Abstract
The formation of the venation pattern in leaves is ideal for examining signaling pathways that recognize and respond to spatial and temporal information, because the pattern is two-dimensional and heritable and the resulting veins influence the three-dimensional spatial organization of the surrounding differentiating leaf cell types. We identified a provascular/procambial cell–specific gene that encodes a Leu-rich repeat receptor kinase, which we named VASCULAR HIGHWAY1 (VH1). A change in the expression domain and level of VH1 marks the transition from an uncommitted provascular state to a committed procambial state in early vascular development. The coding sequence, expression pattern, and transgenic phenotypes together suggest that VH1 transduces extracellular spatial and temporal signals into downstream cell differentiation responses in provascular/procambial cells.
INTRODUCTION
The study of pattern formation in development has furthered our understanding of morphogenetic processes and of the mechanisms of signal transduction pathways. For example, genetic studies of the two-dimensional venation pattern in the Drosophila wing found that genes affecting the venation pattern also affect cell proliferation in the wing and that they encode components of the EGF-R, Notch, and dpp signaling pathways (Diaz-Benjumea et al., 1989; de Celis et al., 1994; Segal and Gelbart, 1995). Indeed, intercellular signaling by growth signals is essential for pattern formation in metazoan development, such as the establishment of the left-right body axis in mammals, the body size and tail morphology in nematodes, and the formation of adult appendages in flies (Posakony et al., 1991; Collignon et al., 1996; Savage et al., 1996). This is true as well for plant development, in which such signaling regulates cell division versus cell differentiation in meristems and in lateral organ primordia.
The formation of the venation pattern in leaves is ideal for examining signaling pathways that recognize and respond to spatial and temporal information, because the pattern is two-dimensional and heritable and arises during intercalary growth of the leaf primordium (Nelson and Dengler, 1997). The developing veins of the leaf primordium appear to serve as morphogenetic centers that organize spatially many of the differentiating leaf cell types through an undefined intercellular signaling system. These spatial relationships make functional sense, because many of the cell types must function in cooperation with veins. For example, studies of the differentiation of bundle sheath and mesophyll cells in maize and other C4 monocots suggested the siting of the veins as the first step in the patterning of surrounding photosynthetic cell types (Langdale et al., 1988, 1989; Nelson and Dengler, 1992).
At the cellular level, the formation of the venation pattern begins in the leaf primordium with the siting of procambial (PC) cells among equivalent provascular (PV) cells, which are cells in an uncommitted meristematic state with vascular potential. PC strands are initiated by longitudinal divisions that result in aligned elongate cells. PC cells give rise to differentiated vascular cell types and thus constitute a vascular meristematic tissue that exists transiently at sites that foreshadow the location of the venation network (Nelson and Dengler, 1997). Several genes with potential regulatory or sig-naling roles in venation patterning also exhibit PC expression patterns. Certain homeodomain-leucine zipper genes appear to be PC cell specific, including the Arabidopsis genes Athb8 and IFL1/REV, the rice gene Oshox1, and the tomato gene Vahox1 (Baima et al., 1995; Tornero et al., 1996; Zhong and Ye, 1999; Ratcliffe et al., 2000; Scarpella et al., 2000). Some authors do not distinguish the PC stage and describe all early vascular stages as PV.
Here, we describe the identification of a PV/PC cell–specific gene that encodes a putative Leu-rich repeat (LRR) receptor kinase, which we named VASCULAR HIGHWAY1 (VH1). Ectopic expression of VH1 in transformed wild-type Arabidopsis plants affects both venation patterning and cell proliferation and differentiation of many cell types in juvenile leaves. A loss-of-function allele causes premature senescence and cell death in leaves, possibly as a consequence of a defect in transport among PC and/or mature vascular cells. VH1's coding sequence, expression pattern, and transgenic phenotypes suggest that VH1 transduces extracellular spatial and temporal signals into downstream cell differentiation responses in PV/PC cells.
RESULTS
VH1 Is Associated with a PV/PC Enhancer
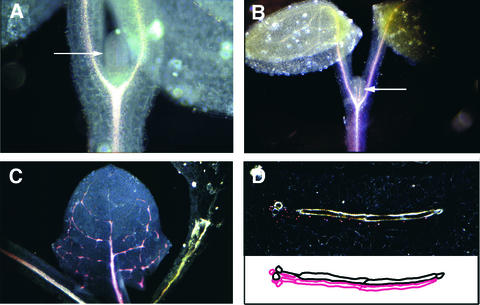
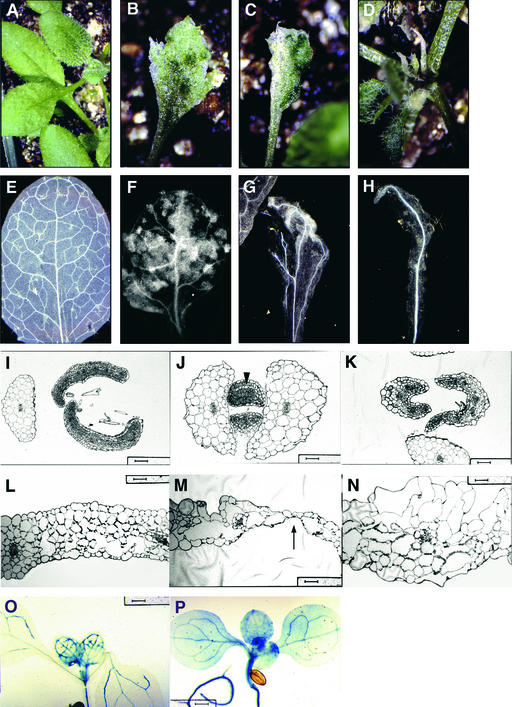
In the course of screening an Arabidopsis enhancer trap collection, we identified a line in which reporter expression was observed in PC cells throughout the seedling as well as in PV cells of young leaf primordia before the onset of vascularization. At a later stage of leaf organogenesis, the broad expression domain seen in leaf primordia (Figure 1A) becomes restricted to the PC cells of the primary midvein (Figure 1B); subsequently, it is limited to the paths of the formation of higher order veins, which proceeds from leaf tip to leaf base through a hierarchical series (Figure 1C). At this stage, reporter expression continues to mark PC cells (Figure 1D), which now exist transiently at many locations that correspond to the growing tips of veins and to vein junctions where cell proliferation occurs. We cloned the genomic region immediately upstream from the minimal promoter of the reporter gene via inverse PCR and found the enhancer trap T-DNA to be ∼2.6 kb upstream of a gene on chromosome 2, 9 to 10 centimorgan on the genetic map (data not shown). We subsequently confirmed that this gene, VH1, exhibits an expression pattern identical to that of the reporter in the enhancer trap line.
Figure 1.
VH1 Is Associated with a PV/PC Enhancer.
(A) and (B) Seedlings from an enhancer trap line at successive stages of leaf development were stained for GUS expression and viewed under dark-field illumination (GUS staining appears pink). Arrows point to GUS-stained leaf primordium (A) and to a GUS-stained PC strand of the midvein (B).
(C) GUS staining is specific to PC cells, and when those cells mature, they no longer stain for GUS activity.
(D) A transverse section through an expanding leaf shows a single file of GUS-stained PC cells underneath the xylem strand (top). Below is the corresponding tracing of xylem cells (black) and PC cells (pink).
VH1 Encodes an LRR Receptor Kinase
The nucleotide sequence of VH1 revealed one large, intronless open reading frame of 3431 bp that encodes a predicted protein of 1143 amino acids with an estimated molecular mass of 124 kD. There is a typical TATA box sequence (TAATAAAT) at positions −137 to −144 and no other ATGs downstream. A canonical poly(A) addition site (AATAAA) (Joshi, 1987) was found 150 bp downstream from the stop codon.
