Abstract
We demonstrate a new method, microarray-assisted bulk segregant analysis, for mapping traits in yeast by genotyping pooled segregants. We apply a probabilistic model to the progeny of a single cross and as little as two microarray hybridizations to reliably map an auxotrophic marker, a Mendelian trait, and a major-effect quantitative trait locus.
MAPPING the genes responsible for traits remains one of the most basic goals of genetics. There are several techniques available for reaching this goal, especially in model organisms such as the yeast Saccharomyces cerevisiae. The dense and well-characterized genetic map of S. cerevisiae supports traditional tetrad analysis, which is still widely used in spite of its requirements for time- and labor-intensive crosses and dissections. More recently developed genomics techniques take advantage of parallel genotyping of single feature polymorphisms (SFPs) and other small differences between two strain backgrounds (Winzeler et al. 1998, 1999, 2003; Brem et al. 2002; Steinmetz et al. 2002; Deutschbauer and Davis 2005). Other approaches use the strains created by the yeast deletion project as a dense collection of genetic markers that can be crossed in parallel to a query strain (Jorgensen et al. 2002). Finally, a new approach looks for mutations directly by hybridizing DNA from a strain of interest to a reference sequence microarray (Gresham et al. 2006).
All of these approaches have limitations that reduce their value as routinely applied methods for mapping suppressors or other mutations. Most highly parallel methods are still resource intensive, requiring 10 or more array-typed segregants. Mapping strategies that use the yeast deletion collection are appropriate only for limited classes of phenotypes and are tedious and time-consuming without substantial robotic automation. Methods that find DNA sequence changes directly are unable to link these changes to particular phenotypes without extensive follow-up work.
Here we describe a microarray-based method—microarray-assisted bulk segregant analysis—that applies a highly parallel genotyping strategy to mapping genes in pooled populations of yeast. Bulk segregant analysis measures the variation present in pools of segregants that have been sorted according to phenotype and uses the correlation between these measurements and the pool phenotype to assign a likely map location. This is an improvement over methods that require individual genotyping, as it simultaneously measures the average genotype of progeny with a given phenotype. The approach of bulk segregant analysis was first developed (Michelmore et al. 1991) and adapted for microarray-based genotyping (Borevitz et al. 2003; Hazen et al. 2005) in plants. We have adapted the method for use in yeast and have also developed an analytical model that takes advantage of the linkage that is expected to occur between loci at similar map locations. Incorporating this cosegregation into the mapping model greatly improves the accuracy of the method and allows the calculation of confidence limits for derived map locations. Our method requires the isolation of ∼100 segregants, either individually dissected or randomly isolated from populations of tetrads, and hybridization to as few as two microarrays.
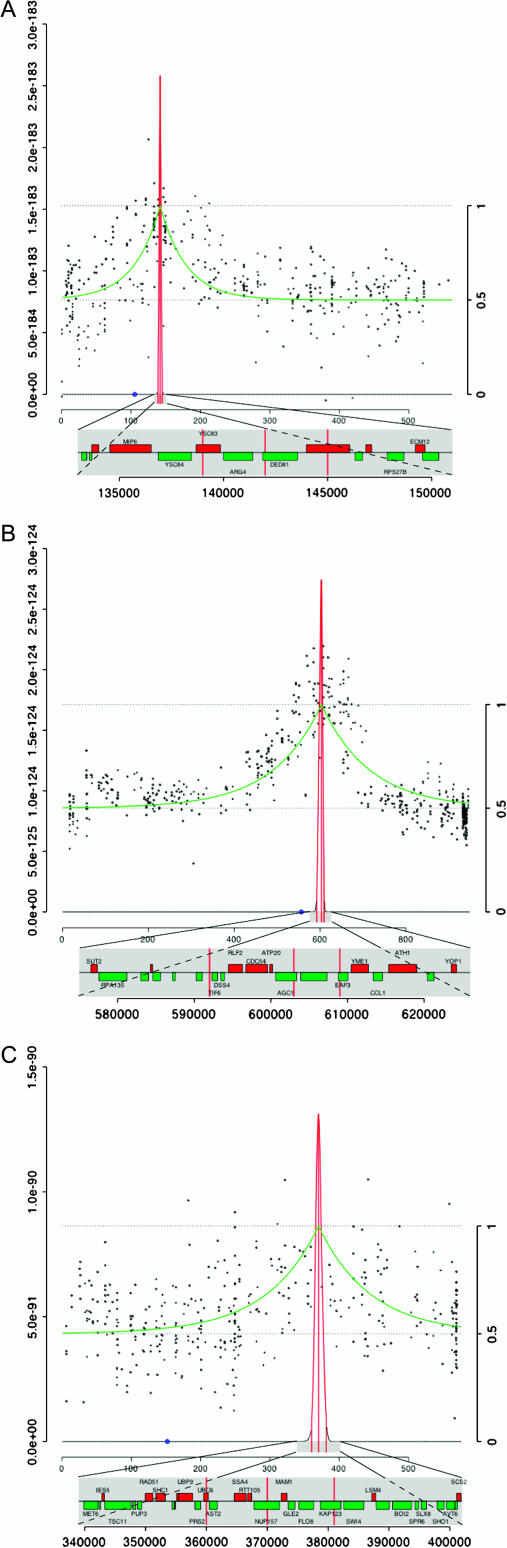
As a demonstration of the technique, we mapped three traits of varying difficulty and complexity (Figure 1). For each experiment, the basic layout was the same. Two strains differing both by genetic background and by the trait of interest were crossed. A total of 30–60 tetrads were dissected from the diploid and the segregants from these dissections were grown individually and pooled according to phenotype. Genomic DNA from each pool was hybridized to an Affymetrix S98 microarray, consisting of 25mer probes designed from the sequence of a reference strain at an average density of 16 probes/gene over 6400 genes.
