Abstract
Isolated mesophyll cells from Zinnia elegans are induced by auxin and cytokinin to form tracheary elements (TEs) in vitro with high synchrony. To reveal the changing patterns of gene expression during the 48 h of transdifferentiation from meso-phyll to TE cell fate, we used a cDNA–amplified fragment length polymorphism approach to generate expression profiles of >30,000 cDNA fragments. Transcriptional changes of 652 cDNA fragments were observed, of which 304 have no previously described function or sequence identity. Sixty-eight genes were upregulated within 30 min of induction and represent key candidates for the processes that underlie the early stages of commitment and differentiation to a TE cell fate.
INTRODUCTION
A central goal in plant developmental biology is to understand how embryonic cells differentiate into the 40 or so cell types that constitute plants. Meristems of shoot and root are a source of cells that must be maintained in a proliferative state, often regarded as undifferentiated, so one approach has been to study mutants in which meristematic function has been compromised (Haecker and Laux, 2001). A second approach has been to study mutants in which a clear developmental phenotype for a particular cell type can be identified, for example, root hairs (Parker et al., 2000), trichomes (Hulskamp and Kirik, 2000), or xylem (McCann and Roberts, 2000). Together, these two approaches have led to the identification of key regulatory genes involved in either the maintenance of an undifferentiated state or the promotion of a differentiated state. In the future, global gene expression technologies may permit the dissection of downstream events through comparisons of mutants in these pathways; however, to date, only a few genes have been identified that are specific to particular cell types. Many genes involved in vascular cell fates have been identified in two large-scale cDNA-sequencing projects using material derived from young xylem tissue of loblolly pine (Allona et al., 1998) and poplar (Sterky et al., 1998). These EST databases are derived from many vascular cell types and from asynchronously differentiating cells; consequently, the cell specificity of each EST and the order of differentiation-related events are difficult to establish. However, tissue-specific transcript profiles have been obtained using DNA microarray analysis of 3000 ESTs of poplar (Hertzberg et al., 2001).
The use of a remarkable synchronized in vitro cell system, the Zinnia elegans mesophyll cell system (Fukuda and Komamine, 1980), allows us to establish the chronology of molecular and biochemical events during the commitment and differentiation to a specific cell fate (McCann and Roberts, 2000; Milioni et al., 2001). Cells isolated from the leaves of Zinnia cv Envy, an ornamental plant, are put into liquid culture and supplied with two plant growth factors, auxin and cytokinin. Cells that were already differentiated as photosynthetic mesophyll cells in the leaf now transdifferentiate—change cell fate—to become tracheary elements (TEs) (Fukuda and Komamine, 1980). Sheets of epidermal cells also can be induced to transdifferentiate to TE fate (Church and Galston, 1989). TEs are cells, dead at maturity, that in the plant form a series of connected tubes that transport water and dissolved minerals from the root to the shoot.
During the formation of TEs, hoops of secondary cell wall material are deposited and subsequently stiffened and waterproofed by the deposition of lignin. Finally, the end wall of the cell is broken down and the cell contents are autolysed. The transdifferentiation of mesophyll or epidermal cells in vitro to a TE fate is analogous to the ability of cortical cells in wounded stems to transdifferentiate into xylem elements and reestablish vascular continuity (Sinnott and Bloch, 1945). Thus, TE fate is terminal. The Zinnia mesophyll cell system has several key advantages for studying xylogenesis in vitro rather than in planta. First, the entry into a new developmental pathway is induced by adding plant growth factors that act as a molecular switch to start the process of transdifferentiation. Second, the cells that form TEs do so with remarkable synchrony, making it possible to stage more precisely the events involved in building a TE.
We have shown that the exogenous growth factors, auxin and cytokinin, are not required in the first 48 h after isolation of Zinnia mesophyll cells; furthermore, as little as 10 min of exposure to the growth factors at 48 h is both necessary and sufficient to commit cells to the TE differentiation pathway (Milioni et al., 2001). These findings suggest that the first 48 h of culture represents a time in which the cells adapt to liquid culture and acquire the competence to respond to the inductive signals (McCann, 1997; Milioni et al., 2001). At 48 h, the auxin and cytokinin act as a switch to start the developmental pathway of TE formation. Other signals then are required for cells to progress through the subsequent stages of differentiation leading ultimately to cell death. By delaying the addition of auxin and cytokinin for 48 h, we have improved the synchrony of the culture significantly, so that up to 60% of the cells transdifferentiate to TEs in just 48 h after the addition of growth factors (Milioni et al., 2001). The precise start point, good synchrony, and compressed time scale of the transdifferentiation process provide a new and improved context in which to discover the earliest genes involved in switching on the developmental program.
