Abstract
The YABBY (YAB) genes specify abaxial cell fate in lateral organs in Arabidopsis. Loss-of-function mutants in two early-expressing YAB genes, FILAMENTOUS FLOWER (FIL) and YAB3, do not exhibit vegetative phenotypes as a result of redundancy. Mutations in these genes result in the derepression of the KNOX homeobox genes SHOOTMERISTEMLESS (STM), BREVIPEDICELLUS, and KNAT2 in the leaves and in the partial rescue of stm mutants. Here, we show that fil yab3 double mutants exhibit ectopic meristem formation on the adaxial surfaces of cotyledons and leaf blades. We propose that in addition to abaxial specification, lateral organ development requires YAB function to downregulate KNOTTED homeobox genes so that meristem initiation and growth are restricted to the apex.
INTRODUCTION
In seed plants, lateral organs are initiated from the peripheral regions of the shoot apical meristem (SAM), a dome of self-perpetuating and self-organizing cells. Genes of the KNOTTED1 homeodomain, or KNOX, family are required for the maintenance and growth of the SAM (Long et al., 1996; Kerstetter et al., 1997; Bowman and Eshed, 2000; Vollbrecht et al., 2000). Initiation of lateral organs from the apex coincides with the downregulation of KNOX genes as organ primordia develop. Subsequently, KNOX gene expression remains off as lateral organs differentiate, ensuring that these organs display determinate growth, as opposed to the indeterminate growth of the shoot apex.
Recently, it was demonstrated that the YAB family of abaxially expressed genes, which encode presumptive transcription factors with high-mobility group and zinc-finger domains, promote abaxial cell fates in the lateral organs of Arabidopsis (Eshed et al., 1999; Sawa et al., 1999; Siegfried et al., 1999; Villanueva et al., 1999; Bowman, 2000). One member of the YAB gene family, FILAMENTOUS FLOWER (FIL) (Chen et al., 1999; Kumaran et al., 1999; Sawa et al., 1999; Siegfried et al., 1999), is required for normal flower development. FIL is thought to act redundantly with YAB2 and YAB3 because of their overlapping expression pattern and sequence homology (Siegfried et al., 1999), consistent with the absence of aberrant vegetative phenotypes in fil loss-of-function mutants. Siegfried et al. (1999) reported the analysis of the fil-5 yab3-1 double mutant in which an enhanced floral phenotype and a vegetative phenotype were observed. However, the yab3-1 allele, in which a T-DNA insertion is located upstream of the YAB3 promoter, is not a null mutation; rather, it results in a low level of delocalized YAB3 expression. Thus, some functions of FIL and YAB3 may have been obscured.
Here, we report the isolation of a null allele of yab3 (yab3-2) and the generation of double-mutant plants with fil-8. We show that SHOOTMERISTEMLESS (STM), BREVIPEDICELLUS (BP; also known as KNAT1), and KNAT2, which are members of the KNOX gene family, are expressed ectopically in the leaves of fil-8 yab3-2 double mutants. Furthermore, the loss of YAB activity is able to compensate partially for the lack of meristematic activity in stm-1 mutants. In addition, ectopic vegetative and inflorescence meristems are produced on the margins of cotyledons, rosettes, and cauline leaves in fil-8 yab3-2 double mutants. These results suggest that FIL and YAB3 act, directly or indirectly, to downregulate meristematic genes during lateral organ development in wild-type plants in addition to promoting abaxial cell fate in the lateral organs.
RESULTS
Identification of yab3 Mutant Alleles
We reported previously the cloning of the Abnormal Floral Organs (AFO) gene, which is required for normal flower development in Arabidopsis (Kumaran et al., 1999). Because AFO is identical to FIL and YAB1, we renamed the afo-1 mutant allele fil-8. fil-8 results in partially radialized floral organs but has no effect on vegetative development, as is the case with other fil alleles. To address the possibility of overlapping functions of FIL with other YAB genes during vegetative development, we screened for more knockouts in this family from the Ds transposant insertion lines that we generated (Sundaresan et al., 1995; Parinov et al., 1999). Based on the flanking sequence analysis, we isolated two alleles of YAB3 called yab3-2 and yab3-3, which correspond to Ds insertions in the coding region (Figure 1). Both mutants are indistinguishable from wild-type plants under normal growth conditions. For this study, we used the yab3-2 allele, which carries a gene-trap Ds insertion in the first exon, resulting in undetectable levels of YAB3 transcript by reverse transcriptase–mediated (RT) PCR (data not shown) and in β-glucuronidase (GUS) reporter gene expression. In seedlings heterozygous for yab3-2, GUS expression was observed in the young leaves, and as the leaf matured, expression was restricted to the abaxial tissues of leaves (Figure 2A). Apart from the abaxial tissue expression pattern, we observed GUS expression on either side of the leaf margin in the younger tissues of leaf blades (Figure 2B). YAB3::GUS also had GUS expression in floral organs (data not shown). These data are consistent with the results of previous in situ hybridization studies (Siegfried et al., 1999).