Figure 1.—
Mapping of the locus for (A) a known arginine auxotrophy, (B) an unknown acetate growth defect, and (C) a major QTL involved in flocculation. For each experiment, 10 μg of DNA from each parent strain or pool was labeled as described (Winzeler et al. 1999) and hybridized to Affymetrix (Santa Clara, CA) YG-S98 GeneChip microarrays. Hybridized arrays were washed using the Affymetrix fluidics station, scanned with a GeneChip Scanner 3000, and analyzed with Affymetrix software to obtain the signal intensity for each feature (the “CEL” file). A detailed protocol is available as supplemental data at http://genomics-pubs.princeton.edu/snappy. Signal variation caused by location-specific artifacts was calculated for each feature by computing the mean intensity within a 37 × 37-feature window centered on that feature. This variation was subtracted from each feature on the array. The resulting normalized hybridization values from each feature were log transformed, averaged over hybridization replicates, and stored for further analysis. Using the parental strain hybridizations, 7236 SFPs were identified between SK1 and FY, and 5787 between Y55 and FY. Hybridization intensity data for these probes were scaled for each pool so that the reference parent had a value of 1.0, while the divergent parent's value was 0.0. Scaled and normalized data (shown as black dots) were applied to a likelihood model of recombination (best fit shown in green) to more precisely map the region of the chromosome most tightly linked to the gene of interest. The interval marked by the outer red vertical lines corresponds to the region containing loci with likelihood ratios of >20. Raw and processed data are available as supplemental data at http://genomics-pubs.princeton.edu/snappy, and analysis scripts are available upon request.
Each parent strain was also hybridized by itself to an array. Approximately 6000 probes had significantly different signal intensities between the divergent and reference strain hybridizations. These variable probes correlate with interparent strain sequence polymorphisms (Winzeler et al. 1998, 1999, 2003; Brem et al. 2002; Steinmetz et al. 2002; Deutschbauer and Davis 2005). By comparing the hybridization intensity of each pool to each parent, the variable probes were used as markers to estimate the genotype frequency of each pool at each probe location. Most markers, inherited in equal frequencies from each parent, were unlinked to the trait and yielded an intermediate intensity value. Markers that were linked to the trait showed a bias in intensity toward the parent with the corresponding phenotype.
These intensity data were scaled to represent relative genotype frequencies and analyzed using a derivation of Haldane's recombination model (see supplemental data at http://genomics-pubs.princeton.edu/snappy). The parameters estimated by the fit to the model were used to define a region of linkage to the trait of interest. Genes in this region were then tested for association with the trait.
As a test case, we first mapped a known arginine auxotrophy that segregated 2:2 in the cross of reference strain FY (ARG4) and divergent strain SK1 (arg4). Fifty-six arginine prototrophs and 62 arginine auxotrophs were pooled and processed as described above. The ARG4 locus was contained in a 6-kb region with three other genes in the only interval found to have a location estimate likelihood ratio of at least 20 (Figure 1A). Increasing the number of segregants in each pool did not significantly improve the mapping resolution (data not shown).
We next mapped a Mendelian trait for which the causative gene was not known. The strain CP1AB, derived from S288C for use in chemostat evolutions (Paquin and Adams 1982), has a slight growth defect relative to other S288C strains when acetate is supplied as the sole carbon source (“acetate −”). When CP1AB was crossed to FY, the trait segregated 2:2 in tetrads, indicating that a single gene was responsible. Since the phenotype was a subtle one, we were unable to clone the gene by complementation with a transformed library. Neither could we map the gene by crossing with the ordered deletion set (Jorgensen et al. 2002). We mated isolates of CP1AB with the divergent strains SK1 and Y55. From each cross, we pooled ∼30 acetate+ and 30 acetate− segregants and hybridized DNA from each of the four pools to an array. Both mapping crosses indicated that the gene was close to the centromere of chromosome 16 (Y55 cross shown in Figure 1B), which we confirmed by traditional mapping using crosses with strains from the yeast deletion collection containing G418 resistance cassettes in the place of nearby genes. Due to the reduced levels of recombination near the centromere, we were unable to locate the gene more precisely. Nevertheless, the localization of the gene to this region allowed us to identify the relevant mutation from among those found subsequently using a tiling microarray-based genomewide polymorphism screen (Gresham et al. 2006). We confirmed a point mutation in the CEN16-linked AEP3, which, when deleted, causes respiratory defects (Ellis et al. 2004). Since many possible mutations were identified, the mapping data were essential for associating this mutation with the acetate growth phenotype.