To determine the range of genes involved in this complex process, we require sequence information on both high- and low-abundance transcripts. We selected five time points at which to sample mRNA populations: they reflect early changes in gene expression (30 min) and immediate downstream events (4 h), the time at which secondary wall synthesis begins (24 h), and the time at which TEs begin to deposit lignin and autolyse their cellular contents (48 h). We then performed an extensive cDNA–amplified fragment length polymorphism (cDNA-AFLP) study (Bachem et al., 1996; Durrant et al., 2000) in which we systematically characterized the amplification products whose abundance changed during the 48 h in which the cells transdifferentiate. Of the ∼30,000 cDNA fragments inspected, 652 (∼2.1%) exhibited altered expression profiles. A subset of 68 of these 652 ESTs was accumulated within 30 min of adding the auxin and cytokinin, and these represent a set of candidate genes whose expression is involved in the very earliest events in cell commitment and differentiation.
RESULTS
A cDNA-AFLP Screen Identified 652 Differentially Accumulated ESTs during the Time Course of TE Formation
The addition of auxin and cytokinin to Zinnia mesophyll cells, after they have been cultured in maintenance medium for 48 h, is followed by TE differentiation of ∼60% of the cell population between 96 and 120 h (Milioni et al., 2001). Poly(A+) RNA was extracted from Zinnia cells at 48 h without growth factors (regarded as time 0) and then at 30 min, 4 h, 24 h, and 48 h after the addition of the growth factors at time 0 and used for cDNA-AFLP analysis, as described in Methods. The cDNA expression profiles were determined by PCR selective amplification using 512 different primer combinations, and we screened ∼30,000 cDNA fragments. Comparison of fingerprints obtained from these five cell populations identified transcript-derived fragments (TDFs) from genes that were accumulated differentially (Figure 1). As anticipated, the majority of the TDFs showed similar levels of accumulation in all five samples. To examine the reproducibility of the fragment banding patterns, reaction products that derived from two sets of independently prepared samples of mRNA were compared. Using 12 combinations of primers, we detected ∼800 cDNA fragments. The differential amplification of the TDFs was reproducible (see supplemental data online).
Figure 1.
A cDNA-AFLP Autoradiogram Showing the Accumulation Patterns of TDFs from Zinnia Cultured Cells during TE Formation.
(A) Templates were derived from Zinnia cells cultured for 48 h in medium with no growth factors (lane 0) and 0.5, 4, 24, and 48 h after the addition of auxin and cytokinin. Lanes are in groups of five: each group was amplified using a different primer combination with two selective nucleotides.
(B) Enlarged view of the boxed region in (A). Various expression patterns can be detected across the time course: constitutive expression (i), downregulation (ii), transient expression (iii), and upregulated expression (iv).
A total of 652 differentially accumulated TDFs, ranging in length from 50 to 450 bp, were recovered from gels and reamplified, subcloned, and sequenced. Each sequence was identified by homology search using the Basic Local Alignment Search Tool (BLAST) program (Altschul et al., 1997) against the GenBank nonredundant public sequence database. A total of 349 fragments (53.5%) of the differentially expressed genes showed close matches (BLASTX expectation values [E] of <10−5) to database entries with assigned identities. We classified these sequences into 13 groups based on functional categories established for Arabidopsis (Arabidopsis Genome Initiative, 2000), and the proportion of genes in each category is shown in Figure 2. The major group is involved in primary and secondary metabolism and energy generation (19.2%), whereas a slightly higher proportion (8%, compared with 5.6% in Arabidopsis) is cell wall related, reflecting the restructuring of the primary wall and the deposition of secondary cell wall during TE formation. An additional 9.7% of the TDFs are involved in information processing and constitute genes involved in transcriptional control and signal transduction. Interestingly, 12.4% of the sequences share significant similarity to unknown or hypothetical genes with no assigned function from various genome projects (Figure 2). These fragments represent new candidate proteins involved in cell fate determination, differentiation, cell wall remodeling, and cell death. Furthermore, no function could be assigned to 34% of the clones, because they showed no or only poor sequence similarity to any database entries. They may represent either previously uncharacterized genes or fragments too short to reveal any significant identity. The distribution of our 652 sequences by putative function is similar to that for the loblolly pine and poplar EST databases (Allona et al., 1998; Sterky et al., 1998).
Figure 2.
