Abstract
The auxin indole-3-acetic acid is a key plant hormone essential for a broad range of growth and developmental processes. Here, we show that auxin activates Rac-like GTPases (referred to as Rac/Rop GTPases), and they in turn stimulate auxin-responsive gene expression. In particular, we show that overexpressing a wild-type tobacco Rac/Rop GTPase, NtRac1, and its constitutively active mutant form activates auxin-responsive gene expression. On the other hand, overexpressing dominant-negative NtRac1 and Rac-negative regulators, or reducing the endogenous NtRac1 level, suppresses auxin-induced gene expression. Furthermore, overexpression of NtRac1 activity or suppression of its expression in transgenic seedlings induces phenotypes that are similar to auxin-related defects. Together, our results show that a subset of plant Rac/Rop GTPases functions in mediating the auxin signal to downstream responsive genes.
INTRODUCTION
The ability of plants to perceive, transduce, and respond to the hormone auxin (indole-3-acetic acid [IAA]) and to regulate its homeostasis intracellularly is critical to plant growth and development (Davies, 1995; Normanly and Bartel, 1999). Auxin regulates cellular processes such as cell division, elongation, and differentiation and organismal responses such as phototropism, gravitropism, and apical dominance, which together give rise to the ultimate architecture of the whole plant. It also plays key roles in developmental processes, from embryogenesis to reproduction and senescence. Induction of these diverse auxin responses is believed to involve multiple divergent and intersecting signal transduction pathways (Moller and Chua, 1999; McCarty and Chory, 2000). Protein kinases, in particular the mitogen-activated protein kinase (MAPK) cascades and ubiquitin-regulated proteolysis, are known to play important roles in auxin signaling (Rogg and Bartel, 2001). However, the receptors and initial signaling components that relate the auxin stimulus to these intracellular pathways remain to be elucidated.
Among the best-characterized auxin responses is its regulation of a set of early response genes (Abel and Theologis, 1996; Guilfoyle et al., 1998a, 1998b). Expression of these genes is controlled by a family of transcription factors, the auxin response factors (ARFs) (Ulmasov et al., 1997a), which bind directly to the auxin-responsive elements in the promoter region of auxin-responsive genes. In Arabidopsis, specific ARFs either activate or repress auxin-responsive gene expression (Ulmasov et al., 1999). Aux/IAA proteins constitute another family of important regulators for auxin-responsive gene expression. They are rapidly auxin-induced, short-lived proteins that act as repressors for ARF-activated gene expression (Ulmasov et al., 1997b; Gray et al., 1999; Worley et al., 2000; Reed, 2001; Rogg and Bartel, 2001; Tiwari et al., 2001). Auxin promotes the degradation of these proteins, derepressing ARF-mediated gene expression.
Heterotrimeric G proteins and Ras-related small GTPases in animals and yeast are prominent signaling molecules that mediate a wide variety of external stimuli to intracellular signaling pathways (Bourne et al., 1991; Bar-Sagi and Hall, 2000; Hur and Kim, 2002). Previous biochemical studies have suggested the involvement of GTP binding proteins in the transduction of the auxin signal in rice coleoptiles (Zaina et al., 1990) and wheat mesophyll protoplasts (Bossen et al., 1991). Recent genetic analysis of a heterotrimeric G-protein mutant in Arabidopsis has suggested a role for this protein in modulating several hormonal signals, including auxin (Ullah et al., 2001; Wang et al., 2001; Assmann, 2002). In animals, Rho, Rac, and Cdc42 are members of the Rho subfamily of the Ras superfamily of GTP binding proteins (Mackey and Hall, 1998; Bar-Sagi and Hall, 2000). They respond to extracellular stimuli and modulate the activity of downstream signaling pathways by alternating between the inactive GDP-bound form and the active GTP-bound form. The GDP-GTP exchange itself is controlled by a number of regulatory proteins (Van Aelst and D'Souza-Schorey, 1997; Aspenstrom, 1999). In plants, the Rho family of GTPases is represented by a large number of proteins that most resemble the mammalian Rac GTPases; they have been referred to variably as Racs and Rops (Winge et al., 1997, 2000; Arabidopsis Genome Initiative, 2000; Valster et al., 2000; Yang, 2002). We refer to this class of proteins as Rac/Rop GTPases; for specific members of the family, we use the nomenclature in the original literature that describes these proteins.
Like their counterparts in other organisms, Rac/Rop GTPases mediate a variety of stimuli to multiple cellular and developmental responses. Several Rac/Rop GTPases have been shown to regulate polar cell growth in pollen tubes and root hairs (Kost et al., 1999; Fu et al., 2001; Molendijk et al., 2001; Jones et al., 2002). These small GTPases also play prevalent roles in processes that involve oxidative reactions. For instance, the rice OsRac1 has been shown to regulate pathogen-elicited and hydrogen peroxide–mediated programmed cell death (Kawasaki et al., 1999; Ono et al., 2001), and the cotton Rac13 mediates oxidative reactions involved in secondary wall formation (Potikha et al., 1999). Moreover, inactivation of a Rac/Rop GTPase negative regulator, RopGAP4, results in decreased tolerance to oxygen deprivation in Arabidopsis, implying an important role for these small GTPases in oxidative stress responses (Baxter-Burrell et al., 2002). Rac/Rop GTPases also have been implicated in regulating hormonal responses. For instance, downregulation of Rac/Rop GTPases is important for abscisic acid–induced stomatal closure (Lemichez et al., 2001) and altering Rop2At signaling capacity in transgenic Arabidopsis results in defects in several developmental processes regulated by different hormones, including auxin, brassinosteroid, and abscisic acid (Li et al., 2001). A protein related immunologically to pea Rop1 has been detected in the CLAVATA signaling complex, suggesting the possible involvement of a Rac/Rop protein in the regulation of meristem development (Trotochaud et al., 1999).
To examine the biological roles of Rac/Rop GTPases, we overexpressed a tobacco Rac/Rop GTPase, NtRac1, and a constitutively active mutant form of this protein, NtRac1(CA), in transgenic tobacco. These transgenic plants produced progeny seedlings with a range of phenotypes, complicating a mechanistic dissection of the functional role of NtRac1 in plant growth and development. However, some of the phenotypes in the transformed tobacco seedlings were suggestive of defects in auxin-related processes (see below). This led us to examine specifically whether NtRac1 plays a role in auxin signaling. Here, we provide biochemical evidence from whole-plant studies that auxin activates Rac/Rop GTPases. We also describe gene expression studies in transfected protoplasts and in transgenic plants showing that several of these small GTPases from Arabidopsis and tobacco, in particular the tobacco NtRac1, signal auxin-inducible gene expression.
RESULTS
Tobacco NtRac1 Stimulates Auxin-Responsive Gene Expression
GH3 and DR5 are well-characterized natural and synthetic auxin-inducible promoters, respectively (Liu et al., 1994; Ulmasov et al., 1997b). GH3-GFP (green fluorescent protein) expression was induced by auxin in transfected protoplasts (Figure 1) (Kovtun et al., 1998). Similarly, GH3-GUS (β-glucuronidase) and DR5-GUS were induced by naphthalene acetic acid (NAA) (Figures 2A and 2B) (Ulmasov et al., 1997b). To examine the effect of NtRac1 on auxin-responsive gene expression, we cotransfected protoplasts with 35S-NtRac1 or 35S-NtRac1(CA) [NtRac1(CA) has a G15V conversion rendering it constitutively in the active GTP-bound form] and one of these model auxin-responsive genes or the Arabidopsis auxin-responsive IAA2-GUS (Luschnig et al., 1998). As shown in Figures 1, 2A, 2B, and 2D, coexpression of NtRac1 or NtRac1(CA) activated expression from these auxin-responsive genes in the absence of exogenous auxin and enhanced their expression when auxin was provided in the medium. N-terminal GFP fusion to various forms of NtRac1 had little effect on their auxin-responsive gene expression stimulating activity (Figure 2C). These fusion proteins provided a molecular tag with which to quantitate expression levels from the transgenes. Immunoblot analysis showed that the different forms of GFP-NtRac1 were expressed to similar levels in transfected protoplasts (Figure 2C), confirming that their differential effects on auxin-responsive gene expression were not attributable to different levels of transgene expression.
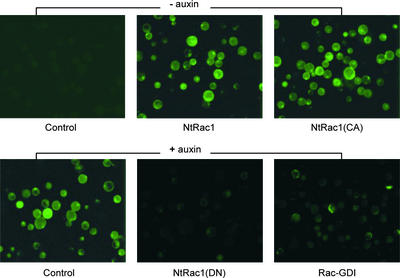
Figure 1.
GH3-GFP Is Stimulated by Auxin and Regulated by NtRac1.
All samples were cotransfected with the reporter gene GH3-GFP and the transgene indicated. Protoplasts were cultured in the absence (− auxin) or the presence (+ auxin) of 0.1 μM NAA. Results from Rac-GAP (data not shown) were similar to those for Rac-GDI.