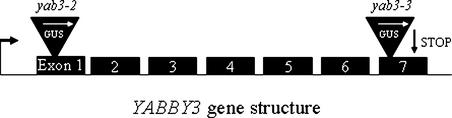
Figure 1.
Different Alleles of YAB3 Generated by Ds Transposable Elements.
The Ds gene-trap insertions in the yab3-2 and yab3-3 alleles are at nucleotide positions 191 and 755, respectively, with reference to the YAB3 mRNA sequence (ATG at 109).
Figure 2.
YAB3::GUS Expression in Heterozygotes, and Phenotype of fil-8 yab3-2 Plants.
(A) YAB3::GUS in a leaf showing abaxial expression. ab, abaxial surface; ad, adaxial surface.
(B) Strong YAB3::GUS expression at the leaf margins.
(C) Bifurcated leaf of a fil-8 yab3-2 seedling (arrow).
Generation and Phenotypic Analysis of Double-Mutant Plants for fil yab3
The single-mutant alleles of fil-8 and yab3-2 do not have any visible aberrant vegetative phenotype under normal growth conditions. They produce leaves that are similar to wild-type leaves in shape, size, and epidermal cell types on both adaxial and abaxial surfaces (data not shown). By contrast, fil-8 yab3-2 double-mutant plants show an abnormal vegetative phenotype, including thin, elongated cotyledons and partially radialized leaves, suggesting a partial loss of leaf polarity. In addition, leaves are sometimes bifurcated near the base (Figure 2C). The floral organs show stronger defects than in fil mutants and exhibit a much greater degree of radialization. These phenotypes are very similar to those of the fil-5 yab3-1 double mutants (Siegfried et al., 1999). We also examined the adaxial and abaxial cell types of fil-8 yab3-2 mutant leaves by scanning electron microscopy and found that the cells on the abaxial and adaxial leaf surfaces were clearly distinguishable, indicating that polarity had not been eliminated (data not shown).
The absence of abaxial trichomes on the first rosette leaves is a useful marker for the correct specification of abaxial cell types and the maintenance of leaf polarity. For example, seedlings homozygous for the kanadi1 mutation, which affects abaxial specification, have trichomes on the abaxial leaf surfaces of the first two rosette leaves (Kerstetter et al., 2001). We examined the number of trichomes on the abaxial leaf surfaces of wild-type, fil-8, yab3-2, and fil-8 yab3-2 seedlings. The first four rosette leaves were selected for analysis from 15 10-day-old seedlings from each mutant class. Neither the wild-type control nor any of the fil-8, yab3-2, and fil-8 yab3-2 leaves produced trichomes on their abaxial surfaces. Subsequently, all seedlings produced three to five abaxial trichomes on the fifth leaf, with none of the fil-8 yab3-2 seedlings producing more than three abaxial trichomes at this stage, perhaps as a result of their smaller leaves. These results indicate that abaxial polarity is not abolished in the early rosette leaves of the mutants.
Formation of Ectopic Meristems on the Lateral Organs of Double-Mutant Plants
A striking phenotype observed in fil-8 yab3-2 double-mutant plants was the production of ectopic vegetative and inflorescence meristems on the adaxial surfaces of the cotyledons and leaf blades (Figure 3). Ectopic meristem emergence on cotyledons and leaves occurred in 5 to 7% of double-mutant plants. Ectopic inflorescence meristems arose mainly from the rosette leaves (Figures 3A to 3E), with a few exceptions. This finding could be attributable to the fact that these double-mutant plants produced very few cauline leaves. The ectopic inflorescence meristems had mutant cauline leaves and flowers with the same phenotype as the primary inflorescence meristems of fil-8 yab3-2 plants (Figure 3A and data not shown).
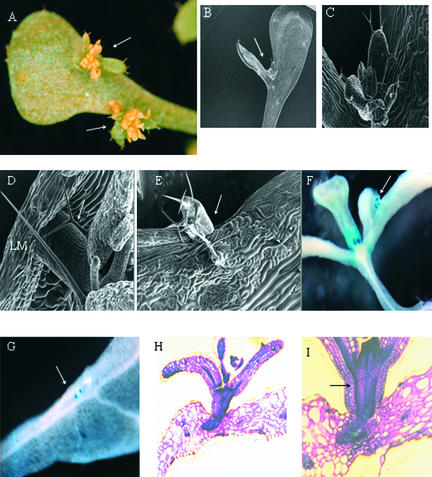
Figure 3.
