Abstract
The intronic sequences flanking exon–intron junctions (i.e., exon flanks) are important for splice site recognition and pre-mRNA splicing. Recent studies show a higher degree of sequence conservation at flanks of alternative exons, compared to flanks of constitutive exons. In this article we performed a detailed analysis on the evolutionary divergence of exon flanks between human and chimpanzee, aiming to dissect the impact of mutability and selection on their evolution. Inside exon flanks, sites that might reside in ancestral CpG dinucleotides evolved significantly faster than sites outside of ancestral CpG dinucleotides. This result reflects a systematic variation of mutation rates (mutability) at exon flanks, depending on the local CpG contexts. Remarkably, we observed a significant reduction of the nucleotide substitution rate in flanks of alternatively spliced exons, independent of the site-by-site variation in mutability due to different CpG contexts. Our data provide concrete evidence for increased purifying selection at exon flanks associated with regulation of alternative splicing.
THE intronic sequences that flank exon–intron junctions play a critical role in exon recognition and pre-mRNA splicing (Fairbrother and Chasin 2000; Sun and Chasin 2000; Zhang et al. 2003). These intronic sequences, referred to as “exon flanks” (Zhang et al. 2005), contain important splicing regulatory signals such as intronic splicing enhancers and silencers. Zhang and colleagues performed a genomewide study on the splicing signals in 50-nt exon flanks. They showed that exon flanks displayed a bimodal distribution of GC content, and high-GC and low-GC exon flanks might be associated with different mechanisms of exon recognition (Zhang et al. 2005). Some important splicing regulatory motifs are phylogenetically conserved in exon flanks (Kol et al. 2005; Minovitsky et al. 2005). Mutations at exon flanks frequently cause disruption of normal splicing patterns, such as the recognition of a cryptic splice site or the skipping of an entire exon (Eng et al. 2004; Costa et al. 2005; Coutinho et al. 2005). Such splicing mutations are one of the major causes for various human diseases (Garcia-Blanco et al. 2004). Therefore, although noncoding, exon flanks experience negative selection during evolution due to constraints of splicing regulation.
Recent comparative genomic analyses indicate a reduced nucleotide substitution rate at flanks of alternatively spliced exons. Sorek and Ast analyzed the 100-nt flanks of exons that were alternatively spliced in human and mouse transcripts. They observed a nearly 20% increase of percentage of sequence identity at 30-nt exon flanks of alternatively spliced exons, compared to flanks of constitutive exons (Sorek and Ast 2003). The average lengths of conserved sequences were 103 nt for upstream flanks of alternative exons and 94 nt for downstream flanks of alternative exons (Sorek and Ast 2003). Other studies reported similar results (Sugnet et al. 2004, 2006; Xing and Lee 2005). In fact, the increased sequence identity at flanks of alternative exons has been successfully used to predict alternative splicing from raw genomic sequences (Philipps et al. 2004; Sorek et al. 2004b; Yeo et al. 2005). It has been suggested that such a higher sequence conservation reflects stronger negative selection associated with regulation of alternative splicing (Sorek and Ast 2003; Itoh et al. 2004).
However, there are other factors that affect the nucleotide substitution rate in a genomic region. One important factor is the mutation rate. It is well known that the mutation rate varies significantly across the entire genome, leading to mutational hot spots and cold spots (Chuang and Li 2004). Even within the same gene, there can be remarkable site-by-site variations of the mutation rate. For instance, CpG dinucleotides are highly mutable due to methylation effects (Li et al. 2002). Keightley et al. analyzed the substitution rate in rodent genomes. They found that the substitution rate was significantly higher in CpG-susceptible sites (i.e., sites preceded by C or followed by G, which thus were likely to be parts of ancestral CpG dinucleotides) compared to non-CpG-susceptible sites (Keightley and Gaffney 2003; Gaffney and Keightley 2005). A similar observation was made through genomewide human–chimpanzee comparisons (Ebersberger et al. 2002; Hellmann et al. 2003) and more recently at distant intronic sites and fourfold redundant synonymous sites using human–chimpanzee genome alignments (Kondrashov et al. 2006). These data indicate the heterogeneity of mutation rate even within a small region of a gene, depending on CpG contexts.