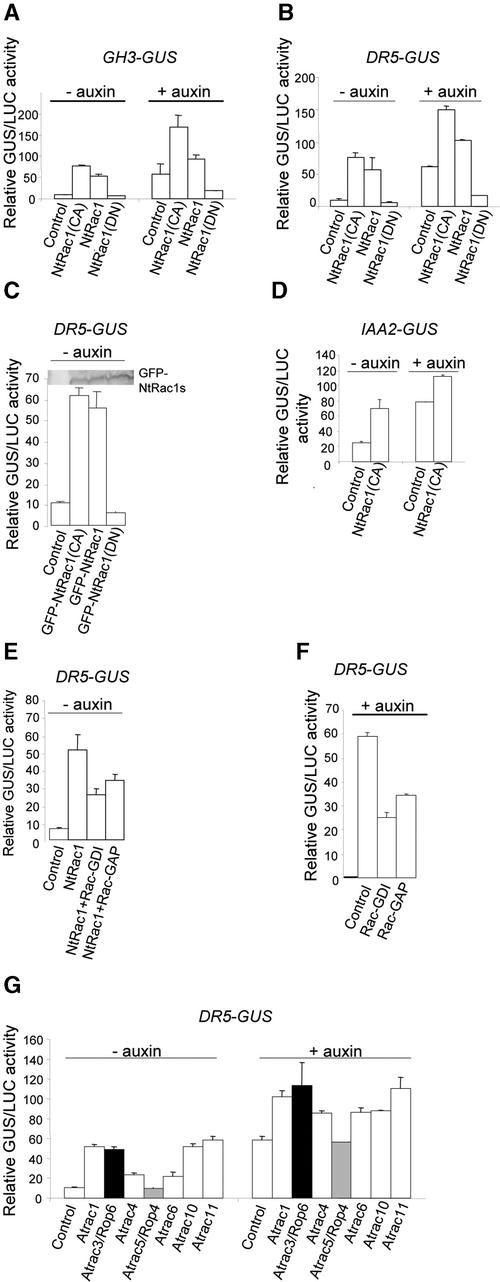
Figure 2.
NtRac1 Regulates Auxin-Responsive Gene Expression in Transfected Protoplasts.
The reporter gene (GH3-GUS, DR5-GUS, or IAA2-GUS) used for each experiment is indicated above each histogram. The cotransfecting transgene(s) used is as indicated. Data for NtRac1(DN) were from the D121A mutant gene. Results from the NtRac1(DN) T20N mutation were qualitatively similar (data not shown). Samples cultured in the absence of exogenous auxin are indicated (− auxin). Auxin-supplemented cultures (+ auxin) were maintained in 0.1 μM NAA (except in [D], for which 10 μM NAA was used) before GUS reactions. Transfections were performed in triplicate for each condition. GUS and luciferase (LUC) assays were performed in duplicate for each transfection. GUS activity was expressed relative to the internal reference luciferase activity. Each experiment was repeated at least three times with similar results. The data presented are averaged from one representative experiment. Where no error bar is shown, the standard deviation was negligible.
(A) Response of GH3-GUS to various forms of NtRac1.
(B) Response of DR5-GUS to various forms of NtRac1.
(C) Response of DR5-GUS to various forms of GFP-NtRac1. The protein blot shows that the level of GFP-tagged NtRac1 expressed in each of the transfected cultures was comparable.
(D) Response of Arabidopsis IAA2-GUS to NtRac1(CA).
(E) The negative Rac regulators Rac-GDI and Rac-GAP suppressed NtRac1-activated DR5-GUS expression in auxin-free medium. These negative Rac regulators had no effect on 35S-GUS expression (data not shown).
(F) Rac-GDI and Rac-GAP suppressed auxin-activated DR5-GUS expression in the presence of exogenous auxin. These negative Rac regulators may target multiple endogenous Rac/Rop GTPases, including NtRac1, so the data reflect the sum total of their inactivation.
(G) Multiple Arabidopsis Atracs stimulate auxin-responsive gene expression. Protoplasts were cotransfected with DR5-GUS and each of the 35S-Atracs as indicated. Black and gray bars refer to Atrac3 and Atrac5, respectively (equivalent to Rop6 and Rop4, respectively), which have been shown to have cell membrane and perinuclear predominant localizations, respectively (Bischoff et al., 2000).
The Rac-GTPase regulators Rac-GAP (GTPase-activating protein) and Rac-GDI (guanine nucleotide dissociation inhibitor) downregulate the activity of Rac GTPases by maintaining them predominantly in the GDP-bound inactive state (Hancock and Hall, 1993; LaMarche and Hall, 1994; Sasaki and Takai, 1998; Borg et al., 1999; Bischoff et al., 2000; Wu et al., 2000). Expression of these negative regulators in transfected protoplasts counteracted considerably the ability of coexpressed NtRac1 to activate auxin-responsive promoters (Figure 2E). Together, these observations suggest that active NtRac1 upregulates at least a subset of auxin-responsive promoters in transfected protoplasts.
Rac/Rop GTPase–Mediated Signaling Is Essential for Auxin-Induced Gene Expression
The effect of dominant-negative mutant forms of NtRac1 [NtRac1(DN), with either a D121A or a T20N conversion rendering them constitutively in the inactive GDP-bound state] on auxin-responsive gene expression was also examined. Unlike their wild-type and constitutively active counterparts, NtRac1(DN)s and their GFP-tagged counterparts did not activate auxin-responsive promoters in the absence of auxin (Figures 2A to 2C), but they suppressed the auxin-induced expression of cotransfected auxin-responsive genes to basal levels in the presence of exogenous auxin (Figures 1, 2A, and 2B). This observation excludes the possibility that the NtRac1- and NtRac1(CA)-induced expression from auxin-responsive promoters described above had resulted from increased levels of endogenous auxin.
Dominant-negative mutations in signaling molecules presumably act by interfering with other components of the endogenous signaling machinery. Therefore, the ability of NtRac1(DN)s to suppress auxin-induced expression from responsive promoters (Figures 1 and 2A to 2C) suggests that endogenous NtRac1 and/or closely related Rac/Rop GTPases (collectively called NtRacs herein) are necessary to mediate the auxin signal to responsive genes. Further support for this notion was provided by the observation that the expression of Rac-GAP and Rac-GDI also reduced auxin-induced DR5-GUS expression in transfected protoplasts (Figure 2F). These negative regulators would act via downregulation of endogenous target NtRacs, presumably including endogenous NtRac1, providing compelling evidence that these small GTPases play important signaling roles in auxin-regulated gene expression.
NtRac1 Mediates the Signaling of Auxin-Responsive Gene Expression in Transgenic Plants
The effect of NtRac1 on auxin-responsive gene expression was also examined in whole plants. A single copy of the DR5-GUS reporter gene was crossed to tobacco plants that had been transformed by 35S-NtRac1, 35S-NtRac1(CA), 35S-NtRac1(DN)s, or their GFP-tagged counterparts. Constitutive expression of NtRac1 and NtRac1(CA) stimulated DR5-GUS expression in the double-transformed seedlings in the absence or presence of exogenously supplied auxin (Figure 3A). On the other hand, overexpression of NtRac1(DN)s inhibited auxin-induced DR5-GUS expression in the presence of exogenous auxin (Figure 3A).
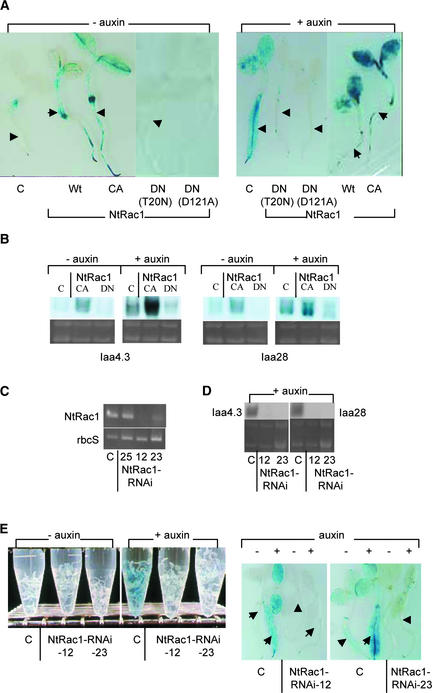
Figure 3.
NtRac1 Mediates Auxin-Responsive Gene Expression in Whole Plants.
(A) GUS expression in control DR5-GUS–transformed seedlings (C) and seedlings that were double transformed with DR5-GUS and various forms of 35S-NtRac1 [wild type (Wt), CA, DN (T20N), or DN (D121A)]. Seedlings were incubated either with 15 μM NAA (+ auxin) or without auxin (− auxin) before GUS reaction. Arrowheads point to individual seedlings.
(B) RNA gel blot analysis of auxin-responsive expression of tobacco Iaa4.3 and Iaa28 in control (C) and 35S-NtRac1(CA)– and 35S-NtRac1(DN)–transformed seedlings. Seedlings were treated with 50 μM NAA (+ auxin) or without auxin (− auxin) for 6 h before RNA isolation. RNA gel blots were probed with either 32P-labeled Iaa4.3 or Iaa28 cDNA. rRNA bands from the corresponding RNA gels are shown below each autoradiogram.
(C) RT-PCR for NtRac1 mRNA (top) and rbcS mRNA (bottom; a control for mRNA quantitation) from control (C) and 35S-NtRac1-RNAi–transformed lines 25, 12, and 23. Line 25 produced kanamycin-resistant but otherwise normal seedlings. Lines 12 and 23 produced progeny seedlings that showed strong developmental defects (see Figure 6G).