Induction of Ectopic Meristems on the Lateral Organs of fil-8 yab3-2 Plants.
Arrows indicate the positions of ectopic meristems.
(A) Ectopic inflorescence meristems on both sides of the leaf margin.
(B) Ectopic meristem at the bifurcation point on a leaf.
(C) Higher magnification of the meristem shown in (B).
(D) Ectopic inflorescence meristem on a rosette leaf margin (LM).
(E) Emergence of an ectopic meristem on the adaxial surface of a rosette leaf.
(F) and (G) A YAB3::GUS marker was used to identify the emergence of ectopic meristems from cotyledon (F) and leaf (G).
(H) Section of a leaf that has ectopic meristems reveals that they emerge from the vasculature tissues.
(I) Differentiation of the procambium strand from the vein of a leaf producing ectopic meristems.
Even though the emergence of ectopic meristems can occur anywhere on the leaf blade, they developed most frequently toward the base and near the leaf margins. It is interesting that YAB3 expression also was more intense near the leaf margins (Figure 2B). Similar to the previous results with the fil-5 yab3-1 mutants (Siegfried et al., 1999), these double-mutant plants also produced bifurcated leaves and cotyledons, with the bifurcation often induced near the margins of the lateral organs. As noted by Siegfried et al. (1999), in some cases bifurcated leaves and cotyledons gave rise to ectopic meristems at the point of bifurcation (Figures 3B and 3F). Here, the YAB3::GUS fusion in the yab3-2 allele also served as a marker to identify the emerging lateral organs arising from ectopic meristems on cotyledons and leaves (Figures 3F and 3G). The ectopic meristems that we observed in fil yab3 plants arose only from the adaxial surface of the mutant leaves, consistent with the retention of some polarity in the leaves of fil yab3 plants. As seen in Figure 2, although the leaves appeared partially radialized, they retained leaf lamina, with distinguishable cell types on the leaf surfaces and trichomes only on the adaxial surface of the early rosette leaves.
The vasculature of the ectopic shoots arose from a single vein of the leaf, and the shoots appeared to be associated with the vasculature of the leaf of origin (Figure 3H). The leaves originating from the ectopic shoots had well-differentiated procambial strands that were linked with the vein of the leaf from which the ectopic meristem arose (Figure 3I). Therefore, it seems that a normal pattern of cell differentiation occurs within the tissues and organs derived from the ectopic meristems, despite their abnormal positions.
Derepression of KNOX Genes in fil-8 yab3-2 Plants
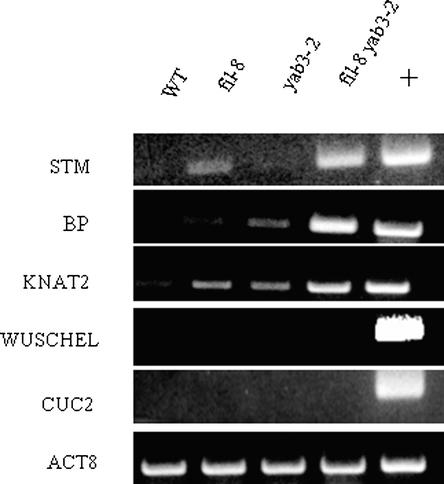
fil-8 yab3-2 plants appear to phenocopy plants overexpressing the KNOX homeobox genes in that ectopic meristems arise from the leaves. In transgenic lines that overexpress BP (known previously as KNAT1) using the 35S promoter, ectopic meristems emerge from the sinuses of severely lobed leaves (Chuck et al., 1996). To test for the misexpression of genes that are expressed normally only in meristems, we performed RT-PCR with primers specific for STM, BP, KNAT2, WUSCHEL (WUS), and CUP-SHAPED COTYLEDONS2 (CUC2) on mRNA isolated from lateral organs of fil-8, yab3-2, and fil-8 yab3-2 plants (Figure 4). Importantly, the leaves selected for these experiments had no detectable ectopic meristems. We observed the expression of STM in the leaves of double mutants but not in wild-type plants. Interestingly, STM leaf expression was detectable even in fil-8 and yab3-2 single-mutant plants that did not show any vegetative abnormality. Similarly, BP was expressed in leaves of the fil-8 and yab3-2 single mutants as well as in fil-8 yab3-2 double mutants, but not in wild-type plants. We detected low-level expression of KNAT2 in the leaves of wild-type plants, and an enhanced level of expression was detected in the leaves of both the fil-8 and yab3-2 single mutants as well as the fil-8 yab3-2 double mutants. We did not detect the expression of WUS and CUC2 in any of these genotypes, confirming that the leaves selected for analysis did not contain ectopic meristems.
