0052 | 26.5 | 9 | 0.82 |
| 4 | MCM2 | 10.53 | 22.5 | 31.5 | 555 | 502 | 7 | 0.791 | 0.796 | 0.771 | 0.0038 | 39.4 | 11 | 0.8 |
| 4 | MAF70 | 1.7 | 28.6 | 54 | 381 | 263 | 6 | 0.743 | 0.785 | 0.749 | 0.1455 | 61.4 | 17 | 0.9 |
| 4 | L168685 | 7.76 | 12.9 | 82.6 | 462 | 220 | 4 | 0.545 | 0.559 | 0.503 | 0.0268 | 83.3 | 9 | NA |
| 4 | ILSTS62 | 28.98 | 7 | 95.5 | 517 | 332 | 5 | 0.617 | 0.602 | 0.543 | 0 | 91.3 | 16 | 0.88 |
| 4 | CP26 | 18.59 | 14.4 | 102.5 | 560 | 422 | 5 | 0.738 | 0.723 | 0.674 | 0.0054 | 98.7 | 6 | 0.74 |
| 4 | HH35 | 21.57 | 13.3 | 116.9 | 548 | 408 | 4 | 0.695 | 0.696 | 0.643 | 0.0125 | 114.9 | 7 | 0.7 |
| 4 | BPGM | 15.12 | 12.5 | 130.2 | 545 | 394 | 6 | 0.653 | 0.641 | 0.611 | 0 | 126.1 | 7 | 0.7 |
| 4 | MCM73 | 142.7 | 545 | 316 | 3 | 0.486 | 0.489 | 0.402 | 0.0851 | 143.6 | 13 | 0.88 | ||
| 5 | WNT3K13 | 14.56 | 9.3 | 0 | 550 | 191 | 4 | 0.438 | 0.415 | 0.386 | 0 | 0 | 8 | 0.76 |
| 5 | TGLA176 | 9.37 | 12.3 | 9.3 | 549 | 292 | 3 | 0.574 | 0.538 | 0.464 | 0 | 17.8 | 8 | NA |
| 5 | MCM380 | 9.63 | 0 | 21.6 | 559 | 70 | 4 | 0.114 | 0.116 | 0.111 | 0 | 26.6 | 10 | 0.78 |
| 5 | CSRD138 | 0 | 50 | 21.6 | 535 | 383 | 5 | 0.6 | 0.577 | 0.519 | 0 | 38.7 | 10 | 0.76 |
| 5 | BMS2258 | 5 | 22.7 | 71.6 | 551 | 266 | 2 | 0.454 | 0.479 | 0.364 | 0 | 89.6 | 8 | NA |
| 5 | TGLA137 | 26.63 | 2.2 | 94.3 | 516 | 288 | 3 | 0.523 | 0.517 | 0.462 | 0 | 113.2 | 8 | NA |
| 5 | EPCDV26 | 32.74 | 1.9 | 96.5 | 553 | 302 | 5 | 0.582 | 0.58 | 0.542 | 0 | 117.4 | 7 | NA |
| 5 | MCM527 | 27.7 | 12.1 | 98.4 | 397 | 324 | 6 | 0.783 | 0.758 | 0.727 | 0.0152 | 125.5 | 6 | 0.67 |
| 5 | MCM108 | 30.28 | 8.8 | 110.5 | 545 | 503 | 7 | 0.811 | 0.784 | 0.753 | 0 | 135.2 | 11 | 0.82 |
| 5 | CSRD134 | 5.36 | 18.8 | 119.3 | 501 | 281 | 4 | 0.559 | 0.613 | 0.557 | 0.0286 | 140.8 | 6 | 0.55 |
| 5 | BMS1247 | 138.1 | 545 | 370 | 3 | 0.662 | 0.629 | 0.552 | 0.0086 | 157 | 7 | NA | ||
| 6 | BM9058 | 22.59 | 5 | 0 | 452 | 312 | 5 | 0.615 | 0.624 | 0.578 | 0 | 12.9 | 10 | NA |
| 6 | MCM204 | 20.7 | 6.8 | 5 | 541 | 318 | 4 | 0.54 | 0.516 | 0.465 | 0 | 18.2 | 8 | 0.79 |
| 6 | MCM53 | 6.29 | 10.7 | 11.8 | 558 | 378 | 5 | 0.67 | 0.664 | 0.623 | 0.0064 | 29.7 | 9 | 0.76 |
| 6 | MCMA14 | 2.09 | 14.5 | 22.5 | 543 | 88 | 3 | 0.179 | 0.178 | 0.17 | 0 | 45 | 8 | 0.67 |
| 6 | JP27 | 25.46 | 11.9 | 37 | 526 | 449 | 8 | 0.793 | 0.798 | 0.768 | 0.0084 | NA | NA | NA |
| 6 | BM143 | 10.29 | 20.7 | 48.9 | 544 | 332 | 6 | 0.563 | 0.549 | 0.506 | 0 | 59 | 10 | NA |
| 6 | BMS360 | 20.64 | 8.3 | 69.6 | 536 | 388 | 4 | 0.688 | 0.671 | 0.609 | 0 | 80.8 | 12 | NA |
| 6 | MCM140 | 5.28 | 22.3 | 77.9 | 447 | 317 | 5 | 0.609 | 0.599 | 0.533 | 0.0123 | 95.8 | 10 | 0.79 |
| 6 | BM4311 | 10.78 | 10.7 | 100.2 | 447 | 355 | 5 | 0.996 | 0.705 | 0.647 | 0 | 111.6 | 9 | 0.79 |
| 6 | CSRD93 | 5.77 | 27.3 | 110.9 | 537 | 427 | 4 | 0.695 | 0.7 | 0.646 | 0 | 122.7 | 11 | 0.73 |
| 6 | MCM214 | 138.2 | 552 | 417 | 4 | 0.679 | 0.658 | 0.597 | 0 | 147.1 | 7 | 0.72 | ||
