Abstract
A global analysis of gene expression events during shoot development in Arabidopsis was conducted using oligonucleotide array analysis. Shoots can be induced in tissue culture by preincubating root explants on an auxin-rich callus induction medium (CIM) and by transferring explants to a cytokinin-rich shoot induction medium (SIM), during which time explants become committed to shoot formation and ultimately form shoots. Oligonucleotide array data obtained during shoot development from ∼8000 Arabidopsis genes were subjected to principal component analysis, which demonstrated that the major components of variation in gene expression during shoot development can be represented by groups of genes, each group of which is upregulated at only one developmental stage. Two percent to three percent of the ∼8000 Arabidopsis genes monitored in this study were upregulated by fourfold or more at any one stage during shoot development. When upregulated and downregulated genes were categorized by function, it was observed that numerous hormone response genes were upregulated during preincubation on CIM. Groups of genes involved in signaling and/or transcription were induced at or before the time of shoot commitment, and genes that encode components of the photosynthetic apparatus were upregulated later in development before shoot emergence. Primary hormone response genes, such as Aux/IAA genes, were upregulated during preincubation on auxin-rich CIM, and cytokinin-responsive response regulator genes were upregulated during incubation on cytokinin-rich SIM. The expression of ARABIDOPSIS RESPONSE REGULATOR5, a type-A response regulator gene, was upregulated at the time of shoot commitment, and its expression was localized to sites of presumptive shoot formation. Two “hybrid” His kinases involved in cytokinin responses, CRE1, which encodes a cytokinin receptor, and CKI1, a gene that is capable of conferring cytokinin-independent shoot development, were upregulated during incubation on SIM.
INTRODUCTION
The power of plant hormones to promote development in tissue culture has amazed and baffled plant scientists for years. In pioneering experiments, Skoog and Miller (1957) demonstrated that the developmental fate of regenerating tobacco pith cells in tissue culture could be directed by the balance of cytokinin and auxin added to the growth medium: high cytokinin/auxin ratios promoted shoot formation, and low ratios favored the formation of roots. To this day, the findings of Skoog and Miller remain largely unexplained, as do the mechanisms by which cytokinin and auxin promote shoot or root development.
The regeneration of organs from explanted vegetative tissue in culture is a form of organogenesis or adventitious shoot and root development. Regeneration in various species can be classified by whether organogenesis is direct or indirect (Hicks, 1994; Bhojwani and Razdan, 1996). In direct organogenesis, explants respond to inductive hormones and do not require previous callus growth, as is the case in shoot organogenesis from detached pine cotyledons (Flinn et al., 1988; Thorpe, 1993). Shoot or root organogenesis from Convolvulus leaf explants or tobacco pith, on the other hand, requires callus production before shoot or root induction and so is an example of indirect organogenesis (Thorpe, 1993). Typically, procedures for the production of shoots from root and or hypocotyl explants in Arabidopsis involve indirect organogenesis (Valvekens et al., 1988). Explants are preincubated on an auxin-rich callus induction medium (CIM) and then are transferred to a cytokinin-rich shoot induction medium (SIM) for shoot formation.
The hormone requirements for shoot regeneration in tissue culture have been investigated extensively in various species; however, the developmental events that occur during regeneration and the mechanisms by which the hormones promote shoot development are less well known. Recently, it was found that genes related to those encoding components of the cytokinin signaling pathway influence the formation of shoots (Hwang and Sheen, 2001). Cytokinin signaling is mediated by a “two-component” phosphorelay transfer system, which in Arabidopsis is composed of sensor His kinases (AHKs), His-containing phosphotransmitters, and response regulators (ARRs). The AHKs are “hybrid” His kinases composed of His kinase and receiver domains and, in some cases, an extra receiver-like domain.
CRE1/AHK4/WOL (CRE1) encodes a sensor His kinase that was the first to be reported as a cytokinin receptor (Inoue et al., 2001; Yamada et al., 2001). The locus was identified in Arabidopsis through a loss-of-function mutation (cre1-1) that required higher levels of exogenous cytokinin for shoot regeneration (Inoue et al., 2001). Inoue et al. (2001) demonstrated that CRE1 complements a His kinase defect in the budding yeast Saccharomyces cerevisiae in the presence of cytokinin and that the same cre1-1 mutation (a missense mutation in the His kinase domain) inactivated the complementing activity. Similarly, Suzuki et al. (2001) demonstrated that the same gene (which they called AHK4) functions as a cytokinin receptor in Schizosaccharomyces pombe. CRE1 is related to CYTOKININ INDEPENDENTI (CKI1), AHK2, and AHK3 (Ueguchi et al., 2001a). AHK2 and AHK3 also appear to encode cytokinin receptors, although the case is not as clear as it is for CRE1. Cotransfection of Arabidopsis protoplasts with AHK2 or AHK3 and ARR6 constructs activates ARR6 expression in the presence of cytokinin (Hwang and Sheen, 2001), although AHK2 fails to complement His kinase defects in yeast (Ueguchi et al., 2001a). AHK3 complements these defects in yeast, but it can do so in the absence of added cytokinin (Ueguchi et al., 2001a). However, in fission yeast and Escherichia coli, AHK2 and AHK3 complement two-component system mutations in a cytokinin-dependent manner (Suzuki et al., 2001; Ueguchi et al., 2001b; Yamada et al., 2001). CKI1 also encodes a hybrid His kinase and was once a candidate for a cytokinin receptor (Kakimoto, 1996). This idea emerged from activational tagging experiments in Arabidopsis in which Kakimoto (1996) showed that overexpression of CKI1 overcame the requirement for cytokinin in shoot development. However, cytokinin does not enhance CKI1 activity in an Arabidopsis protoplast system (Hwang and Sheen, 2001), except at high cytokinin concentrations (I. Hwang, personal communication).
The effectors in the cytokinin signaling pathway in Arabidopsis are ARRs. The Arabidopsis genome encodes >30 ARRs or ARR-like proteins (Hwang et al., 2002), and they can be classified in three subgroups: type B, containing a myb-like domain; type A without the domain (Imamura et al., 1999); and pseudo-RRs, which have a phospho-accepting receiver-like domain but lack the critical phospho-accepting Asp site (Makino et al., 2000).
Some of the type-B ARRs function as transcriptional activators of cytokinin-induced gene expression, whereas the type-A ARRs may serve as repressors (Hwang and Sheen, 2001). Some of the type-A ARRs are “primary response genes” that can be induced rapidly by cytokinin in Arabidopsis (Brandstatter and Kieber, 1998; Imamura et al., 1998), and the induction of ARR5 is controlled, at least in part, transcriptionally (D'Agostino et al., 2000). The transcription of ARR5 and other type-A ARRs can be transactivated in an Arabidopsis protoplast system by type-B ARRs (Sakai et al., 2000, 2001; Hwang and Sheen, 2001). For example, coexpression of ARR1, ARR2, and ARR10 activated ARR6 expression, and in the presence of cytokinin, the effect of ARR2 was most dramatic, enhancing ARR6 expression by >1000-fold (Hwang and Sheen, 2001). Sakai et al. (2001) engineered transgenic Arabidopsis to express ARR1 under the control of a steroid-inducible promoter and demonstrated enhanced expression of ARR6 in seedlings after treatment with the inducer dexamethasone. The gene products of the type-A, ARR4, ARR5, ARR6, and ARR7, inhibit transcription and act as negative feedback inhibitors of cytokinin-induced gene expression (Hwang and Sheen, 2001; Sheen, 2002).
Auxin also plays a critical role in shoot development in tissue culture, serving as the principal hormone in CIM to promote callus formation and, seemingly, as a counterpart to cytokinin in SIM. It is known that auxin activates the expression of three major families of primary response genes: the Aux/IAA (auxin/indoleacetic acid) genes, the GH3 gene family, and the SAUR gene family (Abel and Theologis, 1996). The Aux/IAA genes are the best characterized, encoding short-lived nuclear proteins that are capable of homodimerization and heterodimerization with themselves and with members of the ARF gene family (Kim et al., 1997; Ulmasov et al., 1997; Guilfoyle et al., 1998). The Aux/IAA proteins appear to act as repressors of ARF function (Ulmasov et al., 1997; Tiwari et al., 2001), and their repression is countermanded by the protein degradation machinery activated by auxin (Dharmasiri and Estelle, 2002). The Aux/IAA gene family in Arabidopsis consists of ∼30 members that play a variety of roles in auxin signaling and plant development (Liscum and Reed, 2002). A number of auxin mutants, such as the semidominant mutant axr3-1, have been mapped to loci encoding Aux/IAA proteins (Rouse et al., 1998). axr3-1 does not interfere with the dimerization function or nuclear localization of the protein, but it dramatically increases the stability of this rapidly turned-over protein (Ouellet et al., 2001). The phenotypes of axr3-1—enhanced apical dominance, reduced root elongation, increased adventitious rooting, no root gravitropism, and ectopic expression from the SAUR-AC1 promoter—suggest an increased auxin response (Leyser et al., 1996). The mutant phenotype can be reversed partially by cytokinin, a treatment that could normalize the balance between auxin and cytokinin responses (Leyser et al., 1996).