In this article, we performed an in-depth analysis on the evolutionary divergence of exon flanks. We are interested in the following questions. First, how does the local CpG context affect substitution rates at flanks of constitutive and alternative exons? Second, what is the major cause for the increased sequence conservation at flanks of alternative exons? Is it reduced mutability or increased selection or both? Since multiple substitutions at a single site can obscure the impact of ancestral CpG dinucleotides, it is important to compare closely related species, where multiple substitutions are unlikely to occur (Ebersberger et al. 2002). The human and chimpanzee exon flanks are the most suitable for our comparison, because these two species diverged only ∼5 million years ago (Hedges 2002). The goal of our analysis is to dissect the contribution of mutability and selection to the evolutionary rate of exon flanks. We divided sites within exon flanks into distinct types according to their local CpG contexts. Windows ranging from 50 to 350 nt were used in previous analyses of exon flanks (Sorek and Ast 2003; Kaufmann et al. 2004; Sugnet et al. 2004, 2006; Xing and Lee 2005; Yeo et al. 2005). Throughout this article, following the consensus choice in the current literature (Sorek and Ast 2003; Sugnet et al. 2004, 2006), we defined our exon flanks as the 100-nt intronic sequences immediately upstream (upstream exon flanks) or downstream (downstream exon flanks) of the exon–intron junctions. A different definition (e.g., 50-nt exon flanks) produced very similar results (data not shown).
MATERIALS AND METHODS
We identified alternatively spliced exons in the human and mouse genome, by aligning expressed sequences (mRNA/EST) to the genomic sequences (Modrek et al. 2001). We matched orthologous exons as previously described (Modrek and Lee 2003). We compiled a set of ancestral alternatively spliced exons between human and mouse, i.e., exons whose inclusion and skipping forms were observed in both human and mouse transcripts (Xing and Lee 2005). The independent observation of alternative splicing in two species is widely adopted as a criterion for functional alternative splicing events (Sorek et al. 2004a; Yeo et al. 2005). For such an exon, alternative splicing is likely to be present in the common ancestor of human and mouse, therefore affecting the rate of nucleotide substitutions throughout subsequent evolution. We also used the data of Pan and colleagues to compile an independent set of ancestral alternatively spliced exons (Pan et al. 2004). Pan and colleagues identified mouse alternative exons that were also alternatively spliced in human, using EST–genome alignments and microarray data. For each mouse exon, we mapped it to the human genome (hg17, http://hgdownload.cse.ucsc.edu/goldenPath/hg17/bigZips/) using BLAST (Altschul et al. 1997), requiring a match to the exon over its entire length and an expectation value of <1e−4. Last, we compiled a set of exons that were constitutively spliced in human and mouse.
For each ancestral alternative or constitutive exon, we calculated the evolutionary divergence of its flanks between human and chimpanzee. We extracted the alignment of its flanks between human and chimpanzee using the UCSC human–chimpanzee pairwise genome alignments (http://hgdownload.cse.ucsc.edu/goldenPath/hg17/vsPanTro1/axtNet/) (Kent et al. 2003; Schwartz et al. 2003). We divided the nucleotide sites at exon flanks into four distinct types on the basis of the local CpG context: sites preceded by a C and followed by a G (postCpreG), sites followed by a G but not preceded by a C (preG), sites preceded by a C but not followed by a G (postC), and sites neither preceded by a C nor followed by a G (nonCpG) (Keightley and Gaffney 2003; Kondrashov et al. 2006). The first three types of sites were CpG susceptible according to the definition of Keightley and Gaffney, i.e., they were likely to be parts of ancestral CpG dinucleotides (Keightley and Gaffney 2003). We required each site to be flanked by two conserved nucleotides in the alignment and excluded sites that corresponded to insertions or deletions. For each nucleotide site, we checked the alignment to see whether it was conserved or substituted between human and chimpanzee. For each type of CpG sites, we counted the number of substituted sites and the total numbers of sites. The alignment data can be accessed at http://www.bioinformatics.ucla.edu/yxing/cpg/.
RESULTS AND DISCUSSION
We investigated the evolutionary divergence of exon flanks between human and chimpanzee. The complication of multiple substitutions at single sites is minimal because of the close relationship of human and chimpanzee (Ebersberger et al. 2002; Hedges 2002), allowing an accurate inference for the ancestral CpG dinucleotides. We extracted the alignments of 141 upstream flanks of ancestral alternatively spliced exons from the UCSC human–chimpanzee pairwise alignment. We also extracted the alignments of 40,638 upstream flanks of constitutive exons. We divided all nucleotide sites into four distinct types according to the local CpG contexts (see materials and methods).