| 7 | BM3033 | 5.02 | 28.2 | 0 | 541 | 379 | 3 | 0.632 | 0.634 | 0.562 | 0.0084 | 0 | 17 | NA |
| 7 | BMS528 | 1.7 | 38.2 | 28.2 | 533 | 348 | 3 | 0.612 | 0.618 | 0.536 | 0.0097 | 32.6 | 6 | NA |
| 7 | INRA107 | 3.82 | 26.6 | 66.4 | 517 | 386 | 3 | 0.598 | 0.574 | 0.49 | 0.0358 | 75.4 | 17 | 0.79 |
| 7 | MCM139 | 1.2 | 42.6 | 93 | 518 | 236 | 5 | 0.542 | 0.555 | 0.524 | 0.0203 | 97.6 | 11 | 0.81 |
| 7 | MCM156 | 135.6 | 534 | 294 | 4 | 0.614 | 0.632 | 0.572 | 0.0756 | 133.4 | 8 | 0.77 | ||
| 8 | MNS50A | 10.11 | 13.3 | 0 | 547 | 463 | 5 | 0.779 | 0.737 | 0.688 | 0.0165 | 5 | 11 | 0.85 |
| 8 | INRA127 | 0 | 54.6 | 13.3 | 503 | 211 | 3 | 0.342 | 0.366 | 0.301 | 0 | 17.2 | 8 | 0.81 |
| 8 | BMS434 | 62.99 | 2.4 | 67.9 | 515 | 394 | 6 | 0.724 | 0.692 | 0.652 | 0.0069 | 61.3 | 9 | NA |
| 8 | KD101 | 16.03 | 16.8 | 70.3 | 552 | 452 | 6 | 0.766 | 0.758 | 0.716 | 0.0048 | 71.1 | 12 | 0.83 |
| 8 | CSRD129 | 2.99 | 27.2 | 87.1 | 541 | 328 | 5 | 0.636 | 0.627 | 0.566 | 0 | 86 | 11 | 0.82 |
| 8 | URB024 | 9.86 | 23.5 | 114.3 | 528 | 270 | 4 | 0.532 | 0.549 | 0.49 | 0 | 117.5 | 12 | 0.65 |
| 8 | BMS1967 | 6.09 | 16.1 | 137.8 | 535 | 503 | 6 | 0.776 | 0.769 | 0.731 | 0 | 132.8 | 11 | 0.71 |
| 8 | PLG | 153.9 | 277 | 144 | 2 | 0.596 | 0.5 | 0.375 | 0 | NA | NA | NA | ||
| 9 | BM757 | 26.84 | 10.9 | 0 | 550 | 364 | 4 | 0.675 | 0.67 | 0.615 | 0.0069 | 16.2 | 7 | NA |
| 9 | CSSM66 | 12.42 | 15.4 | 10.9 | 550 | 325 | 4 | 0.578 | 0.569 | 0.507 | 0.0309 | 24.2 | 10 | NA |
| 9 | ILSTS11 | 9.6 | 18.2 | 26.3 | 533 | 290 | 3 | 0.465 | 0.449 | 0.401 | 0 | 40.1 | 10 | 0.68 |
| 9 | BM4630 | 6.18 | 2.6 | 44.5 | 540 | 368 | 6 | 0.548 | 0.611 | 0.555 | 0.0451 | 60 | 8 | 0.78 |
| 9 | MAF33 | 0 | 24.4 | 47.1 | 193 | 36 | 4 | 0.285 | 0.302 | 0.286 | 0.2015 | 60 | 9 | 0.7 |
| 9 | MCM138 | 1.12 | 23.1 | 71.5 | 539 | 222 | 2 | 0.341 | 0.418 | 0.33 | 0.1234 | 78.5 | 9 | 0.64 |
| 9 | BMS1304 | 3.31 | 0.4 | 94.6 | 45 | 33 | 6 | 1 | 0.782 | 0.737 | 0 | 89.6 | 12 | NA |
| 9 | MCM63 | 29.71 | 8 | 95 | 546 | 362 | 5 | 0.632 | 0.645 | 0.602 | 0.016 | 93.1 | 14 | 0.87 |
| 9 | BM4513 | 103 | 429 | 403 | 7 | 0.811 | 0.806 | 0.776 | 0 | 100.3 | 10 | 0.82 | ||
| 10 | SRCRS25 | 7.22 | 21.1 | 0 | 553 | 253 | 4 | 0.483 | 0.498 | 0.463 | 0.0515 | 5.3 | 11 | 0.82 |
| 10 | AGLA226 | 10.64 | 15.5 | 21.1 | 481 | 312 | 6 | 0.636 | 0.68 | 0.646 | 0.0078 | 31.5 | 8 | 0.78 |
| 10 | HH41 | 9.83 | 11.7 | 36.6 | 545 | 366 | 7 | 0.615 | 0.634 | 0.585 | 0 | 45.9 | 11 | 0.8 |
| 10 | CSRD87 | 14.02 | 9.8 | 48.3 | 538 | 291 | 4 | 0.61 | 0.585 | 0.504 | 0 | 56.2 | 10 | 0.78 |
| 10 | ILSTS56 | 22.31 | 14.7 | 58.1 | 500 | 359 | 6 | 0.682 | 0.72 | 0.687 | 0.0621 | 61.2 | 9 | 0.7 |
| 10 | VH117 | 19.16 | 10.7 | 72.8 | 541 | 451 | 6 | 0.726 | 0.734 | 0.693 | 0.0108 | 68.2 | 11 | 0.75 |
| 10 | INRA5 | 11.47 | 17.8 | 83.5 | 403 | 314 | 8 | 0.695 | 0.679 | 0.635 | 0.0181 | 78 | 13 | 0.87 |
| 10 | BMS1316 | 5.93 | 21.9 | 101.3 | 547 | 452 | 5 | 0.682 | 0.673 | 0.609 | 0 | 93.5 | 13 | 0.81 |
| 10 | INRA209 | 123.2 | 546 | 179 | 2 | 0.264 | 0.297 | 0.253 | 0.043 | 108 | 10 | 0.58 | ||