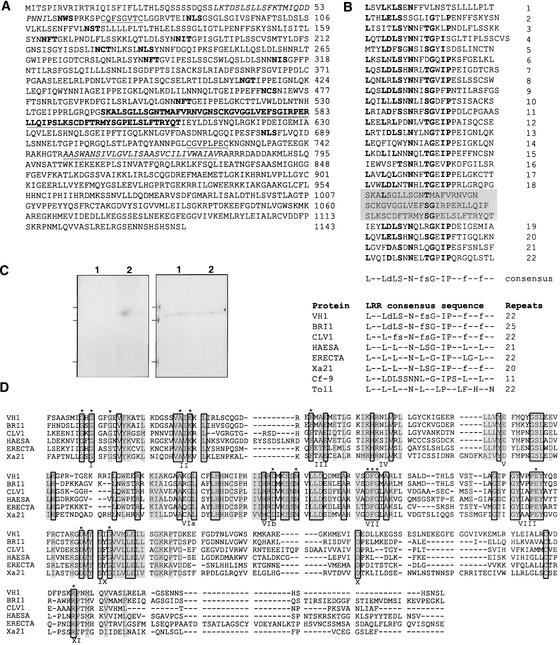
The VH1 protein is a member of a large family of plant LRR receptor kinases. Its highest protein sequence identity is to the Arabidopsis genes BRI1 (45% identity; Li and Chory, 1997), HAESA (32% identity; Jinn et al., 2000), ERECTA (33% identity; Torii et al., 1996), and CLV1 (32% identity; Clark et al., 1997), all of which are involved in signaling pathways that affect plant development. Like the other family members, the predicted VH1 protein has several distinct domains: a signal peptide, an extracellular LRR domain, a transmembrane domain, and an intracellular Ser/Thr kinase domain (Figure 2A).
Figure 2.
VH1 Encodes a LRR Receptor Kinase.
(A) The predicted VH1 protein contains the following regions: a signal peptide (amino acids 1 to 31); a putative Leu zipper (italicized); an extracellular LRR domain (amino acids 105 to 708) that contains a buried 68–amino acid island (underlined and in boldface) and is flanked by two conservatively spaced Cys-pair regions (underlined); a transmembrane domain (italicized and underlined) that is flanked by stop transfer sequences; and an intracellular kinase domain (amino acids 795 to 1143) with Ser/Thr specificity. Potential N-glycosylation sites are shown in boldface.
(B) The extracellular domain contains 22 LRRs with a unit length of 24 amino acids. Numbers at right of the LRR domain indicate the specific LRR number, and the bottom line is the consensus sequence for the VH1 LRR. Dashes stand for any amino acid, and φ indicates an aliphatic amino acid residue. Amino acid residues that match the deduced consensus sequence are shown in boldface. Lowercase letters in the consensus sequence indicate residues identical in at least half of the repeats. A 68–amino acid island that does not fit the consensus sequence is buried between the 18th and 19th LRRs (black box). In the table at bottom, the LRR consensus sequence of VH1 is compared with those of other LRR-containing signaling proteins: Arabidopsis BRI1, CLV1, HAESA, and ERECTA; rice Xa21; tomato Cf-9; and Drosophila Toll.
(C) Maltose binding protein (MBP) fusions to an active and an inactive version of the VH1 kinase domain (MBP::VH1 and MBP::K866N, respectively) were affinity purified from isopropylthio-β-galactoside–induced E. coli cells and used in an in vitro kinase assay. The gel at right is a Coomassie blue–stained 10% SDS-PAGE gel, and the gel at left is the corresponding autoradiogram, which shows 32P-labeled MBP::VH1 (lane 2) as a product of an autophosphorylation reaction. MBP::K866N is in lane 1. Lines at left indicate molecular mass markers (107, 76, and 52 kD from top to bottom).
(D) Alignment of the kinase domain among putative LRR receptor kinases in plants. Residues that are conserved among all of the compared sequences are boxed, and those that are conserved among at least four sequences are shaded. The 12 conserved protein kinase domains are indicated I to XI (Hanks and Quinn, 1991). The 15 invariant amino acids present in all protein kinases are indicated by asterisks.
The N terminus of the VH1 protein has a potential 31–amino acid signal peptide that directs secretion (von Heijne, 1990), which is followed by a putative extracellular domain consisting of 22 tandem copies of a 24–amino acid LRR with 11 N-linked glycosylation consensus sites (N-X-S/T). The LRRs resemble repeats found in animal hormone receptors and plant disease resistance receptors, such as the rice Xa21 (Song et al., 1995), the tomato Cf-9 (Jones et al., 1994), and the Arabidopsis FLS2 (Gomez-Gomez and Boller, 2000) (Figure 2B). They are believed to play a role in protein–protein interactions (Kobe and Deisenhofer, 1994). The LRR region is thought to form a semibarrel structure (Jiang et al., 1995). In addition, the region is interrupted by a 68–amino acid “island” between the 18th and 19th LRRs. Islands also have been found in the LRR domains of BRI1, Cf-9, and the Arabidopsis gene CLV2 (Jeong et al., 1999). In the case of BRI1, the island was shown to be essential for its function. As with many LRR-containing proteins, the LRR region of VH1 is flanked by pairs of conservatively spaced Cys residues (Jones and Jones, 1996).
The transmembrane and kinase domains of the VH1 protein are typical. The transmembrane domain is flanked by two stop-transfer sequences. The putative intracellular kinase domain has Ser/Thr specificity and contains all of the subdomains and invariant amino acid residues found among Ser/Thr protein kinases (Hanks and Quinn, 1991) (Figure 2D). Escherichia coli–expressed recombinant VH1 has kinase activity (Figure 2C).
VH1 Expression Is Associated with PV/PC Sites
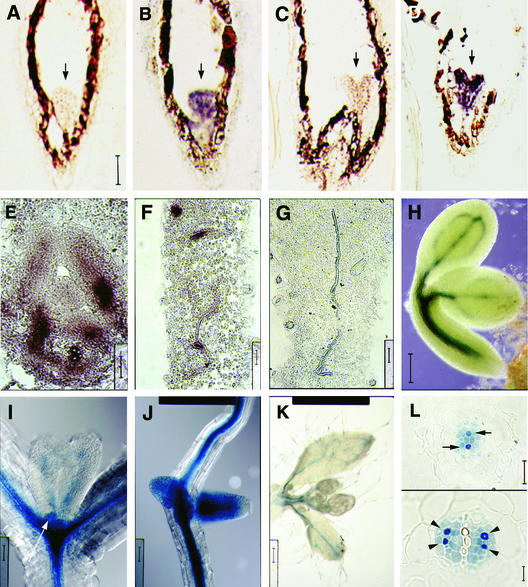
To determine the function of VH1 in Arabidopsis, we first determined the developmental profile of its expression pattern by in situ RNA hybridization and by staining transgenic plants containing 2.6 kb of VH1's 5′ sequences driving the β-glucuronidase (GUS) reporter gene (VH1::GUS). This construct proved to include all regions needed to produce the VH1 expression pattern. VH1 mRNA is detected throughout globe- to heart-staged embryos (Figures 3B and 3D, respectively). This broad expression domain becomes restricted to PC cells by the late torpedo stage, and this pattern persists throughout the duration of embryo development (Figure 3H). After germination, VH1 is expressed not only in PC cells throughout the plant (Figures 3F, 3I, and 3K) but also in all lateral organ primordia before the onset of vascularization (Figures 3E, 3I, and 3J). Thus, VH1 expression is associated with PV/PC sites throughout plant development.