We observed a systematic difference of nucleotide substitution rates among different types of sites. In upstream flanks of the constitutive exons, the nucleotide substitution rate averaged over all four types of sites was 0.0113. postCpreG sites had the highest rate of nucleotide substitution at 0.0172, followed by postC (0.0134) and preG (0.0124) sites. nonCpG sites had the lowest rate of nucleotide substitution at 0.0097, a nearly twofold reduction compared to postCpreG sites (see Table 1). The nucleotide composition at different types of sites also showed a systematic difference: postC sites had a very low frequency of G; preG sites had a very low frequency of C; while postCpreG sites were strongly depleted of both C and G. We observed a similar trend at upstream flanks of ancestral alternatively spliced exons. The overall nucleotide substitution rate was 0.0075. The rate was the lowest (0.0063) for non-CpG-susceptible sites (nonCpG sites, compared to CpG-susceptible sites including postCpreG, postC, and preG sites) (see Table 1). Analyses of downstream exon flanks yielded very similar results (see Table 2). Our results were consistent with human–chimpanzee analyses on distant intronic sites and fourfold redundant synonymous sites in exons (Kondrashov et al. 2006). These data indicate increased mutability at CpG-susceptible sites compared to non-CpG-susceptible sites in exon flanks, regardless of constitutive splicing or alternative splicing.
TABLE 1.
The nucleotide substitution rates at upstream exon flanks
| All sites | PostCpreG | PostC | PreG | NonCpG | |
|---|---|---|---|---|---|
| Constitutive | |||||
| No. of sites | 3,825,522 | 231,695 | 701,611 | 620,871 | 2,271,345 |
| No. of substituted sites | 43,207 | 3,996 | 9,412 | 7,685 | 22,114 |
| Frequencies | |||||
| A | 22.9% | 41.1% | 21.6% | 25.3% | 20.8% |
| T | 31.3% | 44.7% | 33.3% | 32.5% | 29.0% |
| C | 24.2% | 7.4% | 40.4% | 4.9% | 26.1% |
| G | 21.5% | 6.7% | 4.7% | 37.3% | 24.0% |
| Proportion of nucleotide difference | 0.0113 | 0.0172 | 0.0134 | 0.0124 | 0.0097 |
| Alternative | |||||
| No. of sites | 13,412 | 749 | 2,801 | 2,078 | 7,784 |
| No. of substituted sites | 101 | 7 | 26 | 19 | 49 |
| Frequencies | |||||
| A | 21.1% | 36.8% | 20.1% | 24.8% | 19.1% |
| T | 32.4% | 44.3% | 33.5% | 34.0% | 30.4% |
| C | 26.2% | 9.6% | 41.7% | 6.1% | 27.5% |
| G | 20.3% | 9.2% | 4.7% | 35.1% | 23.0% |
| Proportion of nucleotide difference | 0.0075 | 0.0093 | 0.0093 | 0.0091 | 0.0063 |
TABLE 2.
The nucleotide substitution rates at downstream exon flanks
| All sites | PostCpreG | PostC | PreG | NonCpG | |
|---|---|---|---|---|---|
| Constitutive | |||||
| No. of sites | 3,831,232 | 222,572 | 614,689 | 759,625 | 2,234,346 |
| No. of substituted sites | 44,107 | 4,501 | 8,806 | 9,185 | 21,615 |
| Frequencies | |||||
| A | 24.2% | 39.5% | 24.2% | 26.4% | 21.9% |
| T | 28.3% | 44.3% | 30.0% | 28.6% | 26.1% |
| C | 22.1% | 7.6% | 40.6% | 4.6% | 24.4% |
| G | 25.4% | 8.6% | 5.2% | 40.4% | 27.6% |
| Proportion of nucleotide difference | 0.0115 | 0.0202 | 0.0143 | 0.0121 | 0.0097 |
| Alternative | |||||
| No. of sites | 13,682 | 830 | 2,394 | 2,640 | 7,818 |
| No. of substituted sites | 97 | 16 | 19 | 19 | 43 |
| Frequencies | |||||
| A | 22.5% | 35.8% | 22.3% | 24.7% | 20.3% |
| T | 28.6% | 45.2% | 30.7% | 29.3% | 26.0% |
| C | 23.8% | 9.2% | 41.4% | 5.2% | 26.2% |
| G | 25.1% | 9.9% | 5.6% | 40.8% | 27.5% |
| Proportion of nucleotide difference | 0.0071 | 0.0193 | 0.0079 | 0.0072 | 0.0055 |
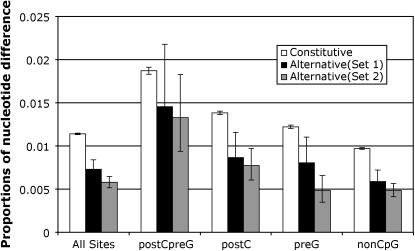
Comparing upstream flanks of alternative and constitutive exons, we observed a significant reduction of nucleotide substitution rate associated with alternative splicing (see Table 1). The overall nucleotide substitution rate at upstream flanks was 0.0075 for alternative exons, a 33.6% reduction compared to constitutive exons (0.0113). Remarkably, all four types of sites experienced a reduced substitution rate at flanks of alternative exons (see Table 1), ranging from a reduction of 45.9% (at postCpreG sites) to 26.6% (at preG sites). We also compared the substitution rate at downstream flanks of alternative and constitutive exons and observed a similar trend. The overall substitution rate had a 38.3% reduction at downstream flanks of alternative exons. All four types of CpG sites had reduced substitution rates (see Table 2). These data suggest that alternative splicing is associated with a universal increase in the strength of negative selection at exon flanks, independent of the site-by-site variation of mutation rates due to different CpG contexts (see Figure 1, solid bars vs. open bars).