| 11 | HEL10 | 4.2 | 27.7 | 0 | 550 | 349 | 5 | 0.645 | 0.624 | 0.588 | 0.0077 | 22.4 | 12 | 0.85 |
| 11 | CSSME70 | 4.33 | 10.7 | 27.7 | 550 | 261 | 4 | 0.58 | 0.574 | 0.498 | 0 | 47.9 | 9 | 0.78 |
| 11 | SRCRSP6 | 10.56 | 5.8 | 38.4 | 537 | 394 | 4 | 0.68 | 0.663 | 0.612 | 0 | 59.9 | 9 | 0.73 |
| 11 | FCB193 | 7.63 | 2.3 | 44.2 | 564 | 146 | 3 | 0.229 | 0.226 | 0.203 | 0 | 65.4 | 12 | 0.41 |
| 11 | THRA | 12.8 | 8.9 | 46.5 | 545 | 258 | 3 | 0.499 | 0.497 | 0.387 | 0.0138 | 67.2 | 8 | 0.73 |
| 11 | EPCDV23 | 61.23 | 3 | 55.4 | 546 | 371 | 7 | 0.689 | 0.665 | 0.624 | 0 | 79.9 | 10 | NA |
| 11 | MCM120 | 27.89 | 12.2 | 58.4 | 550 | 490 | 8 | 0.773 | 0.76 | 0.725 | 0 | 87.7 | 16 | 0.85 |
| 11 | ETH3 | 3.57 | 20.6 | 70.6 | 536 | 319 | 3 | 0.565 | 0.557 | 0.491 | 0 | 99.4 | 5 | NA |
| 11 | CSSM08 | 91.2 | 541 | 229 | 4 | 0.447 | 0.43 | 0.404 | 0 | 112.4 | 5 | 0.53 | ||
| 12 | HUJ614 | 1.99 | 10.5 | 0 | 543 | 65 | 2 | 0.145 | 0.141 | 0.131 | 0 | 7.6 | 9 | 0.72 |
| 12 | TGLA53 | 15.23 | 9.9 | 10.5 | 400 | 308 | 7 | 0.64 | 0.651 | 0.621 | 0.039 | 39.3 | 8 | NA |
| 12 | BM4025 | 5.29 | 22.4 | 20.4 | 445 | 319 | 5 | 0.712 | 0.714 | 0.659 | 0.0074 | 24 | 9 | NA |
| 12 | CSSM03 | 12.42 | 19.8 | 42.8 | 535 | 229 | 3 | 0.393 | 0.396 | 0.36 | 0 | 54.5 | 10 | NA |
| 12 | MCMA52 | 3.5 | 27.4 | 62.6 | 536 | 387 | 5 | 0.674 | 0.678 | 0.624 | 0.0069 | 68.8 | 10 | 0.78 |
| 12 | INRA35 | 90 | 523 | 212 | 5 | 0.512 | 0.477 | 0.436 | 0 | 92.1 | 9 | 0.68 | ||
| 13 | BMC1222 | 19.35 | 15.5 | 0 | 508 | 416 | 5 | 0.734 | 0.706 | 0.67 | 0 | 12.3 | 15 | 0.82 |
| 13 | SCYAMS | 14.79 | 20.4 | 15.5 | 535 | 407 | 5 | 0.693 | 0.683 | 0.642 | 0.0197 | 37.4 | 20 | 0.89 |
| 13 | MCM152 | 23.42 | 10.9 | 35.9 | 527 | 356 | 4 | 0.651 | 0.635 | 0.56 | 0.0087 | 52.1 | 10 | 0.79 |
| 13 | HUJ616 | 13.51 | 10.3 | 46.8 | 546 | 365 | 5 | 0.615 | 0.609 | 0.538 | 0.0092 | 65 | 15 | NA |
| 13 | URB058 | 4.59 | 17.5 | 57.1 | 548 | 254 | 4 | 0.5 | 0.508 | 0.387 | 0.0261 | 74.4 | 13 | 0.78 |
| 13 | CTSBJ12 | 5.63 | 8.9 | 74.6 | 552 | 154 | 4 | 0.252 | 0.249 | 0.222 | 0 | 98 | 9 | 0.77 |
| 13 | ADA | 11.85 | 8.1 | 83.5 | 456 | 217 | 2 | 0.382 | 0.38 | 0.307 | 0 | NA | NA | NA |
| 13 | MMP9 | 8.94 | 11.4 | 91.6 | 542 | 359 | 5 | 0.646 | 0.635 | 0.585 | 0 | 115.4 | 9 | 0.79 |
| 13 | MAF18 | 103 | 335 | 171 | 3 | 0.573 | 0.559 | 0.488 | 0.019 | 125.8 | 5 | 0.41 | ||
| 14 | TGLA357 | 41.95 | 5.1 | 0 | 525 | 315 | 6 | 0.651 | 0.657 | 0.606 | 0 | 11.7 | 8 | 0.8 |
| 14 | INRA38 | 15.8 | 11 | 5.1 | 536 | 398 | 6 | 0.668 | 0.69 | 0.643 | 0 | 17.6 | 13 | 0.82 |
| 14 | CSRD70 | 25.28 | 7.2 | 16.1 | 530 | 358 | 5 | 0.675 | 0.656 | 0.608 | 0 | 25.5 | 14 | 0.78 |
| 14 | BMS2213 | 27.13 | 5.9 | 23.3 | 530 | 325 | 4 | 0.587 | 0.59 | 0.52 | 0 | 33.8 | 10 | 0.82 |
| 14 | LS29 | 17.05 | 7.2 | 29.2 | 536 | 367 | 6 | 0.631 | 0.614 | 0.575 | 0 | 46.5 | 14 | 0.84 |
| 14 | MCM133 | 11.53 | 11.2 | 36.4 | 532 | 185 | 3 | 0.361 | 0.362 | 0.31 | 0.0272 | 56.8 | 9 | 0.75 |