Figure 3.
VH1 Expression Is Associated with PV/PC Sites.
(A) to (G) Digoxigenin-labeled VH1-specific RNA probes were hybridized to serial sections of tissue from wild-type plants. Transverse sections of young leaves ([F] and [G]) as well as longitudinal sections of globe-stage ([A] and [B]) and heart-stage ([C] and [D]) embryos (arrows) and leaf primordia (E) were hybridized to antisense ([B] and [D] to [F]) and sense ([A], [C], and [G]) probes. (A) to (D) are at the same magnification; bar in (A) = 50 μm. Bars in (F) to (G) = 25 μm.
(H) to (L) Transgenic plants containing the VH1::GUS reporter construct were stained for GUS activity, and whole-mounts ([H] to [K]) and serial transverse sections through a young root (K) were viewed using Nomarski optics. Whole-mount tissues include bent-cotyledon-stage embryos (H), leaf primordia (arrow) and very young leaves (I), lateral root primordia (J), and young cauline leaves that subtend floral buds (K). Bars = 50 μm for (H) and 40 μm for (I) to (K). In (L), arrows indicate PC cells that will differentiate as xylem, and arrowheads indicate PC cells that will differentiate as phloem. Bars = 8 μm.
To determine the developmental fate of VH1-expressing PV/PC cells and any corresponding change in VH1 expression levels, we examined transverse sections of stained roots of VH1::GUS reporter transgenic plants, starting from the apical meristem. In the root, the vascular column develops continuously as a single axial unit behind the growing apical meristem. The resulting correlation between cell position along the main axis of the root and the degree of cell type differentiation makes it possible to describe the developmental history and fate of each of the vascular cells based on its original location (van den Berg et al., 1998). Mature Arabidopsis roots have a simple diarch vascular system consisting of two phloem poles and two xylem poles (Dolan et al., 1993). We observed that all of the PV cells in the vascular column immediately behind the apical meristem express VH1, but cells that will differentiate as xylem express VH1 more strongly (Figure 3L, top). Farther away from the apical meristem, cells that will differentiate as phloem also express VH1 more strongly, whereas the fully differentiated xylem cells have undergone cell death and thus no longer express VH1 (Figure 3L, bottom). Together, these expression data indicate that both the domain and the level of VH1 expression change during the early stages of vascular development.
Ectopic Expression of VH1 Results in Reduced Venation and Premature Cell Differentiation of Nonvein Cell Types
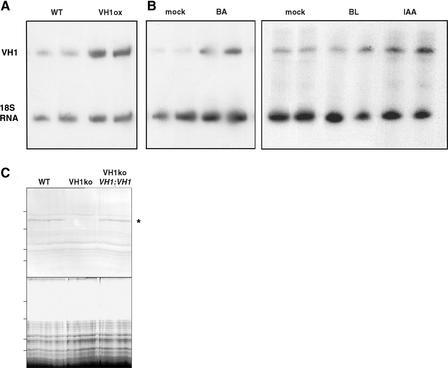
We generated transgenic plants that contained the constitutive 35S promoter of Cauliflower mosaic virus (CaMV) driving the VH1 gene (35S CaMV::VH1). Multiple transgenic lines were selected, and two independent transgenic lines in which the transgene inserted at a single locus were characterized further. Relative quantitative reverse transcriptase–mediated PCR results confirm that VH1 expression levels in these lines were increased more than twofold (Figure 4A).
Figure 4.
VH1 Expression Data from Transgenic Lines and from Exogenous Application of Hormones.
(A) and (B) Both VH1 (425-bp fragment) and 18S RNA (315 bp) were amplified by PCR from cDNA generated from total RNA for each sample and loaded on a denaturing acrylamide gel in duplicate. The VH1 band was quantified relative to the 18S RNA.
(A) One-week-old wild-type (WT) and 35S CaMV::VH1 (VH1ox) transgenic seedlings.
(B) Etiolated seedlings were incubated for 1 h in a salt solution alone (mock) or with the addition of benzyladenine (BA), 24-epibrassinolide (BL), or indoleacetic acid (IAA).
(C) The gel at top shows a protein blot of whole-cell extracts from 5-day-old seedlings probed with VH1 antiserum. Both the wild type (WT) and the vh1 insertion line containing a wild-type VH1 transgene driven by its own promoter (VH1ko + VH1::VH1) produced an ∼150-kD band, whereas the vh1 insertion line alone (VH1ko) did not. The gel at bottom shows the corresponding Coomassie blue–stained 7.5% SDS-PAGE gel demonstrating that the samples were loaded equally. The asterisk indicates the VH1 protein band. Lines at left indicate molecular mass markers (184, 121, 86, and 69 kD from top to bottom).
Consistent with VH1's normal expression pattern, the 35S CaMV::VH1 transgenic lines exhibited a defect in the cotyledons and juvenile leaves. However, with successive leaves, the phenotype gradually disappeared, suggesting that an activating ligand is present only during the earlier stages of development, that VH1 expression is irrelevant in other stages of development, or that the transgene is gradually silenced. However, we observed no or very subtle phenotype in the roots and hypocotyls. Normally, VH1 is highly expressed there, so additional increases in expression may be negligible. The juvenile leaves of the transgenic lines exhibit dramatic alterations in shape, color, and pattern of leaf cell types (Figures 5B to 5D). They also differ in severity of the mutant phenotype; the most severe case involves the formation of pin-like leaves (Figure 5D).
Figure 5.
Ectopic VH1 Expression Results in Premature Leaf Cell Differentiation.
Juvenile leaves from wild-type plants were used for comparison ([A], [E], [I], and [L]).
(A) to (H) Fully expanded juvenile leaves were viewed with a dissecting microscope ([A] to [D]) and then were cleared and viewed using dark-field optics ([E] to [H]) to visualize the venation pattern and the mesophyll cell layers.
(I) to (N) Transverse sections through leaf primordia ([I] and [J]) and expanding (K) and fully expanded ([L] to [N]) juvenile leaves were stained with toluidine blue and viewed using bright-field optics. The arrowhead in (J) points to primordial ground cells (normally identified by their dense cytoplasmic staining) that have prematurely ceased cell division and have differentiated, resulting in leaves with fewer mesophyll cells that are unusually sparse in interveinal regions (K). (L) to (N) are oriented so that the adaxial surface of the leaf section is at top. The adaxial surface is above the xylem (light blue) portion of the midvein. The arrow in (M) points to a region composed of a single epidermal layer. Bars = 50 μm for (I) and (K) to (M) and 40 μm for (J) and (N).
(O) and (P) Plants containing both the 35S CaMV::VH1 and the VH1::GUS constructs (P) expressed GUS constitutively, expanding the GUS expression domain seen in plants containing only the VH1::GUS reporter construct (O). Bars = 400 μm.
The range in leaf pigmentation from mottled green to white suggests that the photosynthetic cell types are affected. Indeed, examination of cleared leaves revealed that the palisade layer, which normally is composed of tightly packed chloroplast-bearing mesophyll cells, is missing or depleted in mainly interveinal regions of the leaf (Figures 5F and 5G), particularly in the white, pin-like leaves, in which few or no mesophyll cells were present (Figure 5H). However, we observed no histological evidence of cell death among mesophyll cells. Instead, transverse sections of leaf primordia show that the cells appear to cease cell division and to differentiate prematurely even while the organ continues to expand (Figure 5J), accounting for the fewer numbers of mesophyll cells and their pulled-apart appearance in interveinal regions (Figure 5K). Moreover, in otherwise fully expanded leaves, small fields of cells or individual cells appear to remain meristematic or proliferative (as indicated by their small size) and very photosynthetically active (as indicated by the high starch production in their chloroplasts; data not shown).