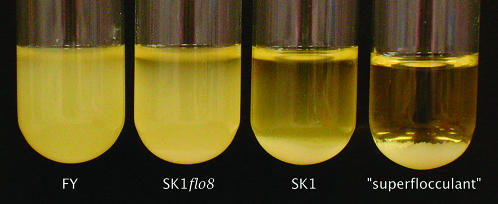
Finally, we mapped a major-effect quantitative trait locus (QTL), showing that our technique is robust to some amount of genetic complexity. In the cross between SK1 and FY used to map the ARG4 locus, we found a segregating flocculation phenotype. Cells of the FY parent strain were not clumpy and settled slowly, while those of SK1 grew in small, granular clumps that settled quickly. Segregants were grouped into three pools: FY like (76 segregants), SK1 like (38 segregants), and a pool of “superflocculent” cultures (23 segregants) that formed one large clump that was poorly dispersed by vortexing (see Figure 2, for example). The mapping data yielded a strong peak when the very flocculent and FY-like pools were compared (Figure 1C). The 21-kb interval defined by the peak contained the flocculation transcription factor FLO8, which is known to be mutated in S288C strains (Liu et al. 1996). We deleted FLO8 in SK1 and reproduced the FY-like phenotype (Figure 2). Crosses between SK1flo8 and FY resulted in many fewer flocculent segregants with no obvious pattern, indicating that several small-effect QTL probably remain.
Figure 2.—
Flocculation phenotype of FY, SK1flo8, SK1, and a “superflocculent” segregant. Saturated cultures in rich media were vortexed and allowed to settle for 10 min. FLO8 was deleted and replaced with URA3 by a standard one-step transformation procedure and then checked by PCR.
With bulk segregant analysis we have successfully mapped two traits to windows of 6 and 21 kb, and a third trait to a particular centromere. These results are comparable to applications of other mapping techniques. An example of a mapping strategy using crosses to the yeast deletion collection yielded a map interval of 90 kb, which was further reduced only with the additional step of random spore analysis (Jorgensen et al. 2002). Mapping using array-genotyped individual segregants has achieved intervals between 11 and 64 kb for Mendelian traits (10 typed segregants) (Winzeler et al. 1998) and 52 kb for a major-effect QTL (19 typed segregants), which was narrowed to 32 kb using custom genotyping on more segregants (Steinmetz et al. 2002). A more recent article mapped three QTL to intervals of 12–30 kb through a combination of segregant genotyping, backcrossing to isolate the individual QTL, and custom-targeted SNP typing across candidate intervals (Deutschbauer and Davis 2005). All of these approaches have required additional mapping and functional assays to further define the causative region. Overall, our method attains comparable accuracy but with less expense and in a shorter time.
Acknowledgments
The authors thank David Botstein and members of the Botstein lab for critical guidance during development of the method. We also thank Leonid Kruglyak and Mark Rose for help with the mapping model. This work was supported in part by grants from the National Institutes of Health to M.J.B. (F32 HG02649-01) and to David Botstein (R01 GM046406 and P50 GM071508).
References
- Borevitz, J. O., D. Liang, D. Plouffe, H. S. Chang, T. Zhu et al., 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A. M., and R. W. Davis, 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Ellis, T. P., K. G. Helfenbein, A. Tzagoloff and C. L. Dieckmann, 2004. Aep3p stabilizes the mitochondrial bicistronic mRNA encoding subunits 6 and 8 of the H+-translocating ATP synthase of Saccharomyces cerevisiae. J. Biol. Chem. 279: 15728–15733. [DOI] [PubMed] [Google Scholar]
- Gresham, D., D. M. Ruderfer, S. C. Pratt, J. Schacherer, M. J. Dunham et al., 2006. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311: 1932–1936. [DOI] [PubMed] [Google Scholar]
- Hazen, S. P., J. O. Borevitz, F. G. Harmon, J. L. Pruneda-Paz, T. F. Schultz et al., 2005. Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 138: 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., B. Nelson, M. D. Robinson, Y. Chen, B. Andrews et al., 2002. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics 162: 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., C. A. Styles and G. R. Fink, 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R. W., I. Paran and R. V. Kesseli, 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin, C., and J. Adams, 1982. Isolation of sets of a, α, a/α, a/a and α/α isogenic strains in Saccharomyces cervisiae. Curr. Genet. 6: 21–24. [DOI] [PubMed] [Google Scholar]
- Steinmetz, L. M., H. Sinha, D. R. Richards, J. I. Spiegelman, P. J. Oefner et al., 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. R. Richards, A. R. Conway, A. L. Goldstein, S. Kalman et al., 1998. Direct allelic variation scanning of the yeast genome. Science 281: 1194–1197. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., B. Lee, J. H. McCusker and R. W. Davis, 1999. Whole genome genetic-typing in yeast using high-density oligonucleotide arrays. Parasitology 118(Suppl.): S73–S80. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., C. I. Castillo-Davis, G. Oshiro, D. Liang, D. R. Richards et al., 2003. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]