| 14 | CSRD32 | 8.34 | 23 | 47.6 | 527 | 293 | 4 | 0.545 | 0.511 | 0.459 | 0 | 64.6 | 15 | 0.82 |
| 14 | LS30 | 21.14 | 6.5 | 70.6 | 532 | 308 | 4 | 0.609 | 0.635 | 0.568 | 0 | 94.4 | 11 | 0.79 |
| 14 | RM128 | 36.86 | 2.2 | 77.1 | 556 | 303 | 4 | 0.482 | 0.473 | 0.436 | 0 | 104.6 | 11 | 0.81 |
| 14 | MCMA19 | 79.3 | 519 | 189 | 3 | 0.374 | 0.354 | 0.326 | 0 | 109.3 | 6 | NA | ||
| 15 | MCMA16 | 15.86 | 12.9 | 0 | 551 | 372 | 4 | 0.633 | 0.631 | 0.575 | 0.0081 | 0 | 9 | 0.63 |
| 15 | BR3510 | 3.04 | 12.9 | 12.9 | 515 | 289 | 5 | 0.561 | 0.542 | 0.502 | 0 | 19.3 | 8 | NA |
| 15 | BMS1004 | 6.33 | 0.98 | 25.8 | 532 | 99 | 2 | 0.195 | 0.189 | 0.171 | 0 | 27.2 | 13 | NA |
| 15 | ADCYC | 21.26 | 17 | 26.78 | 539 | 379 | 5 | 0.651 | 0.65 | 0.585 | 0 | 35.3 | 7 | 0.74 |
| 15 | JAB1 | 23.14 | 1.2 | 43.78 | 533 | 388 | 8 | 0.683 | 0.703 | 0.655 | 0.0061 | 46.5 | 19 | 0.82 |
| 15 | MAF65 | 0 | 29.8 | 44.98 | 328 | 172 | 4 | 0.518 | 0.512 | 0.453 | 0 | 47 | 8 | 0.62 |
| 15 | HAEM | 2.29 | 9.8 | 74.78 | 216 | 57 | 2 | 0.528 | 0.501 | 0.375 | 0 | NA | NA | NA |
| 15 | POTCHA | 30.64 | 5.3 | 84.58 | 510 | 413 | 5 | 0.716 | 0.717 | 0.668 | 0 | 85.1 | 11 | NA |
| 15 | BMS1660 | 13.4 | 10 | 89.88 | 532 | 282 | 4 | 0.515 | 0.484 | 0.442 | 0 | 96.5 | 10 | 0.77 |
| 15 | BMS2076 | 4.04 | 19.1 | 99.88 | 531 | 301 | 3 | 0.605 | 0.606 | 0.537 | 0 | 105.4 | 14 | 0.83 |
| 15 | MCM105 | 118.98 | 534 | 500 | 8 | 0.785 | 0.784 | 0.75 | 0.0044 | 123.8 | 10 | 0.81 | ||
| 16 | RM106 | 12.64 | 0 | 0 | 327 | 130 | 4 | 0.474 | 0.48 | 0.447 | 0 | 3.8 | 10 | NA |
| 16 | BM1225 | 0 | 16.5 | 0 | 543 | 133 | 3 | 0.274 | 0.267 | 0.249 | 0 | 13.2 | 9 | 0.74 |
| 16 | TGLA126 | 6.98 | 18.5 | 16.5 | 525 | 212 | 4 | 0.371 | 0.383 | 0.342 | 0.0247 | 34.3 | 18 | 0.84 |
| 16 | AGLA29 | 30.04 | 6.7 | 35 | 502 | 308 | 6 | 0.631 | 0.637 | 0.581 | 0.0083 | 46.9 | 16 | NA |
| 16 | CSRD69 | 6.28 | 3.7 | 41.7 | 519 | 361 | 6 | 0.667 | 0.681 | 0.636 | 0.0073 | 55.3 | 10 | 0.67 |
| 16 | MCM506A | 4.85 | 6.6 | 45.4 | 533 | 73 | 3 | 0.126 | 0.12 | 0.116 | 0 | 63.2 | 17 | 0.75 |
| 16 | SRCRS27 | 4.58 | 5.2 | 52 | 509 | 385 | 6 | 0.68 | 0.688 | 0.631 | 0 | 69.4 | 8 | 0.72 |
| 16 | MCM150 | 57.2 | 544 | 42 | 2 | 0.131 | 0.147 | 0.136 | 0 | 83.9 | 9 | 0.68 | ||
| 17 | MCM4 | 0 | 16.9 | 0 | 550 | 404 | 5 | 0.596 | 0.662 | 0.616 | 0 | 0 | 13 | 0.85 |
| 17 | VH98 | 9.19 | 6 | 16.9 | 553 | 71 | 4 | 0.179 | 0.178 | 0.172 | 0 | 19.7 | 9 | 0.67 |
| 17 | CP49 | 22.83 | 12.6 | 22.9 | 543 | 449 | 7 | 0.703 | 0.687 | 0.649 | 0.0063 | 28.5 | 7 | 0.76 |
| 17 | BMS2780 | 3.23 | 8.8 | 35.5 | 517 | 289 | 4 | 0.509 | 0.516 | 0.439 | 0.0147 | 38.4 | 8 | 0.75 |
| 17 | FCB48 | 7.12 | 7.4 | 44.3 | 302 | 109 | 4 | 0.45 | 0.457 | 0.405 | 0.1119 | 42.5 | 11 | 0.76 |
| 17 | MAF209 | 5.31 | 34 | 51.7 | 402 | 332 | 7 | 0.756 | 0.738 | 0.692 | 0.0515 | 48 | 8 | 0.79 |
| 17 | MCMA20 | 85.7 | 507 | 489 | 6 | 0.984 | 0.806 | 0.777 | 0 | 89.2 | 11 | 0.73 | ||
| 18 | BM3413 | 6.73 | 15.5 | 0 | 555 | 237 | 4 | 0.373 | 0.378 | 0.327 | 0 | 22.4 | 8 | 0.7 |