Classification of Differentially Accumulated Zinnia ESTs during in Vitro TE Formation.
On the basis of BLASTX expectation (E) values of <10−5, 348 sequences were assigned to functional categories, 80 sequences shared homology with hypothetical proteins or proteins of unknown function, and 224 sequences showed no or low similarity to existing sequences.
Validation of Expression Patterns by Reverse Transcriptase–PCR Analysis in the Zinnia System
To validate the cDNA-AFLP expression profiles, reverse transcriptase (RT)–PCR and/or RNA gel blot analysis were used to study 41 differentially accumulated TDFs using mRNA populations harvested from cells at the same time points used for the cDNA-AFLP analysis. Of these TDFs, 12 were selected for their similarity to genes implicated previously in the formation of vascular tissues (Figure 3). The cDNA-AFLP sequences were named Ze (for Zinnia elegans) followed by the number corresponding to the AFLP fragment. Zinnia cDNA sequences Ze203, Ze349, and Ze622 were similar to MONOPTEROS (E < 10−24) (Berleth et al., 2000), PIN1 (E < 10−26) (Gälweiler et al., 1998), and REVOLUTA/IFL1 (E < 10−10) (Talbert et al., 1995; Ratcliffe et al., 2000) from Arabidopsis, respectively. All of them were accumulated or induced within 4 h and have been identified in Arabidopsis mutant screens with vascular phenotypes. Two TDFs, Ze452 and Ze414, similar to CESA (E < 10−31) (Richmond, 2000) and CSL (E < 10−16) (Richmond and Somerville, 2001), respectively, were shown to be accumulated differentially in differentiating TEs of Zinnia (data not shown).
Figure 3.
Comparison of Expression Patterns of Selected Genes by Both cDNA-AFLP and RT-PCR.
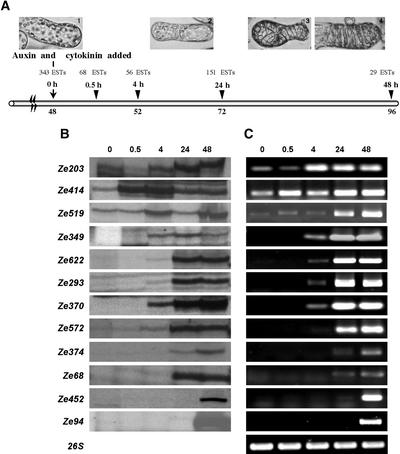
(A) Time course of Zinnia mesophyll cells transdifferentiating into TEs, and the corresponding number of ESTs first accumulated at each time point.
(B) and (C) Comparison of expression patterns of 12 cDNA-AFLP fragments, similar to genes known to be involved in vascular differentiation, by cDNA-AFLP (B) and RT-PCR (C). RNA templates were derived from Zinnia cells cultured in noninductive medium for 48 h (lane 0) and cells left for 30 min, 4 h, 24 h, and 48 h (lanes 0.5, 4, 24, and 48, respectively) after the addition of auxin and cytokinin. Ze203 (MONOPTEROS-like), Ze519 (ZCAD1-like), and Ze414 (CSL-like) transcripts were present already before the addition of auxin and cytokinin. Ze349 (PIN1-like), Ze622 (REVOLUTA/IFL1-like), Ze572 (TED3-like), Ze370 (PEL1-like), and Ze293 (EXP1-like) were upregulated at the 4-h time point. The mRNAs for genes similar to Ze452 (CESA-like), Ze374 (TED4-like), Ze68 (TED2-like), and Ze94 (LACCASE-like) were upregulated at later stages of the developmental pathway.
Marker genes isolated previously from the Zinnia system also were identified in our screen, demonstrating that the cDNA-AFLP results are fully comparable with data obtained from alternative methods of differential display. The TDFs Ze68, Ze572, and Ze374 were most similar to the molecular markers TED2 (E < 10−8), TED3 (E < 10−20), and TED4 (E < 10−8), of which TED3 is expressed preferentially in cells that transdifferentiate into TEs (Demura and Fukuda, 1994). Ze370 was similar to ZePEL1 (E < 10−12), which is involved in remodeling the cell wall (Domingo et al., 1998), whereas Ze293 was similar to ZeEXP1 (E < 10−14), which is expressed in developing TEs in vitro (Im et al., 2000). Ze519 and Ze94 were similar to ZCAD1 (E < 10−36) (Sato et al., 1997) and LACCASE-like (E < 10−8) (LaFayette et al., 1999), respectively, which are known to be expressed during lignification. The accumulation patterns observed by cDNA-AFLP for these selected genes were confirmed by semiquantitative PCR (Figure 3).