Figure 4.
RT-PCR Analysis of Meristematic Genes in Mutant Leaves.
RT-PCR analysis using gene-specific primers for STM, BP, KNAT2, WUS, and CUC2 on RNA from wild-type (WT), fil-8, yab3-2, and fil-8 yab3-2 leaves and control wild-type whole seedlings (+). ACTIN8 (ACT8) primers were used as an internal control.
We also confirmed the derepression of KNOX gene expression by crossing to plants carrying a BP::GUS fusion. In a wild-type background, BP::GUS expression is detected in the meristematic zone and in lateral root primordia (Ori et al., 2000). As lateral organs emerged from the apical meristem, the expression of BP::GUS was downregulated (Figure 5A). Because the yab3-2 allele itself had reporter GUS activity, the fil-8 yab3-2 mutants could not be used in combination with BP::GUS to study the expression of the BP gene. Instead, we looked for BP::GUS expression in the emerging lateral organs of fil-8 mutant plants, because the RT-PCR results showed that they also misexpressed KNOX genes in the lateral organs. In most of the fil-8 seedlings, BP::GUS was detectable in emerging leaves and was especially prominent in the leaf blade margins, corresponding to the positions at which ectopic meristems most often arose in the fil-8 yab3-2 plants (Figures 5B and 5C), even though the fil-8 mutants had normal vegetative development. In some fil-8 seedlings, BP::GUS expression appeared to be restricted to the abaxial domains of the emerging leaves (Figure 5D).
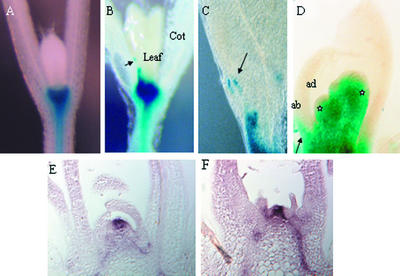
Figure 5.
Misexpression of KNOX Genes in yab Mutants.
(A) A wild-type seedling showing BP::GUS expression restricted to the meristematic regions and downregulated during lateral organ emergence.
(B) BP::GUS expression persists in a fil-8 mutant in the leaf blades near the margins. The arrow indicates GUS expression in the leaf margin.
(C) Enlarged view of the leaf shown in (B).
(D) BP::GUS expression is observed in the abaxial tissues of leaves in a fil seedling. The arrow indicates GUS expression in the abaxial tissue layer of a fil-8 seedling leaf. The asterisks indicate younger leaves emerging from the SAM. ab, abaxial surface; ad, adaxial surface.
(E) STM expression in wild-type plants is limited to the apical meristem and is excluded from leaf anlagen and primordia.
(F) STM expression in fil-8 yab3-2 plants is not detected from leaf anlagen, as in the wild type.
To further characterize the ectopic KNOX gene expression, we examined STM expression in fil-8 yab3-2 mutants during leaf initiation, because ASYMMETRIC LEAVES1 (AS1) expression at this stage can repress BP but not STM (Byrne et al., 2000; Ori et al., 2000). In wild-type plants, STM expression was excluded from leaf anlagen, being restricted to the apical meristem and developing vasculature (Figure 5E) (Long et al., 1996). Likewise, in fil-8 yab3-2 plants, STM expression was excluded from leaf anlagen and was restricted to the apical meristem (Figure 5F). Thus, either STM is downregulated properly at this stage, or if it is expressed ectopically, it is below the level of detection in our experiments.
Because AS1 shows an altered expression pattern in stm mutants (Byrne et al., 2000), we also tested for alterations in the YAB expression pattern in stm mutant plants using the YAB3::GUS insertion. However, we detected no differences in the YAB gene expression pattern in the cotyledons of stm mutants and wild-type plants (data not shown). We also tested, by RT-PCR, the levels of AS1 transcripts in fil yab3 leaves, and these appeared unchanged compared with those in wild-type leaves (data not shown).