The production of shoots from roots must involve considerable genetic reprogramming. The magnitude of gene expression changes during the regeneration of shoots from roots can be estimated from microarray analyses, such as the study by Ruan et al. (1998) that compared gene expression patterns between roots and shoots (leaves) in mature Arabidopsis. Of 1443 cDNAs in their analysis, 34% were expressed differentially in shoots and roots; 443 were expressed at least twofold higher in roots than in leaves, whereas 57 were expressed at least twofold higher in leaves than in roots. A similar study was conducted in Arabidopsis by Zhu et al. (2001) using oligonucleotide microarray analysis. Of ∼8300 genes in their study, 94 were expressed uniquely in leaves and 64 were expressed uniquely in roots. Of the genes expressed preferentially in leaves, most were metabolic or unknown, and of those expressed preferentially in roots, most were involved in defense or were unknown.
Here, we describe the global program of gene expression during shoot development and the changes in expression of genes involved in cytokinin and auxin signaling. In doing so, we identify some important trends in large-scale gene expression patterns and genes that may be key regulators of this process.
RESULTS
Global Gene Expression Patterns
The developmental events involved in shoot formation can be studied during regeneration of shoots from root explants in Arabidopsis tissue culture. To do so, root explants were preincubated on CIM for 4 days and then transferred to SIM for 14 or 15 days. Under these conditions, explants acquire competence to respond to shoot formation signals during preincubation on CIM (Cary et al., 2001). On transfer to SIM, explants become committed to shoot formation after ∼6 days on SIM, and shoots begin to emerge 6 to 8 days later (12 to 14 days on SIM; Cary et al., 2002).
To describe the program of gene expression that underlies shoot development, a global analysis of gene expression was undertaken using oligonucleotide array analysis. Root segments were explanted in bulk from Arabidopsis seedlings and harvested for RNA extraction at seven different time points (time 0, 2 and 4 days on CIM, and 3, 6, 10, and 15 days on SIM). Total RNA was used to generate cDNA probes and hybridized to Affymetrix Arabidopsis GeneChips.
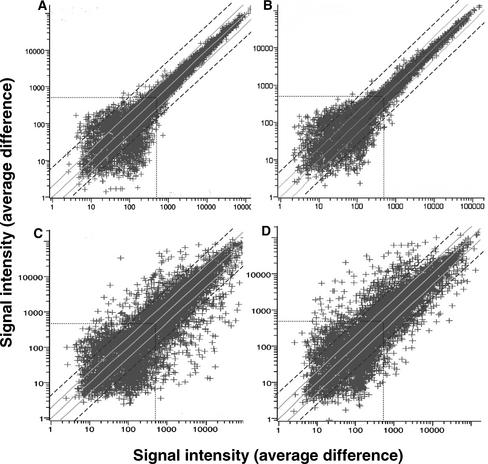
Gene expression changes at different time points in shoot development were compared using scatterplots. The error variation from chip to chip was assessed in scatterplots by hybridizing the time-0 probe to two different chips (Figure 1A). The variation resulting from experimental error from chip to chip increased with decreasing signal intensity. We eliminated from consideration gene expression profiles with maximum signal intensities of <500 (Figure 1A, dotted lines), because signals below that threshold frequently varied by more than twofold (Figure 1A, solid gray lines.) Thus, by focusing on genes with a greater than fourfold change in gene expression and with a maximum signal intensity of >500, we eliminated most false-positive results.
Figure 1.
Scatterplots Comparing Gene Expression Levels of ∼8200 Arabidopsis Genes at Different Times during Shoot Development.
Gene expression levels (represented by signal intensities or average differences) were analyzed by Affymetrix Arabidopsis oligonucleotide arrays. Solid gray lines represent twofold differences in signal intensity (gene expression) from the diagonal (same signal intensity in both hybridizations), and dashed black lines represent fourfold differences in signal intensity. Dotted lines represent the arbitrary signal intensity threshold of 500.
(A) Control scatterplot used to assess chip-to-chip error variation by comparing time 0 (chip 1, x axis) with time 0 (chip 2, y axis). In this control experiment, the same probe was hybridized to two different chips.
(B) Additional control scatterplot to assess experiment-to-experiment error variation by comparing 3 days on SIM (experiment 1, x axis) with 3 days on SIM (experiment 2, y axis). In this control experiment, two different probes derived from different experiments conducted several months apart were hybridized to two different chips.
(C) and (D) Scatterplots comparing time 0 (chip 1, x axis) with 2 days on CIM (y axis) (C) and time 0 (chip 1, x axis) with 15 days on SIM (y axis) (D).
Error variation arising from tissue culture procedures was reduced by replicating and pooling samples within a time point. Root segments were explanted onto 10 to 20 Petri plates. (More plates and root segments were required for earlier time points to obtain 1 g fresh weight of material.) The root segments on each plate were maintained as separate groups during transfer from CIM to SIM plates and were pooled with samples from other plates before RNA extraction. Error variation arising from time point to time point was estimated by comparing the expression of ubiquitin genes at different time points. For ubiquitin 4 (At5g20620), the mean signal intensity and standard error was 6378 ± 338 (±5.2% of the mean). For ubiquitin 11 (At4g05320), there are three sets of oligonucleotides on the Affymetrix 8000 GeneChips. The standard errors for the mean of determinations at various time points ranged from ±3.3 to ±4.9% of the mean. Because we chose to study genes that are upregulated or downregulated by fourfold or more (Figure 1A, dashed lines), the error variation from extraction to extraction appears to lie well within the experimental variation for most genes of interest with signal intensities of >500.
Error variation from experiment to experiment was estimated by conducting another independent experiment (several months later) and comparing one of the time points (3 days on SIM) between the two experiments. (This time point was chosen because, as described below, there are interesting changes in the expression of genes that encode signaling components and transcription factors at this stage.) Scatterplots comparing the data at the single time point between the two experiments (Figure 1B) show no greater error variation than the chip-to-chip variation described above (Figure 1A).
Scatterplots were used to compare gene expression levels (signal intensity) at the first time point, 2 days on CIM (Figure 1C), and the last time point, 15 days on SIM (Figure 1D), with those at time 0 (time of explant). The scatterplots demonstrated that many genes were upregulated and downregulated (with respect to the time-0 control) at both the earliest and latest time points. After 2 days of incubation on CIM, 189 genes (2.3% of the total) were upregulated by more than fourfold (and had signal intensities of >500) compared with time 0. At 15 days of incubation on SIM, 257 genes (3.1% of the total) were upregulated by more than fourfold (and had signal intensities of >500) compared with time 0.
It was surprising that more genes were not upregulated as shoot development progressed. One might expect that if shoot development was the result of a gene cascade stimulated by hormonal signals, more genes might be upregulated with time. Some of this was seen during incubation on SIM. At 3 days on SIM, 141 genes (1.7% of the total) were upregulated by more than fourfold compared with time 0. The number of genes upregulated by more than fourfold approximately doubled at 15 days of incubation on SIM. However, one might expect greater numbers of genes to be upregulated in a gene cascade.
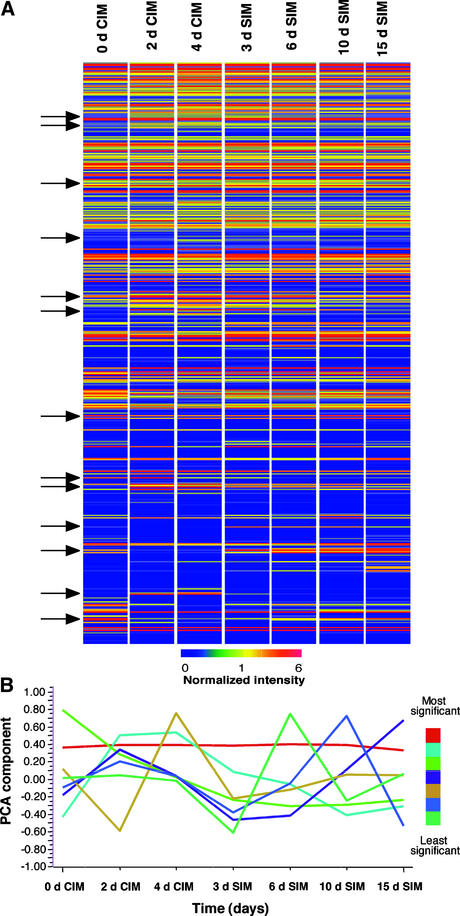
One reason why more genes are not upregulated by 15 days on SIM is that many genes are expressed transiently (i.e., their expression is stage specific). This can be seen in a hierarchical cluster of expression profiles grouped by pattern similarity (Figure 2A). Although not all ∼8200 genes could be resolved, the expression of most genes did not change significantly during shoot development. However, small groups of genes (Figure 2A, arrows) did show different patterns of accumulation, with some groups of genes turning on and then turning off again.