Various numbers of PCR cycles were tested to ensure that the reactions had not reached the plateau, as described in Methods. In addition, RNA gel blot analysis was used to confirm the accumulation patterns of the Ze121, Ze198, Ze48, Ze138, and Ze266 cDNA-AFLP fragments. Ze121, Ze198, and Ze48 showed similarity to PG1 (E < 10−5) and ZeEXP1 (E < 10−14) and to a ripening-related protein–like gene (E < 10−5) from Arabidopsis, respectively. Ze138 and Ze266 were most similar to Arabidopsis putative endo-β-1,4- glucanase (E < 10−36) and peroxidase (E < 10−16) genes, respectively (see supplemental data online). RNA gel blot analysis was performed using the differentially amplified cDNA fragments as digoxigenin-labeled probes. The comparisons described above showed that 32 of the 41 cDNAs examined had the same expression profiles as on the original cDNA-AFLP analysis (data not shown). The remaining nine cDNAs all showed differential expression patterns on subsequent RT-PCR or RNA gel blot analysis, but these differed from the original cDNA-AFLP patterns (see supplemental data online). These results indicate that the original cDNA-AFLP pattern was validated in 78% of the cases, so the general approach is a reliable method for identifying differentially expressed genes in the Zinnia system.
Sixty-Eight ESTs Are Expressed within 30 min of the Addition of Growth Factors to the Zinnia System
The differentially expressed genes identified in this screen can be divided into two main categories: 43% of the 652 ESTs are expressed at 48 h (before the addition of auxin and cytokinin), whereas the remainder are expressed at very low or undetectable levels and then upregulated. From the second category, we identified 68 cDNA-AFLP fragments that correspond to genes that are newly expressed within 30 min of adding auxin and cytokinin. Table 1 lists all of the sequences within that category, together with three genes downregulated within 30 min that show high similarity (E < 10−5) to sequences in the public databases. Some of the induced sequences, such as those for Ze620 (IAA3-like), Ze92 (GH3-like), and Ze235 (NAC4-like), were similar to those of auxin-induced genes. The IAA/AUX and GH3 gene families are known to be induced very early by auxin. The proteins encoded by the IAA/AUX family are short-lived nuclear proteins that are involved in auxin signaling (Ouellet et al., 2001). NAC domain–containing proteins (for petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2) are a family of transcription factors, specific to plants, that play a role in a diverse set of developmental processes. Sequences corresponding to proteins involved in metabolic processes such as ion, nucleotide, and amino acid transportation, energy production, and protein metabolism are induced. Because relatively little is known about basic metabolic processes during xylogenesis, information on the induction of these genes will help our understanding of the coordination of metabolism during vascular development. Furthermore, early changes in the expression profiles of 10 other genes with no previous functional description were observed. These 10 genes, and others, represent new candidate genes involved in the early stages of the TE developmental pathway.
Table 1.
cDNA-AFLP Fragments DIfferentially Expressed within 30 min of Addition of Growth Factors to the Zinnia Systema
Zinnia cell cultures were supplied with auxin and cytokinin for the times (in hours) indicated within the accumulation pattern column.
The Zinnia System Provides a Set of Marker Genes for Xylogenesis
To confirm that the cDNA-AFLP screen provides reliable potential markers for vascular development, we chose three differentially accumulated TDFs representative of unknown proteins predicted from the Arabidopsis sequencing project. These were selected by their cDNA-AFLP expression patterns: Ze469 was expressed very early (30 min), Ze126 was expressed later in the time course (24 h), and Ze86 was expressed at 48 h. The expression of these genes was studied in a more extensive time course by semiquantitative RT-PCR (Figure 4). RNA was reverse transcribed from cells cultured for 48 h in the absence of growth factors (i.e., at time 0) and from cells at six time points (30 min and 4, 24, 48, 60, and 72 h) after the addition of auxin and cytokinin. Additionally, expression profiles were determined in cells exposed for 72 and 96 h to either auxin or cytokinin only. There was no detectable transcript accumulation in Zinnia cells cultured for 48 h in the absence of the plant growth factors (i.e., at time 0). Ze469 mRNA accumulation (first detected at 30 min after the addition of auxin and cytokinin to the culture) reached a peak level 48 h later and started declining thereafter (Figure 4). Transcripts of Ze126 were first observed at high levels 24 h after the addition of auxin and cytokinin. Ze86 was seen at 48 h after the addition of the growth factors and then declined; it was not detected in cells exposed to either auxin or cytokinin alone (Figure 4). The expression of both Ze469 and Ze126 was upregulated by auxin only, whereas the expression of the former also was induced by cytokinin alone.