Partial Suppression of the stm-1 Mutant Phenotype in yab Mutant Plants
In plants homozygous for stm-1, a strong but nonnull allele, a functional SAM, is not produced (Barton and Poethig, 1993). Thus, stm-1 homozygotes lack structures at the site typically occupied by the SAM (Figure 6A). Because fil-8 yab3-2 plants induce ectopic meristems that are correlated with the misexpression of class-1 KNOX genes, we tested for genetic interactions by crossing stm-1 to the fil-8 and fil-8 yab3-2 mutant backgrounds. Analysis of a segregating population from F2 progeny of stm-1/+ crossed with fil-8 yab3-2/fil-8 yab3-2 showed that the loss of SAM activity in the stm-1 mutant was suppressed partially by mutations in YAB genes. For example, leaves were produced from the shoot apex in fil-8 stm-1 plants (Figure 6B, Table 1). However, the morphology of leaves could be abnormal, because lobed and butterfly-shaped leaves were found frequently (Figures 6C and 6D). Vegetative shoots of fil-8 stm-1 plants were distinguishable from those of fil-8 plants in that the former exhibited reduced apical dominance, with the formation of multiple shoots with short internodes that initiated leaves, resulting in an abnormal arrangement of leaves. By contrast, stm-1 plants produced few leaves and no shoots. The fil-8 stm-1 plants failed to form flowers. In place of inflorescence meristems, leaf-like organs developed, indicating that a reduction in FIL activity alone could not rescue the stm-1 phenotype during flower formation, suggesting that the meristems that are initiated cannot be maintained (Figure 6E).
Figure 6.
Suppression of the stm-1 Phenotype in fil and fil yab3 Mutant Backgrounds.
(A) The stm-1 mutant fails to initiate leaves.
(B) stm-1 fil-8 double-mutant plants showing leaf proliferation but failure to produce flowers.
(C) and (D) Lobed (C) and butterfly-shaped (D) leaves of stm-1 fil-8 double-mutant plants.
(E) Proliferation of leaves from the apex of stm-1 fil-8 double-mutant plants.
(F) Scanning electron microscopy analysis showing the emergence of a leaf (arrow) from the meristem in a fil-8 yab3-2 stm-1 triple-mutant seedling.
(G) Formation of radialized leaves in fil yab3 stm triple mutants.
(H) The arrow indicates the filamentous-like organs from the inflorescence apex of fil yab3 stm triple mutants.
Table 1.
Number of Leaves in stm-1 Plants in the yabby Mutant Background
| Genotype | Number of Plants Analyzed |
Number of Plants Producing Leaves |
Number of Leaves after 5 Weeks |
|---|---|---|---|
| stm-1/stm-1 | 12 | 3 | 4 to 6 |
| fil-8 stm-1/fil-8 stm-1 | 10 | 10 | 18 to 24 |
| fil-8 yab3-2 stm-1/fil-8 yab3-2 stm-1 | 4 | 4 | 14 to 17 |
In fil-8 yab3-2 stm-1 triple-mutant plants, the stm-1 phenotype was suppressed to an even greater extent. A single leaf structure was produced initially at the site that normally is occupied by the SAM during germination, and leaf initiation continued such that a large number of leaves were formed before the plants flowered (Figure 6F, Table 1). However, the leaves of fil-8 yab3-2 stm-1 plants differed from those of fil-8 stm-1 plants in that they displayed the fil-8 yab3-2 phenotype, and the leaves produced later often were radialized (Figure 6G). The triple-mutant plants also exhibited reduced apical dominance and excess leaf proliferation, as was observed in fil-8 stm-1 plants. Eventually, flower-like organs developed, indicating that the loss of FIL and YAB3 was able to partially rescue the stm phenotype during flower formation (Figure 6H). The differences in the degree of rescue of the reproductive phenotype may reflect differences in the levels of ectopic KNOX gene expression.