(D) RNA gel blot analysis of auxin-responsive expression of Iaa4.3 and Iaa28 in control (C) and 35S-NtRac1-RNAi–transformed seedlings from lines 12 and 23. Seedlings were treated with 50 μM NAA for 6 h before RNA isolation. rRNA bands from the corresponding RNA gels are shown below each autoradiogram.
(E) GUS expression in 8-day-old control (DR5-GUS–transformed) (C) and DR5-GUS 35S-NtRac1-RNAi (lines 12 and 23) double-transformed seedlings. Seedlings were either treated without auxin (− auxin) or with 15 μM NAA (+ auxin) for 6 h before GUS reaction. Data for groups of seedlings are shown at left; data for individual seedlings (arrowheads) are shown at right.
An RNA interference (RNAi) construct (Chuang and Meyerowitz, 2000), 35S-NtRac1-RNAi, was used to produce transformed plants in which endogenous NtRac1 mRNA (and possibly other related NtRac mRNAs) had been suppressed to very low levels (Figure 3C; see also Figure 4A, right). Like the effect of NtRac1(DN)s on DR5-GUS expression shown in Figure 3A, auxin-induced expression from DR5-GUS in the 35S-NtRac1-RNAi and DR5-GUS double-transformed seedlings also was blocked (Figure 3E). Results from these transgenic plants were in accordance with those obtained from transient expression analyses in transfected protoplasts (Figures 1 and 2), allowing for the conclusion that NtRac1 and possibly other endogenous NtRacs mediate the auxin signal to downstream responsive genes to be extended to the whole-plant level.
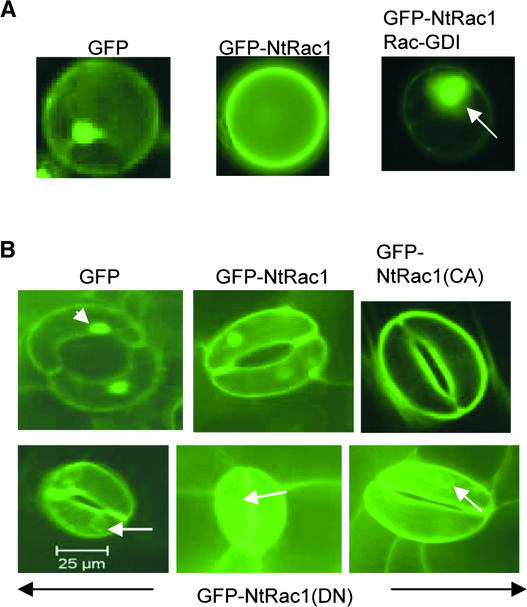
Figure 4.
Cell Membrane Association of Rac/Rop GTPases Correlates with Their Ability to Stimulate Auxin-Responsive Gene Expression.
(A) Left, protoplasts transfected with 35S-GFP typically had strong nuclear fluorescence (arrowhead) along with cytoplasmic signals, as reported previously (Grebenok et al., 1997). Middle, protoplasts transfected with 35S-GFP-NtRac1 typically showed a strong cell membrane localization for GFP-NtRac1. Right, protoplasts cotransfected with 35S-GFP-NtRac1 and 35S-Rac-GDI typically showed a diminished cell membrane association and a strong perinuclear and nuclear localization (arrow) for GFP-NtRac1.
(B) Localization of GFP, GFP-NtRac1, GFP-NtRac1(CA), and GFP-NtRac1(DN) (T20N) in epidermal cells of transformed seedlings. A single guard cell from each of the transgenic plants is shown because the signals in these cells were the strongest. The arrowhead points to the nucleus; arrows point to the perinuclear region.
To confirm that NtRac signaling is not restricted only to introduced transgenes, we also examined the auxin responsiveness of two endogenous auxin-inducible genes, Iaa3.4 and Iaa28 (Dargeviciute et al., 1998) in transgenic seedlings with perturbed NtRac1 signaling activity. RNA gel blot analysis (Figures 3B and 3D) showed that overexpressed NtRac1(CA) stimulated the levels of these two endogenous auxin-responsive mRNAs even in the absence of exogenous auxin. On the other hand, when treated with exogenous auxin, NtRac1(DN)s and NtRac1-RNAi significantly reduced the auxin-induced accumulation of these Iaa mRNAs. These observations indicate that NtRacs, in particular NtRac1, also mediate the auxin signal to at least a subset of endogenous auxin-inducible genes.
Multiple Arabidopsis Rac/Rop GTPases Can Activate Auxin-Inducible Gene Expression
Studies of Rac/Rop GTPases have found considerable structural and functional homology among these proteins from different plants. For example, Arabidopsis Rac/Rop GTPases have been shown to induce similar phenotypes in Arabidopsis and tobacco pollen tubes (Kost et al., 1999; Fu et al., 2001). When members of the Arabidopsis Atrac GTPase family (Winge et al., 2000) were tested in transfected protoplasts, Atrac1, Atrac3, Atrac10, and Atrac11 (most homologous with NtRac1) stimulated expression from DR5-GUS in both the absence and the presence of auxin (Figure 2G). Atrac4 and Atrac6 had detectable but weaker stimulatory activities. The ability of multiple Atracs to stimulate auxin-responsive gene expression suggests that they could play a signaling role for auxin similar to that played by NtRac1. The expression profiles of individual Atracs (Winge et al., 1997; Li et al., 1998; Kost et al., 1999) may determine to what extent each contributes to auxin signaling in different cell and tissue types or at different developmental stages.
Plasma Membrane Association of Rac/Rop GTPases Correlates with Stimulatory Activity toward Auxin-Responsive Expression
It is known that C-terminal sequences in small GTPases have signal sequences for isoprenylation to facilitate membrane association while in the active form (Shields et al., 2000). The stimulatory activity that NtRac1, NtRac1(CA), and their GFP-tagged counterparts exhibited on auxin-respon-sive promoters (Figures 1 and 2A to 2D) could be correlated with a predominant localization of GFP-NtRac1 and GFP-NtRac1(CA) to the plasma membrane in transfected protoplasts and transformed leaf cells (Figure 4). On the other hand, the inability of NtRac1(DN)s to stimulate auxin-responsive promoters (Figures 1, 2A, and 2B) was associated with diminished cell membrane association, as suggested by the enhanced cytosolic and perinuclear localization of GFP-NtRac1(DN)s (Figure 4B). Moreover, the ability of Rac-GDI to suppress the NtRac1- and auxin-induced expression from DR5-GUS (Figures 2E and 2F) was correlated with its ability to induce a more cytosolic location for GFP-NtRac1, especially around the nucleus (Figure 4A). Furthermore, among the Arabidopsis Rac/Rop GTPases, the only Atrac tested that had no effect on auxin-responsive gene expression, Atrac5 (Figure 2G, gray bars), also referred to as AtRop4, has a C-terminal region that confers a preferred perinuclear localization (Bischoff et al., 2000). On the other hand, Atrac3, also referred to as AtRop6, one of the Atracs that showed strong stimulatory activity toward auxin-responsive gene expression (Figure 2G, black bars), has been shown to have strong cell membrane localization (Bischoff et al., 2000). These observations suggest that the cell membrane association of Rac/Rop GTPases (Figure 4) may be an important aspect of their auxin-responsive gene-activating activity. Therefore, it is plausible that these small GTPases act on the plasmalemma level to mediate the incoming auxin signal to downstream cytoplasmic and nuclear processes, perhaps via a yet to be identified auxin-receptor complex.
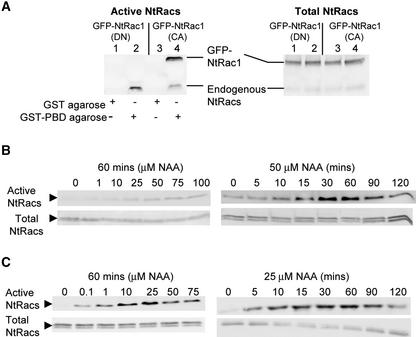
Auxin Stimulates the Activation of Rac-Like GTPases
For Rac/Rop GTPases to mediate the auxin signal, their activity should be regulated by the stimulus itself. To examine this idea, we adopted a pulldown assay that specifically targets the GTP-bound active form of Rac-GTPases. These assays involve the use of the protein binding domain (PBD) from mammalian PAK (p21-activated kinase) that binds specifically to activated Racs (Benard et al., 1999; Ren and Schwartz, 2000). PBD has been shown to distinguish between active and inactive Arabidopsis Rac/Rop GTPases (Lemichez et al., 2001). We show that it can efficiently pull down GFP-NtRac1(CA) but not GFP-NtRac1(DN)s from transgenic seedling proteins (Figure 5A), confirming that it effectively distinguishes between active and inactive forms of NtRacs. When wild-type seedlings that had been grown on solid medium were treated with auxin, the level of the PBD-pulled down, active form of NtRacs increased with increasing auxin concentration, saturating at ∼75 μM NAA (Figure 5B, gels at left). When treated with a subsaturating concentration of NAA (50 μM), the level of active NtRacs became saturated at 30 to 60 min after auxin treatment (Figure 5B, gels at right). When hydroponically grown seedlings were treated with auxin, 0.1 μM NAA was adequate to stimulate NtRacs, and significant NtRac stimulation was observed by 5 min after treatment with 25 μM NAA (Figure 5C).
Figure 5.
Auxin Activates NtRacs.