Figure 4.
Expression of Ze469, Ze126, and Ze86 cDNAs in Cells Cultured in Vitro.
(A) The lanes contain total RNA from cells cultured in noninductive medium for 48 h (lane 1), from cells at 30 min and 4, 24, 48, 60, and 72 h (lanes 2, 3, 4, 5, 6, and 7, respectively) after the addition of auxin and cytokinin at 48 h, from cells in medium containing 1.0 mg/L auxin but no cytokinin for 72 and 96 h (lanes 8 and 9), and from cells in medium containing 1.0 mg/L cytokinin but no auxin for 72 and 96 h (lanes 10 and 11). The corresponding Zinnia accession numbers are presented at the end of Methods.
(B) cDNA-AFLP expression patterns of Ze469, Ze126, and Ze86. The Zinnia cells were cultured with auxin and cytokinin for the times indicated above the gels (in hours) before harvesting.
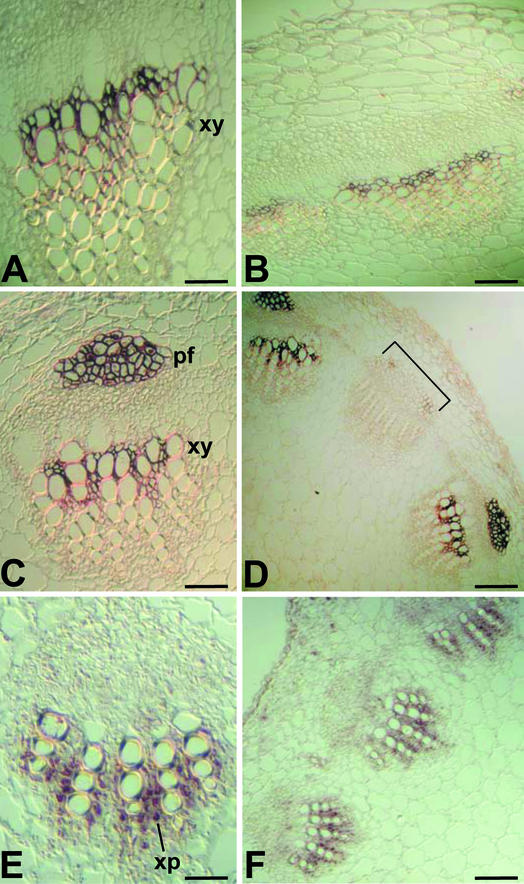
To determine the spatial patterns of expression of Ze469, Ze126, and Ze86, we performed in situ RNA hybridization experiments in transverse sections of stem tissues from the second internodes of 4-week-old Zinnia plants (Figure 5). Sections were hybridized with Ze469, Ze126, and Ze86 antisense and sense probes. No hybridization signals were detected using sense probes (data not shown). Transcripts of Ze469 were localized to young xylem tissue within the vascular bundle (Figures 5A and 5B). The presence of Ze126 transcripts could be seen in both young xylem tissue and phloem fiber cells (Figures 5C and 5D). Because fibers, like vessel elements and TEs, deposit thick lignified secondary walls, it is likely that common genes are involved in secondary wall thickening in both cell types. There was a strong Ze126 signal in alternating vascular bundles around the circumference of the stem, with no signal detected in the leaf trace bundles (Figure 5D). Transcripts of Ze86 (Figures 5E and 5F) were restricted to xylem parenchyma cells. These data demonstrate that Ze469, Ze126, and Ze86 transcripts are under temporal and spatial regulation and are all correlated with vascular cell differentiation.
Figure 5.
Transverse Sections of the Inflorescence Stems of 4-Week-Old Zinnia Plants Probed with Partial Sequences of Ze469, Ze126, and Ze86 by in Situ Hybridization.
The sections were probed with antisense digoxigenin-labeled Ze469 RNA ([A] and [B]), Ze126 RNA ([C] and [D]), and Ze86 RNA ([E] and [F]). The Ze469 transcripts were localized to young xylem tissue (xy). The Ze126 transcript strongly accumulated in the phloem fibers (pf) and in young xylem tissue (xy) of alternate vascular bundles. No signal was detected in vascular bundles (bracket in [D]) going to the leaves. The Ze86 transcripts were detected mainly in older xylem parenchyma cells (xp). Shown are photographs of bright-field images using Nomarski optics. Bars = 70 μm in (A), (C), and (E) and 120 μm in (B), (D), and (F).