DISCUSSION
Derepression of KNOX Genes Is Not a Direct Consequence of the Loss of Dorsoventral Polarity
Previous studies have demonstrated a correlation between adaxialization and the promotion of meristematic growth. For example, the phabulosa (phab) mutation, which results in completely adaxialized leaves, leads to new meristems arising from any position on the leaf axil (McConnell and Barton, 1998; McConnell et al., 2001). Conversely, the phantastica (phan) mutant of Antirrhinum, which results in the complete abaxialization of leaves, also can result in meristem arrest (Waites and Hudson, 1995; Waites et al., 1998). Similarly, the ectopic expression of FIL or YAB3 produces seedlings with abaxialized leaves and arrested meristems (Sawa et al., 1999; Siegfried et al., 1999). Therefore, the ectopic meristems observed in fil yab3 mutants could be interpreted as a consequence of the adaxialization of the lateral organs caused by the loss of genes conferring abaxial cell identity, resulting in the promotion of meristem formation via KNOX gene derepression. However, we do not favor this interpretation, because mutations such as phab and kanadi, which result in more extreme adaxialization than was observed in the fil yab3 double mutants, do not produce ectopic meristems from leaf lamina (McConnell and Barton, 1998; Eshed et al., 2001; Kerstetter et al., 2001). By contrast, the degree of adaxialization observed in fil yab3 double mutants was only partial, as seen in Figure 2. The partial retention of polarity can be explained by the activity of other genes that specify abaxial cell identity, such as YAB2, YAB5, and members of the KANADI gene family, which are still active in these plants. The restriction of the ectopic meristems to the adaxial surface may reflect a requirement for adaxial-specific factors that promote meristematic growth, possibly those encoded by class III homeodomain-zipper genes (e.g., PHB) that are expressed adaxially (McConnell et al., 2001).
Therefore, we propose that FIL/YAB genes may act as regulators of KNOX genes independently of their role in abaxial tissue fate. Consistent with this interpretation is our observation that KNOX gene derepression was present even in fil single mutants (Figures 3 and 4), in which no abnormal vegetative phenotypes were observed. With this interpretation, the ectopic meristems seen in the fil yab3 double mutants are more likely to result from further derepression of KNOX genes than from adaxialization of the lateral organs. Conversely, it is possible that the downregulation of KNOX genes in the shoot meristem could be partially responsible for the arrest of meristem growth observed in plants that overexpress FIL or YAB3 (Sawa et al., 1999; Siegfried et al., 1999).
Both as1 and fil yab3 can partially suppress the apical defect in stm mutants. In the case of as1 (Byrne et al., 2000), this effect is mediated through the ectopic expression of BP, because apical meristems fail to form in as1 stm bp triple mutants (Byrne et al., 2002). Because BP also is expressed ectopically in fil yab3 leaves, the partial rescue of meristem defects in the fil yab3 stm background is likely to be mediated by BP. Because the KNOTTED homeodomain proteins have been shown to move through plasmodesmata (Lucas et al., 1995), a possible mechanism for the suppression of the stm mutation is the misexpression of BP in leaf primordia, which results in the transport of BP proteins into the shoot meristem to rescue the stm phenotype.
YAB Repression of KNOX Genes May Act by a Mechanism Separate from That of the PHAN Myb Genes
Three orthologous Myb genes from different plant species, called AS1, PHAN, and ROUGH SHEATH2 (RS2) in Arabidopsis, Antirrhinum, and maize, respectively, have been shown to downregulate KNOX genes in lateral organs (Schneeberger et al., 1998; Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000). Even though mutations in these genes result in the misexpression of KNOX genes in the lateral organs, the phenotypes that they exhibit are very different for each species. The phan mutation gives rise to radialized leaves in Antirrhinum, rs2 mutants are defective in proximal-distal polarity in maize, and as1 mutants produce lobed leaves in Arabidopsis.
In Arabidopsis, AS1 represses BP and KNAT2, but not STM, during leaf development (Byrne et al., 2000; Ori et al., 2000). The loss of as1 function can partially phenocopy the BP overexpression phenotype, but only in the background of either serrate (se) or pickle (pkl) mutants (Ori et al., 2000). PKL (also called GYMNOS) and SE appear to encode chromatin-remodeling factors (Eshed et al., 1999; Ogas et al., 1999; Prigge and Wagner, 2001). Similar results have been obtained with the as2 mutation (Ori et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002). However, in fil yab3 plants, enhanced levels of KNOX genes are sufficient to induce meristem activity in the leaves despite the presence of wild-type PKL and SE gene products, perhaps because STM also is derepressed in this background.
In contrast to maize, Arabidopsis, and Antirrhinum, in tomato, which forms compound leaves, PHAN alone is insufficient to repress KNOX gene expression as a result of overlapping domains of expression (Koltai and Bird, 2000). Because the observed KNOX gene expression is restricted to the adaxial region, abaxial-specific factors may be more important for the downregulation of KNOX genes in tomato. It is interesting that BP::GUS was misexpressed in the abaxial tissues of fil-8 plants, indicating the role played by YAB genes in the repression of KNOX genes in the abaxial tissues of the lateral organs. The complementary nature of KNOX and AS1 gene expression may not be maintained in Arabidopsis in all contexts. For example, levels of AS1 transcript appeared unchanged in fil yab3 leaves despite the ectopic expression of STM in these tissues. This finding suggests that AS1 is not likely to function downstream of YAB genes and that the repression of AS1 by STM may require other factors that are restricted to the apical meristems.