(A) Glutathione S-transferase (GST)–PBD pulls down GFP-NtRac1(CA) but not GFP-NtRac1(DN). Proteins were isolated from 35S-GFP-NtRac1(CA)– or 35S-GFP-NtRac1(DN) (T20N)–transformed seedlings. Left, the pulled down, active Rac GTPases were immunodetected on protein blots by anti-NtRac1 antibodies, which recognize all related endogenous Rac GTPases (NtRacs) (bottom band) as well as the GFP-tagged NtRac1 fusion proteins (top band). GFP-NtRac1(CA) was pulled down but GFP-NtRac1(DN) was not, indicating that PBD effectively distinguished between the active and inactive forms of tobacco NtRacs. Control binding by GST-agarose did not pull down any NtRac-related proteins. At right, an immunoblot of the original protein extracts used in the pulldown assays shown at left illustrates the relative levels of total endogenous NtRacs and the transgenic GFP-NtRacs in the various samples examined. The numbers (1 to 4) indicate pulldown samples and their corresponding total protein samples.
(B) GST-PBD pulldown assays for active NtRacs in wild-type tobacco seedlings that were cultured on solid medium and treated with increasing concentrations of NAA for 60 min (left) or with 50 μM NAA for increasing lengths of time (right). The relative levels of total NtRacs (active and inactive) were comparable in these samples.
(C) GST-PBD pulldown assays for active NtRacs in wild-type tobacco seedlings that were cultured hydroponically and treated with increasing concentrations of NAA for 60 min (left) or with 25 μM NAA for increasing lengths of time (right). The relative levels of total NtRacs (active and inactive) were comparable in these samples. The faster kinetics of NtRac activation by auxin treatment in these samples and the saturation of this activation by lower concentrations of auxin than those shown in (B) probably were the result of easier access of the exogenously added hormone into the hydroponically grown plants.
Gene expression analyses have shown that early auxin response genes, including GH3 and some Aux/IAAs, are fully activated within 5 to 10 min (Hagen and Guilfoyle, 1985; Theologis et al., 1985; McClure et al., 1989; Abel et al., 1995; Rogg and Bartel, 2001). The kinetics of auxin-activated NtRacs and auxin-induced gene expression, together with the ability of active Rac/Rop GTPases to activate expression from auxin-responsive genes (Figures 1 to 3), suggest that at least a subset of auxin-activated gene expression could be mediated by auxin-stimulated activation of these small GTPases.
Perturbing Endogenous NtRac1 Activity in Transgenic Plants Induces Multiple Phenotypes, Some of Which Are Similar to Auxin-Related Defects
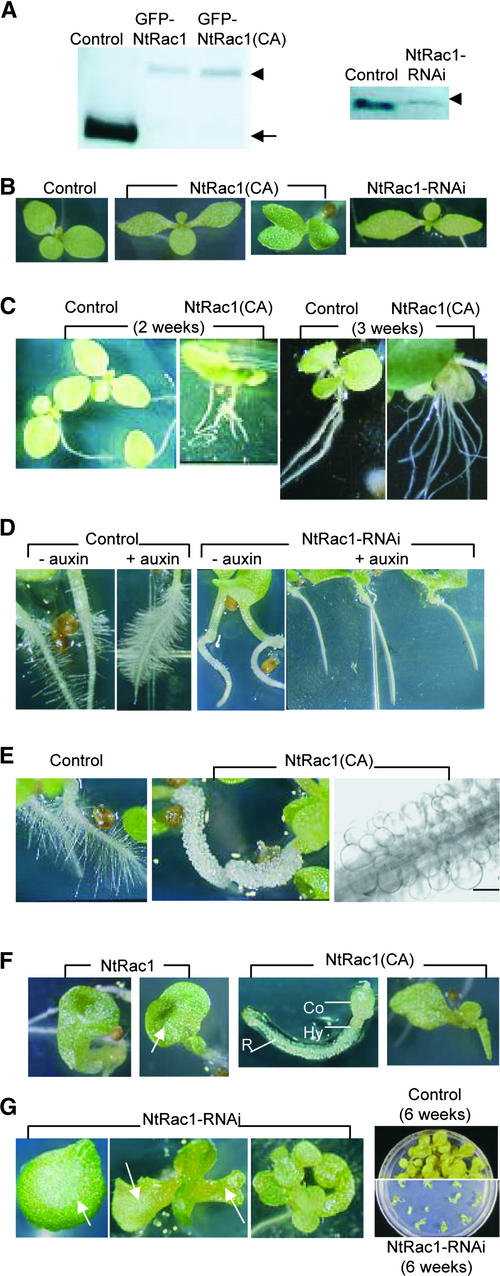
Tobacco plants were transformed by 35S-NtRac1, 35S-NtRac1(CA), or their respective GFP fusion counterparts to confer increases in NtRac1 activity or by 35S-NtRac1-RNAi to confer losses in NtRac1 (and possibly other related NtRacs) activity in transgenic plants. A majority of the recovered T0 transgenic plants (∼60) grew to maturity with little noticeable vegetative phenotype. These transgenic plants showed low levels of transgene expression (Figure 6A, gel at left), but the effects of the transgenes were clearly observable (Figures 3 and 6A, gel at right). The recovery of only relatively normal T0 transgenic plants could have resulted from lethality among transformed plantlets that harbored high levels of transgene expression. Under normal growth conditions, progeny from transformed lines also segregated a variety of phenotypes, ranging from relatively normal with slightly altered cotyledon shapes (Figure 6B) to highly seedling defective (Figures 6F and 6G, Table 1). Except for the highly defective seedlings, most of these T1 plants also reached maturity relatively normally.
Figure 6.
Overexpression of NtRac1 and NtRac1(CA) and Suppression of NtRacs by 35S-NtRac1-RNAi–Induced Seedling Phenotypes, Some of Which Were Similar to Auxin-Related Defects.
Samples shown are representative of progeny from multiple independent lines transformed by 35S-NtRac1, 35S-NtRac1(CA), their GFP-tagged counterparts, or 35S-NtRac1-RNAi. Similar phenotypes were observed in each progeny generation until the T4 generation.
(A) Left, immunoblot analysis of proteins from 2-week-old 35S-GFP–, 35S-GFP-NtRac1–, and GFP-NtRac1(CA)–transformed seedlings as detected by anti-GFP antibodies. The level of overproduced GFP-NtRac1 or GFP-NtRac1(CA) fusion proteins (arrowhead) in a population of seedlings in general was very low relative to the level of GFP (arrow) that accumulated in control seedlings. When observed by epifluorescence, 35S-GFP-NtRac1– and 35S-GFP-NtRac1(CA)–transformed seedlings that showed severely defective phenotypes similar to those shown in (F) had significantly higher levels of green fluorescence relative to their less severely affected counterparts (two middle panels in [B]). Right, immunodetection of NtRacs in control and 35S-NtRac1-RNAi–transformed seedlings (line 12) showing that the endogenous level of NtRacs (arrowhead) was significantly lower than that in control seedlings.
(B) Mild phenotypes of altered cotyledon shapes were prevalent among NtRac1(CA)–overexpressing and 35S-NtRac1-RNAi–transformed seedlings.
(C) Overexpressing NtRac1(CA) induced root branching. The two left panels (2-week-old seedlings) are of identical magnification; the two right panels (3-week-old seedlings) also are of identical magnification. Seedlings shown in the two right panels were representative of 30 3-week-old seedlings measured (see Table 1).
(D) 35S-NtRac1-RNAi suppressed root hair development. Control seedlings responded to auxin treatment (0.1 μM NAA) and showed increased root hair proliferation (two left panels). 35S-NtRac1-RNAi–transformed seedlings did not develop root hairs even in the presence of exogenous auxin (two right panels).
(E) Overexpressing NtRac1(CA) induced ballooned (and club-shaped; data not shown) root hairs (middle and right panels). The left and middle panels show 5-day-old seedlings. The right panel shows root hairs from a 2-week-old seedling. Bar = 100 μm.
(F) Overexpressing NtRac1 and NtRac1(CA) induced severe seedling defects that were similar to auxin-related mutant phenotypes such as the pin-like fused cotyledons (two left panels), the gnom-like seedlings arrested in both root and shoot development (middle panel), and the mp-like seedling lacking primary root development (right panel). These mutant phenotypes appeared less frequently among NtRac1-overexpressing seedlings. The most severely defective phenotype, gnom-like, occurred mostly among NtRac1(CA)-overexpressing seedlings. The arrow points to fused cotyledons. Co, cotyledon; Hy, hypocotyl; R, root.
(G) 35S-NtRac1-RNAi induced severe seedling defects. Most of these seedlings were arrested for further development (right panel, bottom). Arrows point to fused or abnormal cotyledons. Observation of more lethality among 35S-NtRac1-RNAi–transformed seedlings could be attributable to the downregulation of multiple Rac/Rop GTPases, because NtRac1-RNAi may suppress the expression of NtRac1 and other related small GTPases.
Table 1.