Figure 2.
Gene Expression Patterns during Shoot Development in Arabidopsis.
(A) Hierarchical cluster analysis of the expression patterns of ∼8200 genes (arrayed along the vertical axis) at different times during shoot development (horizontal axis). Normalized signal intensities are color coded according to the scale at bottom, and genes are grouped according to similarities in expression profiles. Arrows highlight groups of genes with changing expression patterns during development.
(B) Principal component analysis (PCA) of gene expression profiles. The seven profiles accounting for most of the variation in gene expression during the time course are ranked in order of significance (see color scale at right). Data were analyzed using GeneSpring software.
Principal Component Analysis of Gene Expression Patterns
Because the data sets in the scatterplots and cluster analysis were large, we attempted to reduce their dimensionality to seven or eight components using principal component analysis. Principal component analysis showed that the most significant component contributing to the variation in gene expression profiles was a pattern representing “no change” in gene expression (Figure 2B). The second most significant component was one that peaked at two time points, 2 and 4 days on CIM. However, the next most significant components were profiles that peaked at single time points.
To understand the gene expression changes that gave rise to these principal components, expression profiles were sorted into groups of genes with stage-specific patterns that peaked at different time points. The groups were defined by selecting genes with prototypic expression profiles peaking at each time point and using the profiles of these genes to search for other genes with similar profiles. The identification of some of the highly upregulated genes in each group gave a snapshot of the changes in the gene expression program at different developmental stages.
The gene AIR1A (At4g12550), which encodes the wall–plasma membrane disconnecting protein, was selected as the most highly upregulated gene with a profile peaking at 2 days on CIM (Table 1). This is a critical time because it is when root explants acquire nearly full competence to respond to shoot induction signals (Cary et al., 2001). The profile for AIR1A was used to identify other genes with similar profiles. The most highly upregulated genes in this class included a putative pathogenesis-related protein (At2g19970), β-fructosidase (At3g13790), and a peroxidase (At2g38390) (Table 1). Other genes with similar profiles encoding putative transcription factors or signaling components also were identified. They included genes encoding a zinc finger protein (At3g28210), a putative Ser/Thr protein kinase (At1g51170), and a RING zinc finger protein (At3g60220).
Table 1.
Genes with Stage-Specific Expression Patterns
| Stage | 2 Days on CIM | 4 Days on CIM | 3 Days on SIM | 6 Days on SIM | 10 Days on SIM | 15 Days on SIM |
|---|---|---|---|---|---|---|
| Genes with prototypic profiles | AIR1A wall/membrane disconnect protein (At4g12550) | Protodermal factor, PDF1 (At2g42840) |
Cold-regulated protein, KIN1 (At5g15960) | Putative expansin (At2g40610) | Hypothetical protein (At4g27730) | CAB protein CP29 (At5g01530) |
| High performers (most highly expressed) | Putative pathogenesis-related protein (At2g19970) | Nonspecific lipid transfer protein (At2g38540) |
Myrosinase-associated protein (At1g54000) | Aquaporin/MIP-like protein (At3g54820) | Peroxidase-like protein (At4g36430) | Hypothetical protein (At1g20620) |
| Hypothetical protein (At4g15610) | Gly-rich protein (At4g29020) |
Cold-regulated protein, KIN2 (At5g15970) | Putative pectate lyase (At4g24780) | Metallothionein 2b (At5g02380) |
Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit 3b (At5g38410) |
|
| β-Fructosidase (At3g13790) | Nonspecific lipid transfer protein (At2g38530) |
Glutathione S-transferase (At1g78380) |
Expansin (At1g69530) | Putative peroxidase (At2g18150) | Putative Gly-rich protein (At2g05520) | |
| Peroxidase (At2g38390) | Putative protein translocase (At2g37410) |
His kinase (AHK1) (At2g17820) | ARR5 response regulator (At3g48100) | Putative protein (At2g19120) | Calmodulin-like protein (At2g41100) | |
| Genes encoding transcription or signaling factors | Zinc finger protein (At3g28210) | Putative AP2 domain transcription factor (At4g23750) |
Transcription factor BBFa (At3g61850) |
CUP-SHAPED COTYLEDON2 (At5g53950) | Putative protein kinase (At2g32800) | Putative homeodomain transcription factor (At2g35940) |
| Putative Ser/Thr protein kinase (At1g51170) | Putative heat-shock transcription factor (At2g26150) |
Myb-like transcription factor (At5g60890) | Basic domain/Leu zipper transcription factor (At4g34590) | Myb-related transcription factor (At5g57620) | Putative RING-H2 zinc finger protein (At2g15580) | |
| RING-H2 zinc finger protein (At3g60220) | Homeodomain protein (At4g36740) |
PERIANTHIA transcription factor (At1g68640) | Putative protein kinase (At2g18470) | Myb-related transcription factor (At5g23000) | CCAAT binding transcription factor (At4g14540) |
The gene with a prototypical expression profile peaking at 4 days on CIM was PDF1 (At2g42840), which encodes protodermal factor 1 (Table 1). Four days on CIM also is a key time point because explants acquire full competence for shoot formation at this time and are poised to respond to shoot development signals when transferred to SIM (Cary et al., 2001). The high performers at this time point included two genes that encode nonspecific lipid transfer protein isoforms (At2g38540 and At2g38530) (Table 1). Among the putative transcription factors and signaling proteins with similar profiles were those encoding a putative AP2 domain transcription factor (At4g23750), a putative heat shock transcription factor (At2g26150), and a homeodomain-containing protein (At4g36740).
A gene that encodes a cold- and abscisic acid–inducible protein (At5g15960) was chosen as a highly expressed gene with a stage-specific pattern peaking at 3 days on SIM (Table 1). Curiously, other high performers with similar expression profiles were genes, such as KIN1 (At5g15960) and KIN2 (At5g15970), that also encode cold-regulated proteins (Table 1). Genes with similar expression profiles that encode putative transcription factors included the genes encoding transcription factor BBFa (At3g61850), a myb-like transcription factor (At5g60890), and a PERIANTHIA transcription factor (At1g68640).
A prototypic gene selected with an expression profile peaking at 6 days on SIM was an expansin gene (At2g40610) (Table 1). A profile similarity search revealed another expansin gene (At1g69530) as a highly upregulated gene at this stage, along with an aquaporin/MIP-like protein (At3g54820), a putative pectate lyase (At4g247880), and a type-A response regulator, ARR5 (At3g48100) (Table 1). (ARR5 will be discussed further below.) CUP-SHAPED COTYLEDON2 (At5g53950), a gene encoding a NAC domain–containing protein required for shoot meristem formation (Aida et al., 1999), also was upregulated with a similar expression profile, as was a gene encoding a basic domain/Leu zipper transcription factor (At4g34590).
A gene that encodes a hypothetical protein (At4g27730) was chosen as the gene with a prototypical profile peaking at 10 days on SIM (Table 1). This gene was used to cull others with similar profiles, and among the top performers were two genes that encode putative peroxidases (At4g36430 and At2g18150) and one that encodes a metallothionein (At5g02380) (Table 1). Interesting upregulated genes that encode transcription factors and signaling components included a putative protein kinase (At2g32800) and myb-related transcription factors (At5g57620 and At5g23000).
The final time point in our study was 15 days on SIM, a time when shoots emerge. The prototypic gene upregulated at this stage was one that encodes a chlorophyll a/b binding protein (At5g01530) (Table 1). The most highly induced genes with expression profiles similar to the cab gene included a gene encoding a small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (At5g38410) and one encoding a Gly-rich protein (At2g05520) (Table 1). Genes that encode a putative RING-H2 zinc finger protein (At2g15580) and two putative transcription factors, a homeodomain (At2g35940) and a CCAAT binding factor (At4g14540), also were upregulated at 15 days on SIM.