The patterning of the entire leaf is affected. Transverse sections of leaves show that they vary in thickness and in number of cell layers. In the most severe cases, the leaves are thinned in some regions to a single epidermal layer (Figure 5M). In some leaves, the abaxial/adaxial arrangement of the mesophyll cells is lost or inverted. For example, the palisade layer of the transgenic leaf shown in Figure 5N appears to be along the abaxial side (bottom surface) of the leaf rather than along the adaxial side (top surface), inconsistent with the abaxial/adaxial arrangement of xylem and phloem in the veins. Both epidermal and mesophyll cells are irregular in size, and some become unusually large. However, the veins in these leaves are aligned correctly, have the normal arrangement of xylem and phloem, and function in exporting photosynthates, as demonstrated by an in situ starch-staining assay (data not shown).
VH1 appears to positively regulate its own transcription. Plants containing both the 35S CaMV::VH1 and the VH1:: GUS reporter constructs express GUS not only in PC cells but throughout the cotyledons and juvenile leaves (Figure 5P). This finding suggests (1) that the VH1 signaling pathway is activated in those cells, (2) that this activity directs additional VH1 expression, and (3) that the ligand is broadly expressed or available. Alternatively, those cells that normally do not express VH1 have taken on a PV/PC character that permits VH1 expression.
Loss of VH1 Results in Premature Leaf Senescence and Defective Vascular Transport
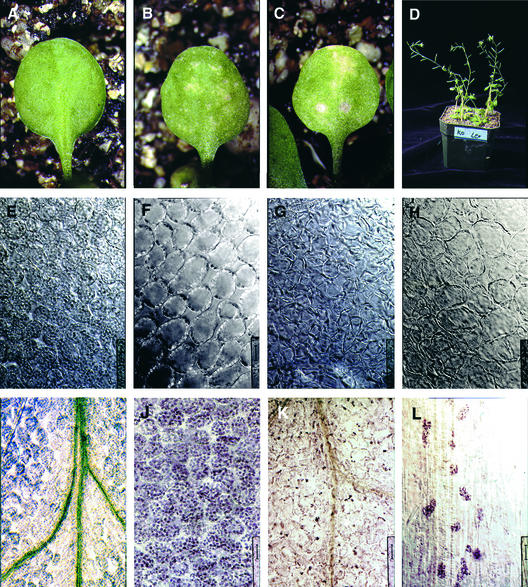
To obtain a loss-of-function allele of VH1, we screened a collection of Ds transposon insertion lines for plants containing an insertion in the coding sequence of VH1. We found a line with a single insertion of the complete 6.4-kb Ds gene trap module (see Methods) in the 15th LRR repeat of VH1. The insertion line was confirmed to be a VH1 null protein by immunoblot analysis (Figure 4C). Transgenic plants homozygous for this insertion allele are wild type in general appearance (Figure 6D). However, their leaves senesce and undergo cell death earlier than wild-type leaves (Figures 6B and 6C). Moreover, leaf senescence is uniform, whereas wild-type leaves do not show a well-defined starting point of senescence and instead undergo differential senescence, usually from the leaf margins inward.
Figure 6.
Loss of VH1 Results in Premature Leaf Senescence and Possible Defect in Vascular Transport.
Juvenile leaves from wild-type (A) and vh1 mutant ([B] and [C]) 3-week-old plants were viewed with a dissecting microscope to visualize senescence. In (D), vh1 mutant plants are at left and wild-type plants are at right. Expanding ([E] and [G]) and fully expanded ([F] and [H]) leaves of wild-type ([E] and [F]) and vh1 mutant ([G] and [H]) plants were cleared and viewed with Nomarski optics to visualize the chloroplast-bearing mesophyll cells. Bars = 25 μm for (A) to (H). Cleared leaves of wild-type ([I] and [J]) and vh1 mutant ([K] and [L]) plants were stained with I/KI for starch accumulation. Bars = 50 μm for (I) and (K) and 25 μm for (J) and (L).
The premature leaf senescence phenotype segregates as a single recessive trait and is complemented in two independent lines by the introduction of 2.6-kb VH1 promoter sequences driving the VH1 transgene (data not shown). We generated a vh1 excision allele by crossing mutant plants with a line containing an Ac transgene to mobilize the Ds insertion. This excision line retains an 8-bp footprint (TCA-ACAAG) 7 bp upstream of the site of the original insertion, which causes a shift in the reading frame. Plants homozygous for the excision allele also exhibited the premature leaf senescence phenotype.
The first microscopically visible changes in leaf senescence are in the chloroplasts (Thimann, 1980; Gepstein, 1988). We examined chloroplasts in expanding and fully expanded juvenile leaves of plants homozygous for the vh1 insertion allele. The chloroplasts were fewer in number per mesophyll cell, flattened, and lost many of their granal stacks of thylakoids (part of the photosynthetic machinery) (Figures 6G and 6H) as well as many of their characteristic transitory starch deposits in the stroma between thylakoids (Figure 6K). These traits can be detected as early as in the cotyledons of 1-week-old seedlings and are consistent with a decrease in photosynthetic capability, which is associated with early leaf senescence (Badenhuizen, 1969; Thimann, 1980). There were regional differences in the integrity of the chloroplasts; mesophyll cells along the main veins and leaf margins contained some wild-type-looking and starch-accumulating chloroplasts (Figure 6L), suggesting that those cells received some transported factor that delayed their senescence.
Leaf senescence can be induced artificially by a block in vascular transport (Thimann, 1980), in which case the induced leaf senescence is distributed uniformly. We tested whether the veins in the loss-of-function mutant were functioning properly in transporting photosynthates out of the leaf to the rest of the plant by staining the leaves for starch. We observed in the vh1 mutant unusual starch precipitation in the chloroplasts of the mesophyll cells along large veins (Figure 6K). These chloroplasts had most likely converted into starch-storing organelles, the amyloplasts. Furthermore, there were no starch drainage zones around veins, as in the wild type.
This apparent defect in the vascular transport of photosynthates and/or the transport of some antisenescence signal may prematurely induce leaf senescence, including the observed chloroplastic defects. This apparent transport defect likely is phloem specific, because the xylem appears to function normally in transporting water to the leaf, as indicated by the turgidity of the leaves. However, we cannot exclude the possibility that the transport defect may be PC cell specific, because the observed chloroplastic defects occur early in leaf development, when photosynthetic activity is normally high and venation pattern is still formative. The veins in these leaves are anatomically normal, with aligned cell files and a wild-type arrangement of xylem and phloem (data not shown), but apparently they are functionally abnormal or have an abnormal association with mesophyll cells.
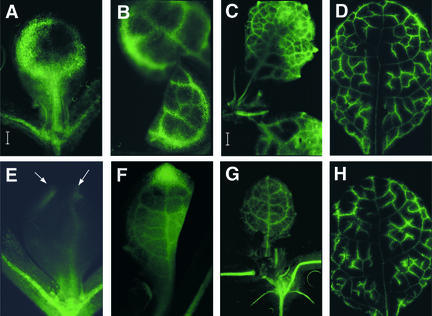
To assay phloem transport, we used a phloem-mobile fluorescent probe, carboxyfluorescein (CF), to image the phloem transport of CF along with the flow of photoassimilates from expanded source leaves to developing sink leaves. The pattern of dye transport in wild-type Arabidopsis leaves mimicked that reported for tobacco leaves (Roberts et al., 1997). In developing leaves that have formed one or two pairs of secondary veins (Figures 7A and 7B, respectively), the CF dye was seen first in the midvein and in the secondary veins that branch from the midvein and interconnect to form loops, but not in the minor veins. By the time the leaf has formed the third pair of secondary veins, the CF dye was observed to enter the minor veins and unload into the mesophyll (Figure 7C). However, in developing vh1 mutant sink leaves that have formed one or two pairs of secondary veins, only the major veins at the tip of the leaf were fluorescent (Figures 7E and 7F), suggesting that phloem transport occurred at much reduced levels. Furthermore, this pattern of dye transport persisted during early leaf expansion, such that the vasculature of the sink leaves fluoresced less (were darker) than the other vasculature of the plant (Figure 7G). Interestingly, in fully expanded leaves of both the wild type and the vh1 mutant, CF unloading was reduced and the dye was restricted to the minor veins (Figures 7D and 7H), suggesting that by the time the leaves undergo the sink-to-source transition, phloem transport in the vh1 mutant occurred at nearly wild-type levels.