| 18 | VH54 | 49.16 | 1.5 | 15.5 | 554 | 320 | 4 | 0.605 | 0.625 | 0.555 | 0 | 41.9 | 7 | 0.7 |
| 18 | BP33 | 25.55 | 13.9 | 17 | 549 | 438 | 6 | 0.719 | 0.715 | 0.675 | 0 | 43.4 | 12 | 0.85 |
| 18 | UWCA4 | 14.29 | 18.5 | 30.9 | 551 | 401 | 4 | 0.641 | 0.634 | 0.584 | 0 | 56.1 | 6 | 0.72 |
| 18 | BMC5221 | 31.87 | 1.4 | 49.4 | 551 | 409 | 5 | 0.713 | 0.715 | 0.666 | 0 | 77 | 10 | NA |
| 18 | HH47 | 2.5 | 16 | 50.8 | 379 | 205 | 5 | 0.623 | 0.658 | 0.607 | 0.0108 | 77 | 10 | 0.77 |
| 18 | ILSTS54 | 0 | 16.8 | 66.8 | 552 | 89 | 2 | 0.239 | 0.248 | 0.217 | 0.1071 | 91.7 | 3 | NA |
| 18 | IDVGA30 | 0 | 29.5 | 83.6 | 533 | 150 | 2 | 0.257 | 0.344 | 0.285 | 0.3782 | 110.5 | 2 | NA |
| 18 | CSAP28E | 113.1 | 558 | 261 | 4 | 0.461 | 0.439 | 0.393 | 0.0169 | 121.6 | 5 | 0.62 | ||
| 19 | PZ963 | 12.91 | 16.9 | 0 | 549 | 350 | 6 | 0.65 | 0.618 | 0.574 | 0 | 10.6 | 22 | NA |
| 19 | AE119 | 3.98 | 21.6 | 16.9 | 523 | 298 | 3 | 0.535 | 0.513 | 0.459 | 0 | 27.7 | 8 | 0.76 |
| 19 | CSSM41 | 9.33 | 0 | 38.5 | 551 | 151 | 2 | 0.385 | 0.39 | 0.314 | 0.0219 | NA | NA | NA |
| 19 | BM3628 | 2.47 | 40.1 | 38.5 | 499 | 264 | 5 | 0.515 | 0.511 | 0.47 | 0 | 43.3 | 4 | NA |
| 19 | FCB304 | 78.6 | 577 | 352 | 4 | 0.591 | 0.586 | 0.506 | 0 | 66 | 9 | 0.54 | ||
| 20 | BM1815 | 6.78 | 24.8 | 0 | 544 | 255 | 3 | 0.504 | 0.521 | 0.405 | 0.0867 | 26.8 | 6 | NA |
| 20 | OLADRB | 48.16 | 0 | 24.8 | 529 | 478 | 8 | 0.79 | 0.819 | 0.794 | 0.0149 | 52.2 | 13 | NA |
| 20 | OLADRBps | 23.85 | 5.6 | 24.8 | 263 | 203 | 6 | 0.795 | 0.786 | 0.752 | 0.014 | NA | NA | NA |
| 20 | OMHC1 | 4.58 | 22 | 30.4 | 294 | 222 | 5 | 0.568 | 0.597 | 0.564 | 0.0325 | NA | NA | NA |
| 20 | BM1818 | 6.45 | 19.2 | 52.4 | 533 | 388 | 8 | 0.657 | 0.678 | 0.629 | 0.0848 | 64.9 | 10 | NA |
| 20 | BM1905 | 71.6 | 562 | 320 | 2 | 0.528 | 0.488 | 0.369 | 0 | 77.8 | 2 | NA | ||
| 21 | BMS1787 | 7 | 11 | 0 | 524 | 438 | 4 | 0.716 | 0.711 | 0.656 | 0 | 15.5 | 16 | 0.84 |
| 21 | RM044 | 5.07 | 9.8 | 11 | 520 | 125 | 3 | 0.26 | 0.251 | 0.23 | 0 | 22.1 | 10 | 0.83 |
| 21 | CSAP30E | 4.62 | 23.1 | 20.8 | 540 | 269 | 3 | 0.48 | 0.508 | 0.401 | 0 | 29.1 | 15 | 0.79 |
| 21 | MCM135 | 2.84 | 22.9 | 43.9 | 493 | 399 | 5 | 0.996 | 0.676 | 0.618 | 0.0093 | 46 | 13 | 0.85 |
| 21 | BMC1206 | 66.8 | 538 | 212 | 2 | 0.522 | 0.499 | 0.374 | 0 | 58.1 | 6 | 0.67 | ||
| 22 | BMS907 | 48.34 | 10.1 | 0 | 498 | 480 | 7 | 0.815 | 0.838 | 0.817 | 0.0083 | 13.8 | 12 | 0.83 |
| 22 | HEL11 | 24.14 | 9.1 | 10.1 | 531 | 509 | 8 | 0.825 | 0.849 | 0.83 | 0.0154 | 30 | 17 | NA |
| 22 | BM1314 | 34.63 | 4.6 | 19.2 | 333 | 288 | 8 | 0.802 | 0.81 | 0.784 | 0 | 34.5 | 6 | NA |
| 22 | INRA81 | 29.94 | 12.9 | 23.8 | 504 | 420 | 7 | 0.734 | 0.772 | 0.733 | 0 | 35.7 | 22 | 0.85 |
| 22 | BM4505 | 32.57 | 11.9 | 36.7 | 518 | 402 | 7 | 0.707 | 0.734 | 0.69 | 0.0058 | 43.5 | 11 | NA |
| 22 | BMS882 | 5.18 | 27.4 | 48.6 | 545 | 485 | 5 | 0.734 | 0.734 | 0.691 | 0 | 59.7 | 7 | 0.79 |
| 22 | MCM373 | 76 | 531 | 231 | 6 | 0.405 | 0.411 | 0.393 | 0.021 | 82.9 | 13 | 0.84 | ||