These and other gene expression profiles during shoot development can be seen at http://www.bioinformatics.iastate.edu/howell/.
Expression Changes of Genes Grouped by Function
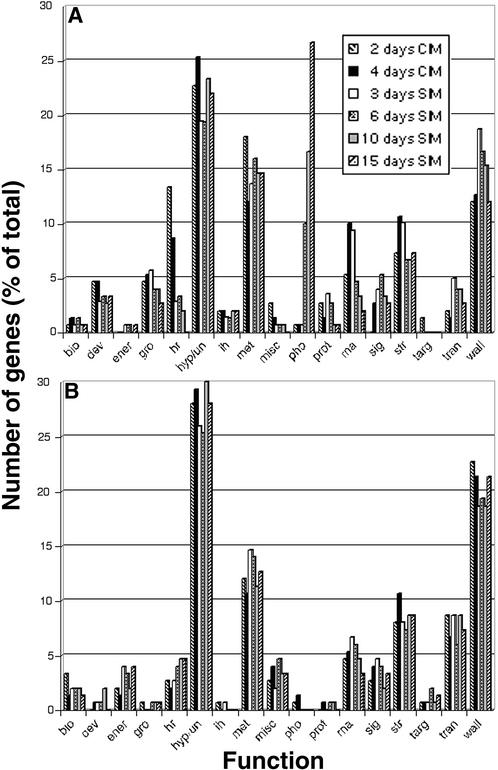
To assess global changes in expression patterns of genes related by function, we sorted the 150 most highly upregulated and downregulated genes into categories based on assigned function. These are genes that showed fourfold or more upregulation or downregulation compared with time 0. The selected upregulated genes had a signal intensity at the maximum time point of >500, and the downregulated genes had a signal intensity at time 0 of >500. This analysis differed from the analysis described above in that genes were not grouped by expression profiles that peaked at single stages. All upregulated or downregulated genes were considered whether they were expressed in a stage-specific manner or not.
Some interesting trends were observed during the time course of development among the upregulated genes. In particular, the number of genes that were involved in hormone responses increased dramatically at 2 days on CIM and decreased rapidly during shoot development (Figure 3A). Most of the hormone response genes in this group were Aux/IAA genes, and these genes will be described more fully below. Another interesting trend late in shoot development was the dramatic increase in genes that encode components of the photosynthetic apparatus (Figure 3A). As stated above, this was not unexpected because explants green and shoots emerge later during incubation on SIM. Another trend was the increase in the numbers of upregulated genes that encode transcription factors and signaling components during the transition from CIM (4 days on CIM) to SIM (3 days on SIM) (Figure 3A). This appears to be a time during shoot development when the gene regulatory machinery undergoes a major transition.
Figure 3.
Most Highly Upregulated and Downregulated Genes during Shoot Development Grouped According to Function.
(A) A total of 150 of the most highly upregulated genes (compared with time 0) at each developmental time point were categorized according to assigned function.
(B) A total of 150 of the most highly downregulated genes (compared with time 0) at each developmental time point were categorized according to assigned function.
Categories are as follows: bio, cell structure; dev, developmental; ener, energy metabolism; gro, cell growth and division; hr, hormone response; hyp/un, hypothetical or unknown; ih, ion homeostasis; met, intermediary metabolism; misc, miscellaneous; pho, photosynthesis; prot, protein synthesis and RNA binding proteins; rna, transcription or DNA binding proteins; sig, signal transduction; str, stress or pathogen response; targ, protein targeting; trans, transport; wall, cell wall metabolism.
Unlike the upregulated genes, there were almost no significant trends during shoot development among the downregulated genes (Figure 3B). In comparing the overall patterns of upregulated to downregulated genes, it is apparent that the downregulated genes are dominated by fewer categories of genes, particularly by the hypothetical/unknown genes and the genes encoding cell wall components or enzymes that are involved in cell wall metabolism. Cell wall metabolism genes are both upregulated and downregulated in greater numbers than most other categories of genes during shoot development (Figures 3A and 3B). It is of interest that the most highly downregulated genes were large numbers of genes encoding peroxidases.
Cytokinin-Related Gene Expression Patterns
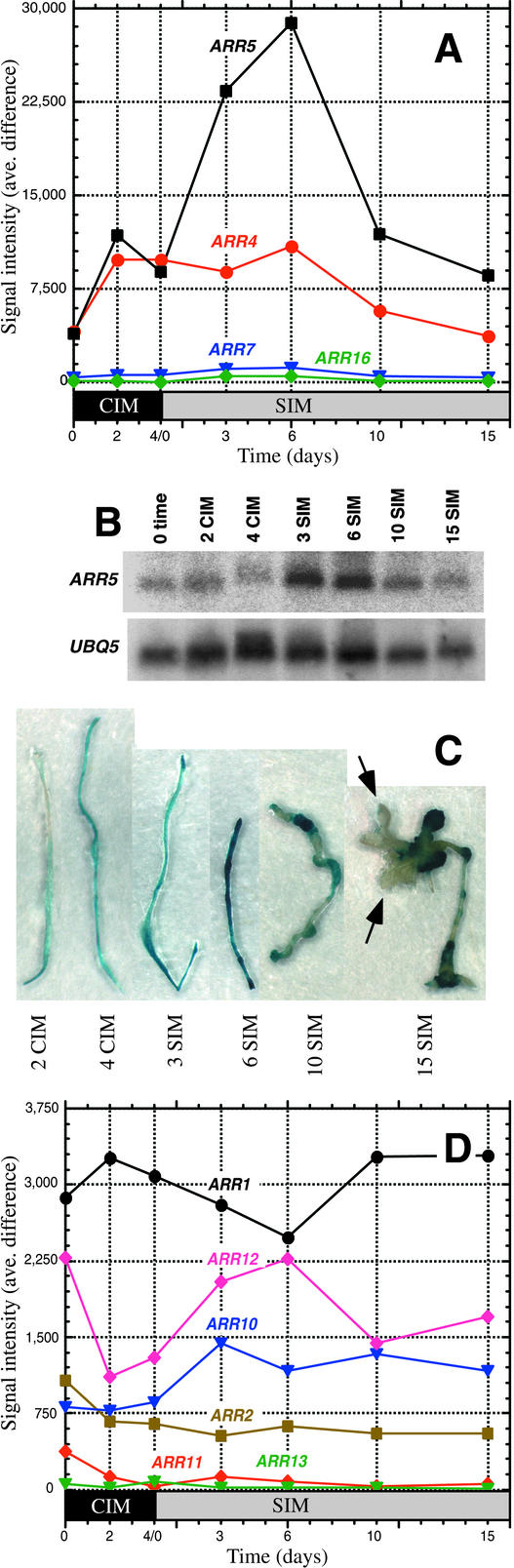
Because cytokinin plays a major role in directing shoot development, expression patterns were examined for genes that encode proteins involved in cytokinin signaling. The effects of cytokinin should be most apparent after transferring explants to cytokinin-rich SIM (containing 5.0 μM isopentenyladenine).
Cytokinins rapidly activate the expression of type-A ARRs (Brandstatter and Kieber, 1998; Imamura et al., 1998), a family of genes encoding proteins that are thought to act as feedback repressors of cytokinin responses (Hwang and Sheen, 2001). Several ARR genes are represented on the 8200-gene Affymetrix Arabidopsis GeneChip, and two of them that encode type-A ARRs are upregulated significantly during incubation on SIM (Figure 4A). One of these genes, ARR5 (At3g48100), was a high performer at 6 days on SIM (Table 1), upregulated by more than sevenfold, and its induction pattern was verified by RNA gel blot analysis (Figure 4B). ARR5 (formerly called IBC6) has been shown by others to be upregulated transcriptionally in seedlings exposed to cytokinin (D'Agostino et al., 2000), so the induction of ARR5 on SIM, a high-cytokinin-containing medium, is consistent with its regulation in seedlings. However, the upregulation observed in seedlings was transient, declining within hours (as assessed by nuclear-off assays or by the presence of transcripts), whereas in our system, the upregulation was longer term, lasting for days.
Figure 4.
Expression Patterns of Arabidopsis Genes That Encode ARRs during Shoot Development.
(A) Expression profiles for type-A ARRs were determined by oligonucleotide array analysis.
(B) RNA gel blot analysis of the expression profile of ARR5, one of the type-A ARRs. The blot was stripped and reprobed with UBQ5, which was used as a loading control.