Figure 7.
Loss of VH1 Results in Defective Vascular Transport of a Phloem-Mobile Dye Early in Leaf Development.
The pattern of phloem transport of CF was imaged in the sink leaves of wild-type ([A] to [D]) and vh1 mutant ([E] to [H]) plants at successive stages of leaf development. Arrows in (E) indicate top portions of major veins that are fluorescing. (A), (B), (E), and (F) are at the same magnification; bar in (A) = 40 μm. (C), (D), (G), and (H) are at the same magnification; bar in (C) = 400 μm.
It would appear that a knockout of VH1 function affects phloem transport early in leaf development, possibly through a slowing of the phloem differentiation process, and that this transport defect occurs before the observed chloroplastic defects, suggesting a causal relationship. Furthermore, the site of action of VH1 corresponds to its gene expression pattern. The subtle defect in the vh1 mutant and the presence of a large number of LRR receptor kinases encoded in the genome of Arabidopsis make it likely that one or more genes may be partially functionally redundant with VH1. The knockout of these genes may permit a more complete account of VH1's role in development.
Hormone Induction Studies
Both auxin and cytokinin have been implicated in leaf senescence and vascular differentiation. Also, VH1 is most similar in sequence among the characterized receptor kinases to BRI1, which is a putative brassinosteroid receptor (Li and Chory, 1997). We tested whether VH1 transcription was induced upon hormone application and found that the auxin indoleacetic acid and the cytokinin benzyladenine induced VH1 transcription 2.5- and 3-fold, respectively, within 1 h of hormone application, whereas 24-epibrassinolide, which is a substrate for brassinosteroid biosynthesis, showed no induction relative to that of the mock control (Figure 4B).
DISCUSSION
Plant development requires the integration of various signaling pathways that recognize and respond to spatial and temporal information. The formation of the leaf venation pattern and its organizing role in leaf morphogenesis are likely to involve signaling pathways that recognize and respond to positional information. In this regard, we identified a gene, VH1, that is PV/PC cell specific and whose expression pattern reflects the progressive formation of the venation pattern in developing juvenile leaves. Furthermore, the protein has all of the canonical features for a potential role in intercellular signaling: an extracellular putative ligand binding domain, a transmembrane domain, and an intracellular kinase domain that we demonstrated to be active.
Although VH1's expression is PV/PC cell specific, its expression domain and level change according to the developmental state of the expressing cells. At present, it is not possible to determine whether a meristematic cell in the vascular system is PC (i.e., that it has actually progressed toward differentiation into xylem or phloem) or whether it is still PV (i.e., an uncommitted but vascular-competent cell) (Nelson and Dengler, 1997). One possible explanation of VH1's expression pattern is that the domain and level of expression may reflect two possible developmental states: PV and PC. A lower but broader expression of VH1 may mark PV cells, whereas a stronger but more restricted expression of VH1 may mark PC cells.
With regard to expression level, VH1 appears to positively regulate its own transcription via the activation of its own signaling pathway. This suggests a mechanism underlying the transition from PV to PC. For example, PV cells that receive the ligand and thus have their VH1 signaling pathways activated express more VH1, thereby reinforcing their reception of the ligand, and the increase in VH1 expression induces the subsequent differentiation of those PV cells into PC cells. This mechanism also could explain the restriction of the expression domain of VH1 from most of the leaf primordium to just PC cells. Indeed, this mechanism does not differentiate the ligand that VH1 perceives from the positional information that may restrict or localize VH1 to PV/PC cells. Rather, the ligand carries positional information. Regardless of the mechanism, it is very likely that the positional information that restricts VH1 expression is transmitted through the files of PC cells in nascent veins.
If VH1 acts as a receptor, what signal molecule activates the VH1 signaling pathway, and where is its source? Whatever the ligand is, it appears to be broadly expressed or available, and given the specificity of VH1 expression, only PV/PC cells can respond to it. Furthermore, the VH1 protein contains a large LRR region, which is believed to be a specific ligand binding site for small peptide(s) or glycoprotein(s) (Kobe and Deisenhofer, 1994) or pathogenic elicitor(s) (Baker et al., 1997). Based on the studies of other plant LRR receptor kinases, the ligand for the VH1 receptor is likely to be a small peptide or a small nonpeptide hormone. Aside from pathogenesis elicitors, the only other peptide ligand known to date is the small, secreted peptide CLV3, which is thought to bind to CLV1 in cooperation with another protein, CLV2 (Clark et al., 1997; Jeong et al., 1999; Trotochaud et al., 2000). CLV3 is a member of a family of genes that encode peptides that share a conserved 14–amino acid motif but otherwise are unrelated at the sequence level (Cock and McCormick, 2001). However, the LRR repeats of VH1 are most closely related in sequence to those in BRI1, a brassinosteroid-responsive receptor kinase, and share with it the presence of an amino acid island that is thought to contribute to ligand specificity (Li and Chory, 1997), although this sequence differs between the two. It is not known whether BRI1 interacts with brassinosteroids directly, but a chimeric receptor approach showed that the extracellular domain of BRI1 could confer brassinosteroid responsiveness to the intracellular kinase domain of a heterologous LRR kinase (He et al., 2000). Furthermore, the number of LRR receptor kinases encoded in the genome of Arabidopsis is large (∼170), and they appear to correspond to a diversity of ligands (McCarty and Chory, 2000). This leaves the nature of VH1's ligand unresolved. Because VH1 is PV/PC cell specific and most likely is involved in vascular differentiation and leaf venation patterning, the VH1 protein might be involved in the perception of a signal associated with the action of either of the two growth-promoting substances, auxin and cytokinin, both of which have been demonstrated to induce VH1 transcription. It also is conceivable that VH1 acts indirectly in cells in which it is activated to confer developmental competence for the hormone response.
Auxin plays a well-documented role in vascular differentiation, which is influenced by auxin production in young leaves and its vascular transport through PC cells to the roots. Polar auxin transport is thought to be essential for the correct spatial pattern and cellular alignment of this differentiation (Nelson and Dengler, 1997; Berleth et al., 2000). According to the “canalization of signal flow” hypothesis (Sachs, 1981, 1991), the formation of a path of auxin flow induces the axial elongation of transporting cells, thereby reinforcing the path, and auxin induces the subsequent differentiation of those transporting cells into vascular elements. Studies of the effects of polar auxin transport inhibitors on the leaf venation pattern suggest that the venation pattern itself may reflect a previous pattern of auxin drainage from sites of synthesis at the margins to the base of the leaf primordium (Mattsson et al., 1999). The restriction of the initially broad expression domain of VH1 throughout the leaf primordium to just PC cells occurs in a pattern predicted by the canalization of signal flow hypothesis. This expression pattern also is shared by the Arabidopsis gene MONOPTEROS, an auxin response transcription factor involved in the axialization of PC cells and possibly in polar auxin transport (Berleth and Jurgens, 1993; Przemeck et al., 1996). Furthermore, the increase in VH1 expression levels from PV cells to PC cells is consistent with the idea of positive feedback making those cells better transporters of auxin through cell differentiation processes. It is conceivable that VH1 interacts with auxin itself, with an auxin binding protein, or with a secondary signal generated by auxin perception, and the signal is transduced through the kinase into effects on PV/PC cell division and/or differentiation.