| 23 | BL6 | 8.13 | 20.8 | 0 | 520 | 310 | 7 | 0.619 | 0.636 | 0.576 | 0.0091 | 15.7 | 12 | NA |
| 23 | CSRD148 | 12.28 | 9.2 | 20.8 | 502 | 319 | 5 | 0.697 | 0.673 | 0.616 | 0 | 33.2 | 15 | 0.82 |
| 23 | BMS2270 | 9.05 | 11.7 | 30 | 557 | 224 | 5 | 0.361 | 0.361 | 0.339 | 0 | 37.7 | 8 | 0.79 |
| 23 | AGLA269 | 13.22 | 15.2 | 41.7 | 508 | 333 | 5 | 0.646 | 0.621 | 0.561 | 0 | 49.1 | 18 | NA |
| 23 | MAF35 | 10.49 | 10.5 | 56.9 | 583 | 369 | 3 | 0.568 | 0.565 | 0.473 | 0 | 59.2 | 5 | 0.61 |
| 23 | MCM136 | 0 | 37.6 | 67.4 | 552 | 438 | 5 | 0.737 | 0.743 | 0.694 | 0 | 67.6 | 8 | 0.83 |
| 23 | URB031 | 105 | 516 | 148 | 4 | 0.347 | 0.369 | 0.347 | 0.0567 | 97 | 7 | 0.62 | ||
| 24 | EPCDV03 | 45.75 | 3.6 | 0 | 529 | 344 | 4 | 0.59 | 0.565 | 0.508 | 0.0108 | 28.9 | 8 | NA |
| 24 | BP28 | 21.85 | 14.8 | 3.6 | 540 | 400 | 4 | 0.67 | 0.664 | 0.611 | 0.0143 | 38.5 | 18 | 0.92 |
| 24 | FIBROSN | 1.9 | 31.4 | 18.4 | 549 | 421 | 4 | 0.727 | 0.724 | 0.674 | 0.0057 | 48.9 | 12 | 0.82 |
| 24 | EPCD152 | 49.8 | 525 | 439 | 5 | 0.985 | 0.656 | 0.587 | 0 | 83.2 | 6 | NA | ||
| 25 | MCMA7 | 12.21 | 8.4 | 0 | 528 | 224 | 3 | 0.422 | 0.391 | 0.337 | 0 | 31 | 12 | 0.9 |
| 25 | PERF3-2 | 16.39 | 6.8 | 8.4 | 525 | 382 | 5 | 0.714 | 0.671 | 0.615 | 0.0077 | 40 | 15 | 0.86 |
| 25 | VH72 | 3.18 | 21.1 | 15.2 | 522 | 171 | 4 | 0.276 | 0.26 | 0.247 | 0 | 45.6 | 7 | 0.76 |
| 25 | AE54 | 3.97 | 15.1 | 36.3 | 402 | 231 | 5 | 0.617 | 0.632 | 0.567 | 0.0442 | 64 | 9 | 0.82 |
| 25 | RBP3 | 51.4 | 332 | 200 | 3 | 0.642 | 0.608 | 0.536 | 0 | 69.9 | 4 | 0.61 | ||
| 26 | BMS629 | 19.53 | 8 | 0 | 527 | 309 | 3 | 0.575 | 0.579 | 0.49 | 0 | 6.9 | 9 | NA |
| 26 | BM6526 | 26.85 | 2.3 | 8 | 541 | 336 | 4 | 0.534 | 0.536 | 0.48 | 0.0121 | 16.4 | 9 | NA |
| 26 | LS41 | 7.1 | 18.1 | 10.3 | 535 | 310 | 4 | 0.624 | 0.617 | 0.542 | 0.0091 | 20.6 | 12 | 0.77 |
| 26 | CSRD163 | 17.18 | 16.9 | 28.4 | 524 | 270 | 3 | 0.578 | 0.614 | 0.531 | 0.0097 | 39 | 8 | NA |
| 26 | JMP23 | 20.7 | 5.3 | 45.3 | 531 | 443 | 8 | 0.727 | 0.731 | 0.681 | 0.0058 | 53.5 | 13 | NA |
| 26 | JMP58 | 9.71 | 8.9 | 50.6 | 401 | 222 | 5 | 0.589 | 0.596 | 0.522 | 0.0263 | 51.4 | 9 | 0.67 |
| 26 | POLBF17 | 11.2 | 14.8 | 59.5 | 497 | 328 | 7 | 0.678 | 0.669 | 0.641 | 0 | 61.5 | 11 | 0.82 |
| 26 | BM203 | 74.3 | 335 | 278 | 9 | 0.773 | 0.79 | 0.759 | 0.0424 | 71.1 | 10 | NA | ||
| X | MCM158 | 4.27 | 34.6 | 0 | 528 | 397 | 7 | 0.714 | 0.648 | 0.599 | 0 | 0.8 | 12 | 0.88 |
| X | MAF45 | 18.21 | 24.7 | 34.6 | 709 | 623 | 6 | 0.779 | 0.742 | 0.701 | 0 | 31.2 | 12 | 0.84 |
| X | ILSTS17 | 66.49 | 5.1 | 59.3 | 530 | 543 | 3 | 0.577 | 0.47 | 0.41 | 0 | 66.8 | 11 | 0.79 |
| X | CP131 | 72.24 | 1.8 | 64.4 | 524 | 537 | 7 | 0.517 | 0.525 | 0.494 | 0 | 80.5 | 9 | 0.83 |
| X | MCM25 | 66.2 | 528 | 464 | 5 | 0.729 | 0.628 | 0.579 | 0 | 90.8 | 15 | NA | ||
| Average | 14.58 | 15.0 | 510 | 310 | 4.58 | 0.58 | 0.58 | 0.52 | 0.01 | 10.06 | 0.75 | |||
| SD | 14.88 | 9.9 | 73 | 113 | 1.58 | 0.17 | 0.16 | 0.16 | 0.03 | 3.49 | 0.11 | |||
Chromosome number.