(C) Localization of pARR5:GUS expression during shoot development. Root segments from 7-day-old transgenic seedlings bearing pARR5:GUS constructs with an ∼1.5-kb ARR5 promoter fragment were explanted and incubated for varying times on CIM and SIM. Explants were stained histologically for GUS expression and decolorized with 70% ethanol. Arrows point to emerging shoots.
(D) Expression profiles for type-B ARRs determined by oligonucleotide array analysis.
Because ARR5 is highly responsive to cytokinin and is induced during shoot development, we attempted to localize ARR5 expression by generating an ARR5 promoter:reporter construct (pARR5:GUS). Several transgenic lines containing the construct showed high-level induction of β-glucuronidase (GUS) in seedlings exposed to cytokinin (20 μM 6-benzyladenine) (data not shown). In root explants, we were interested in whether ARR5 expression, which peaked during incubation on SIM, marked presumptive sites of shoot emergence. We found that pARR5:GUS generally was expressed in the proliferating pericycle/callus tissue along the length of the root segment during preincubation on CIM and early during incubation on SIM (Figure 4C). GUS expression peaked at ∼6 days on SIM; after that, it diminished except at sites of callus outgrowth, from which shoots tended to emerge. Thus, later during incubation on SIM, pARR5:GUS expression was concentrated in regions of callus protuberances and may mark presumptive sites of shoot formation. Emerging shoots showed very little ARR5 expression. In general, ARR5 expression levels tended to increase with the production of undifferentiated callus tissue. ARR5 expression levels remained high in callus destined to give rise to green callus and shoots, but the gene was not expressed in differentiated tissues of the emerging shoot, except in the newly formed shoot meristem.
Type-B ARR genes are thought to be genetic activators in the cytokinin signaling pathway (Hutchison and Kieber, 2002; Hwang et al., 2002). They have been shown to activate the expression of the type-A ARRs described above (Hwang and Sheen, 2001), and the overexpression of ARR2, a type-B ARR, promotes shoot regeneration in Arabidopsis tissue culture in the absence of cytokinin. In our shoot development system, the type-B ARRs in general, and ARR1 (At3g16857) and ARR2 (At5g58080) in particular, were not induced during shoot formation (Figure 4D). However, type-B ARRs are not thought to be regulated transcriptionally; rather, they act on signals from the cytokinin phosphorelay transfer system (Hwang and Sheen, 2001; Hutchison and Kieber, 2002; Hwang et al., 2002).
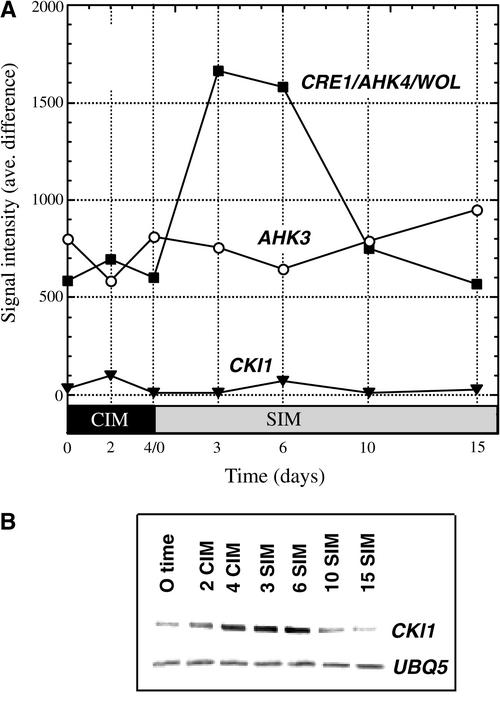
We also examined the expression profiles of genes that encode “hybrid” His kinases involved in cytokinin perception and signaling. CRE1/AHK4/WOL (At2g01830; hereafter referred to as CRE1) was the first hybrid His kinase shown to function as a cytokinin receptor and to be capable of conferring cytokinin responsiveness in a heterologous (yeast) system (Inoue et al., 2001; Yamada et al., 2001). CRE1 expression was upregulated by approximately threefold after transfer to SIM, peaking at 3 to 6 days on SIM and then declining thereafter (Figure 5A). AHK3 (At1g27320), another related His kinase that also has been reported to encode a cytokinin receptor (Suzuki et al., 2001; Ueguchi et al., 2001b; Yamada et al., 2001), showed little change in expression during shoot development in our system. CKI1 (At2g47430) also encodes a hybrid His kinase (Kakimoto, 1996), but its role in cytokinin signaling has not been resolved. CKI1 was thought originally to encode a cytokinin receptor (Kakimoto, 1996); however, CKI does not appear to confer cytokinin responsiveness in various expression systems, or if it does, it does so only at high levels of cytokinin (I. Hwang, personal communication). Nonetheless, we were interested in CKI1 because Kakimoto (1996) showed that CKI1 overexpression stimulates cytokinin-independent shoot formation in tissue culture. In our Affymetrix GeneChip analysis, the expression of CKI1 (At2g47430) was well below the levels that we considered significant (signal intensity of >500) (Figure 5A). Therefore, to assess changes in CKI1 expression, we conducted quantitative reverse transcription PCR analysis using primers specific to CKI1 (Figure 5B). The expression profile for CKI1 was similar to that for CRE1, increasing somewhat earlier, peaking at ∼3 to 6 days on SIM at levels some fourfold greater than at time 0, and declining thereafter. Thus, both CRE1 and CKI1 appear to be upregulated on or before transfer to SIM and downregulated thereafter.
Figure 5.
Expression Profiles of the Genes That Encode Hybrid His Kinases Related to Cytokinin Function during Shoot Development.
(A) Expression profiles for His kinase genes as analyzed by oligonucleotide array analysis. Expression levels at different time points during shoot development are expressed as signal intensity (average difference).
(B) Expression levels of CKI1 and UBI5 (control) were analyzed by quantitative reverse transcription PCR. An ethidium bromide–stained gel of PCR products is shown.
Auxin-Related Gene Expression Patterns
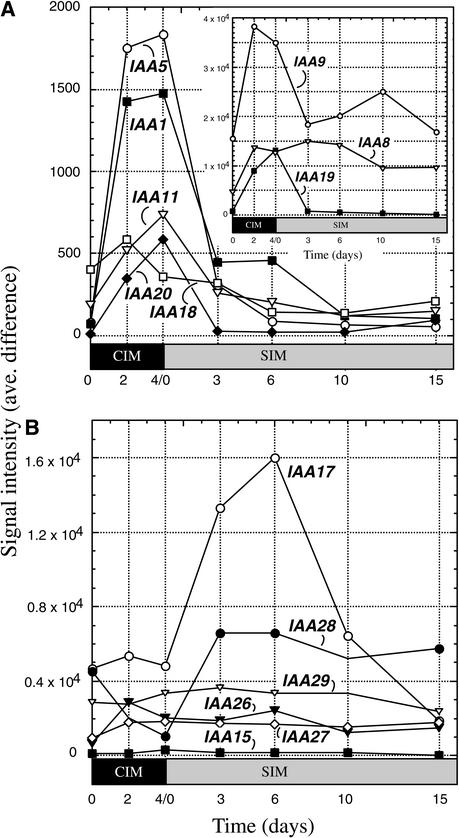
Auxin is a key hormone present in CIM and one that is critical to maintain in balance with cytokinin during shoot development. A number of hormone response genes are upregulated during incubation on CIM, and many of these encode Aux/IAA proteins. The Affymetrix Arabidopsis 8000 GeneChip contains 14 of the ∼30 known Aux/IAA genes. Of those, eight were upregulated on CIM; IAA5 (At1g15580) and IAA1 (At4g14560) were upregulated by >20-fold and IAA19 (At3g15540) was upregulated by >10-fold (Figure 6A). All but one, IAA8 (At2g22670), was downregulated after transfer to SIM. Abel et al. (1995) observed that IAA8 is not highly regulated by auxin, so its pattern of expression may reflect the relative nonresponsiveness of that gene to the high levels of auxin in CIM. The six other Aux/IAA genes have different expression profiles that do not relate strictly to the presence of auxin in CIM (Figure 6B). For example, levels of the IAA17/AXR3 (At1g04250) transcripts increased during incubation on SIM and peaked at 6 days on SIM, declining thereafter. Many of the Aux/IAA genes that were upregulated on CIM were identified originally by their responsiveness to auxin, particularly IAA1 to IAA14 (At4g14550), except for IAA8 (Abel et al., 1995), whereas the others were recognized as members of the Aux/IAA gene family by sequence similarity (Liscum and Reed, 2002).
Figure 6.
Expression Profiles of Genes That Encode Aux/IAA Proteins during Shoot Development.