Alternatively, the signal may be associated with the hormone cytokinin, which moves from the roots to the leaves through the phloem. Not only is cytokinin important for vascular differentiation (Nelson and Dengler, 1997), it also is attributed a regulatory role in leaf senescence because of its production in the roots, its transport to leaves, and its senescence-delaying effects there (van Staden et al., 1988). Furthermore, cytokinin may act in conjunction with auxin to prevent leaf senescence. In detached tobacco leaves, both cytokinin and auxin inhibit yellowing; however, each causes a different and incomplete pattern of chlorophyll retention. Cytokinin treatment retained the green pigmentation in the interveinal regions, whereas auxin treatment kept the area along the main veins green (Engelbrecht and Conrad, 1961; Nooden, 1988). Together, they produced a uniform retention of chlorophyll. The premature leaf senescence in the vh1 loss-of-function alleles, particularly in the interveinal regions, may be attributed to a defect in the phloem transport and possible perception of cytokinin. With regard to VH1 function, the effects of transgenic alteration of its expression suggest that VH1 acts in the juvenile leaf as a signal transduction component in the communication of cell division and/or cell differentiation signals.
The ectopic expression of VH1 causes a number of developmental abnormalities, which can be grouped into three types of defects: (1) a shortened cell proliferation phase, particularly for mesophyll cells; (2) abnormal expansion of some leaf cell types; and (3) a reduction and alteration of the venation pattern. Because much of the venation pattern is determined during the intercalary growth of the leaf blade, venation patterning and cell proliferation in the leaf must be coordinated. For instance, PC cells at the tips of growing veins are recruited from the PV or ground cells of the leaf, and cell proliferation is active in this region. It is predictable that defects in leaf cell proliferation are accompanied by defects in venation patterning and vice versa. Also, the abnormally sized leaf cell types in the 35S CaMV::VH1 transgenic lines have been observed previously in plants under a variety of circumstances that require compensation for defects in cell proliferation (Clark and Schiefelbein, 1997). Moreover, the defects can be summarized as resulting from activity that either inhibits cell proliferation or promotes cell differentiation. The inclusion of most leaf cell types in the phenotype suggests that a family of LRR receptor kinases phosphorylate common signal transduction intermediates to control leaf cell proliferation and cell differentiation, and the specific expression pattern of VH1 may serve to limit its activity to PV/PC cells. Highly similar and potentially redundant LRR receptor kinases may act in other leaf cell types, and ectopic VH1 expression may activate their pathways.
Plants containing either the insertion or excision null allele of vh1 exhibit premature and uniform leaf senescence, the beginnings of which can be observed in young expanding leaves. The loss of starch production in the stroma of chloroplasts and starch drainage zones around veins and the presence instead of large reserve starch deposits and starch-storage plastids suggest that the premature leaf senescence is caused by a compromised vascular transport system or an abnormal association between the vascular and mesophyll cells. The reduced phloem transport of CF in early leaf development suggests that phloem differentiation may be affected.
Two processes are candidates for the downstream target of VH1: the regulation of cell division, and the regulation of cell differentiation. VH1 could function to repress cell division in PV/PC cells by regulating their meristematic state, or VH1 could function to promote the differentiation of PV to PC cells and/or of PC cells to xylem and phloem in response to a ligand that serves as a temporal or spatial cue. Three lines of evidence suggest that VH1 is involved in the promotion of differentiation. First, the defect in the vascular transport of starch, CF, and possibly some antisenescence factor in the vh1 null alleles may reflect a defect in the vascular differentiation process, particularly that of phloem. Most likely, the rate of vascular differentiation was affected, because the mature veins looked anatomically normal and the phloem transport of CF eventually was restored. Second, as suggested above, the progressive restriction of VH1 expression, the increase in VH1 expression in PC cells, and VH1's positive transcriptional regulation all implicate VH1 as a marker of the transition from PV to PC that occurs during vascular cell development. Third, the effects of the ectopic expression of VH1 (shortened cell proliferation phase and abnormal expansion) also suggest that VH1 promotes cell differentiation generally and that its specific expression pattern restricts its function to that of PV/PC cells.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotypes Columbia (Col-O) and Landsberg erecta (Ler) were used for comparison with mutant plants. Seeds were grown under constant white light (∼300 μE·m−2·s−1) either on 0.75% agar medium consisting of Murashige and Skoog (1962) basal salts (Sigma), Haughn and Somerville (1986) nutrient solution, 0.5 g/L Mes, and 10 g/L Suc or on soil (3:1 mix of Metro-Mix 200 to vermiculite; Scotts, Marysville, OH).
Isolation of the VH1 Gene
A collection of enhancer trap lines was provided by J.E. Malamy and P.N. Benfey (New York University). Construction of these lines has been described by Malamy and Benfey (1997). To recover the genomic region flanking the enhancer trap insertion, genomic DNA was isolated from frozen plant material as described by Dellaporta et al. (1983), digested with BglII or PvuI, and used in genomic DNA gel blot analysis and inverse PCR as described by Ponce et al. (1998).
Histological Analyses
One- to 3-week-old seedlings were stained for β-glucuronidase (GUS) activity overnight at 37°C in 1 × GUS buffer (100 mM Tris [pH 7], 1 mM potassium ferricyanide, and 50 mM NaCl), 20% methanol, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronidase as described by Malamy and Benfey (1997). For observation of whole mounts, both stained and unstained tissues were fixed in ethanol:acetic acid (3:1) overnight, dehydrated in an ethanol series, dehydrated in 100% ethanol overnight, cleared in Hemo-De (Fisher Scientific), and mounted in Permount:xylene (2:1; Fisher Scientific). Specimens were visualized on a Zeiss Axiophot microscope (Oberkochen, Germany). For histological analysis, fixed dehydrated specimens were embedded in Spurr's medium (Electron Microscopy Sciences, Fort Washington, PA) or in Paraplast Plus (Fisher Scientific). Plastic sections (2 μm) were cut on a Sorvall MT2-B ultramicrotome (Newtown, CT) and stained briefly with 1% toluidine blue. Paraffin sections (8 μm) were cut on a model 820 microtome (Arthur H. Thomas Co., Philadelphia, PA).
In Situ Hybridizations
The region of the VH1 gene corresponding to the amino acid island region (nucleotides 1615 to 1831) was used to generate digoxigenin-labeled antisense and sense mRNA probes. In situ hybridizations and labeling reactions were performed as described by Jackson (1991) with some modifications. Before hybridization, the sections were soaked in Hemo-De (Fisher Scientific) to remove the paraffin, hydrated with an ethanol/water series, incubated with 15.6 μg/mL proteinase K for 30 min at 37°C, acetylated with acetic anhydride, dehydrated with an ethanol series, and allowed to air dry. Then, they were hybridized with digoxigenin-labeled probes overnight at 42°C. After hybridization, the sections were rinsed in 0.2 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), incubated with 20 μg/mL ribonuclease A for 20 min at 37°C, and washed for 1 h in 0.2 × SSC. Sections were blocked in 1% blocking agent (Boehringer Mannheim), and the signal was detected with an anti-digoxigenin alkaline phosphatase reaction according to the manufacturer's protocol.