LOD score for linkage between adjacent markers.
Intermarker spacing.
Chromosomal position.
Number of sheep genotyped.
Number of informative meioses detected at the marker locus.
Number of alleles.
Observed marker heterozygosity.
Estimated marker heterozygosity.
Polymorphism information content.
Estimated error rate from mother–offspring pairs using CERVUS.
IMF map characteristics (Australian Sheep Gene Mapping website at http://rubens.its.unimelb.edu.au/∼jillm/jill.htm).
Acknowledgments
We thank J. G. Pilkington and many volunteers for collecting field data and genetic samples; the National Trust for Scotland for granting permission to work on St. Kilda and QinetiQ for logistical support; and two anonymous reviewers for their comments on the first version of this article. The long-term data collection on St. Kilda has been supported by Natural Environment Research Council (NERC) and Wellcome Trust grants to T. H. Clutton-Brock, B. T. Grenfell, L. E. B. Kruuk, M. J. Crawley, and J.M.P. This study was funded by NERC through its Environmental Genomics thematic program (grant no. NER/T/S/2002/00189).
References
- Abecasis, G. R., D. Ghosh and T. E. Nichols, 2005. Linkage disequilibrium: ancient history drives the new genetics. Hum. Hered. 59: 118–124. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and P. D. Keightley, 2002. Understanding quantitative genetic variation. Nat. Rev. Genet. 3: 11–21. [DOI] [PubMed] [Google Scholar]
- Bennett, D. C., and M. L. Lamoreux, 2003. The color loci of mice—a genetic century. Pigment Cell Res. 16: 333–344. [DOI] [PubMed] [Google Scholar]
- Brem, R. B., and L. Kruglyak, 2005. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 102: 1572–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. N., 1974. St. Kilda and Its Sheep. Island Survivors: The Ecology of the Soay Sheep of St. Kilda. The Athlone Press, London.
- Carlborg, O., and C. S. Haley, 2004. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618–625. [DOI] [PubMed] [Google Scholar]
- Clutton-Brock, T. H., and J. M. Pemberton, 2004. Soay Sheep Dynamics and Selection in an Island Population. Cambridge University Press, Cambridge, UK.
- Clutton-Brock, T. H., K. Wilson and I. R. Stevenson, 1997. Density-dependent selection on horn phenotype in Soay sheep. Philos. Trans. R. Soc. Lond. B Biol. Sci. 352: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock, T. H., B. T. Grenfell, T. Coulson, A. D. C. MacColl, A. W. Illius et al., 2004. a Population dynamics in Soay sheep, pp. 52–88 in Soay Sheep Dynamics and Selection in an Island Population, edited by T. H. Clutton-Brock and J. M. Pemberton. Cambridge University Press, Cambridge, UK.
- Clutton-Brock, T. H., J. M. Pemberton, T. Coulson, I. R. Stevenson and A. D. C. MacColl, 2004. b The Soay sheep of St. Kilda, pp. 17–51 in Soay Sheep Dynamics and Selection in an Island Population, edited by T. H. Clutton-Brock and J. M. Pemberton. Cambridge University Press, Cambridge, UK.
- Coltman, D. W., and J. M. Pemberton, 2004. Inheritance of coat colour and horn type in Soay sheep, pp. 321–327 in Soay Sheep Dynamics and Selection in an Island Population, edited by T. H. Clutton-Brock and J. M. Pemberton. Cambridge University Press, Cambridge, UK.
- Coltman, D. W., J. A. Smith, D. R. Bancroft, J. Pilkington, A. D. MacColl et al., 1999. Density-dependent variation in lifetime breeding success and natural and sexual selection in Soay rams. Am. Nat. 154: 730–746. [DOI] [PubMed] [Google Scholar]
- Conner, J. K., 2002. Genetic mechanisms of floral trait correlations in a natural population. Nature 420: 407–410. [DOI] [PubMed] [Google Scholar]
- Coulson, T., E. A. Catchpole, S. D. Albon, B. J. Morgan, J. M. Pemberton et al., 2001. Age, sex, density, winter weather, and population crashes in Soay sheep. Science 292: 1528–1531. [DOI] [PubMed] [Google Scholar]
- Dolling, C. H. S., 1961. Hornedness and polledness in sheep. IV. Triple alleles affecting horn growth in the Merino. Aust. J. Agric. Res. 12: 353–361. [Google Scholar]
- Doney, J. M., M. L. Ryder, R. G. Gunn and P. Grubb, 1974. Colour, Conformation, Affinities, Fleece and Patterns of Inheritance of the Soay Sheep. Island Survivors: The Ecology of the Soay Sheep of St. Kilda. The Athlone Press, London.
- Erickson, D., 2005. Quantitative trait loci: mapping the future of QTL's. Heredity 95: 417–418. [DOI] [PubMed] [Google Scholar]
- Erickson, D. L., C. B. Fenster, H. K. Stenoien and D. Price, 2004. Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 13: 2505–2522. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1958. The Genetical Theory of Natural Selection. Dover, New York.
- Green, P., K. Falls and S. Crooks, 1990. Documentation for CRI-MAP. Washington University, St Louis.
