Abstract
A T-DNA–tagged population of Arabidopsis was screened for mutations in AtOPT3, which encodes a member of the oligopeptide (OPT) family of peptide transporters, and a recessive mutant allele, opt3, was identified. Phenotypic analysis of opt3 showed that most homozygous embryos were arrested at or before the octant stage of embryo development and that none showed the usual periclinal division leading to the formation of the protoderm. This defective phenotype could be reversed by complementation with the full-length, wild-type AtOPT3 gene. A β-glucuronidase (GUS) fusion to DNA sequences upstream of the putative AtOPT3 ATG start codon was constructed, and the expression pattern was assayed in transgenic plants. AtOPT3 was expressed in the vascular tissues of seedlings and mature plants as well as in pollen. Consistent with the function of AtOPT3 in embryogenesis, AtOPT3::GUS expression also was detected in developing embryos and in the maternal tissues of seeds. These data suggest a critical role for peptide transport in early embryo development.
INTRODUCTION
Peptide transport is a widely observed phenomenon in both prokaryotes and eukaryotes. The process is characterized by the ability of cells to transport peptides across membranes in a carrier-mediated, energy-dependent manner. Transported peptides are hydrolyzed, and the resulting amino acids are used for protein synthesis or as alternative sources of nitrogen and carbon (Perry et al., 1994; Steiner et al., 1995). Peptide transport systems usually are restricted to small peptides of two to six amino acids, exhibit high stereoselectivity, have no strict side chain specificity, and prefer α-peptide bonds and the l-stereoisomers of amino acids (Becker and Naider, 1980; Payne and Smith, 1994). In addition to a nutritional role, peptide transport systems also are involved in other cellular processes, such as bacterial quorum sensing (Swift et al., 1996), yeast mating (Kuchler et al., 1989; McGrath and Varshavsky, 1989), and mammalian immune response (Neefjes et al., 1993; Shepherd et al., 1993; Uedel et al., 1995). Peptide transport systems also can take up physiologically active peptide derivatives. For example, peptide transport systems in animals are pharmacologically important for the uptake of peptide-derived antibiotics (e.g., cephalosporins [Okano et al., 1986a, 1986b]) and anticancer agents (e.g., bestatin [Inui et al., 1990]).
The ABC (ATP binding cassette) superfamily of transporters uses ATP hydrolysis for the transmembrane translocation of a large variety of substances (including peptides), and members have been identified in organisms ranging from Escherichia coli to human (Blackmore et al., 2001; reviewed by Detmers et al., 2001). In contrast to the ABC family, the PTR (peptide transport) family uses the transmembrane proton gradient to energize transport. Members of the PTR family transport dipeptides and tripeptides as well as amino acids and nitrate (reviewed by Williams and Miller, 2001; Stacey et al., 2002). PTR proteins have been identified in yeast (Perry et al., 1994; Basrai et al., 1995), human (Liang et al., 1995), rabbit (Fei et al., 1994, 2000), mouse (Ganapathy et al., 1995), barley (West et al., 1998), and Arabidopsis (Frommer et al., 1994; Steiner et al., 1994; Song et al., 1997). The Arabidopsis AtPTR2 transporter, when expressed in yeast, transported dipeptides and tripeptides but not larger peptides (Steiner et al., 1995; Song et al., 1996). AtPTR2 antisense plants exhibited a delay in flowering time and arrest in seed development (Song et al., 1997). Database comparisons with the Arabidopsis genome sequence identified 51 additional PTR family members, most of which were predicted to transport peptides (Stacey et al., 2002).
The OPT (oligopeptide transport) family likely uses the proton motive force to drive transmembrane transport. The OPT family was first described in the pathogenic yeast Candida albicans (Lubkowitz et al., 1997) and subsequently in Schizosaccharomyces pombe (Lubkowitz et al., 1998) and Saccharomyces cerevisiae (Hauser et al., 2000). OPT proteins are genetically and physiologically distinct from the ABC and PTR proteins and predominantly transport tetrapeptides and pentapeptides (Lubkowitz et al., 1997, 1998). OPT transporters are predicted to have 12 to 14 transmembrane domains, and sequence homology indicates several conserved motifs (Hauser et al., 2001; Koh et al., 2002). In addition to short peptides, the S. cerevisiae Opt1p was shown to transport glutathione (Bourbouloux et al., 2000) as well as enkephalins, which are endogenous opioid pentapeptides in the central nervous system (Hauser et al., 2000). Database searches for homology with the C. albicans CaOpt1p revealed nine putative Arabidopsis OPT orthologs that form a distinct subfamily compared with the fungal OPTs (Koh et al., 2002; Stacey et al., 2002). Full-length cDNAs for seven Arabidopsis OPT transporters (AtOPT1 to AtOPT7) were cloned and tested for their ability to take up various peptides when expressed in yeast (Koh et al., 2002). Five (AtOPTs 1, 4, 5, 6, and 7) of the seven AtOPTs were functional transporters in yeast, transporting at least one of the synthetic peptides KLLG, KLGL, and KLLLG. Peptide transport by AtOPT3 or AtOPT2 was not observed, perhaps as a result of the limited number of substrates available for testing. Using gene-specific primers, the expression of the cloned AtOPTs was detected by quantitative reverse transcriptase–mediated PCR in Arabidopsis, and each was found to exhibit a distinct tissue-specific expression pattern (Koh et al., 2002). The Arabidopsis OPT transporters were the first nonfungal orthologs to be identified.
Plants can assimilate nitrogen in a variety of forms, such as ammonium, nitrate, amino acids, complex insoluble nitrogen-containing compounds, and soluble peptides (reviewed by Williams and Miller, 2001). In contrast to the information available on plant transport of ammonium, nitrate, and amino acids, very little is known about peptide transporters and their physiological substrates. The presence of multiple peptide transporters in the Arabidopsis genome, as well as the known function of peptide transport systems in bacteria, fungi, and animals, supports the idea that plant peptide transporters play an important role in plant growth and development. Our previous reports on the cloning and characterization of PTR and OPT transporters in Arabidopsis further support this notion. To address the functional role of peptide transport in plants, we used a reverse genetics approach to identify a loss-of-function mutation in one of the Arabidopsis oligopeptide transporter homologs, AtOPT3. Phenotypic analysis of plants carrying a T-DNA insertion in AtOPT3 showed that the opt3 allele is recessive and that embryos homozygous for opt3 are arrested very early in embryogenesis. In addition, we also determined the tissue-specific expression pattern of an AtOPT3 promoter–β-glucuronidase (GUS) fusion in transgenic Arabidopsis. We found that AtOPT3 is expressed preferentially in the vascular tissues of seedlings and mature plants as well as in pollen and in developing embryos.