(A) Expression profiles for Aux/IAA genes upregulated during preincubation on auxin-rich CIM. The inset shows profiles for three genes with higher expression profiles (note the difference in scale).
(B) Expression profiles for Aux/IAA genes not upregulated specifically during preincubation on CIM. Expression levels at different time points during shoot development are expressed as signal intensity (average difference).
DISCUSSION
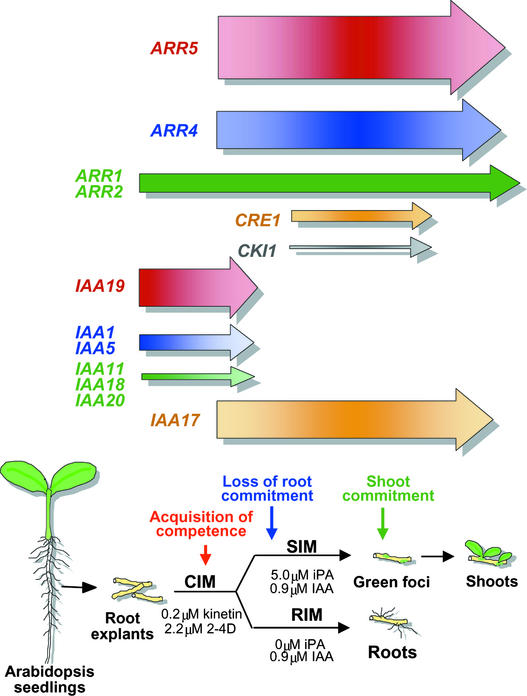
In companion articles, we identified developmental events, such as the “acquisition of competence” and “shoot commitment,” that occur during shoot formation (Cary et al., 2001, 2002). Explants acquire the competence to respond to shoot formation signals during preincubation on CIM (Figure 7). Explants do not form shoots on SIM unless they are preincubated on CIM. Explants also become committed to producing roots during CIM preincubation, so that if explants are transferred to basal medium, they continue to develop roots. Explants rapidly lose their commitment to form roots when transferred to SIM (Cary et al., 2002). After that, they become committed to the formation of shoots (6 to 8 days on SIM). Green foci (centers of green callus) begin to form at approximately that time, and shoots emerge.
Figure 7.
Summary of Developmental Events and Gene Expression Profiles of Selected Genes or Groups of Genes.
Arrow thickness represents maximum expression level. The color gradient within each arrow represents changes in signal intensity at different times. The schedule for developmental events was from Cary et al. (2002).
The general patterns in gene expression during shoot development provide insights into the mechanisms that may underlie this developmental process. First, a number of hormone response genes, largely Aux/IAA genes, were upregulated during preincubation on CIM. Second, many genes that encode signaling and/or transcription components were highly induced before shoot emergence at approximately the time of shoot commitment. This finding suggests that there may be substantial changes in signaling and gene regulatory activities at these stages. Third, as shoots emerged, genes that encode products of differentiated cells were the most highly induced—mostly genes that encode components of the photosynthetic apparatus.
We found little evidence in the overall phenomenon of gene expression changes for a simple cascade of events during shoot development. Based on such a model, one might expect exponentially increasing numbers of genes to be upregulated with time. Instead, many genes were upregulated throughout the time course of shoot development, and most genes that were upregulated subsequently were downregulated. Principal component analysis demonstrated that the major components that contribute to the variation in the overall pattern of gene expression represent groups of genes, each group of which is upregulated at a single developmental stage. However, the total picture may miss many important subpatterns that take the form of gene cascades.
One concern that we have had about this analysis is that early events leading to shoot development might be obscured by the presence of other tissues in the root explants that do not participate in the organization of the shoot primordium. We have tried to minimize this problem by using root explants from seedlings of the Columbia ecotype, which are fairly robust in forming shoots, usually producing one or two shoots per explant. However, that leaves much of the explant uninvolved in shoot formation. If the early events in shoot formation engage only a few progenitor cells, it is unlikely that one would see gene expression changes associated with a minority of cells.
On the other hand, it is possible that much tissue in the root explants engage in the early events of shoot formation and that cells progressively drop out of the process as development proceeds. One gains that impression by comparing green callus formation with shoot development. Shoots generally develop from broader areas of green callus, usually from protuberances in the green callus patches (green foci) that develop on explants. Not all green calli and protuberances give rise to shoots. However, if green callus formation is a step in the production of shoots, then shoot development may be a selective process whereby fewer and fewer cells reach later milestones in the shoot formation process as development proceeds.
In another study (Cary et al., 2002), we observed that a green fluorescent protein enhancer trap marker reporting on the gene CUP-SHAPED COTYLEDON1 (CUC1), which is required for shoot meristem formation, was expressed in a manner consistent with the concept described above. CUC1, which is expressed in the shoot apex of seedlings, was expressed widely throughout the pericycle-derived tissue in root explants at early stages in shoot development and was expressed more locally at protuberances later as shoots emerged. The expression pattern in root explants was in contrast to that in zygotic embryos, in which the enhancer trap was expressed only in the region of the presumptive shoot meristem early in embryo development.
Response Regulators
Cytokinin is a major driver of shoot regeneration, and two cytokinin primary response genes, ARR4 and ARR5, were highly upregulated during shoot development. ARR5, in particular, was upregulated by more than sevenfold. In seedlings, ARR5 is induced transiently within hours of the addition of cytokinin (Brandstatter and Kieber, 1998; Imamura et al., 1998). In our system, ARR5 was not expressed with the same kinetics as it is in seedlings. ARR5 was expressed in the callus produced by root explants, and expression levels increased over days, rising during preincubation on CIM. It also increased during the early stages of incubation on SIM, peaking at 6 days, and declined thereafter at approximately the time of shoot commitment (Figure 7). ARR5 reporter gene expression declined throughout the explants except at callus protuberances, which are the presumptive sites of shoot formation. The ARR5 reporter gene was not expressed in emerging shoots, except at the site of the new shoot apical meristem. Thus, ARR5 expression does not appear to be a simple reporter of cytokinin signaling in this system; rather, ARR5 expression accompanies the early stages of callus accumulation and later appears to mark presumptive sites of shoot formation.
By contrast, type-B ARR genes such as ARR1 and ARR2 were not regulated demonstrably during shoot development in our system, despite the fact that these genes have extraordinary powers in shoot development. For example, ARR2 overexpression can drive cytokinin-independent shoot formation (Hwang and Sheen, 2001). However, the mechanism by which type-B ARRs act and activate gene expression is not known. It has been argued that the transcriptional activation capacity of ARR1 is inhibited by its receiver domain and that phosphorylation of the receiver domain by cytokinin-induced phosphorelay signaling may overcome this inhibition. In support of this idea is the finding that truncation of the N-terminal receiver domain increases transcriptional activation by ARR1 in protoplast transient assay systems (Sakai et al., 2000, 2001). However, it was found that overexpression of full-length ARR1 or ARR2 (even with the N-terminal receiver domain intact) in protoplasts or in transgenic plants activates cytokinin responses even in the absence of cytokinin (Hwang and Sheen, 2001; Sakai et al., 2001). Additionally, it was observed that a substitution mutation in the presumed phospho-accepting Asp in ARR2 did not abolish its transcriptional activation of ARR6 in a protoplast assay system (Hwang and Sheen, 2001). From these findings, Hwang and Sheen (2001) proposed that the type-B ARRs normally may be tied up by repressors and that cytokinin-induced phosphorylation may liberate the ARRs in the active form. In any case, the mechanism of action of the type-B ARRs appears to depend on post-translational events and not on the upregulation or downregulation of their genes.
His Kinases
Some of the hybrid His kinases serve as cytokinin receptors, and we found that CRE1/AHK4/WOL, the first described cytokinin receptor (Inoue et al., 2001), was induced by approximately threefold on transfer to SIM. CRE1 functions as a cytokinin receptor in a yeast heterologous signaling system and is essential for shoot regeneration in tissue culture (Inoue et al., 2001). CKI1 is expressed at very low levels in our system but is induced in a pattern that is somewhat similar to that of CRE1. The function of CKI1 is not understood. CKI1 is similar to other cytokinin receptor His kinases, although it lacks a C-subterminal receiver-like domain (Hwang et al., 2002), and it can activate shoot development and the expression of cytokinin primary response genes even in the absence of cytokinin (Kakimoto, 1996; Hwang and Sheen, 2001). The upregulation of CRE1 and CKI1 in the transition from CIM to SIM may be related to the acquisition of competence. During preincubation on CIM, explants acquire the competence to respond to cytokinin as a shoot induction signal (Figure 7). The upregulation of CRE1 early during incubation on SIM may be a consequence of previous competence-building events that occurred during preincubation on CIM. On the other hand, CRE1 upregulation might simply be a response to cytokinin and the transfer to SIM.