- Hansson, B., M. Akesson, J. Slate and J. M. Pemberton, 2005. Linkage mapping reveals sex-dimorphic map distances in a passerine bird. Proc. Biol. Sci. 272: 2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne, D. J., and S. Via, 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412: 904–907. [DOI] [PubMed] [Google Scholar]
- Heath, S. C., 1997. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am. J. Hum. Genet. 61: 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann, J., and T. Mitchell-Olds, 2005. Epistasis and balanced polymorphism influencing complex trait variation. Nature 435: 95–98. [DOI] [PubMed] [Google Scholar]
- Lander, E., and L. Kruglyak, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- Laurie, C. C., S. D. Chasalow, J. R. LeDeaux, R. McCarroll, D. Bush et al., 2004. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 168: 2141–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer, C., M. E. Welch, J. L. Durphy and L. H. Rieseberg, 2003. Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol. Ecol. 12: 1225–1235. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Maddox, J. F., K. P. Davies, A. M. Crawford, D. J. Hulme, D. Vaiman et al., 2001. An enhanced linkage map of the sheep genome comprising more than 1000 loci. Genome Res. 11: 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, T. C., J. Slate, L. E. Kruuk and J. M. Pemberton, 1998. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 7: 639–655. [DOI] [PubMed] [Google Scholar]
- McRae, A. F., and D. Beraldi, 2006. Examination of a region showing linkage map discrepancies across sheep breeds. Mamm. Genome 17: 346–353. [DOI] [PubMed] [Google Scholar]
- McRae, A. F., J. M. Pemberton and P. M. Visscher, 2005. Modeling linkage disequilibrium in natural populations: the example of the Soay sheep population of St. Kilda, Scotland. Genetics 171: 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J. M., J. M. Pemberton, S. Brotherstone and S. D. Albon, 2000. Estimating variance components and heritabilities in the wild: a case study using the ‘animal model’ approach. J. Evol. Biol. 13: 804–813. [Google Scholar]
- Milner, J. M., S. D. Albon, L. E. B. Kruuk and J. M. Pemberton, 2004. Selection on phenotype, pp. 190–216 in Soay Sheep Dynamics and Selection in an Island Population, edited by T. H. Clutton-Brock and J. M. Pemberton. Cambridge University Press, Cambridge, UK.
- Montgomery, G. W., H. M. Henry, K. G. Dodds, A. E. Beattie, T. Wuliji et al., 1996. Mapping the Horns (Ho) locus in sheep: a further locus controlling horn development in domestic animals. J. Hered. 87: 358–363. [DOI] [PubMed] [Google Scholar]
- Moorcroft, P. R., S. D. Albon, J. M. Pemberton, I. R. Stevenson and T. H. Clutton-Brock, 1996. Density-dependent selection in a fluctuating ungulate population. Proc. R. Soc. Lond. B Biol. Sci. 263: 31–38. [DOI] [PubMed] [Google Scholar]
- Morton, N. E., 1955. Sequential tests for the detection of linkage. Am. J. Hum. Genet. 7: 277–318. [PMC free article] [PubMed] [Google Scholar]
- O'Connell, J. R., and D. E. Weeks, 1998. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall, A. D., K. A. Byrne, J. G. Pilkington and J. M. Pemberton, 2005. Heterozygosity, inbreeding and neonatal traits in Soay sheep on St. Kilda. Mol. Ecol. 14: 3383–3393. [DOI] [PubMed] [Google Scholar]
- Parsons, Y. M., M. R. Fleet and D. W. Cooper, 1999. The Agouti gene: a positional candidate for recessive self-colour pigmentation in the Australian merino. Aust. J. Agric. Res. 50: 1099–1103.
- Roff, D. A., and A. M. Simons, 1997. The quantitative genetics of wing dimorphism under laboratory and “field” conditions in the cricket Gryllus pennsylvanicus. Heredity 78: 235–240. [Google Scholar]
- Slate, J., 2005. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 14: 363–379. [DOI] [PubMed] [Google Scholar]
- Slate, J., P. M. Visscher, S. MacGregor, D. Stevens, M. L. Tate et al., 2002. A genome scan for quantitative trait loci in a wild population of red deer (Cervus elaphus). Genetics 162: 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, I. R., P. Marrow, B. T. Preston, J. M. Pemberton and K. Wilson, 2004. Adaptive reproduction strategies, pp. 243–275 in Soay Sheep Dynamics and Selection in an Island Population, edited by T. H. Clutton-Brock and J. M. Pemberton. Cambridge University Press, Cambridge, UK.
- Terwilliger, J. D., and J. Ott, 1994. Handbook of Human Genetic Linkage. Johns Hopkins University Press, Baltimore.
- Wilson, A. J., D. W. Coltman, J. M. Pemberton, A. D. Overall, K. A. Byrne et al., 2005. a Maternal genetic effects set the potential for evolution in a free-living vertebrate population. J. Evol. Biol. 18: 405–414. [DOI] [PubMed] [Google Scholar]
- Wilson, A. J., J. G. Pilkington, J. M. Pemberton, D. W. Coltman, A. D. Overall et al., 2005. b Selection on mothers and offspring: Whose phenotype is it and does it matter? Evolution Int. J. Org. Evolution 59: 451–463. [PubMed] [Google Scholar]
- Wilson, K., B. T. Grenfell, J. G. Pilkington, H. E. G. Boyd and F. M. D. Gulland, 2004. Parasites and their impact, pp. 52–88 in Soay Sheep Dynamics and Selection in an Island Population, edited by T. H. Clutton-Brock and J. M. Pemberton. Cambridge University Press, Cambridge, UK.
- Zhang, Y. M., Y. Mao, C. Xie, H. Smith, L. Luo et al., 2005. Mapping quantitative trait loci using naturally occurring genetic variance among commercial inbred lines of maize (Zea mays L.). Genetics 169: 2267–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]