RESULTS
The opt3 Insertion Allele Is Lethal
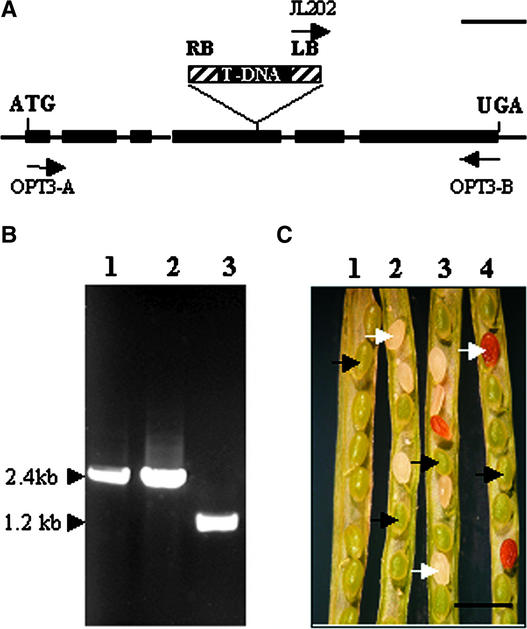
Using a PCR-based approach, we identified an Arabidopsis line harboring a T-DNA insertion in the predicted fourth exon of the AtOPT3 gene (Figure 1A). The mutant line, designated N4, is heterozygous at the AtOPT3 locus (Figure 1B). DNA gel blot analysis using the T-DNA–encoded NPTII (neomycin phosphotransferase) gene as a probe showed a single band (data not shown), consistent with a single T-DNA copy inserted within AtOPT3. The N4 plant showed wild-type phenotypes with regard to plant size and leaf and flower development. However, a seed-defective phenotype was observed. Before the greening stage, developing seeds appeared approximately similar in color and size (data not shown). After the greening stage (i.e., late heart to torpedo stages of embryo development), abnormal seeds become readily distinguishable as white seeds compared with the wild-type, green seeds (Figure 1C). The mutant seeds then became brownish and eventually dried out. This seed-defective phenotype is consistent with that observed for tagged embryo-lethal mutants (Meinke and Sussex, 1979; Errampalli et al., 1991).
Figure 1.
Isolation of a T-DNA Insertional Mutation in AtOPT3 and the Embryo-Lethal Phenotype Associated with the Mutation.
(A) T-DNA insertion site in the predicted fourth exon of AtOPT3. Black boxes represent exons, and black arrows indicate the relative locations and orientations of pertinent primers. The T-DNA insert and primers are not drawn to scale. LB, left border; RB, right border. Bar = 500 bp.
(B) The isolated N4 line is heterozygous for the opt3 allele. PCR amplification of the AtOPT3 wild-type allele with primers OPT3-A and OPT3-B (lane 2) and amplification of the mutant opt3 allele with primers JL202 and OPT3-B (lane 3) using genomic DNA isolated from the N4 (T2) plant. As a control, AtOPT3 was amplified (lane 1) from the wild type.
(C) Embryo-lethal phenotype associated with opt3. An immature silique from a wild-type plant showed only green seeds (silique 1), whereas heterozygous siliques from the N4 plant segregated green and white seeds, with the latter siliques turning purplish/brown in older siliques (siliques 2 to 4). Black arrows, wild-type seeds; white arrows, mutant seeds. Bar = 1.2 mm.
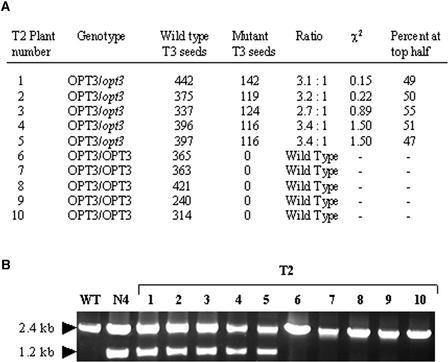
To determine the frequency and locations of mutant seeds, T3 plants derived from N4 were allowed to self-pollinate, and siliques from each plant were scored for the number and locations of mutant and wild-type T4 seeds. The genotype of each T3 plant also was determined to examine the cosegregation of the defective seed phenotype with the T-DNA insertion within AtOPT3. Genotype determination and segregation analyses for 10 selected T3 plants are presented in Figure 2. The five plants heterozygous for opt3 (plants 1 to 5) gave segregation ratios of wild-type to mutant seeds that were not significantly different from 3:1. By contrast, all nontagged plants (plants 6 to 10) produced wild-type seeds. In the heterozygous plants, 47 to 55% of mutant seeds were distributed in the upper half of each silique, indicating that the opt3 mutation has no apparent effect on the development of the male gametophyte (Meinke, 1982). In addition, 25 more T3 and T4 plants also were examined for cosegregation of the defective seed phenotype with the opt3 allele. None of these plants was homozygous for opt3, and plants heterozygous at the AtOPT3 locus produced wild-type and defective seeds (data not shown). Except for the defective seed phenotype, all viable T3 and T4 plants examined showed normal vegetative and reproductive development. Together, these results clearly indicate that the observed embryo-lethal phenotype is linked to the T-DNA insertion within AtOPT3 and that opt3 is recessive and results in seed abortion of the homozygotes.
Figure 2.
Segregation Analysis of Embryo Lethality Associated with the opt3 Mutation.
(A) Segregation and distribution of defective seeds in 10 T3 plants.
(B) Genotype determination of the 10 T3 plants by PCR amplification of wild-type (WT) AtOPT3 and mutant opt3 alleles. Similar primers were used as in Figure 1B, except that a single PCR procedure containing OPT3-A, OPT3-B, and JL202 was performed for each of the T3 plants.
To confirm that the defective seed phenotype is the result of the insertion mutation within AtOPT3, genetic complementation was performed by transforming plants heterozygous at the AtOPT3 locus with a genomic DNA encoding the AtOPT3 gene and promoter region (promoter::AtOPT3). As a control, transformation also was performed with a DNA encoding the GUS gene expressed from the native AtOPT3 promoter (promoter::GUS). Four independent transgenic plants heterozygous for AtOPT3 and hemizygous for the promoter::AtOPT3 transgene were selfed and scored for seed lethality. All four lines showed a reduction in the number of defective seeds, giving an average ratio of wild-type to mutant seeds that was not significantly different from 15:1 (Table 1). By contrast, control plants transformed with promoter::GUS showed no genetic complementation (Table 1). Therefore, the seed-lethal phenotype genetically complemented by AtOPT3 confirmed that the mutant phenotype is the result of the opt3 insertion allele.
Table 1.
Genetic Complementation of the opt3 Mutation
| Genotype | Plant Number | Wild Type | Mutant | Ratio | Hypothesized Ratio | χ2 |
|---|---|---|---|---|---|---|
| AtOPT3/opt3 | 1 | 579 | 42 | 13.8:1 | 15:1 | 0.26 |
| Genomic AtOPT3/-a | 2 | 530 | 47 | 11.3:1 | 15:1 | 3.56 |
| 3 | 477 | 35 | 13.6:1 | 15:1 | 0.28 | |
| 4 | 523 | 33 | 15.8:1 | 15:1 | 0.14 | |
| AtOPT3/opt3 | 1 | 252 | 75 | 3.4:1 | 3:1 | 0.80 |
| AtOPT3 promoter–GUS/-b | 2 | 218 | 85 | 2.6:1 | 3:1 | 1.55 |
| 3 | 315 | 107 | 2.9:1 | 3:1 | 0.03 | |
| 4 | 388 | 132 | 2.9:1 | 3:1 | 0.04 |
Heterozygous for the opt3 allele and hemizygous for the complementing AtOPT3 genomic DNA.