Reporter, Overexpression, and Complementation Vector Construction
The VH1 reporter construct (VH1::GUS) was generated by adding restriction enzyme sites (HindIII and BamHI) to the 5′ and 3′ ends, respectively, of VH1's promoter sequence (2.6 kb) via PCR and cloning the digested PCR product into the similarly digested pBI101 plant transformation vector (Clontech, Palo Alto, CA) upstream of the GUS reporter gene.
The VH1 overexpression construct (35S CaMV::VH1) was generated by adding restriction enzyme sites (XbaI and SstI) to the 5′ and 3′ ends, respectively, of the VH1 structural gene (3.4 kb) via PCR from Col-O genomic DNA and cloning the digested PCR product into the similarly digested pBI121 plant transformation vector (Clontech) downstream of the constitutive 35S promoter of Cauliflower mosaic virus. Both the reporter and overexpression constructs were used to transform wild-type Arabidopsis plants (Col-O) by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998).
The VH1 complementation construct was generated by PCR amplification from Ler genomic DNA of the VH1 promoter sequence flanked by HindIII and XbaI sites and the VH1 structural gene flanked by XbaI and SstI sites and digesting and cloning them into the HindIII and SstI sites of the pBI101 plant transformation vector. This construct was used to transform vh1 plants (Ler background) as described above.
Total RNA Isolation and Relative Quantitative Reverse Transcriptase–Mediated PCR Analyses
Total RNA was isolated from 1-week-old etiolated seedlings with Trizol reagent (Gibco BRL) according to the manufacturer's protocol. One microgram of total RNA was reverse transcribed with random decamers (Ambion, Austin, TX), and the cDNA product served as template in multiplex relative quantitative reverse transcriptase–mediated PCR with QuantumRNA 18S internal standards (Ambion), 2 μCi/μL α-32P-dCTP, and primers to the 3′ end of the VH1 gene (415 bp). PCR products were resolved on an 8% denaturing acrylamide gel and quantified with a Fuji BAS-2000 phosphorimaging system (Tokyo, Japan).
Recombinant VH1 Protein Construction, Site-Directed Mutagenesis, and Kinase Activity Assays
The VH1 kinase domain (positions 2357 to 3352) and the last 318 bp of the kinase domain were amplified by PCR with BamHI sites at both ends, and the digested products were cloned into the similarly digested pMALcR1 expression vector (New England Biolabs, Beverly, MA) downstream of the maltose binding protein (MBP) gene. Similarly, the last 318 bp of the kinase domain was cloned into the pGEX expression vector (Pharmacia) downstream of the glutathione S-transferase (GST) gene.
Site-directed mutagenesis of Lys-866, which is invariant and necessary for phosphotransfer in all protein kinases (Hanks et al., 1988; Hanks and Quinn, 1991; Goring and Rothstein, 1992), to Asp-866 was performed as follows. Regions corresponding to the 5′ half (positions 2357 to 2533) and the 3′ half (positions 2502 to 3352) of the VH1 kinase domain were amplified by PCR with primers that substituted a G with a C at position 2518. The two PCR products overlapped for 32 nucleotides and were introduced as templates in a second PCR procedure. The final PCR product is an inactive mutant version of the catalytic domain of VH1 and was cloned into the pMALcR1 vector as described above.
MBP fusion proteins (MBP::VH1, MBP::VH1-TAIL, and MBP:: K866N) and the GST::VH1-TAIL protein were expressed in Escherichia coli (BL21) and purified with amylose-agarose (New England Biolabs) and glutathione-agarose (Pharmacia) affinity columns, respectively. MBP::VH1 and MBP::K866N were assayed for protein kinase activity as described by Horn and Walker (1994).
Isolation of vh1 Insertion and Excision Alleles
The vh1 insertion allele was a gift of A. Groover and R. Martienssen (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) and was generated by a random insertion of a Ds gene trap transposon into the VH1 gene. This gene trap strategy and construct have been described by Sundaresan et al. (1995). The exact insertion site was determined by amplifying the flanking sequences by PCR and sequencing the product. The number of Ds insertions in this line was determined by genomic DNA gel blot analysis with a Ds probe.
vh1 mutant plants (Ler) were crossed to plants containing the Ac transgene (Ler) to mobilize the Ds transposon. Genomic DNA from individual F2 plants of each F1 line was analyzed for excision of Ds from the VH1 gene. In one such F1 line, the Ds was excised from the VH1 gene and left an 8-bp DNA footprint. This DNA polymorphism was used to genotype F2 individuals by PCR amplification and electrophoresis and was sequenced from F2 individuals homozygous for the excision allele.
Protein Extraction, Antibodies, and Protein Gel Blot Analysis
Protein extraction was performed as described by He et al. (1996) with some modifications. Approximately 200 5-day-old seedlings were ground in liquid N2, thawed in 1 mL of 1 × protein sample buffer (50 mM Tris-Cl, pH 6.8, 50 mM DTT, 4% SDS, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride), and boiled for 5 min. The low-speed supernatants of the denatured samples were resolved by 7.5% SDS-PAGE and electroblotted to polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked for 2 h with PBS containing 5% nonfat milk.
Affinity-purified MBP::VH1-TAIL was used to immunize rabbits. The VH1 antiserum was affinity purified on NHS-activated Hi-Trap columns (Pharmacia), cross-linked with GST::VH1-TAIL fusion protein, and used in dilution (1:5000) for protein gel blot analysis. Protein bands that cross-reacted with the VH1 antibody (diluted 1:5000) were identified by reaction with alkaline phosphatase–conjugated anti-rabbit IgG (Sigma).
In Situ Starch Staining
One-week-old seedlings and fully expanded young leaves were fixed in FAA (10% formalin, 5% acetic acid, 50% ethanol) overnight at 4°C, cleared in 50% ethanol, and stained with I/KI solution for 1 to 5 min as described by Fukaki et al. (1998). They were then rinsed in 50% ethanol for 5 s and mounted in chloral hydrate:glycerol:water solution (8:1:2, w/v/v) for examination by light microscopy.
Phloem Transport and Imaging of Carboxyfluorescein
The imaging of phloem transport in sink leaves was performed as described by Roberts et al. (1997) with some modifications. The adaxial surface of a source leaf was abraded gently with a toothbrush, and 10 μL of 6(5)-carboxyfluorescein diacetate (60 μg/mL; Sigma) was applied to the abraded leaf surface. To image phloem transport in leaves undergoing sink-to-source transition, 10 μL of 6(5)-carboxyfluorescein diacetate (60 μg/mL) was applied to the surface just below the hypocotyl-root junction. After translocation in the light in a humid chamber for 1 to 3 h, the leaves were detached from the plant and mounted on slides in 50% glycerol. Emitted fluorescence was photographed with Kodak Ektachrome 400 film using a blue filter.
Hormone Induction Analyses
One-week-old etiolated seedlings were incubated for 1 h in solution consisting of half-strength Murashige and Skoog (1962) basal salts with one of the following additions: 20 μM indoleacetic acid, 20 μM benzyladenine, or 1 μM 24-epibrassinolide (CIDtech Research, Mississauga, Ontario, Canada). Total RNA isolation and relative quantitative reverse transcriptase–mediated PCR analyses were performed as described above.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Number
The accession number for the VH1 gene reported in this article is AAD20088.
Acknowledgments
We thank Jocelyn Malamy and Philip Benfey (New York University) for giving us access to their enhancer trap collection, Andrew Groover and Robert Martienssen (Cold Spring Harbor Laboratory) for providing us with the Ds gene trap line, and Robert Turgeon (Cornell University) for suggesting the iodine stain assay. Thanks to Joanne Chory (Salk Institute) and Jianming Li (University of Michigan) for sharing information before publication. We also thank Andrew Cary for affinity purifying the VH1 antiserum. This work was supported by grants to T.N. from the National Science Foundation (IDN 9808295) and the U.S. Department of Agriculture (2001 35304 09992).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005884.