Because CKI1 overexpression promotes cytokinin-independent shoot development (Kakimoto, 1996), it is tempting to speculate that the upregulation of CKI1 expression in our system may be related to shoot commitment, which also occurs at approximately the same stage (Figure 7). Commitment to shoot formation is defined as the developmental stage at which explants continue to form shoots even when transferred from SIM to basal medium. Thus, the induction of CKI1 may be a transitional step in shoot development during which shoot development changes from a hormone-dependent to a hormone-independent process.
Aux/IAA Genes
As discussed above, during preincubation on CIM, root explants acquire the competence to respond to the subsequent shoot-forming signals on SIM (Cary et al., 2001). CIM contains the potent auxin 2,4-D, and during preincubation on CIM, many of the Aux/IAA genes are upregulated and then subsequently downregulated when transferred to SIM. The downregulation of auxin-responsive IAA genes after transfer to SIM corresponds to the loss of root commitment that occurs rather precipitously at that time (Figure 7) (Cary et al., 2002). Aux/IAA genes that are known to be auxin responsive, such as IAA1, IAA5, IAA9, and IAA11 (Abel et al., 1995), are upregulated and downregulated during shoot development, depending on the presence of auxin in the medium. However, some of the Aux/IAA genes have expression profiles that increase and decrease independent of exogenous auxin. For example, levels of IAA17 (AXR3) increase dramatically during incubation on SIM, peaking at ∼6 days, and decrease rapidly thereafter. We find the expression profile of IAA17 (AXR3) to be intriguing because gain-of-function mutations in this gene have rather profound developmental effects (Leyser et al., 1996). However, it remains to be determined whether perturbation in the expression of IAA17 (AXR3) affects shoot development in this system.
METHODS
Shoot Development Conditions
Arabidopsis thaliana ecotype Columbia seedlings were grown to 7 days of age on plant nutrient solution medium [5 mM KNO3, 2.5 mM KH2PO4, 2 mM MgSO4, 2 mM Ca(NO3)2, 0.05 mM Fe/EDTA, 70 μM H3BO3, 14 μM MnSO4, 0.5 μM CuSO4, 1.0 μM ZnSO4, 0.2 μM Na2MoO4, 10 μM NaCl, 0.01 μM CoCl2, 5 g/L Suc, pH 5.5, and 0.7% Bacto-agar]. Root segments (5 mm) were cut and transferred to callus induction medium (CIM), which consists of Gamborg's B5 medium (Gamborg et al., 1968) with 5 g/L Mes, 2.2 μM 2,4-D, 0.2 μM kinetin, and 0.8% agarose. Explants were incubated on CIM for 4 days under constant light conditions and then transferred to shoot induction medium (SIM). SIM is prepared as CIM except that SIM contains the hormones isopentenyladenine (5.0 μM) and 3-indoleacetic acid (0.9 μM). Root reduction medium contains 0.9 μM 3-indoleacetic acid. After transfer to SIM, explants were incubated under constant-light conditions to induce shoot formation. Shoot development was recorded with a Nikon CoolPix 990 digital camera (Tokyo, Japan).
RNA Procedures
Total RNA was isolated from plant tissues by TRIzol (Life Technologies, Gibco BRL) extraction. Tissue was ground and homogenized in TRIzol solution (∼100 mg of tissue per 1 mL of TRIzol) and extracted and precipitated as described by the manufacturer. Precipitated RNA was solubilized in water with 0.1% (v/v) diethylpyrocarbonate, purified using the RNeasy kit according to the manufacturer (Qiagen, Valencia, CA), and quantified by 260/280-nm UV light absorption.
RNA samples (20 μg), isolated from various hormone- and inhibitor-treated tissues, were denatured at 95°C and subjected to electrophoresis in 1% (w/v) agarose and 1 × Mops [0.2 M 3-(N-morpholino)-propanesulfonic acid, 0.5 M sodium acetate, and 0.01 M EDTA] on 13.3% formaldehyde gels. RNA was blotted to Hybond-N membranes (Amersham Pharmacia). Probes specific for ARR5 and UBQ5 were generated by PCR and labeled by random-hexamer labeling with a DNA-bead labeling kit, as described by the manufacturer (Amersham Life Sciences). The primers used for UBQ5 amplification were UBQ5F (5′-CTTGAAGACGGCCGTACCCTC-3′) and UBQ5R (5′-CGCTGAACCTTTCAAGATCCATCG-3′). ARR5-specific primers were IBC6F (5′-CTGAGGTTTTGCGTCCCGAGATG-3′) and IBC6R (5′-GCGCGTTTTAGCTGCGAGTAGATATC-3′). Probes were hybridized to the membranes overnight in Church buffer (7% SDS, 1% BSA, 1 mM EDTA, and 250 mM NaH2PO4). The membranes were washed and imaged using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Signals were quantified with ImageQuant software (Molecular Dynamics).
Quantitative Reverse Transcription PCR
Total RNA (2 μg) was reverse transcribed using TaqMan reverse transcription reagent (Applied Biosystems, Foster City, CA) in a 100-μL reaction. PCR was performed using 2 μL of the reverse transcription reaction as a template. Cycle numbers were optimized for each sample to obtain data in the exponential range. Amplified DNA fragments were separated on 2% agarose gels and stained with ethidium bromide. Signal intensities were quantified using NIH Image software (http://rsb.info.nih.gov/nih-image). UBQ5, amplified with the primers described above, was used as an internal control. The primers used for CKI1 amplification were CKI1L (5′-AAAGCTTGTGGCTTCACG-TC-3′) and CKI1R (5′-GAGATCCAAATGCACTCGAAA-3′).
Probe Synthesis and Hybridization to Affymetrix GeneChips
TRIzol-extracted total RNA (10 μg) was treated with RNase-free DNase (Life Technologies) and purified further using the RNeasy total RNA cleanup protocol (Qiagen). Purified RNA was assessed for integrity by formaldehyde/agarose gel electrophoresis and quantified by absorbance at 260 nm.
RNA samples were submitted to the DNA facility at the University of Iowa (http://dna-9.int-med.uiowa.edu/microarrays.htm) and were handled as follows. An aliquot of copy RNA (cRNA) control mixture was added to 10 μg of purified total RNA. The cRNA control mixture is composed of a set of four in vitro transcripts generated by T3 RNA polymerase, encoding the Bacillus subtilis genes thr, trp, phe, and lys. Probes corresponding to these bacterial transcripts are represented on all GeneChips (including test chips) from Affymetrix (Santa Clara, CA). Each synthetic transcript is quantified and represented at copy numbers of 5 × 106 to 5 × 108, corresponding approximately to the expected dynamic range of detection for the GeneChip system. This set of control transcripts allows for the monitoring of cDNA synthesis and the in vitro transcription of biotinylated targets and also provides a reference sample for normalizing between experiments.
RNA was converted to double-stranded cDNA using the SuperScript Choice system (Life Technologies) according to the supplier's protocol, except that an HPLC-purified T7-dT24 primer (Genosys) was used for first-strand synthesis. After synthesis, double-stranded cDNA was purified by phenol/chloroform extraction and ethanol precipitation. The purified cDNA was used to generate biotinylated cRNA target using a Bioarray High-Yield RNA Transcript-Labeling Kit (Enzo, Farmingdale, NY) according to the supplier's protocol. The labeled cRNA then was purified using the RNeasy total RNA cleanup protocol (Qiagen) and quantified by absorbance at 260 nm.
cRNA (20 μg) was fragmented by heating at 94°C for 35 min in fragmentation buffer (40 mM Tris-acetate, pH 8.1, 125 mM KOAc, and 30 mM MgOAc). An aliquot of fragmented and unfragmented cRNA was analyzed by formaldehyde/agarose gel electrophoresis to ensure appropriate size distribution (average size, 700 bp of unfragmented cRNA and 100 bp after fragmentation).