Heterozygous for the opt3 allele and hemizygous for the promoter–GUS fusion.
opt3 Embryo Development Arrests at the Preglobular Stage
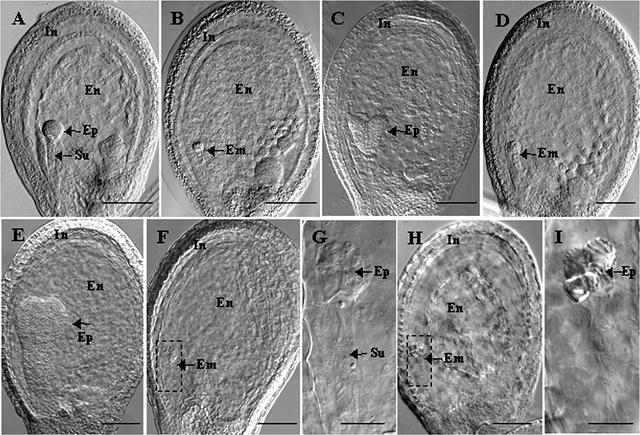
To characterize the nature of the seed lethality associated with opt3, wild-type and mutant embryos from siliques of heterozygous T3 plants were analyzed at various stages of seed development. A comparison of wild-type and mutant embryos at the globular (Figures 3A and 3B), heart (Figures 3C and 3D), and torpedo (Figures 3E to 3G) stages of embryo development showed that mutant embryos remained small and arrested very early in embryogenesis. At the torpedo stage of normal development, mutant embryos became slightly shriveled (Figures 3F and 3G), and most of them disintegrated by the curled cotyledon stage, although a few seeds retained embryos that were clearly disintegrating (Figures 3H and 3I). Despite aberrant embryo development, endosperm development of seeds harboring opt3 embryos appeared normal until the torpedo stage of normal development (Figures 3A to 3F), after which the endosperm and integument layers also started to disintegrate (Figure 3H). In the mature silique, mutant seeds eventually turned brownish and dried out.
Figure 3.
Seed Development in Wild-Type and opt3 Homozygotes.
Siliques from heterozygous opt3 plants were examined at various stages of seed development, and the phenotypes of seeds harboring wild-type embryos ([A], [C], and [E]) were compared with those of the opt3 mutant ([B], [D], and [F] to [I]). For each developmental stage, wild-type and mutant seeds from the same silique were compared. Developmental stages are as follows: (A) and (B), globular; (C) and (D), heart; (E) and (F), torpedo; and (H), late curled cotyledon. (G) and (I) show higher magnifications of boxed mutant embryos in (F) and (H), respectively. In, integument layers; Em, opt3 embryo; En, endosperm; Ep, embryo proper; Su, suspensor. Bars in (A) to (F) and (H) = 80 μm; bars in (G) and (I) = 20 μm.
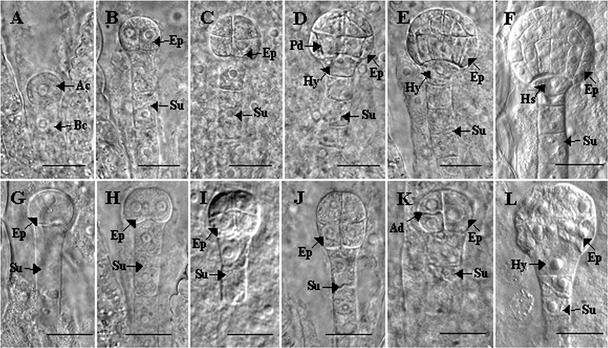
During normal embryo development, the initial division of the zygote is transverse, generating a small, spherical apical cell and a large, elongated basal cell (Figure 4A). The apical cell undergoes two longitudinal divisions to produce a four-celled embryo (Figure 4B), followed by a transverse division to form an octant embryo proper (Figure 4C). A periclinal division of each of the 8 cells gives a 16-celled embryo. The resulting outer cell layer constitutes the protoderm, the first histologically detectable tissue, which gives rise to the epidermis (Figure 4D). The basal cell, on the other hand, undergoes a series of transverse divisions to form the suspensor, which is composed of a single file of 6 to 11 cells, and the hypophysis, the uppermost derivative of the basal cell (Figures 4B to 4E). Asymmetric division of the hypophysis gives rise to the hypophyseal cell, the lens-shaped cell at the base of the embryo proper that is the precursor of the root cap and the root quiescent center (Figure 4F) (Mansfield and Briarty, 1991; West and Harada, 1993).
Figure 4.
Terminal Phenotypes of opt3 Embryos.
Terminal phenotypes of opt3 embryos were determined in heterozygous siliques at the globular to early heart stages of normal embryo development. Early stages of embryogenesis in wild-type seeds are shown in (A) to (F), and representative terminal phenotypes of opt3 embryos are shown in (G) to (L). Developmental stages are as follows: (A), single cell; (B), (G), and (H), 2 or 4 cell; (C), (I), and (J), 8 cell; (D), 16 cell; (E) and (F), globular; and (K), 6 cell. (L) shows a mutant embryo in which cells from the octant stage divided longitudinally without previous periclinal division and the hypophysis was enlarged but did not divide to form the hypophyseal cell. Ac, apical cell; Ad, asynchronous division; Bc, basal cell; Ep, embryo proper; Hs, hypophyseal cell; Hy, hypophysis; Pd, periclinal division; Su, suspensor. Bars = 20 μm.
To determine the stage at which opt3 embryos are arrested during embryogenesis, 86 mutant embryos contained in heterozygous siliques at the globular to early heart stages of normal embryo development were scored for their terminal phenotypes. These developmental stages were selected because younger siliques (single to octant stages) contained mutant embryos that were difficult to distinguish from the wild type, whereas older siliques (torpedo to curled cotyledon stages) contained mutant embryos that already were disintegrating or had fully disintegrated. Of the 86 mutant embryos scored, 12% were arrested at the single or two-celled stage (Figure 4G), 51% were arrested at the two- or four-celled stage (Figure 4H), and 23% were arrested at the octant stage (Figure 4I). A small percentage of mutant embryos (1%) showed an elongated embryo proper (Figure 4J). In addition, 8% of mutant embryos showed asynchronous cell division, in which only two of the cells in the quadrant stage had undergone transverse division, giving a six-celled embryo (Figure 4K). In wild-type seeds, the 16-celled embryo underwent anticlinal and longitudinal divisions of the outer and inner cells, respectively, resulting in a radially symmetrical globular embryo (Figures 4E and 4F). In the case of opt3 embryos, a small percentage of the seeds examined (5%) showed embryos in which the cells that constitute the octant embryo proper underwent further cell divisions without previous periclinal division (Figure 4L), giving rise to embryos lacking the protoderm. Consistent with the early arrest in the development of the embryo proper, the hypophysis of opt3 embryos failed to undergo asymmetric cell division and thus lacked a differentiated hypophyseal cell (Figures 4G to 4L). The average suspensor length of 30 randomly selected opt3 embryos (arrested at the single-celled to octant stages) was comparable to that of wild-type embryos at the two-celled to octant stages of normal development (data not shown).