Sample Segregation of Mutant Phenotypes among Tobacco Plants Transformed by the Gain-of-Function Transgene 35S-NtRAc1(AC) or the Loss-of-Function Transgene 35S-NtRac1-RNAi
| Transgene (Plant Line) | Severely Defective Seedlingsa |
Mild Cotyledon Phenotypeb |
Root Defects |
Number of T1 Seedlings Examined |
Severely Defective Seedlings in the T2 Generation |
|---|---|---|---|---|---|
| Control | <0.017% | 0% | <0.05%c | 586 | – |
| 35S-NtRac1(CA) (71-5) | 4.2% | 95.8% | 100%c,d | 96 | 20% |
| 35S-NtRac1-RNAi (125-12) | 7% | 23% | 100%e | 202 | 50% |
The two transgenic lines were selected for presentation because they produced the highest levels of seedlings with severely defective phenotypes (see Figures 6F and 6G) among all T0 plants analyzed. Both of these lines harbor a single T-DNA insert. The average level was 1.85% (among 1014 T1 seedlings from 16 T0 lines analyzed) for the gain-of-function transgenic plants and 0.42% (among 834 T1 seedlings from 10 T0 lines analyzed) for the loss-of-function transgenic plants. Although low, these percentages were significantly higher than that observed among control seedlings (<0.017% among ∼600 seedlings analyzed). T2 and subsequent-generation progeny from the different classes of transgenic plants segregated a higher level of severely defective seedlings, reaching 50% in the strongest 35S-NtRac1-RNAi line and 20% in the 35S-NtRac1(CA) line. The mild cotyledon and root defects were observed among all of the transgenic lines in frequencies similar to those shown here.
Includes all phenotypes shown in Figures 6F and 6G.
Phenotypes shown in Figure 6B.
Phenotypes shown in Figure 6E.
Phenotypes shown in Figure 6C.
Phenotypes shown in Figure 6D.
The most prevalent phenotypes among these transgenic plants were related to root or root hair development, processes that are known to be highly sensitive to auxin (Pitts et al., 1998; Xie et al., 2000; Rogg and Bartel, 2001). A large number of 35S-NtRac1(CA)–transformed seedlings showed increased root proliferation (Figure 6C, Table 1). The observed increased root branching was similar to the development of excess adventitious and lateral roots observed in auxin-overproducing mutants (Boerjan et al., 1995; King et al., 1995), suggesting that auxin signaling was upregulated in these 35S-NtRac1(CA)–transformed plantlets. When NtRac1 and related NtRacs were downregulated by the expression of NtRac1-RNAi (Figures 3C and 6A, gel at right), a majority of these transformed seedlings developed roots with little or no root hairs (Figure 6D). Furthermore, exogenous auxin stimulated root hair development in wild-type seedlings (Figure 6D, cf. two panels at left) but failed to induce roots hairs in these 35S-NtRac1-RNAi–transformed seedlings (Figure 6D, two panels at right). The insensitivity to auxin-stimulated root hair development is consistent with downregulated auxin signaling activity in these transgenic plantlets with reduced NtRac function.
Another striking and commonly observed root phenotype among 35S-NtRac1(CA)–transformed seedlings was the development of balloon-tipped (Figure 6E) and club-shaped (data not shown) root hairs. Although not as exaggerated as the phenotype shown here, swollen or club-shaped root hairs have been observed in transformed Arabidopsis that overexpress one of a number of AtRop GTPases (Molendijk et al., 2001; Jones et al., 2002). These depolarized root hair growth defects have been attributed to altered AtRop-induced disorganization of the actin cytoskeleton in these cells (Molendijk et al., 2001; Jones et al., 2002), similar to their effect on tip growth pollen tubes (Kost et al., 1999; Fu et al., 2001). We also observed that overexpressing NtRac1 in pollen tubes induced isotropic growth and that the actin cytoskeleton in these cells became highly disorganized (C. Chen, A.Y. Cheung, and H.-m. Wu, unpublished results). Therefore, it is most likely that the balloon-tipped root hairs in NtRac1(CA)-overproducing plants are a consequence primarily of increasing NtRac1 signaling activity to their actin cytoskeleton.
Embryo development is highly dependent on auxin, and many auxin-related defects are manifested at the cotyledon and seedling levels (Mayer et al., 1991; Hadfi et al., 1998; Berleth et al., 2000). A large number of the 35S-NtRac1(CA)–transformed seedlings showed mild cotyledon phenotypes ranging from altered shapes to asymmetrically spaced and/or slightly fused cotyledons (Figure 6B, middle panels). Some of the 35S-NtRac1-RNAi–transformed seedlings, especially those in which NtRac1 expression was suppressed most effectively (Figures 3C and 3D), also showed altered cotyledon shapes (Figure 6B, panel at right). Further development was normal in these plants, suggesting that they had averted the effect of upregulation or downregulation of NtRac1 activity. However, in approximately half of the 35S-NtRac1–, 35S-NtRac1(CA)–, 35S-GFP-NtRac1–, and 35S-GFP-NtRac1(CA)–transformed lines (26 T0 lines), a low percentage of the progeny showed severe seedling defects (Figure 6F, Table 1). Presumably, these were individuals among the progeny population with higher levels of transgene expression during embryogenesis or germination than those expressed in siblings with mild phenotypes. Some of the severe seedling phenotypes shown among progeny of NtRac1- or NtRac1(CA)-overexpressing lines were similar to auxin-related defects seen in embryo developmental mutants. These included the pin-like phenotype (Galweiler et al., 1998) of fused and cup-shaped cotyledons (Figure 6F, two panels at left), the monopteros (mp)-like phenotype (Hardtke and Berleth, 1998) of lacking the primary root axis (Figure 6F, panel at right), and the gnom-like phenotype (Steinmann et al., 1999) of lacking shoot and root development (Figure 6F, third panel). If the shoot apical meristem burst through the fused cotyledons, or adventitious roots developed from the hypocotyls of mp-like seedlings, these plants often developed to maturity.
Reduction of endogenous NtRacs by NtRac1-RNAi also induced severe seedling phenotypes (Table 1), some with unopened or grossly malformed cotyledons or nondiscernible structures in the place of the hypocotyl and cotyledons (Figure 6G). These defective transgenic seedlings were arrested for further development (Figure 6G, panel at right). Both the frequency of severely defective seedlings and their severity were higher among the 35S-NtRac1-RNAi–transformed seedlings. This resulted most likely from reduced levels of not only NtRac1 mRNAs but other closely related NtRac mRNAs as well. Thus, the gross seedling defects and lethality may reflect the reduced signaling capacity of more than one NtRac.
DISCUSSION
How auxin regulates plant growth and development is one of the most extensively studied areas in plant biology. Rac/Rop GTPases are emerging as important molecular switches that regulate the signaling of diverse cellular processes in plants. The results presented here show that Rac/Rop GTPases, in particular the tobacco NtRac1, play a pivotal role in auxin signaling.
Rac/Rop GTPases as Molecular Links in Auxin Signaling
Although known to be important to a variety of cellular processes, signaling relationships between specific extracellular stimuli and downstream pathways are known for only a few Rac/Rop GTPases (Kawasaki et al., 1999; Lemichez et al., 2001; Ono et al., 2001). The observations that NtRac1 and NtRac1(CA) activate auxin-responsive gene expression in the absence of auxin suggest a signaling role for auxin-regulated gene expression for this small GTPase (Figures 1 to 3). The ability of NtRac1(DN) and the negative Rac-GTPase regulators Rac-GAP and Rac-GDI to block auxin-induced gene expression in the presence of exogenous auxin (Figures 1 to 3) provides compelling evidence that endogenous active NtRacs are important for mediating the auxin signal to responsive genes. The observation that auxin activates NtRacs (Figures 5B and 5C) further supports the existence of a signaling pathway whereby auxin-activated NtRacs stimulate downstream responsive gene expression.
Auxin enhances the degradation of auxin-induced, short-lived Aux/IAA proteins that interact with ARFs to modulate auxin-responsive gene expression (del Pozo and Estelle, 1999; Reed, 2001; Rogg and Bartel, 2001). Overexpression of Aux/IAA proteins has been shown to repress ARF-mediated expression from auxin-responsive promoters in transient expression analyses in protoplasts (Ulmasov et al., 1997b; Tiwari et al., 2001). We also observed that, similar to auxin action, overexpression of NtRac1 stimulates the in-stability of an Aux/IAA17-luc fusion protein. Furthermore, overexpression of Aux/IAA17 or Aux/IAA7 suppressed the NtRac1-induced expression from auxin-responsive promoters similar to their suppression of auxin-induced gene expression (our unpublished data). These observations also are consistent with NtRac1 playing an integral role in the auxin-induced signaling pathway of gene derepression via proteolysis.
It is known that individual Rho-type GTPases in animals and yeast relate multiple signals to elicit multiple downstream responses (Bar-Sagi and Hall, 2000). Plant Rac/Rop GTPases are known to be involved broadly in various cellular and developmental processes (Valster et al., 2000; Yang, 2002), and an individual Rac/Rop, AtRop2 (Li et al., 2001; Fu et al., 2002; Jones et al., 2002), may participate in multiple signaling pathways. We observed that NtRac1, like several other Rac/Rop GTPases, also signals cellular activities that regulate the actin cytoskeleton in pollen tubes (our unpublished results), suggesting that NtRac1 participates in multiple signaling pathways. These results allow a signaling relationship between auxin, NtRac1, and downstream responsive gene expression to be defined. However, revealing an auxin signaling activity for Rac/Rop GTPases does not preclude the participation of these signaling molecules as a class or of individual Rac/Rop GTPases such as NtRac1 in mediating stimuli other than auxin. Further work will be needed to dissect the complex biological roles played by Rac/Rop GTPases during plant growth and development.