Fragmented cRNA (15 μg) was added to a hybridization “cocktail” containing 50 pM of control oligonucleotide B2 (Genosys, The Woodlands, TX), 1 × control cRNA mixture, 0.1 mg/mL herring sperm DNA, and 1 × Mes hybridization buffer in a volume of 300 μL. Control oligonucleotide B2 (5′-biotin:GTCGTCAAGATGCTACCG-TTCAGGA-3′) was designed to hybridize to structural features on the chip to allow for proper scanning and grid alignment. The control cRNA mixture was composed of a second set of four biotinylated in vitro antisense transcripts of cDNAs encoding the Escherichia coli biotin synthesis genes bioB, bioC, and bioD and the P1 bacteriophage cre recombinase gene. Probes corresponding to these bacterial transcripts also are represented on all Affymetrix GeneChips (including test chips). Each synthetic transcript was quantified and represented at copy numbers of 2 × 108 to 2 × 1010, corresponding approximately to the expected dynamic range of detection for the GeneChip. This set of control cRNAs allows for the monitoring of hybridization, washing, and staining conditions and also provides a second set of reference samples for normalizing between experiments. The hybridization cocktail was heated to 95°C and then cooled to 45°C, and then 100 μL was applied to a Test3 GeneChip array. The remainder of the cocktail was stored at −20°C, and the test chip was hybridized at 45°C for 16 h.
After hybridization, the hybridization cocktail was removed from the chip and stored at −70°C. The chip was placed immediately in the Affymetrix GeneChip Fluidics Station 400 and washed using the preprogrammed step “mini_euk2.” This wash program involves a low-stringency wash, a high-stringency wash, a steptavidin/phycoerythrin stain, a low-stringency wash, an anti-streptavidin antibody stain, a second steptavidin/phycoerythrin stain, and a final low-stringency wash. After washing and staining, the test chip was placed in the Affymetrix GeneChip array scanner, and image data were captured and converted to numerical output using Microarray Analysis Suite version 5.0 (Scanalytics, Fairfax, VA).
For the test chip, the levels of bioB, bioC, bioD, cre, phe, trp, lys, and thr were assessed. When the labeled cRNA target passed all metrics of the Test3 chip, it was again heated to 95°C, cooled to 45°C, and hybridized to the GeneChip of interest. Hybridization, washing, staining, and scanning were repeated as described above except that the automated fluids program “euk2” was used.
Experimental data acquired from the chip were analyzed using the GeneChip Microarray Analysis Suite version 5.0. For comparison between two chips, the “baseline chip” intensity values were measured and normalized to the average signal intensity. Intensity values of the “experimental chip” then were compared with baseline chip values and a “difference change” was calculated. Data were analyzed using GeneSpring version 4.1.5 (Silicon Genetics, Redwood City, CA).
ARR5 Promoter/Reporter Gene Plasmid Construction
BAC T17F15, containing an Arabidopsis genomic fragment with the full ARR5 gene sequence, was obtained from the ABRC (Columbus, OH). ARR5 5′ flanking sequence was amplified from T17F15 by PCR. Primers used to generate the full-length promoter were (1) the antisense downstream primer ARR52kbR (5′-GCGGATCCAAGAAGA-GTAGGATCGTGAC-3′), which matches the −23 to −2 region (relative to the translation start codon) of the ARR5 5′ untranslated region and contains a BamHI restriction site, and (2) the upstream primer ARR5p3′F (5′-GAGAGGTAAAAACCGAGACCATTAGG-3′), in the sense orientation, which contains the HindIII restriction site. The ∼1.5-kb promoter fragment was cloned into vector pBI221 using the BamHI-HindIII restriction sites in this vector, placing the fragment 21 bp upstream of the uidA reporter gene translational start site. The ARR5 promoter::uidA gene cassette was removed from the pBI221 vector by HindIII-EcoRI digestion and cloned into the Agrobacterium tumefaciens binary plasmid vector pBI121, which had been digested with the same restriction enzymes.
Plant Transformation and Culture Conditions
Four-week-old plants were transformed using the floral-dip method (Clough and Bent, 1998). Agrobacterium strain C58 was transformed with the ARR5 promoter::uidA pBI121 construct. These strains were grown to saturation overnight. They were centrifuged and resuspended in 10% Suc and 0.05% Silwett L-77 to an OD600 of 0.6 to 0.8. The plants were dipped into this medium so that the aboveground tissues were saturated. The plants were covered in plastic wrap overnight and then moved into the growth room. Plants were grown for an additional 2 to 4 weeks, and seeds were collected.
Seeds from the dipped plants were surface-sterilized with 50% bleach and 0.02% Triton X-100. Seeds were resuspended in 0.1% sterile agar and plated on kanamycin–plant nutrient solution selection plates with 50 μg/mL kanamycin. Transformants were identified and transferred to plant nutrient solution plates to recover for 7 days in the growth room and then planted in soil. Transformants were grown for an additional 4 to 6 weeks and allowed to self-pollinate, and seeds representing the T1 generation were collected.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Acknowledgments
D.J.G. and S.H.H. acknowledge support from the National Science Foundation/Cornell Plant Cell and Molecular Biology Program and from the Boyce Thompson Institute. This work was supported in part by the Plant Sciences Institute at Iowa State University.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006668.
References
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570. [DOI] [PubMed] [Google Scholar]
- Bhojwani, S.S., and Razdan, M.K. (1996). Plant Tissue Culture: Theory and Practice. A Revised Edition. (New York: Elsevier Press).
- Brandstatter, I., and Kieber, J.J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary, A.J., Che, P., and Howell, S.H. (2002). Development events and shoot meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J., in press. [DOI] [PubMed]
- Cary, A., Uttamchandani, S.J., Smets, R., Van Onckelen, H.A., and Howell, S.H. (2001). Arabidopsis mutants with increased organ regeneration in tissue culture are more competent to respond to hormonal signals. Planta 213, 700–707. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- D'Agostino, I.B., Deruere, J., and Kieber, J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri, S., and Estelle, M. (2002). The role of regulated protein degradation in auxin response. Plant Mol. Biol. 49, 401–419. [PubMed] [Google Scholar]
- Flinn, B.S., Webb, D.T., and Newcomb, W. (1988). The role of cell clusters and promeristemoids in determination and competence for caulogenesis by Pinus strobus cotyledons in vitro. Can. J. Bot. 66, 1556–1565. [Google Scholar]
- Gamborg, O.J., Miller, R.A., and Ojima, K. (1968). Nutrient requirement of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J., Ulmasov, T., and Hagen, G. (1998). The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 54, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, G.S. (1994). Shoot induction and organogenesis in vitro: A developmental perspective. In Vitro Cell. Dev. Biol. 30P, 10–15. [Google Scholar]
- Hutchison, C.E., and Kieber, J.J. (2002). Cytokinin signaling in Arabidopsis. Plant Cell 14 (suppl.), S47.–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., Chen, H.C., and Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Kiba, T., Ueguchi, C., Sugiyama, T., and Mizuno, T. (1999). Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 40, 733–742. [DOI] [PubMed] [Google Scholar]
- Imamura, A., Hanaki, N., Umeda, H., Nakamura, A., Suzuki, T., Ueguchi, C., and Mizuno, T. (1998). Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 2691–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kakimoto, T. (1996). CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274, 982–985. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Ueguchi, C., Sugiyama, T., and Mizuno, T. (2000). Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41, 791–803. [DOI] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruan, Y., Gilmore, J., and Conner, T. (1998). Towards Arabidopsis genome analysis: Monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15, 821–833. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., and Oka, A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24, 703–711. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., and Oka, A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294, 1519–1521. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (2002). Phosphorelay and transcription control in cytokinin signal transduction. Science 296, 1650–1652. [DOI] [PubMed] [Google Scholar]
- Skoog, F., and Miller, C.O. (1957). Chemical regulation of growth and organ formation in plant tissue cultured in vitro. Symp. Soc. Exp. Biol. 11, 118–131. [PubMed] [Google Scholar]
- Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H., and Mizuno, T. (2001). The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol. 42, 107–113. [DOI] [PubMed] [Google Scholar]
- Thorpe, T.A. (1993). In vitro organogenesis and somatic embryogenesis: Physiological and biochemical aspects. In Morpho-genesis in Plants: Molecular Approaches, K.A. Roubelakis-Angelakis and K. Tran Thanh Van, eds (New York: Plenum Press), pp. 19–38.
- Tiwari, S.B., Wang, X.J., Hagen, G., and Guilfoyle, T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi, C., Koizumi, H., Suzuki, T., and Mizuno, T. (2001. a). Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 42, 231–235. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Sato, S., Kato, T., and Tabata, S. (2001. b). The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42, 751–755. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Lijsebettens, M.V. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., and Mizuno, T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42, 1017–1023. [DOI] [PubMed] [Google Scholar]
- Zhu, T., Budworth, P., Han, B., Brown, D., Chang, H.-S., Zou, G., and Wang, X. (2001). Toward elucidating the global gene expression patterns of developing Arabidopsis: Parallel analysis of 8300 genes by a high-density oligonucleotide probe array. Plant Physiol. Biochem. 39, 221–242. [Google Scholar]