AtOPT3 Is Expressed in Vascular Tissues, Pollen, and Embryos
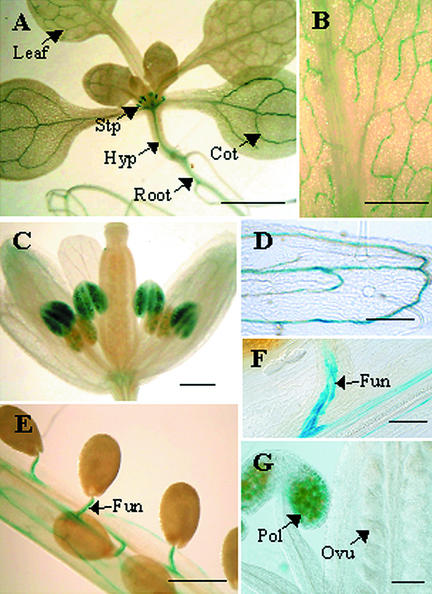
Five independent transgenic lines expressing an AtOPT3 promoter–GUS fusion were analyzed histochemically for GUS activity to assess the temporal and spatial patterns of AtOPT3 expression during plant growth and development. In light-grown seedlings, AtOPT3 was highly expressed in the vascular tissues of cotyledons, hypocotyls, rosette leaves, roots, and stipules (Figure 5A). Likewise, adult plants showed AtOPT3 expression in the vascular tissues of rosette leaves (Figure 5B), sepals (Figures 5C and 5D), and developing siliques and funiculi (Figures 5E and 5F). Weaker AtOPT3 expression also was detected in the vasculature of stamens and petals (data not shown). Strong expression of AtOPT3 was observed in pollen, but none was detected in unfertilized ovules (Figures 5C and 5G). The ubiquitous expression of AtOPT3 in various organs of both seedling and adult plants is consistent with a previous report on the expression pattern of AtOPT3 as determined by reverse transcriptase–mediated PCR (Koh et al., 2002).
Figure 5.
Representative AtOPT3 Promoter–GUS Expression Patterns in Seedlings and Tissues of Mature Plants.
5-Bromo-4-chloro-3-indolyl-β-d-glucuronide staining for AtOPT3 promoter–GUS expression in seedlings and mature plants is shown. Cot, cotyledon; Fun, funiculus; Hyp, hypocotyl; Ovu, ovule; Pol, pollen; Stp, stipules. Bar in (A) = 2 mm; bars in (B) to (E) = 0.5 mm; bars in (F) and (G) = 0.1 mm.
(A) A 10-day-old seedling showing strong GUS staining in the vascular tissues of cotyledons, rosette leaves, hypocotyls, roots, and stipules.
(B) A rosette leaf of a mature plant showing preferential GUS staining in vascular tissues.
(C) and (D) An unfertilized flower showing GUS staining in pollen (C) and in the vascular tissues of sepals (C) and (D).
(E) and (F) Vascular staining in siliques and funiculi.
(G) Anther and carpel of an unfertilized flower showing stained pollen and unstained ovules.
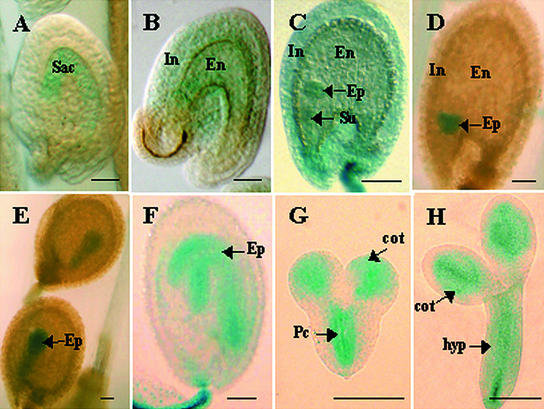
Immediately after fertilization (2 to 4 h after fertilization), AtOPT3 expression was detected in the embryo sac (Figure 6A); subsequently, AtOPT3 was expressed in developing maternal tissues as well (Figure 6B). By the globular stage, prominent GUS staining was observed in the developing endosperm, integument layers, the embryo proper, and the suspensor of the developing embryo (Figure 6C). At the heart stage, prominent staining of the developing embryo, but not the suspensor, was observed (Figure 6D). Although prominent GUS staining was observed in the embryo from the heart stage onward, very weak or no detectable staining was observed in the endosperm and integument tissues at these later stages (Figures 6D to 6F). The transition from globular to heart stage represents a change from radial to bilateral symmetry and marks the formation of the procambium, the precursor of vascular tissue, and the ground meristem (Mansfield and Briarty, 1991; West and Harada, 1993). Consistent with the expression of AtOPT3 in vascular tissues of seedlings and adult plants, strong GUS expression was observed in the developing vasculature of embryos as early as the heart stage (Figures 6G and 6H), with cotyledons showing stronger GUS staining in the developing veins at the curled cotyledon stage.
Figure 6.
AtOPT3 Promoter–GUS Expression in Developing Seeds.
5-Bromo-4-chloro-3-indolyl-β-d-glucuronide staining for AtOPT3 promoter–GUS expression in seeds is shown at different stages of embryo development. cot, developing cotyledon; En, endosperm; Ep, embryo proper; hyp, developing hypocotyl; In, integument layers; Pc, procambium; Sac, embryo sac; Su, suspensor. Bars = 60 μm.
(A) Two to 4 h after fertilization.
(B) Four to 8 h after fertilization.
(C) Globular stage.
(D) and (G) Heart stage.
(E) Torpedo stage.
(F) and (H) Curled cotyledon stage.
DISCUSSION
Characteristics of opt3 Embryos
During plant embryogenesis, the zygote undergoes a series of highly orchestrated developmental changes involving cell division, elongation, and differentiation (West and Harada, 1993; Souter and Lindsey, 2000; Jürgens, 2001). None of the opt3 embryos examined showed the characteristic periclinal division of cells that constitute the octant embryo proper, either because they were blocked before this stage (86%) or because cells in the octant stage divided further without previous periclinal division (5%). Moreover, the hypophysis did not undergo the asymmetric cell division necessary for the formation of the hypophyseal cell. Thus, opt3 embryos are compromised in cell division very early in development and are not able to establish the earliest precursor cells for embryonic organ systems: the protoderm cells for the epidermis and the hypophyseal cell for the embryonic root cap and the root quiescent center. Arrested opt3 embryos then undergo cellular degradation starting at the torpedo stage of normal development, coinciding with the time when the suspensor cells of wild-type embryos undergo programmed cell death (Marsden and Meinke, 1985; Mansfield and Briarty, 1991). Although AtOPT3 is clearly essential for embryo growth, it is not required for normal endosperm development, as shown by the apparently normal endosperm in seeds harboring opt3 embryos.