The apparatus that relates the auxin signal to NtRac1 and the signaling pathway(s) regulated by auxin-activated NtRacs remain to be revealed. Rho-related GTPases are known to activate MAPK cascades to regulate downstream protein or gene expression activities in animals and yeast (Bar-Sagi and Hall, 2000; Roberts et al., 2000), and auxin has been shown to activate MAPKs (Mizoguchi et al., 1994; Mockaitis and Howell, 2000; Rogg and Bartel, 2001). Therefore, it seems plausible that in response to auxin stimulation, NtRac1 may activate one or more MAPK cascades that regulate auxin-mediated gene expression. Studies that biochemically link NtRac1 and other Rac/Rop GTPases upstream to auxin or auxin-activated regulators and downstream to their effectors should reveal components of the Rac/Rop-mediated auxin signaling pathway(s).
NtRac1 Is Important for Auxin Signaling in Plant Growth and Development
Our results from gene expression studies in protoplasts and transgenic plants (Figures 1 to 3), together with the observation that auxin stimulates the activation of NtRacs (Figure 5), provide compelling evidence that NtRac1 mediates the auxin signal to regulate the expression of downstream responsive genes. Multiple root and shoot phenotypes commonly are associated with many auxin-related mutants, and parallel phenotypes often are observed among different classes of auxin-related mutants (Berleth et al., 2000; Rogg and Bartel, 2001). Increased root proliferation has been observed in Arabidopsis mutants with increased auxin levels (Boerjan et al., 1995; King et al., 1995) or in transgenic plants with upregulated auxin signaling activities (Xie et al., 2000). In tobacco, overproduction of IAA also increases adventitious rooting (Romano et al., 1991). Therefore, increased root branching induced by overexpressing NtRac1(CA) (Figure 6B, Table 2) is consistent with upregulated auxin signaling in these plants, supporting a role for NtRac1 in auxin signaling. Overexpression of CA-Rop2At (equivalent to Atrac4) also results in increased lateral root formation in transformed Arabidopsis (Li et al., 2001). This and the observation that Atrac4 is capable of stimulating auxin-responsive gene expression (Figure 2G) support an in planta role for this Arabidopsis small GTPase in auxin signaling as well.
Table 2.
Increased Root Proliferation in 35S-NtRac1(CA)–Transformed Plants
| Transgene (Plant Line) | Primary Root Length (mm)a | Number of Lateral Rootsb | Number of Adventitious Rootsc |
|---|---|---|---|
| Control | 8.06 ± 2.35 | 0 | 1 |
| 35S-NtRac1(CA) (71-5) | 36 ± 13.7 | 2.43 ± 0.85 | 5.76 ± 2.1 |
A majority of these T0 lines produced T1 seedlings that showed increased root proliferation. Among the T1 and subsequent generation seedlings, a majority of them (often up to 100%) showed this phenotype.
Twenty-one-day-old seedlings were scored.
Developed from the primary root.
Developed from the hypocotyl.
Seedlings with fused cotyledons or lack of both root and shoot development, like those induced by increased NtRac1 signaling activity shown in Figure 6F, are prevalent among many auxin-related mutants (Berleth et al., 2000). For instance, fused cotyledons and the lack of a fully developed embryonic axis are characteristics of pin and gnom mutants, respectively, both of which are defective in polar auxin transport (Galweiler et al., 1998; Steinmann et al., 1999). Transgenic tobacco seedlings with altered polar auxin transport also showed cup-shaped fused cotyledons (Naderi et al., 1997). The lack of the primary root axis in transgenic seedlings with upregulated NtRac1 activity is similar to the phenotype of mp mutant seedlings, which are defective in an auxin response transcription factor, ARF5 (Hardtke and Berleth, 1998; Ulmasov et al., 1999). The occurrence of some overlapping phenotypes among the auxin-signaling-upregulated NtRac1(CA)-overexpressing plants described here and the auxin-response-downregulated mp mutants is intriguing. Rac/Rop GTPases have been shown to mediate multiple upstream signals, including those from several hormones, to multiple downstream response systems (Kost et al., 1999; Fu et al., 2001; Lemichez et al., 2001; Li et al., 2001; Baxter-Burrell et al., 2002). Therefore, the phenotypes induced by manipulating the endogenous Rac/Rop signaling capacity could be the sum total of multiple affected pathways, masking an absolute correlation with altered auxin signaling capacity.
It is possible that the signaling capacity of Rac/Rop GTPases itself is modulated by feedback regulation among these small GTPases and their negative regulators, as was proposed recently (Baxter-Burrell et al., 2002). These also may underlie the overlapping phenotype of altered cotyledon shapes among NtRac1(CA)- and NtRac1-RNAi–overexpressing seedlings (Figure 6B). Nevertheless, transgenic seedlings with altered NtRac1 activity are not unlike many of the auxin-related mutants in that a range of phenotypes differing in severity often is observed. For instance, mp mutations are known to induce the milder, variably fused cotyledons, and polar auxin transport is affected in these plants (Przemeck et al., 1996; Hardtke and Berleth, 1998). Moreover, a single mutation, emb30-3 (Emb30 is equivalent to Gnom), induces defective embryos with total lack of organ definition, seedlings with fully fused, partially fused, asymmetrically positioned, or unequally sized cotyledons (Shevell et al., 1994). Therefore, together with the evidence that NtRac1 mediates the auxin signal to responsive genes, altered auxin signaling must have contributed significantly, although not exclusively, to the phenotypes observed in the transgenic seedlings described here.
Auxin is known to be important for root hair development (Pitts et al., 1998). The absence of root hair development in seedlings transformed by 35S-NtRac1-RNAi and their resistance to auxin-stimulated root hair development (Figure 6D) are consistent with reduced auxin signaling capacity resulting from the reduced levels of NtRacs in these plants. On the other hand, Rac/Rop GTPases are important for regulating the actin cytoskeleton organization in the two types of tip growth cells in plants: root hairs and pollen tubes (Kost et al., 1999; Fu et al., 2001; Molendijk et al., 2001; Jones et al., 2002). Therefore, the observed reduced root hair phenotype in transgenic plants with downregulated NtRac signaling activity is likely to be the combined results of at least these two signaling pathways involving NtRacs. Furthermore, ample evidence suggests that auxin and the actin cytoskeleton have interacting relationships (Wang and Nick, 1998; Vissenberg et al., 2000; Geldner et al., 2001; Kandasamy et al., 2001; Muday and DeLong, 2001), including auxin-stimulated expression from a specific actin gene (Kandasamy et al., 2001). Therefore, it is possible that in response to auxin, activated Rac/Rop GTPases activate a pathway that turns on auxin-responsive genes and another that regulates actin cytoskeletal activity. These two pathways provide the biochemical and cellular bases that support the polar cell growth process.
The Functional Role of Rac/Rop GTPases in the Diverse Auxin Signaling Network
The case for multiple auxin receptors and diverse signaling pathways has been made (Gee et al., 1991; Rogg and Bartel, 2001). Different auxin-responsive gene expression properties provide substantive support for the suggestion of diverse perception, signaling, and responsive components in different organs and tissue types and for the notion that multiple pathways may exist within a single cell or tissue type (Gee et al., 1991). The presence of ∼20 MAPK, 25 Aux/IAA, and 23 ARF genes in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000; Rogg and Bartel, 2001; Tiwari et al., 2001) already suggests a complicated network for auxin signaling. The demonstration that multiple Rac/Rop GTPases may relate the auxin signal to downstream responsive genes (Figure 2G) introduces additional complexity to that network. As molecular switches, they may each mediate multiple signals and elicit multiple responses. Therefore, with Rac/Rop GTPases, it is possible to achieve tremendous versatility for cross-talk between auxin and other signaling pathways, plausibly facilitating the fine-tuning of the diverse auxin response. However, this complexity also makes defining the molecules involved in any individual pathway in auxin signaling and their relationships with each other and with other intersecting pathways a formidable task.
Given the importance of auxin to diverse aspects of plant growth and development and the involvement of Rac/Rop GTPases in mediating a variety of stimuli, it is intriguing that single Rac/Rop gene mutations that define their functional roles have not been reported to date. The functional redundancy demonstrated here for multiple Arabidopsis Atracs to signal auxin-responsive gene expression (Figure 2G) could be one of the reasons for the lack of loss-of-function mutations among these genes that would implicate their role in auxin signaling. The actual contribution from individual Atracs to auxin signaling in a particular cell or tissue type or at different developmental stages may depend to a large extent on their expression profiles. The effects of inactivating the primary responsible auxin signaling Atrac in a particular cell or tissue type or at a particular developmental stage may be masked by other Atracs with similar activities, preventing obvious mutant phenotypes from being displayed. The redundancy among Atracs in mediating the auxin signal would ensure that auxin signaling is not impaired easily.
Transient gene expression analysis in transfected protoplasts provides a clear and quantitative assay with which to analyze individual participants in a complex signaling pathway. For instance, it has allowed different ARFs to be assigned auxin-responsive gene-activating or gene-repressing activities (Ulmasov et al., 1999) and has allowed specific MAPKs a role in repressing auxin-responsive gene expression (Kovtun et al., 2000). By examining how individual Rac/Rop GTPases affect the activity of auxin-responsive promoters in transfected protoplasts, we obtained direct evidence that a subset of them mediate the auxin signal to regulate responsive gene expression (Figures 1 and 2). By demonstrating similar signaling activity for NtRac1 in transgenic plants (Figure 3) and the fact that auxin activates endogenous NtRacs in whole plants (Figures 5B and 5C), we show the use of NtRacs in auxin signaling in planta. The established signaling relationship for NtRac1 between auxin and downstream responsive genes provides a molecular basis by which auxin-related defects induced by altered NtRac signaling can be interpreted to reflect the role of these small GTPases in auxin signaling during plant growth and development.