References
- Badenhuizen, N.P. (1969). The Biogenesis of Starch Granules in Higher Plants. (New York: Appleton-Century-Crofts).
- Baima, S., Nobili, F., Sessa, G., Luchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Jurgens, G. (1993). The role of the monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development 118, 575–587. [Google Scholar]
- Berleth, T., Mattsson, J., and Hardtke, C.S. (2000). Vascular continuity and auxin signals. Trends Plant Sci. 5, 387–393. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., and Schiefelbein, J.W. (1997). Expanding insights into the role of cell proliferation in plant development. Trends Cell Biol. 7, 454–458. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cock, J.M., and McCormick, S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126, 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon, J., Varlet, I., and Robertson, E. (1996). Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381, 155–158. [DOI] [PubMed] [Google Scholar]
- de Celis, J.F., Gray, S., and Garcia-Bellido, A. (1994). Roles of the Notch gene in Drosophila wing morphogenesis. Mech. Dev. 46, 109–122. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Diaz-Benjumea, J., Gonzalez-Gaitan, M.A.P., and Garcia-Bellido, A. (1989). Developmental genetics of the wing vein pattern of Drosophila. Genome 31, 612–619. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organization of the Arabidopsis root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Engelbrecht, L., and Conrad, K. (1961). Vergleichende Untersuchungen zur Wirkung von Kinetin and Auxin. Ber. Dtsch. Bot. Ges. 74, 42–46. [Google Scholar]
- Fukaki, H., Fujisawa, H., and Tasaka, M. (1998). The endodermis is essential for shoot gravitropism. Plant J. 14, 425–430. [DOI] [PubMed] [Google Scholar]
- Gepstein, S. (1988). Photosynthesis. In Senescence and Aging in Plants, L. Nooden and A.C. Leopold, eds (San Diego, CA: Academic Press), pp. 85–109.
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Goring, D.R., and Rothstein, S.J. (1992). The S-locus receptor kinase in a self-incompatible Brassica napus line encodes a functional serine/threonine kinase. Plant Cell 4, 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks, S.K., and Quinn, A.M. (1991). Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200, 38–61. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., Quinn, A.M., and Hunter, T. (1988). The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52. [DOI] [PubMed] [Google Scholar]
- Haughn, G.W., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204, 430–434. [Google Scholar]
- He, Z., Wang, Z.-Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- He, Z.-H., Masaaki, F., and Kohorn, B.D. (1996). A cell-wall associated, receptor-like protein kinase. J. Biol. Chem. 271, 19789–19793. [DOI] [PubMed] [Google Scholar]
- Horn, M.A., and Walker, J.C. (1994). Biochemical properties of the autophosphorylation of RLK5, a receptor-like protein kinase from Arabidopsis thaliana. Biochim. Biophys. Acta 1208, 65–74. [DOI] [PubMed] [Google Scholar]
- Jackson, D. (1991). In-situ hybridisation in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, J.S.J. Gurr, and M. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Dreano, M., Buckler, D.R., Cheng, S., Ythjer, A., Wu, H., Hendrickson, W.A., and El Tayar, N. (1995). Structural predictions for the ligand-binding region of glycoprotein hormone receptors and the nature of hormone-receptor interactions. Structure 3, 1341–1353. [DOI] [PubMed] [Google Scholar]
- Jinn, T.-L., Stone, J.M., and Walder, J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., and Jones, J.D.G. (1996). The roles of the leucine-rich repeat proteins in plant defenses. Adv. Bot. Res. Including Adv. Plant Pathol. 25, 89–167. [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Joshi, C.P. (1987). Putative polyadenylation signals in nuclear genes of higher plants: A compilation and analysis. Nucleic Acids Res. 15, 9637–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 19, 415–421. [DOI] [PubMed] [Google Scholar]
- Langdale, J.A., Lane, B., Freeling, M., and Nelson, T. (1989). Cell lineage analysis of maize bundle sheath and mesophyll cells. Dev. Biol. 133, 128–139. [DOI] [PubMed] [Google Scholar]
- Langdale, J.A., Zelitch, I., Miller, E., and Nelson, T. (1988). Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. EMBO J. 7, 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- McCarty, D.R., and Chory, J. (2000). Conservation and innovation in plant signaling pathways. Cell 108, 201–209. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nelson, T., and Dengler, N. (1992). Photosynthetic tissue differentiation in C4 plants. Int. J. Plant Sci. 153, S93–S105. [Google Scholar]
- Nelson, T., and Dengler, N. (1997). Leaf vascular pattern formation. Plant Cell 9, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooden, L.D. (1988). Abscisic acid, auxin, and other regulators of senescence. In Senescence and Aging in Plants, L.D. Nooden and A.C. Leopold, eds (San Diego, CA: Academic Press), pp. 85–109.
- Ponce, M.R., Quesada, V., and Micol, J.L. (1998). Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J. 14, 497–501. [DOI] [PubMed] [Google Scholar]
- Posakony, L.G., Raftery, L.A., and Gelbart, W.M. (1991). Wing formation in Drosophila requires decapentaplegic function along the anterior-posterior compartment boundary. Mech. Dev. 33, 69–82. [DOI] [PubMed] [Google Scholar]
- Przemeck, G.K.H., Mattsson, J., Hardtke, C.S., Sung, Z.R., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Reichmann, J.L., and Zhang, J.Z. (2000). INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA. Plant Cell 12, 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, A.G., Santa Cruz, S., Roberts, I.M., Prior, D.A.M., and Turgeon, R. (1997). Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, T. (1981). The control of patterned differentiation of vascular tissues. Adv. Bot. Res. 9, 151–262. [Google Scholar]
- Sachs, T. (1991). Pattern Formation in Plant Tissues. (Cambridge, UK: Cambridge University Press).
- Savage, C., Das, P., Finelli, A., Townsend, S., and Sun, C. (1996). C. elegans genes sma-2, sma-3, and sma-4 define a conserved family of TGF-β pathway components. Proc. Natl. Acad. Sci. USA 93, 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E., Rueb, S., Boot, K.J.M., Hoge, J.H.C., and Meijer, A.H. (2000). A role for the rice homeobox gene Oshox1 in provascular cell fate commitment. Development 127, 3655–3669. [DOI] [PubMed] [Google Scholar]
- Segal, D., and Gelbart, W.M. (1995). Shortvein, a new component of the dpp gene complex in Drosophila. Genetics 109, 119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.-Y., Wang, G.-L., Chen, L.-L., Kim, H.-S., Pi, L.-Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.-X., Zhu, L.-H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Thimann, K.V. (1980). The senescence of leaves. In Senescence in Plants, K.V. Thimann, ed (Boca Raton, FL: CRC Press), pp. 85–115.
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Cornejero, V., and Vera, P. (1996). Phloem-specific expression of a plant homeobox gene during secondary phase of vascular development. Plant J. 9, 639–648. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Jeong, S., and Clark, S.E. (2000). CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289, 613–617. [DOI] [PubMed] [Google Scholar]
- van den Berg, C., Weisbeek, P., and Scheres, B. (1998). Cell fate and cell differentiation status in the Arabidopsis root. Planta 205, 483–491. [DOI] [PubMed] [Google Scholar]
- van Staden, J., Ackermann, C., and Cooke, E.L. (1988). Manipulating carnation petal senescence. I. The interaction and transport of benzyladenine and IAA. Scientia-Horticulturae 35, 143–156. [Google Scholar]
- von Heijne, G. (1990). The signal peptide. J. Membr. Biol. 115, 195–201. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]