A wide variety of Arabidopsis genes involved in embryo development have been identified. These include genes in biotin biosynthesis (BIO1 [Schneider et al., 1989] and BIO2 [Patton et al., 1998]), cell division (KNOLLE [Lukowitz et al., 1996] and FACKEL [Jang et al., 2000]), intron splicing (SUS2 [Meinke, 1995]), translation (TWN2 [Zhang and Sommerville, 1997] and EDD1 [Uwer et al., 1998]), and hormone signaling (AXR6 [Hobbie et al., 2000] and ABP1 [Chen et al., 2001]). A summary of information on genes known to be involved in seed development, including descriptions of defective embryo development in various mutants, can be found at www.seedgenes.org. In a majority of the known embryo-defective mutants, embryos were arrested after the dermatogen stage, usually at the transition from the globular to the heart stage (Errampalli et al., 1991; Castle et al., 1993; McElver et al., 2001). Examples of Arabidopsis mutants arrested very early in embryogenesis (i.e., before the dermatogen stage) include twn2, a mutation in the valRS gene (Zhang and Sommerville, 1997), and titan (ttn) mutants (i.e., ttn1, ttn2, ttn4, ttn5, ttn7, ttn8, and ttn9), which are disrupted in genes likely involved in modulating chromosome integrity or microtubule assembly during mitosis (Liu and Meinke, 1998; McElver et al., 2000; Liu et al., 2002; Tzafrir et al., 2002). In these mutants, developmental arrest is accompanied by gross abnormalities such as the formation of basal cell–derived multiple embryos, in the case of twn2 (Zhang and Sommerville, 1997), and abnormal endosperm tissue with giant polyploid nuclei (Tzafrir et al., 2002), in the case of the ttn mutants.
By contrast, a notable phenotype of opt3 embryos, in addition to arrest very early in development, is aberrant cell division, which is observed in a small percentage of mutant seeds examined. These include embryos lacking the protoderm layer as a result of the absence of the periclinal division that gives rise to this tissue (5%), as well as embryos that undergo asynchronous cell division (8%). These abnormalities in cell division are similar to those found in certain pattern-formation mutants. For example, a mutation in the Arabidopsis KNOLLE (KN) gene, which encodes a protein related to syntaxins, resulted in embryos that did not exhibit periclinal divisions (Lukowitz et al., 1996). Likewise, asynchronous cell division was observed in axr6 embryos, which are defective in a gene involved in auxin response (Hobbie et al., 2000). Unlike opt3 embryos, kn and axr6 embryos exhibited sustained cell division throughout embryogenesis, giving rise to mutant seedlings with no morphologically distinct epidermis layer (Mayer et al., 1991) and abnormal root, hypocotyl, and vascular tissues (Hobbie et al., 2000), respectively.
Function of AtOPT3 in Plant Growth and Development
In Arabidopsis, nine OPT orthologs were identified that exhibit 61 to 85% sequence similarity (Koh et al., 2002). When tested for the ability to transport peptides and peptide derivatives, AtOPTs were able to take up only selected tetrapeptides and pentapeptides but not dipeptides and tripeptides. Therefore, although the exact nature of the physiological substrate(s) for AtOPT3 remains unanswered, the data suggest that this substrate(s) is likely a small peptide, larger than a tripeptide, or a modified peptide. The expression of AtOPT3 in the vascular tissues of seedlings and mature plants suggests its possible role in the long-distance translocation of nutritionally important peptides necessary for plant growth and development. Significant levels of peptides were reported in phloem and xylem exudates, including non-protein-derived peptides such as alanylaminobutyric acid and glycylketoglutaric acid (Higgins and Payne, 1982). There are certain advantages in plant cells transporting peptides as opposed to individual amino acids. Transported peptides can provide a wide variety of amino acids to cells and may be a more efficient means of long-distance transport of protein degradation products during leaf senescence and seed germination (Higgins and Payne, 1982). Transport of peptides also can protect amino acids from catabolism by enzymes known to be present in the phloem (Higgins and Payne, 1980).
AtOPT3 expression is induced in the embryo sac immediately after fertilization. By the globular stage, AtOPT3 is expressed in all of the maternal and filial tissues of the developing seed. This expression pattern very early in embryogenesis is consistent with the arrest of opt3 embryos at the preglobular stage. Nutrient and signal exchanges between the developing embryo, the endosperm, and the maternal tissues are an important part of seed development. The early developmental arrest of opt3 embryos, as well as the temporal and spatial expression patterns of AtOPT3 in the filial and maternal tissues, suggests the crucial role of peptide transport during the earliest stages of embryogenesis. The induction of AtOPT3 expression after fertilization is coincident with the formation of extensive embryo sac wall ingrowths and the degeneration of the three antipodal cells (Mansfield and Briarty, 1991). These changes in the embryo sac are believed to be indicative of an increased flux of biomolecules from the maternal tissues into the embryo sac. No specific function during reproduction has been attributed to the antipodal cells, although they may function in the transport of nutrients and growth substances for the embryo sac or they may serve as an additional nutrient source upon their degeneration after fertilization (Mansfield and Briarty, 1991; Mansfield et al., 1991). Therefore, peptide transport by AtOPT3 appears to be among the earliest physiological events occurring during embryogenesis, namely, the import of peptide(s) from the maternal tissues and/or the degenerated antipodal cells immediately after fertilization.
Although not much is known about the role of peptides in Arabidopsis during seed development, the importance of peptides in other plants has been reported. For example, in kidney bean seeds, 34% of nonprotein amino nitrogen is present as a γ-glutamyl peptide (γ-glutamyl-S-methyl-l-Cys), which is degraded upon seed germination and thus likely functions as a storage form for nitrogen and/or sulfur (Goore and Thompson, 1967). In barley, a peptide transport system that can transfer dipeptides and tripeptides across the scutellum layer, a specialized absorptive tissue abutting the endosperm, was reported (Higgins and Payne, 1978; West et al., 1998; Waterworth et al., 2000). It was proposed that storage proteins present in barley endosperm are hydrolyzed and transferred as peptides to the growing embryo. Therefore, the embryo-lethal phenotype of opt3 embryos, as well as the expression of AtOPT3 in the endosperm and the embryo, is consistent with the possibility that peptides or modified peptides serve as an important nitrogen source for the developing embryo. Specifically, peptides could be an important form of storage nitrogen in the endosperm and cotyledons as well as a form of imported nitrogen from the plant into the developing seed. The expression of amino acid permeases (AAP1 and AAP2) was demonstrated in Arabidopsis seeds, and these transporters were suggested to provide the developing seeds with amino acids for storage protein synthesis (Hirner et al., 1998). However, the physiological function of various nitrogenous compounds and their transporters in seed development remains poorly characterized.