DISCUSSION
The main advantage of using the Zinnia system and the cDNA-AFLP approach over the existing EST databases is in being able to establish the precise chronology of changing gene expression events during transdifferentiation. The improvements that we made previously to the synchrony of the time course (Milioni et al., 2001) permit a precise mapping of cytological, biochemical, and molecular markers, whereas the cDNA-AFLP screen gives a reasonable indication of the temporal expression profiles of relevant genes. The formation of TEs involves several processes fundamental to plant development, including cell division, local cell signaling, cell elongation, cell fate specification, cell wall synthesis and deposition, lignification, and programmed cell death, involving the activity of hundreds of genes (McCann, 1997).
In this article, we report the isolation of 652 differentially accumulated ESTs across the time course of TE formation in the Zinnia mesophyll cell system using cDNA-AFLP analysis. We estimate that we have sampled two-thirds of the original mRNA populations using one pair of restriction enzymes and all possible primer combinations and that the total number of genes involved in TE formation could be ∼1000. We inspected the sequences of 47 Zinnia mRNAs and found that 66% contained suitable ApoI-MseI fragments of between 50 and 450 bp. This finding suggests that 66% of the genes expressed could be visualized using this enzyme combination. Some genes may be expressed constitutively and yet play a role in the transdifferentiation process. Others may show differential expression and yet not be involved directly in xylogenesis. The reliability of the expression patterns as displayed by the cDNA-AFLP approach was verified. The reproducibility was high even when samples of mRNA prepared from independent batches of starting material were used. A more detailed analysis of a subset of the 652 genes, by RT-PCR and RNA gel blotting, showed that all members of the subset displayed differential expression patterns and that 78% of them matched exactly the cDNA-AFLP pattern. Therefore, we conclude that the cDNA-AFLP screen reliably detects differential patterns of gene expression in the Zinnia system.
Twelve of the differentially accumulated cDNA-AFLP sequences to which we can assign an identity by comparison with the public databases are from genes already known to be involved in processes related to TE formation (McCann and Roberts, 2000). Studies indicate that the various members of the CESA gene family have specialized roles in primary and secondary wall formation (Taylor et al., 1999; Williamson et al., 2001). The Ze452 (CESA-like) gene reported here has highest sequence similarity to Arabidopsis, cotton, and poplar sequences implicated in secondary wall formation. The REVOLUTA/IFL1 gene encodes a homeodomain-Leu zipper protein essential for the normal differentiation of interfascicular fibers and secondary xylem in the inflorescence stem (Zhong and Ye, 1999). Local increased levels of auxin may control cell specification in cambial cells in poplar (Uggla et al., 1996) and in the basal embryo and in the cells surrounding the initials of the seedling root in Arabidopsis (Sabatini et al., 1999). The PIN1 gene encodes an auxin efflux-carrier protein that establishes polar auxin transport in the embryo by the polar localization of PIN1, later becoming localized on the basal side of xylem parenchyma cells (Gälweiler et al., 1998). Ze203 (MONOPTEROS-like) and Ze349 (PIN1-like) TDFs are accumulated or induced within 4 h of the addition of auxin and cytokinin in the Zinnia system. Auxin transport inhibitors can phenocopy the mutant monopteros, which fails to form an interconnected vascular network. MONOPTEROS is a transcription factor similar to ARF1, an auxin response factor that binds to auxin-responsive elements in the promoters of auxin-responsive genes (Berleth and Sachs, 2001).
Auxin and cytokinin have long been implicated in the process of vascular development. Recent genetic and molecular analyses have confirmed the role of auxin as a coordinating signal across the plant (Berleth and Sachs, 2001) and implicated auxin as a positional signal in pattern formation (Uggla et al., 1996). Cytokinin is known to promote the induction of TE differentiation in many tissue culture systems (Blee et al., 2001; Milioni et al., 2001), including differentiation of xylem and phloem tissues in tobacco pith explant cultures in vitro (Boucheron et al., 2002) and in Eucalyptus tumors (Azmi et al., 2001).