METHODS
Protoplast Transfection
Tobacco (Nicotiana tabacum var Petit Havana [SR1]) plants were grown under standard tissue culture conditions. Protoplast isolation and transfection were performed as described (Chiu et al., 1996). For analytical transfections, 0.125 mL of protoplasts (1.6 × 106 cells/mL) was used. Plasmid DNA (10 μg) containing one of the auxin-responsive reporter genes was used for each transfection. The constitutive 35S promoter of Cauliflower mosaic virus (35S) was used to express various forms of NtRac1, Atracs, the negative regulators Rac-GAP and Rac-GDI, and the Aux/IAA proteins. Combinations of 10 μg of these plasmids as indicated in the figures were used in each transfection, which also included 5 μg of an internal reference gene (35S-LUC). In control transfections, an amount of the mock effector plasmid 35S-Nos3′ (which contains only the 35S promoter and the nopaline synthase transcription termination region) was used to ensure that equal amounts of DNA were introduced for each transfection. After transfection, protoplasts were incubated in K3 medium supplemented with 0.4 M Suc, with 0.1 μM naphthalene acetic acid (NAA) (unless stated otherwise), or without auxin at 25°C for 16 h in the dark. Protoplasts were either observed directly by epifluorescence microscopy or used for luciferase (LUC) and β-glucuronidase (GUS) assays. For each experiment, all transfections were performed in triplicate, and GUS assays were performed in duplicate for each transfection. Similar results were obtained in at least three independent experiments.
Plant Transformation and Analysis
Tissue culture–grown SR1 plants were transformed by Agrobacterium tumefaciens Ti plasmid. Transformed T0 plants were grown to maturity under greenhouse conditions. T1 seedlings of all transformed lines and T2 seedlings from a few representative T1 plants that showed seedling phenotypes were used for analyses. Transformed plants homozygous for a single DR5-GUS insertion were used as a pollen donor in genetic crosses. T1 and T2 seedlings were germinated under selective conditions. Root hair sensitivity to auxin was observed for seedlings grown in the presence of various concentrations of NAA.
cDNA Isolation, Mutagenesis, and Plasmid Constructs
Identical NtRac1 cDNA was isolated from tobacco leaf and pollen λZapII (Stratagene) cDNA libraries. Constitutively active (CA) and dominant negative (DN) mutations of NtRac1 were constructed using PCR mutagenesis (Ho et al., 1989; Ito et al., 1991). The GH3 promoter was obtained by PCR of soybean DNA (Liu et al., 1994). The DR5 promoter was composed of seven tandem copies of AuxREs and the minimum 35S promoter (Ulmasov et al., 1997b). IAA2-GUS (Luschnig et al., 1998) was an Arabidopsis auxin-responsive reporter gene. Atrac cDNAs (Winge et al., 2000) were obtained by reverse transcriptase–mediated (RT) PCR of mixed Arabidopsis leaf and flower mRNAs. Arabidopsis Rac-GDI and Rac-GAP (Bischoff et al., 2000; Wu et al., 2000) were obtained by PCR. Tobacco Iaa-4.3 and Iaa-28 (Dargeviciute et al., 1998) cDNAs were obtained by RT-PCR from seedling mRNAs. GFP-NtRac1 designates green fluorescent protein (GFP) fusion to the N terminus of NtRac1. 35S-NtRac1-RNAi was constructed with inverted repeats of the NtRac1 coding region between amino acid 62 and the C terminus, separated by a 100-bp stuffer DNA, and cloned behind the 35S promoter.
GUS and LUC Assays
Fluorometric GUS assays were performed as described (Jefferson, 1987). GUS reactions were performed at 37°C for 2 h. LUC assays were performed as described (Luehrsen et al., 1992). The GUS activity in each sample was expressed relative to its LUC activity to standardize data for variation in transformation efficiency and cell viability. Histochemical staining of seedlings for GUS activity was as described (Jefferson, 1987). Seedlings were incubated in sterile distilled water or in 15 μM NAA, followed by reactions for GUS histochemical staining as described (Ulmasov et al., 1997b).
Protein Gel Blot Analysis
Protoplast transfection as described above was scaled up 10 times for each transfection. One-tenth of the transfected protoplasts was used in GUS and LUC assays; the remaining protoplasts were ruptured by vortexing in 0.1% Triton X-100. Proteins were precipitated at −20°C in 80% acetone for 2 h. The acetone precipitate was resuspended in 100 μL of protein extraction buffer (30 mM Tris-HCl, pH 7.5, 1 mM Na2EDTA, 1% SDS, 0.15% β-mercaptoethanol, 10 mM DTT, and 1 mM phenylmethylsulfonyl fluoride). The entire sample was subjected to 15% SDS-PAGE and electrotransferred to nitrocellulose membranes. Proteins from plants were prepared as described (Kawata and Cheung, 1990). Immunodetection was performed with anti-GFP or anti-NtRac1 antibodies raised against Escherichia coli–produced His-tagged recombinant proteins and horseradish peroxidase–conjugated secondary antibodies raised against rabbit IgG. Proteins were detected by chemiluminescence (Perkin-Elmer).
RNA Gel Blots and RT-PCR for Gene Expression Analysis
Two-week-old seedling RNA was used. RNA gel blots were probed with 32P-labeled tobacco Iaa-3.4 or Iaa-28 cDNAs (Dargeviciute et al., 1998). RT-PCR was performed to monitor the effectiveness of 35S-NtRac1-RNAi suppression of endogenous NtRacs. Primers for NtRac1 and a tobacco rbcS cDNA (as an internal control) were used after first-strand cDNA synthesis.
Detection of Active NtRacs by Protein Binding Domain Pulldown Assays
Ten-day-old seedlings grown on solid medium or 7-day-old seedlings grown in liquid medium were used. After soluble and Triton X-100–extractable protein isolation, pulldown assays (Benard et al., 1999; Ren and Schwartz, 2000; Lemichez et al., 2001) were performed by incubating plant proteins with resins bound to either glutathione S-transferase (control) or glutathione S-transferase–protein binding domain (Benard et al., 1999). Protein binding domain was used as bait for GTP-bound Rac-GTPases. After binding and washing, bound proteins were eluted by boiling in SDS-PAGE loading buffer. Solubilized proteins were examined by immunoblot analysis after SDS-PAGE. A small aliquot (2% of the amount used for pulldown assays) from the original extracts was examined for analyses of total proteins and to ensure that comparable amounts of proteins were used in the pulldown assays. Immunodetection was performed with anti-NtRac1 antibodies and horseradish peroxidase–conjugated secondary antibodies followed by chemiluminescent detection.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession numbers for the genes and proteins mentioned in this article are as follows: Rac-GAP (AAF21198), Rop2-GAP (AAC79102), Rac-GDI (AAF21198), and NtRac1 (AY029330).
Acknowledgments
We thank Jen Sheen and Wan-ling Chiu for sharing the protoplast transfection protocol; Tom Guilfoyle and Jane Murfett for the gift of the DR5 promoter; Christine Chen and Richard Glaven for the use of NtRac1; and Jennifer Normanly for the use of the IAA2-GUS construct. We are grateful to Barbara Osborne and Maurice Fournier for the use of their luminometers. We thank Bernard Rubinstein and Jennifer Normanly for their critical reading of and thoughtful comments on the manuscript. We thank members of our laboratory for helpful discussions. We thank Winslow Briggs for his generous encouragement on this work, and we are most grateful to an anonymous reviewer who persistently provided generous guidance to improve the thoroughness and precision of this study.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006320.