Although it is possible that AtOPT3 plays a solely nutritional role during embryogenesis, one can argue that the opt3 embryos may be blocked in the transport of an important developmental regulator. Indeed, there is a growing body of information on biologically active peptides or peptide derivatives and their importance in plant growth and development (Bisseling, 1999; Matsubayashi et al., 2001; Lindsey et al., 2002). For example, plant growth factors such as auxin and gibberellin are bound frequently to small peptides, and these peptide-hormone conjugates are present in many tissues, including the vascular system and the endosperm of plant seeds (Salisbury and Ross, 1992). Phytosulfokines are disulfated pentapeptides or tetrapeptides that stimulate plant cell division in tissue culture (Matsubayashi and Sakagami, 1996). Regardless of whether AtOPT3 plays a nutritional or a signaling role, the data presented here provide evidence for the critical importance of the OPT family of transporters in seed development. Isolation of less severe opt3 alleles would be useful in further defining the function of AtOPT3. The observed phenotype of the opt3 lesion clearly suggests a specific function of AtOPT3 in embryogenesis. The observed differences in the tissue-specific and subcellular expression patterns of the various AtOPTs, as well as differences in their substrate specificity, suggest that each of the AtOPT transporters plays a unique role in plant metabolism (Koh et al., 2002). Further elucidation of the timing of expression and localization of AtOPTs and the identification of their cognate substrates will add significantly to our understanding of plant growth and development.
METHODS
Isolation of the opt3 Allele and Genetic Analysis
A total of 60,480 T-DNA–tagged Arabidopsis thaliana seeds generated at the University of Wisconsin Knockout Arabidopsis facility (Madison, WI) were screened for opt3 insertion alleles by PCR (see www.biotech.wisc.edu/Arabidopsis for methods). Insertions within AtOPT3 were identified using a primer specific for the T-DNA left border (JL202, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) in tandem with OPT3-specific primers (OPT3-A, 5′-AAGACTAATGTCCATCTCTCCTCGGACCA-3′; OPT3-B, 5′-TCGAGCGCTGCAGAGAGT-ACGTAATTGTA-3′). The locations and orientations of the primers are shown in Figure 1A. Appropriate PCR bands were identified by DNA gel blot analysis using the AtOPT3 cDNA (Koh et al., 2002) as a probe. One line carrying an opt3 allele, designated N4, was identified and sequenced with a T-DNA–specific primer (JL270, 5′-TTTCTCCATATTGACCATTCATACTCATTTG-3′). T-DNA copy number was determined by DNA gel blot analysis using a 546-bp NPTII cDNA fragment as a probe. For seed amplification, the N4 plant was allowed to self.
The genotype of the N4 line with regard to the opt3 allele, as well as subsequent T3 and T4 plants, was determined using a PCR-based approach. Briefly, genomic DNA was isolated from each plant analyzed and used as a template for PCR amplification of DNA fragments corresponding to the wild-type AtOPT3 allele and the opt3 insertion allele. The wild-type allele was amplified with the AtOPT3 gene-specific primers OPT3-A and OPT3-B to give a 2.4-kb PCR product. The opt3 mutant allele was amplified with OPT3-B and the T-DNA–specific primer JL202 to give a 1.2-kb PCR product. PCR products were detected by agarose gel electrophoresis. Partial sequencing using JL202 or OPT3-A confirmed that the observed PCR products corresponded to wild-type AtOPT3 or mutant opt3 sequences.
Plant Growth Conditions and Transformation
Seedlings were germinated aseptically on agar medium containing half-strength Murashige and Skoog (1962) salts (Sigma, St. Louis, MO) and 1% Suc (w/v) supplemented as required with 50 μg/mL kanamycin or 20 μg/mL hygromycin. For seed amplification and analysis of mature plants, 10-day-old seedlings were transferred to Pro-Mix soil (Premier Horticulture, Red Hill, PA) and grown at 22°C under constant fluorescent white light. Stable transformation of Arabidopsis was performed after the vacuum infiltration procedure for Agrobacterium tumefaciens–mediated T-DNA gene transfer (Bechtold and Pelletier, 1998).
Phenotypic Analysis
The growth phenotypes of plants harboring the opt3 allele were observed carefully, making note of plant size, shape, and growth rate during vegetative and reproductive growth phases. Leaf and flower phenotypes were observed for any gross defect using a stereoscope (SZX12; Olympus, Tokyo, Japan). Defective seed development was noted by dissecting siliques of self-pollinated plants and enumerating the number of wild-type and aborted seeds present in each silique. At least eight siliques were scored per plant.
To determine the terminal phenotypes of opt3 embryos, seeds from heterozygous siliques were cleared in Hoyer's solution (7.5 g of gum arabic, 100 g of chloral hydrate, and 5 mL of glycerol in 30 mL of water) and observed for defects in embryogenesis, taking note of the terminal phenotypes of arrested embryos. Abnormalities in endosperm and integument development also were observed. Observation and documentation of morphology were performed using an Eclipse E600 microscope (Nikon Tokyo, Japan) equipped with Nomarski optics or a Leica SP2 laser scanning confocal microscope (Wetzlar, Germany) attached to a photo multiplier.
Construction of the AtOPT3 Promoter–β-Glucuronidase Fusion and Analysis of Transgenic Plants
A 3.5-kb fragment containing sequences upstream of the predicted ATG start codon of AtOPT3 was amplified by PCR using OPT3pFor (5′-ACTGGATCCGGTCCAGTAGGCCATTTCACAT-3′) and OPT3pRev (5′-AGCGAATTCCTGGCAGAAAGTGAATGCTGTT-3′) primers. The amplified AtOPT3 promoter was cloned as an EcoRI-BamHI fragment into the binary vector pCAMBIA 1391Z (Hajdukiewicz et al., 1994) upstream of a promoterless β-glucuronidase (GUS) gene. Transformed Arabidopsis plants carrying the promoter–GUS fusion were selected based on hygromycin resistance encoded in the T-DNA and were stained for GUS using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as a substrate according to published protocols (Jefferson et al., 1987). Stained seedlings and tissues were fixed in 50% ethanol, 5% acetic acid, and 3.7% formaldehyde and destained in 70% ethanol overnight. Additional overnight clearing with lactophenol (water:glycerol:lactate:phenol, 1:1:1:2 [v/v]) was performed for seeds after the ethanol treatment to visualize the embryos. Staining patterns were observed and documented using a stereomicroscope (Olympus SZX12) or an Eclipse E600 microscope (Nikon) equipped with Nomarski optics.
Genetic Complementation
A 6.2-kb fragment encompassing the AtOPT3 genomic DNA and upstream sequences was cloned into the BamHI and BstEII sites of pCAMBIA 1391Z (Hajdukiewicz et al., 1994). Transformed Arabidopsis plants carrying the opt3 allele were selected for hygromycin and kanamycin resistance. Selected plants were allowed to self and analyzed for the segregation of the embryo-lethal phenotype. PCR was used to distinguish transformed plants heterozygous at the opt3 locus. As a control, plants heterozygous at the opt3 locus that were transformed with the AtOPT3 promoter–GUS fusion described above also were analyzed for genetic complementation.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Acknowledgments
The authors gratefully acknowledge the helpful suggestions and comments of Albrecht von Arnim during the course of this study and the helpful review of the manuscript by Walter Gassmann. This research was supported by National Research Initiative Grant 99-35304-8194 from the U.S. Department of Agriculture.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005629.