Zinnia cells can be committed to a TE fate even by 10 min of exposure to auxin and cytokinin; they then differentiate to TEs even when the growth factors have been removed. Thus, differentiation events after 10 min clearly are independent of exogenous auxin and cytokinin (Milioni et al., 2001). Sixty-eight TDFs are switched on within 30 min of the addition of auxin and cytokinin to Zinnia cultures; the expression of these genes is the earliest known event in the signaling networks that result in TE formation. The sequences of Ze620, Ze92, and Ze235 were similar to those of known auxin-induced genes such as the IAA/AUX and GH3 gene families and the NAC domain–containing proteins. The CmNACP gene is expressed in vascular tissues and is involved in shoot-to-meristem signaling (Ruiz-Medrano et al., 1999). Several lines of evidence indicate that NAC1, a transcription activator, mediates auxin signaling to promote lateral root development. An increase of NAC1 transcript level was detected after only 30 min of exposure of Arabidopsis wild-type root cultures to auxin (Xie et al., 2000). These 68 TDFs provide the opportunity for further studies needed to delineate the relationship between the auxin- and/or cytokinin-regulated signaling networks and the functions of both the known and the novel genes that are expressed differentially during the first stages of the developmental pathway of TE formation. A next step is to distinguish genes that are regulated directly by auxin and/or cytokinin from those that are secondary targets. However, additional signals, including ethylene, jasmonate, brassinosteroids, gibberellic acid, and Ca2+, are required to complete TE formation (Fukuda, 1996). Cell commitment triggered by auxin and cytokinin is rapid, but the final restriction to TE fate depends on responses to a network of other signal pathways.
Many plant cells with primary cell walls are capable of growth and division and are pluripotent, which means that some already differentiated cells may be respecified and transdifferentiate to a new cell fate. However, the formation of tracheids is a process of progressive restriction to a single cell fate and is intrinsically irreversible. It is not clear whether the response to auxin and cytokinin is sufficient to commit the cell to a TE fate or if there is a series of intermediate cell fate options, with the final commitment to a TE fate potentially occurring as late as cell autolysis. The idea of the commitment to a TE fate being a multistep process is based on the expression patterns of three TED transcripts (Demura and Fukuda, 1994). It was proposed that these genes are expressed in a temporal order during in vitro culture that corresponds to a spatial distribution in the living plant associated with the progressive restriction of cell fate. Although we did not find these particular genes expressed in the same temporal order in our more synchronous system, either by RNA gel blot analysis (Milioni et al., 2001) or by cDNA-AFLP pattern, this remains an attractive hypothesis. There are two possibilities. The first is that these intermediate stages are simply stages in TE formation that the cell, once committed, must transit. The second is that fate is restricted progressively—for example, to vascular fate, then to xylem, and finally to TE, each step depending on further signaling inputs. If the latter possibility is the case, then it should be possible to provide other signals that would divert the cell to a different developmental pathway (e.g., to induce transdifferentiation to phloem sieve elements). Because TE formation is not cell autonomous until a late stage in the process, it seems more likely that the cells are not committed solely to a TE cell fate by the action of auxin and cytokinin.
From our results, two lines of evidence provide strong support for the idea that the Zinnia system is a robust model for normal plant vascular development rather than for a special pathway of wound-induced vascular connectivity. The first is the presence in the cDNA-AFLP screen of several genes (e.g., Ze203 [MONOPTEROS-like]) that are involved in the establishment of normal vasculature. The second is the in situ results, which confirm that three genes of unknown function from the screen are expressed in plants in different cell types within the vascular bundle, not simply those forming TEs. The patterns of gene expression, when the formation of new vascular strands is taking place, is consistent with the auxin regulation of Ze126. It was shown that Ze469, a gene whose expression is regulated by both auxin and cytokinin in culture, is localized to young xylem tissue. The combination of auxin and cytokinin increases its expression, suggesting that they act in a synergistic manner. Ze86 transcripts are localized conspicuously to the xylem parenchyma cells of vascular bundles, yet they are accumulated late in culture. These patterns of accumulation suggest that each induced cell may adopt one of at least three different cell fates: TEs, xylem parenchyma, and phloem fibers (McCann et al., 2001). The final determination of which fate each cell will adopt probably is the result of complex cell-to-cell signaling interactions. This notion is consistent with progressive cell fate restriction following from an initial commitment to a more general vascular cell fate. It also raises the possibility that other intermediate cell states may be present in culture and that these might be identified by some of the new molecular markers described here.
Probably of greatest significance for understanding the molecular basis of xylogenesis is the fact that 40% of the sequences, including those that accumulate within 30 min, are unknowns: they are either hypothetical proteins in the Arabidopsis database or not represented in any of the plant or animal databases. We have demonstrated that a gene of previously unknown function, Ze469, is expressed within 30 min and is a marker of vascular cell fate in the stem, confirming that the Zinnia system is a useful engine of gene discovery with respect to the very earliest steps in the process of xylogenesis.