References
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–813. [DOI] [PubMed] [Google Scholar]
- Aspenstrom, P. (1999). Effectors for the Rho GTPases. Curr. Opin. Cell Biol. 11, 95–102. [DOI] [PubMed] [Google Scholar]
- Assmann, S.M. (2002). Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14 (suppl.), S355.–S373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi, D., and Hall, A. (2000). Ras and Rho GTPases: A family reunion. Cell 103, 227–238. [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell, A., Yang, Z., Springer, P.S., and Bailey-Serres, J. (2002). RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296, 2026–2028. [DOI] [PubMed] [Google Scholar]
- Benard, V., Bohl, B., and Bokoch, G.M. (1999). Characterization of Rac and Cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 274, 13198–13204. [DOI] [PubMed] [Google Scholar]
- Berleth, T., Mattsson, J., and Hardtke, C.S. (2000). Vascular continuity and auxin signals. Trends Plant Sci. 5, 387–393. [DOI] [PubMed] [Google Scholar]
- Bischoff, F., Vahlkamp, L., Molendijk, A., and Palme, K. (2000). Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol. Biol. 42, 515–530. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Onckelen, H., Van Montagu, M., and Inze, D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg, S., Podenphant, L., Jensen, T.J., and Poulsen, C. (1999). Plant cell growth and differentiation may involve GAP regulation of Rac activity. FEBS Lett. 453, 341–345. [DOI] [PubMed] [Google Scholar]
- Bossen, M., Tretyn, A., Kendrick, R.E., and Vredenberg, W.J. (1991). Comparison between swelling of etiolated wheat (Triticum aestivum L.) protoplasts induced by phytochrome and α-naphthaleneacetic acid, benzylaminopurine, gibberellic acid, abscisic acid and acetylcholine. J. Plant Physiol. 137, 706–710. [Google Scholar]
- Bourne, H.R., Sanders, D.A., and McCormick, F. (1991). The GTPase superfamily: Conserved structure and molecular mechanisms. Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargeviciute, A., Roux, C., Decreux, A., Sitbon, F., and Perrot-Rechenmann, C. (1998). Molecular cloning and expression of the early auxin-responsive Aux/IAA gene family in Nicotiana tabacum. Plant Cell Physiol. 39, 993–1002. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- del Pozo, J.C., and Estelle, M. (1999). Function of the ubiquitin-proteosome pathway in auxin response. Trends Plant Sci. 4, 107–112. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The Rop2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14, 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Gee, M.A., Hagen, G., and Guilfoyle, T.J. (1991). Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.D., Jurgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenok, G.J., Lambert, G.M., and Galbraith, D.W. (1997). Characterization of the targeted nuclear accumulation of GFP within the cells of transgenic plants. Plant J. 12, 685–696. [Google Scholar]
- Guilfoyle, T., Hagen, G., Ulmasov, T., and Murfett, J. (1998. a). How does auxin turn on genes? Plant Physiol. 118, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T., Ulmasov, T., and Hagen, G. (1998. b). The ARF family of transcription factors and their role in plant hormone responsive transcription. Cell Mol. Life Sci. 54, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfi, K., Speth, V., and Neuhaus, G. (1998). Auxin-induced developmental patterns in Brassica juncea embryos. Development 125, 879–887. [DOI] [PubMed] [Google Scholar]
- Hagen, G., and Guilfoyle, T.J. (1985). Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 5, 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J.F., and Hall, A. (1993). A novel role for RhoGDI as an inhibitor of GAP proteins. EMBO J. 12, 1915–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hur, E.M., and Kim, K.T. (2002). G protein-coupled receptor signalling and cross-talk: Achieving rapidity and specificity. Cell. Signal. 14, 397–405. [DOI] [PubMed] [Google Scholar]
- Ito, W., Ishiguro, H., and Kurosawa, Y. (1991). A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene 102, 67–70. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assay for chimeric genes in plants: The GUS fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jones, M.A., Shen, J.-J., Fu, Y., Li, H., Yang, Z., and Grierson, C.S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14, 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., Gilliland, L.U., McKinney, E.C., and Meagher, R.B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 7, 1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96, 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata, E.E., and Cheung, A.Y. (1990). Molecular analysis of an aurea photosynthetic mutant (Su/Su) in tobacco: LHCP depletion leads to pleiotropic mutant phenotypes. EMBO J. 9, 4197–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7, 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-l., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-l., Zeng, W., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. [DOI] [PubMed] [Google Scholar]
- LaMarche, N., and Hall, A. (1994). GAPs for rho-related GTPases. Trends Genet. 10, 436–440. [DOI] [PubMed] [Google Scholar]
- Lemichez, E., Wu, Y., Sanchez, J.P., Mettouchi, A., Mathur, J., and Chua, N. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15, 1808–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Shen, J.-J., Zheng, Z.-L., Lin, Y., and Yang, Z. (2001). The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 126, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Wu, G., Ware, D., Davis, K.R., and Yang, Z. (1998). Arabidopsis Rho-related GTPases: Differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 18, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.-B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, J.T. (1994). The soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen, R.K., DeWet, R.J., and Walbot, V. (1992). Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 216, 397–414. [DOI] [PubMed] [Google Scholar]
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D.J., and Hall, A. (1998). Rho GTPases. J. Biol. Chem. 273, 20685–20688. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Buttner, G., and Jurgens, G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353, 402–407. [Google Scholar]
- McCarty, D.R., and Chory, J. (2000). Conservation and innovation in plant signaling pathways. Cell 103, 201–209. [DOI] [PubMed] [Google Scholar]
- McClure, B.A., Hagen, G., Brown, C.S., Gee, M.A., and Guilfoyle, T.J. (1989). Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Gotoh, Y., Nishida, E., Yamaguchi-Shinozaki, K., Hayashida, N., Iwasaki, T., Kamade, H., and Shinozaki, K. (1994). Characterization of two cDNAs that encode MAPK kinase homologues in Arabidopsis thaliana and analysis of the possible role of auxin in activating such kinase activities in cultured cells. Plant J. 5, 111–122. [DOI] [PubMed] [Google Scholar]
- Mockaitis, K., and Howell, S.H. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24, 785–796. [DOI] [PubMed] [Google Scholar]
- Molendijk, A.J., Bischoff, F., Rajendrakumar, C.S.V., Friml, J., Braun, M., Gilroy, S., and Palme, K. (2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, S.G., and Chua, N.H. (1999). Interactions and intersections of plant signaling pathways. J. Mol. Biol. 293, 219–234. [DOI] [PubMed] [Google Scholar]
- Muday, G.K., and DeLong, A. (2001). Polar auxin transport: Controlling where and how much. Trends Plant Sci. 6, 535–542. [DOI] [PubMed] [Google Scholar]
- Naderi, M., Caplan, A., and Berger, P.H. (1997). Phenotypic characterization of a tobacco mutant impaired in auxin polar transport. Plant Cell Rep. 17, 32–38. [DOI] [PubMed] [Google Scholar]
- Normanly, J., and Bartel, B. (1999). Redundancy as a way of life: IAA metabolism. Curr. Opin. Plant Biol. 2, 207–213. [DOI] [PubMed] [Google Scholar]
- Ono, E., Wong, H.L., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560. [DOI] [PubMed] [Google Scholar]
- Potikha, T.S., Collins, S.C., Johnson, D.I., Delmer, D.P., and Levine, A. (1999). The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 119, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck, G.K., Mattsson, J., Hardtke, C.S., Sung, Z.R., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Ren, X., and Schwartz, M.L. (2000). Determination of GTP loading on Rho. Methods Enzymol. 325, 264–272. [DOI] [PubMed] [Google Scholar]
- Roberts, C.J., et al. (2000). Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873–880. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., and Bartel, B. (2001). Auxin signaling: Derepression through regulated proteolysis. Dev. Cell 1, 595–604. [DOI] [PubMed] [Google Scholar]
- Romano, C.P., Hein, M.B., and Klee, H.J. (1991). Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev. 5, 438–446. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., and Takai, Y. (1998). The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem. Biophys. Res. Commun. 245, 641–645. [DOI] [PubMed] [Google Scholar]
- Shevell, D.E., Leu, W.-M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.-H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Shields, J.M., Pruitt, K., McFall, A., Shaub, A., and Der, C.J. (2000). Understanding Ras: ‘It ain’t over 'til it's over.' Trends Cell Biol. 10, 147–154. [DOI] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318. [DOI] [PubMed] [Google Scholar]
- Theologis, A., Huynth, T.A., and Davis, R.W. (1985). Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J. Mol. Biol. 183, 53–68. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, Z., Hagen, G., and Guilfoyle, T.J. (2001). Aux/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.-G., Young, J.C., Im, K.-H., Sussman, M.R., and Jones, A.M. (2001). Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292, 2066–2069. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Activation and repression of transcription by auxin-responsive factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valster, A.H., Hepler, P.K., and Chernoff, J. (2000). Plant GTPases: The Rhos in bloom. Trends Cell Biol. 10, 141–146. [DOI] [PubMed] [Google Scholar]
- Van Aelst, L., and D'Souza-Schorey, C. (1997). Rho GTPases and signaling networks. Genes Dev. 11, 2295–2322. [DOI] [PubMed] [Google Scholar]
- Vissenberg, K., Quelo, A.H., Van Gestel, K., Olyslaegers, G., and Vergelen, J.P. (2000). From hormone signal, via the cytoskeleton, to cell growth in single cells of tobacco. Cell Biol. Int. 24, 343–349. [DOI] [PubMed] [Google Scholar]
- Wang, Q.Y., and Nick, P. (1998). The auxin response of actin is altered in the rice mutant Yin-Yang. Protoplasma 204, 22–33. [DOI] [PubMed] [Google Scholar]
- Wang, X.-Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292, 2070–2072. [DOI] [PubMed] [Google Scholar]
- Winge, P., Brembu, T., and Bones, A.M. (1997). Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol. Biol. 35, 483–495. [DOI] [PubMed] [Google Scholar]
- Winge, P., Brembu, T., Kristensen, R., and Bones, A.M. (2000). Genetic structure and evolution of Rac-GTPases in Arabidopsis thaliana. Genetics 156, 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]
- Wu, G., Li, H., and Yang, Z. (2000). Arabidopsis RopGAPs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation. Plant Physiol. 124, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2002). Small GTPases: Versatile signaling switches in plants. Plant Cell 14 (suppl.), S375.–S388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaina, S., Reggiani, R., and Bertani, A. (1990). Preliminary evidence for involvement of GTP-binding protein(s) in auxin signal transduction in rice (Oryza sativa L.) coleoptile. J. Plant Physiol. 136, 653–658. [Google Scholar]