Figure 1.—
The nucleotide substitution rates over the 100-nt upstream and 100-nt downstream exon flanks of constitutive and alternative exons. Error bars represent 95% confidence intervals (calculated by assuming a binomial distribution). Alternative (set 1), ancestral alternatively spliced exons from Xing and Lee (2005); Alternative (set 2), ancestral alternatively spliced exons from Pan et al. (2004). See details of these data sets in materials and methods.
To rule out the possibility of artifacts due to potential sampling biases in our alternative splicing data set, we analyzed an independent set of ancestral alternatively spliced exons. This data set included 289 exons, which were identified using EST–genome alignments and microarray data (Pan et al. 2004). We observed the same trend in this data set (see Figure 1). All four types of CpG sites had a statistically significant reduction in their substitution rates, compared to constitutive exons (see Figure 1, shaded bars vs. open bars). We also observed the same trend when we restricted our analysis to 50-nt exon flanks. These data demonstrate that our reported finding is a confident result.
Our study extended previous comparative analyses of intronic sites flanking alternatively spliced exons (Sorek and Ast 2003; Sugnet et al. 2004; Xing and Lee 2005). Instead of analyzing the evolutionary divergence of the entire exon flanks, we divided all nucleotide sites at exon flanks into distinct types, according to their local CpG contexts. Such treatment allowed us to separate the contribution of mutability and selection, two most important factors that affect the divergence rate of exon flanks. Our study indicates significant site-by-site variations of mutability in the exon flanks, depending on the local CpG context. Such a phenomenon was observed in flanks of both alternative and constitutive exons. However, the site-by-site variation of mutability is not the major contributor to the increased conservation at flanks of alternative exons. The higher sequence conservation is not due to a reduced fraction of hypermutable sites (i.e., CpG-susceptible sites). In fact, at all types of CpG sites (with different propensities for mutation) inside flanks of alternative exons, we observed a significant reduction of the nucleotide substitution rate (see Figure 1). This consistent pattern, independent of the variation in mutability due to different CpG contexts, strongly argues for an increased selection at flanks of alternative exons. Such a strong purifying selection might reflect RNA sequence motif (Itoh et al. 2004) or secondary structure (Meyer and Miklos 2005) requirements for proper controls of alternative splicing. Our study provides concrete evidence for widespread selection pressure in mammalian genomes that is associated with alternative splicing (Sorek and Ast 2003; Baek and Green 2005; Xing and Lee 2005). Understanding selection pressure at exon flanks will shed light on the regulation of alternative splicing and how it evolves.
Acknowledgments
We thank Fyodor Kondrashov for key discussions that led to this analysis and for sharing one manuscript prior to its publication. We thank Sandy Pan and Benjamin Blencowe for sharing mouse splicing microarray data and Xiang Zhang and Alex Alekseyenko for helpful comments and suggestions. This work was supported by National Institutes of Health grant U54-RR021813, a Teacher–Scholar award to C.J.L. from the Dreyfus Foundation, a Department of Energy grant DE-FC02-02ER63421, and a Ph.D. dissertation fellowship to Y.X. from University of California, Los Angeles.