Although KNOX gene overexpression phenotypes are observed in both as1 pkl and fil yab3 plants, their leaf phenotypes differ in many respects. Some differences could be attributed to the differences in the levels of expression of specific KNOX genes (e.g., STM), although most are more likely attributable to the other roles that these genes play in leaf development. For instance, unlike as1 pkl plants, leaf blades do not display serration in fil yab3 plants, and the positions and frequencies of ectopic meristems vary between the two genotypes. In addition, the formation of ectopic stipules is prominent in as1 pkl and BP-overexpressing plants, but it is not observed in fil yab3 plants, perhaps because fil yab3 mutants do not produce stipules, even on leaves initiated at the SAM (Siegfried et al., 1999). Finally, the phenotype observed in the as2-14 fil-8 yab3-2 triple mutant appears to be additive, which is consistent with the independent operation of the AS and YAB pathways (Y. Eshed and J.L. Bowman, unpublished results).
FIL/YAB3 Have Two Distinct Functions in Promoting Lateral Organ Development
The region of the leaf at which ectopic meristems arise in fil yab3 plants appears to correlate with the YAB3::GUS expression pattern. Most of these meristems originate near leaf margins, where YAB3::GUS is expressed in heterozygous plants. In general, leaf margins remain densely cytoplasmic for a longer time than the cells in the center of the leaf and are the last to differentiate. In addition, putative marginal meristems have been proposed to be important for lamina outgrowth in many species (Hagemann and Gleissberg, 1996; Donnelly et al., 1999). In the absence of FIL and YAB3 function, KNOX gene activity may be derepressed in the marginal regions, leading to the formation of ectopic shoot meristems. Waites and Hudson (1995) have proposed that the juxtaposition of abaxial and adaxial cell fate is required for the laminar outgrowth of the leaves, consistent with the phenotypes of the phan and phab mutants. In the context of this model, we may link the proposed two functions of FIL and YAB3 (i.e., the promotion of abaxial cell fate and the repression of KNOX genes) as reflecting dual functions required for proper leaf outgrowth. Thus, the outgrowth of the Arabidopsis leaf blade, and by extension that of other lateral organs, requires not only the proper specification of abaxial and adaxial cell identities through the action of the PHAB, YAB, and KANADI genes but also the action of the YAB genes to repress KNOX genes to allow the maintenance of dividing cells at the margins that will not revert to stem cells.
METHODS
Generation of the yab3 Mutant Allele and Genetic Analysis
The afo-1/fil-8 mutant (Kumaran et al., 1999) and the yab3 mutants used in the study are in the Arabidopsis thaliana ecotype Landsberg erecta. All plants were grown on soil at 22°C under long-day conditions. The mutations in the yab-3 alleles (Figure 1) were identified by homology search of the YAB3 gene sequence with the flanking sequences of Ds in the transposants collection that we published previously (Parinov et al., 1999). fil-8 yab3-2 double-mutant plants were generated by crossing fil-8 as a female plant with pollen from yab3-2 plants.
The stm-1 mutation used in this study also is in the Landsberg erecta ecotype (Barton and Poethig, 1993). The fil-8 stm-1 double-mutant plants were identified in F2 progeny segregating for both mutants by phenotypic and PCR analysis. Among 200 plants tested, 8 were confirmed by PCR as homozygous for fil-8 and stm-1. Crossing fil-8 yab3-2 flowers with pollen from stm-1 heterozygous plants was used to generate triple mutants of fil-8 yab3-2 stm-1. Of 320 F2 plants that were examined, 21, 31, 14, 13, and 4 plants showed stm-1, fil-8, fil-8 yab-2, fil-8 stm-1, and fil-8 yab3-2 stm-1 phenotypes, respectively; the genotypes of the last two classes were confirmed by PCR.
Reverse Transcriptase–Mediated PCR Analysis
Total RNA from cotyledons and leaves of the wild type, single fil-8 and yab3-2 mutants, and the fil-8 yab3-2 double mutant was isolated using the Oligotex mRNA midi kit (Qiagen, Valencia, CA). Reverse transcriptase–mediated PCR was performed on an RNA template using a one-step reverse transcriptase–mediated PCR kit supplied by Qiagen. The primers used for amplification were as follows: STM, 5′-TGTCAGAAGGTTGGAGCACCA-3′ and 5′-TTTGTTGCTCCGAAG-GGTAA-3′; BP, 5′-TCATGGAAGCATACTGTGACA-3′ and 5′-TGA-CTCAGAAGGATATGGCCA-3′; KNAT2, 5′-GATTGCCAAAAGGTG-GGAGC-3′ and 5′-TGTCGCCTTCAGTAGGGTA-3′; ACTIN8, 5′-ATG-AAGATTAAGGTCGTGGCA-3′ and 5′-CCGAGTTTGAAGAGGCTAC-3′; CUC2, 5′-CAGCCAATATCTTCCACCGGG-3′ and 5′-GGAGAGGTG-GGAGTGAGACGGA-3′; and WUS, 5′-CAGTCTTGTTCCATAGAT-CC-3′ and 5′-ATCATCATCATCAAGCCGA-3′.