References
- Basrai, M.A., Lubkowitz, M.A., Perry, J.R., Miller, D., Krainer, E., Naider, F., and Becker, J.M. (1995). Cloning of a Candida albicans peptide transport gene. Microbiology 141, 1147–1156. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Becker, J.M., and Naider, F. (1980). Transport and utilization of peptides by yeast. In Microorganisms and Nitrogen Sources, J.W. Payne, ed (New York: John Wiley & Sons), pp. 257–279.
- Bisseling, T. (1999). The role of plant peptides in intercellular signaling. Curr. Opin. Plant Biol. 2, 365–368. [DOI] [PubMed] [Google Scholar]
- Blackmore, C.G., McNaughton, P.A., and van Heen, H.W. (2001). Multidrug transporters in prokaryotic and eukaryotic cells: Physiological functions and transport mechanisms. Mol. Membr. Biol. 18, 97–103. [DOI] [PubMed] [Google Scholar]
- Bourbouloux, A., Shahi, P., Chakladar, A., Delrot, S., and Bachhawat, A.K. (2000). Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275, 13259–13265. [DOI] [PubMed] [Google Scholar]
- Castle, L.A., Errampalli, D., Atherton, T.L., Franzmann, L.H., Yoon, E.S., and Meinke, D.W. (1993). Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol. Gen. Genet. 241, 504–514. [DOI] [PubMed] [Google Scholar]
- Chen, J.-G., Ullah, H., Young, J.C., Sussman, M.R., and Jones, A.M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmers, F.J.M., Lanfermiejer, F.V., and Poolman, B. (2001). Peptides and ATP-binding cassette peptide transporters. Res. Microbiol. 152, 245–258. [DOI] [PubMed] [Google Scholar]
- Errampalli, D., Patton, D., Castle, L., Mickelson, L., Hansen, K., Schnall, J., Feldman, K., and Meinke, D.W. (1991). Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Y., Sugawara, M., Liu, J., Li, H.W., Ganapathy, V., Ganapathy, M.E., and Leibach, F.H. (2000). cDNA structure, genomic organization, and promoter analysis of the mouse intestinal peptide transporter, PEPT1. Biochim. Biophys. Acta 1492, 145–154. [DOI] [PubMed] [Google Scholar]
- Fei, Y.-J., Kanai, Y., Nussberger, S., Ganapathy, V., Leibach, F.H., Romero, M.F., Singh, S.K., Boron, W.F., and Hediger, M.A. (1994). Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 368, 563–566. [DOI] [PubMed] [Google Scholar]
- Frommer, W.B., Hummel, S., and Rentsch, D. (1994). Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Lett. 347, 185–189. [DOI] [PubMed] [Google Scholar]
- Ganapathy, M.E., Brandsch, M., Prasad, P.D., Ganapathy, V., and Leibach, F.H. (1995). Differential recognition of β-lactam antibiotics by intestinal and renal peptide transporter, PepT1 and PepT2. J. Biol. Chem. 270, 25672–25677. [DOI] [PubMed] [Google Scholar]
- Goore, M.Y., and Thompson, J.F. (1967). Gamma-glutamyl transpeptidase from kidney bean fruit. I. Purification and mechanism of action. Biochim. Biophys. Acta 132, 15–26. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hauser, M., Donhardt, A.M., Barnes, D., Naider, F., and Becker, J.M. (2000). Enkephalines are transported by a novel eukaryotic peptide uptake system. J. Biol. Chem. 275, 3037–3041. [DOI] [PubMed] [Google Scholar]
- Hauser, M., Narita, V., Donhardt, A.M., Naider, F., and Becker, J.M. (2001). Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 18, 105–112. [PubMed] [Google Scholar]
- Higgins, C.F., and Payne, J.W. (1978). Peptide transport by germinating barley embryo: Uptake of physiological di- and oligopeptides. Planta 138, 211–216. [DOI] [PubMed] [Google Scholar]
- Higgins, C.F., and Payne, J.W. (1980). Transport and utilization of amino acids and peptides by higher plants. In Microorganisms and Nitrogen Sources, J.W. Payne, ed (New York: John Wiley & Sons), pp. 609–637.
- Higgins, C.F., and Payne, J.W. (1982). Plant peptides. In Encyclopedia of Plant Physiology, Vol. 14A, D. Boulder and B. Parthier, eds (New York: Springer Verlag), pp. 438–458.
- Hirner, B., Fischer, W.N., Rentsch, D., Kwart, M., and Frommer, W.B. (1998). Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 14, 535–544. [DOI] [PubMed] [Google Scholar]
- Hobbie, L., McGovern, M., Hurwitz, L.R., Pierro, A., Liu, N.Y., Bandyopadhyay, A., and Estes, M. (2000). The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127, 23–32. [DOI] [PubMed] [Google Scholar]
- Inui, K.I., Tomita, T., Katsura, T., Okano, T., Takano, M., and Hori, R. (1990). H+ coupled active transport of bestatin via the dipeptide transport system in rabbit intestinal brush-border membranes. J. Pharmacol. Exp. Ther. 260, 482–486. [PubMed] [Google Scholar]
- Jang, J.-C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14, 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, G. (2001). Apical-basal pattern formation in Arabidopsis embryogenesis. EMBO J. 20, 3609–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, S., Donhardt, A.M., Sharp, J., Naider, F., Becker, J.M., and Stacey, G. (2002). An oligopeptide transporter gene family in Arabidopsis thaliana. Plant Physiol. 128, 21–29. [PMC free article] [PubMed] [Google Scholar]
- Kuchler, K., Sterne, R.E., and Thorner, J. (1989). Saccharomyces cerevisiae STE6 gene product: A novel pathway for protein export in eukaryotic cell. EMBO J. 8, 3973–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, R., Fei, Y.J., Prasad, P.D., Ramammorthy, S., Han, H., Yang, F.R.L., Hediger, M.A., Ganapathy, V., and Leibach, F.H. (1995). Human intestinal H+/peptide co-transporter: Cloning, functional expression, and chromosomal localization. J. Biol. Chem. 270, 6456–6463. [DOI] [PubMed] [Google Scholar]
- Lindsey, K., Casson, S., and Chilley, P. (2002). Peptides: New signaling molecules in plants. Trends Plant Sci. 7, 78–83. [DOI] [PubMed] [Google Scholar]
- Liu, C.-M., McElver, J., Tzafrir, I., Joosen, R., Wittich, P., Patton, D., Van Lemmeren, A.A., and Meinke, D.W. (2002). Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 29, 405–415. [DOI] [PubMed] [Google Scholar]
- Liu, C.-M., and Meinke, D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16, 21–31. [DOI] [PubMed] [Google Scholar]
- Lubkowitz, M.A., Barnes, D., Breslav, M., Burchfield, A., Naider, F., and Becker, J.M. (1998). Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Mol. Microbiol. 28, 729–741. [DOI] [PubMed] [Google Scholar]
- Lubkowitz, M.A., Hauser, L., Breslav, M., Naider, F., and Becker, J.M. (1997). An oligopeptide transport gene from Candida albicans. Microbiology 143, 387–396. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jürgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1991). Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 69, 461–476. [Google Scholar]
- Mansfield, S.G., Briarty, L.G., and Erni, S. (1991). Early embryogenesis in Arabidopsis thaliana. I. The mature embryo sac. Can. J. Bot. 69, 447–460. [Google Scholar]
- Marsden, M.P.F., and Meinke, D.W. (1985). Abnormal development of the suspensor in an embryo-lethal mutant of Arabidopsis thaliana. Am. J. Bot. 72, 1801–1812. [Google Scholar]