References
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, D., and P. Green, 2005. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. USA 102: 12813–12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, J. H., and H. Li, 2004. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2: E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, D. B., L. Lozovatsky, P. G. Gallagher and B. G. Forget, 2005. A novel splicing mutation of the {alpha}-spectrin gene in the original hereditary pyropoikilocytosis kindred. Blood 106: 4367–4369. [DOI] [PMC free article] [PubMed]
- Coutinho, G., J. Xie, L. Du, A. Brusco, A. R. Krainer et al., 2005. Functional significance of a deep intronic mutation in the ATM gene and evidence for an alternative exon 28a. Hum. Mutat. 25: 118–124. [DOI] [PubMed] [Google Scholar]
- Ebersberger, I., D. Metzler, C. Schwarz and S. Paabo, 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70: 1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, L., G. Coutinho, S. Nahas, G. Yeo, R. Tanouye et al., 2004. Nonclassical splicing mutations in the coding and noncoding regions of the ATM gene: maximum entropy estimates of splice junction strengths. Hum. Mutat. 23: 67–76. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W. G., and L. A. Chasin, 2000. Human genomic sequences that inhibit splicing. Mol. Cell. Biol. 20: 6816–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, D. J., and P. D. Keightley, 2005. The scale of mutational variation in the murid genome. Genome Res. 15: 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco, M. A., A. P. Baraniak and E. L. Lasda, 2004. Alternative splicing in disease and therapy. Nat. Biotechnol. 22: 535–546. [DOI] [PubMed] [Google Scholar]
- Hedges, S. B., 2002. The origin and evolution of model organisms. Nat. Rev. Genet. 3: 838–849. [DOI] [PubMed] [Google Scholar]
- Hellmann, I., S. Zollner, W. Enard, I. Ebersberger, B. Nickel et al., 2003. Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 13: 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., T. Washio and M. Tomita, 2004. Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes. RNA 10: 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, D., O. Kenner, P. Nurnberg, W. Vogel and B. Bartelt, 2004. In NF1, CFTR, PER3, CARS and SYT7, alternatively included exons show higher conservation of surrounding intron sequences than constitutive exons. Eur. J. Hum. Genet. 12: 139–149. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and D. J. Gaffney, 2003. Functional constraints and frequency of deleterious mutations in noncoding DNA of rodents. Proc. Natl. Acad. Sci. USA 100: 13402–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W. J., R. Baertsch, A. Hinrichs, W. Miller and D. Haussler, 2003. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. USA 100: 11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol, G., G. Lev-Maor and G. Ast, 2005. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum. Mol. Genet. 14: 1559–1568. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F. A., A. Y. Ogurtsov and A. S. Kondrashov, 2006. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J. Theor. Biol. 240: 616–626. [DOI] [PubMed] [Google Scholar]
- Li, W. H., S. Yi and K. Makova, 2002. Male-driven evolution. Curr. Opin. Genet. Dev. 12: 650–656. [DOI] [PubMed] [Google Scholar]
- Meyer, I. M., and I. Miklos, 2005. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 33: 6338–6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minovitsky, S., S. L. Gee, S. Schokrpur, I. Dubchak and J. G. Conboy, 2005. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 33: 714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., and C. Lee, 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased rate of exon creation / loss. Nat. Genet. 34: 177–180. [DOI] [PubMed] [Google Scholar]
- Modrek, B., A. Resch, C. Grasso and C. Lee, 2001. Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res. 29: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., O. Shai, C. Misquitta, W. Zhang, A. L. Saltzman et al., 2004. Revealing global regulatory features of Mammalian alternative splicing using a quantitative microarray platform. Mol. Cell 16: 929–941. [DOI] [PubMed] [Google Scholar]
- Philipps, D. L., J. W. Park and B. R. Graveley, 2004. A computational and experimental approach toward a priori identification of alternatively spliced exons. RNA 10: 1838–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., W. J. Kent, A. Smit, Z. Zhang, R. Baertsch et al., 2003. Human-mouse alignments with BLASTZ. Genome Res. 13: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., and G. Ast, 2003. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 13: 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., R. Shamir and G. Ast, 2004. a How prevalent is functional alternative splicing in the human genome? Trends Genet. 20: 68–71. [DOI] [PubMed] [Google Scholar]
- Sorek, R., R. Shemesh, Y. Cohen, O. Basechess, G. Ast et al., 2004. b A non-EST-based method for exon-skipping prediction. Genome Res. 14: 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet, C. W., W. J. Kent, M. Ares, Jr. and D. Haussler, 2004. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput, 66–77. [DOI] [PubMed]
- Sugnet, C. W., K. Srinivasan, T. A. Clark, G. O'Brien, M. S. Cline et al., 2006. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., and L. A. Chasin, 2000. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 20: 6414–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., and C. Lee, 2005. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl. Acad. Sci. USA 102: 13526–13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G. W., E. Van Nostrand, D. Holste, T. Poggio and C. B. Burge, 2005. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. USA 102: 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. H., K. A. Heller, I. Hefter, C. S. Leslie and L. A. Chasin, 2003. Sequence information for the splicing of human pre-mRNA identified by support vector machine classification. Genome Res. 13: 2637–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. H., C. S. Leslie and L. A. Chasin, 2005. Dichotomous splicing signals in exon flanks. Genome Res. 15: 768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]