Microscopy
Scanning Electron Microscopy
Fresh material was mounted on silver tape and viewed using a JOEL JSM-5310 LV microscope, and images were captured using JOEL SEMafore software (JOEL, Sundbyberg, Sweden). Further processing was performed using Adobe Photoshop 3.0 (Adobe Systems, Mountain View, CA).
Light Microscopy
Leaves having ectopic meristems were fixed in formaldehyde (5%), acetic acid (5%), and alcohol (EtOH 90%) under vacuum for 20 min, dehydrated in an ethanol series to 95%, and embedded in resin medium. Sections were cut at 5 μm with glass knives on a rotary microtome, mounted on slides, and stained with periodic acid–Schiff reagent and 0.1% toluidine blue.
In situ hybridizations were performed as described by Eshed et al. (1999) with the STM probe as described by Long et al. (1996).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Number
The accession number for the YAB3 mRNA sequence is AF136540.
Acknowledgments
We thank Yuval Eshed, Megan Griffith, and Yang Wei-Cai for their valuable discussions and for a critical reading of the manuscript. We are grateful to S. Hake for providing the BP::GUS transgenic seeds. We also are thankful to D. Ye, S. Mayalagu, and S. Parinov for sharing unpublished materials and results and to C.W. Lin for the scanning electron microscopy analysis. This work was supported by grants from the National Science and Technology Board of Singapore (M.K.K. and V.S.) and the U.S. National Science Foundation (J.L.B.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004911.
References
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- Bowman, J.L. (2000). The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3, 17–22. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., and Eshed, Y. (2000). Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 5, 110–115. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Ross, B., Mark, C., Arroya, J.M., Maitreya, D., Hudson, A., and Martienssen, R.A. (2000). asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorwski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chen, Q., Atkinson, A., Otsuga, D., Christensen, T., Reynolds, L., and Drews, G.N. (1999). The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126, 2715–2726. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251.–1260. [DOI] [PubMed] [Google Scholar]
- Hagemann, W., and Gleissberg, S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199, 121–152. [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467.–478. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollmann, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Laudencia-Chingcuanco, D., Smith, L.G., and Hake, S. (1997). Loss-of-function mutations in the maize homeobox gene knotted1 are defective in shoot meristem maintenance. Development 124, 3045–3054. [DOI] [PubMed] [Google Scholar]
- Koltai, H., and Bird, D.M. (2000). Epistatic repression of PHANTASTICA and class 1 KNOTTED genes is uncoupled in tomato. Plant J. 22, 455–459. [DOI] [PubMed] [Google Scholar]
- Kumaran, M.K., Ye, D., Yang, W.C., Griffith, M.E., Chaudhury, A.M., and Sundaresan, V. (1999). Molecular cloning of ABNORMAL FLORAL ORGANS: A gene required for flower development in Arabidopsis. Sex. Plant Reprod. 12, 118–122. [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the SHOOTMERISTEMLESS gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1943–1944. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodelling factor that regulates transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms of Knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Parinov, S., Mayalagu, S., Ye, D., Yang, W.C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from Dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., and Wagner, D.R. (2001). The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E.M., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger, R., Tsiantis, M., Freeling, M., and Langdale, J.A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125, 2857–2865. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Timmermans, M.C., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284, 151–153. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeberger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize ROUGH SHEATH2 gene and leaf development programs in monocot and dicot plants. Science 284, 154–156. [DOI] [PubMed] [Google Scholar]
- Villanueva, J.M., Broadhvest, J., Hauser, B.A., Meister, R.J., Schneitz, K., and Gasser, C.S. (1999). INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 23, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht, E., Reiser, L., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172. [DOI] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). PHANTASTICA: A gene required for dorsoventrality in leaves of Antirrhinum majus. Development 121, 2143–2154. [Google Scholar]
- Waites, R., Selvadurai, R.N., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. [DOI] [PubMed] [Google Scholar]