- Matsubayashi, Y., and Sakagami, Y. (1996). Phytosulfokine, sulfated peptides that induce proliferation of single mesophyll cell of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93, 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, Y., Yang, H., and Sakagami, Y. (2001). Peptide signals and their receptors in higher plants. Trends Plant Sci. 6, 573–577. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Ruiz, R.A.T., Berleth, T., Misera, S., and Jürgens, G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353, 402–407. [Google Scholar]
- McElver, J., Patton, D., Rumbaugh, M., Liu, C.M., Yang, L.J., and Meinke, D.W. (2000). The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12, 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver, J., et al. (2001). Insertional mutagenesis of genes required in seed development in Arabidopsis thaliana. Genetics 159, 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, J.P., and Varshavsky, A. (1989). The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature 340, 400–404. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. (1982). Embryo-lethal mutants of Arabidopsis thaliana: Evidence for gametophytic expression of the mutant genes. Theor. Appl. Genet. 63, 381–386. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. (1995). Molecular genetics of plant embryogenesis. Annu. Rev. Plant Physiol. 46, 369–394. [Google Scholar]
- Meinke, D.W., and Sussex, I.M. (1979). Embryo-lethal mutants of Arabidopsis thaliana: A model system for genetic analysis of plant embryo development. Dev. Biol. 72, 50–61. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, 12 12F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Neefjes, J.J., Momburg, F., and Hammerling, G.J. (1993). Selective and ATP-dependent translocation of peptides by the MHC-encoded transporter. Science 261, 769–771. [DOI] [PubMed] [Google Scholar]
- Okano, T., Inui, K., Maegawa, H., Takano, M., and Hori, R. (1986. a). H+ coupled uphill transport of aminocephalosporins via the dipeptide transport system in rabbit intestinal brush-border membranes. J. Biol. Chem. 261, 14130–14134. [PubMed] [Google Scholar]
- Okano, T., Inui, K., Takano, M., and Hori, R. (1986. b). H+ gradient-dependent transport of aminocephalosporins in rat intestinal brush-border membrane vesicles: Role of dipeptide transport system. Biochem. Pharmacol. 35, 1781–1786. [DOI] [PubMed] [Google Scholar]
- Patton, D.A., Schetter, A.L., Franzmann, L.H., Nelson, K., Ward, E.R., and Meinke, D.W. (1998). An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin biosynthesis. Plant Physiol. 116, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, J.W., and Smith, M.W. (1994). Peptide transport by microorganisms. Adv. Microb. Physiol. 36, 52–69. [DOI] [PubMed] [Google Scholar]
- Perry, J.R., Basrai, M.A., Steiner, H.Y., Naider, F., and Becker, J.M. (1994). Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol. Cell. Biol. 14, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury, F.B., and Ross, C.W. (1992). Hormones and growth regulators: Auxins and gibberellins. In Plant Physiology, F.B. Salisbury and C.W. Ross, eds (Belmont, CA: Wadsworth Publishing), pp. 357–381.
- Schneider, T., Dinkins, R., Robinson, K., Shellhammer, J., and Meinke, D.W. (1989). An embryo-lethal mutant of Arabidopsis thaliana is a biotin auxotroph. Dev. Biol. 131, 161–167. [DOI] [PubMed] [Google Scholar]
- Shepherd, J.C., Schumacher, T.N., Ashton-Rickardt, P.G., Imaeda, S., Ploegh, H.L., Janeway, C.A., Jr., and Tonegawa, S. (1993). TAP1-dependent peptide translocation in vitro is ATP-dependent and peptide selective. Cell 74, 577–584. [DOI] [PubMed] [Google Scholar]
- Song, W., Koh, S., Czako, M., Marton, L., Drenkard, E., Becker, J.M., and Stacey, G. (1997). Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol. 114, 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W., Steiner, H.Y., Zang, L., Naider, F., Becker, J.M., and Stacey, G. (1996). Cloning of a second Arabidopsis peptide transport gene. Plant Physiol. 110, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter, M., and Lindsey, K. (2000). Polarity and signaling in plant embryogenesis. J. Exp. Bot. 51, 971–983. [DOI] [PubMed] [Google Scholar]
- Stacey, G., Koh, S., Granger, C., and Becker, J.M. (2002). Peptide transport in plants. Trends Plant Sci. 7, 257–263. [DOI] [PubMed] [Google Scholar]
- Steiner, H.Y., Naider, F., and Becker, J.M. (1995). The PTR family: A new group of peptide transporters. Mol. Microbiol. 16, 825–834. [DOI] [PubMed] [Google Scholar]
- Steiner, H.Y., Song, W., Zhang, L., Naider, F., Becker, J.M., and Stacey, G. (1994). An Arabidopsis peptide transporter is a member of a new class of membrane transport proteins. Plant Cell 6, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift, S., Throup, J.P., Williams, P., Salmond, G.P.C., and Stewart, G.S.A.B. (1996). Quorum sensing: A population density component in the determination of bacterial phenotype. Trends Biol. Sci. 21, 214–219. [PubMed] [Google Scholar]
- Tzafrir, I., McElver, J.A., Liu, C.-m., Yang, L.J., Wu, J.Q., Martinez, A., Patton, D.A., and Meinke, D.W. (2002). Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128, 38–51. [PMC free article] [PubMed] [Google Scholar]
- Uedel, S., Meyer, T.H., Kraas, W., Kienle, S., Jung, G., Wiesmuller, K.-H., and Tamp, R. (1995). Requirements for peptide binding to the human transporter associated with antigen processing revealed by peptide scans and complex peptide libraries. J. Biol. Chem. 270, 18512–18516. [DOI] [PubMed] [Google Scholar]
- Uwer, U., Willmitzer, L., and Altmann, T. (1998). Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10, 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth, W.M., West, C.E., and Bray, C.M. (2000). The barley scutellar peptide transporter: Biochemical characterization and localization to the plasma membrane. J. Exp. Bot. 51, 1201–1209. [PubMed] [Google Scholar]
- West, C.E., Waterworth, W.M., Stephens, S.M., Smith, C.P., and Bray, C.M. (1998). Cloning and functional characterization of a peptide transporter expressed in the scutellum of barley grain during the early stages of germination. Plant J. 15, 221–230. [DOI] [PubMed] [Google Scholar]
- West, M.A.L., and Harada, J.J. (1993). Embryogenesis in higher plants: An overview. Plant Cell 5, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L.E., and Miller, A.J. (2001). Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 659–688. [DOI] [PubMed] [Google Scholar]
- Zhang, J.Z., and Sommerville, C.R. (1997). Suspensor-derived polyembryony caused by altered expression of valyl-tRNA synthetase in the twn2 mutant of Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7349–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]