METHODS
Preparation of Cell Cultures
Mesophyll cells were isolated and cultured as described by Domingo et al. (1998) using medium with no growth factors, 1.0 mg/L benzylaminopurine and 1.0 mg/L naphthalene acetic acid (inductive medium), 1.0 mg/L benzylaminopurine alone, or 1.0 mg/L naphthalene acetic acid alone. Cells were transferred from noninductive to inductive media at 48 h from the wells in six-well plates into 30-mL screw-cap tubes (Bibby Sterilin, Stone, UK); the tubes were centrifuged at 800 rpm for 2 min, and the cells were gently suspended in new medium at a density of 106 cells/mL.
cDNA–Amplified Fragment Length Polymorphism
Total RNA was prepared from Zinnia elegans cells at different stages of in vitro tracheary element formation using the RNeasy Plant Kit (Qiagen, Valencia, CA). Poly(A+) RNA was isolated using magnetic oligo(dT)25 Dynabeads according to the manufacturer's instructions (Dynal Biotech, Wirral, UK). Synthesis of cDNA was performed with Expand Reverse Transcriptase, RNase H, and DNA Polymerase I (Roche Diagnostics, Lewes, UK). cDNA–amplified fragment length polymorphism (cDNA-AFLP) analysis was performed according to Bachem et al. (1996) and Durrant et al. (2000). Double-stranded cDNA was digested with the restriction enzymes ApoI and MseI. The adaptor primers 5′-CTCGTAGACTGCGTACC-3′ and 3′-CATCTG-ACGCATGGTTAA-5′ (ApoI) and 5′-GACGATGAGTCCTGAG-3′ and 3′-TACTCAGGACTCAT-5′ (MseI) were ligated to the restriction fragments, and a PCR preamplification step was performed using the adaptor primers without selective nucleotides. The selective PCR amplification step was performed using the primers 5′-GACTGC-GTACCAATT(C/T)NN-3′ (ApoI) and 5′-GATGAGTCCTGAGTAANN-3′ (MseI) (N represents G, C, A, or T). All AFLP reactions were performed as described by Durrant et al. (2000). The bands of interest were cut from the gel with a surgical blade, eluted, and reamplified with the same primer set used for the initial amplification. The reamplified cDNAs were subcloned using the pGEM-T vector system (Promega) and sequenced using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit with fluorescent sequencing AmpliTaq DNA polymerase (Perkin-Elmer). Database searches were performed at the NCBI World Wide Web server using the Basic Local Alignment Search Tool (BLAST) network service (Altschul et al., 1997). Each transcript-derived fragment sequence was compared against all sequences in the nonredundant database using the BLASTX program. Sequences that returned with no significant homology were compared again against the EST databases using the BLASTN program. Program default parameters were used for all analyses.
Reverse Transcriptase–PCR Analysis
Total and poly(A+) RNA were prepared as described above. First-strand cDNA synthesis was performed using Expand Reverse Transcriptase (Roche Diagnostics). The following primers were used for the reverse transcriptase (RT)–PCR experiments: for Ze203 (MONOPTEROS-like), 5′-CAATGCAACCTCCAACTCAAGAGCTTCATGT-3′ and 5′-AGATCGCCGTACTCCCATCGCCGTACTCCCAA-3′; for Ze414 (CSL-like), 5′-CTATCTTGGTGACCTTCAGGTGAAAAGT-3′ and 5′-ATCTCCATCACCATTTTCCTAAACAAATT-3′; for Ze-519 (ZCAD-like), 5′-CCAAGTTTAATGTAGGGGATTGCGTGGGAGTT-3′ and 5′-CTCTCAACATCAGTGCTGAGGAGGTAATCATC-3′; for Ze349 (PIN1-like), 5′-GACGGGAAACTTCATGTTACGGTTCGGAAATC-3′ and 5′-GGTTGAAACTAGACCCTCTAGGTGTTGGGTTTC-3′; for Ze622 (REVOLUTA/IFL-like), 5′-ACATCGCCCGAAGCAGTAACTCTAGCT-CAAT-3′ and 5′-GGCAAATTCTGGAACCAACGCCTTTCGTCCA-3′; for Ze293 (ZeEXP1-like), 5′-ACTTGTACAGTCAAGGTTACGGGG-TGAACACT-3′ and 5′-GGATGTGCGTCGGTCACTGGCTGTGAC-TCTAA-3′; for Ze370 (ZePEL1-like), 5′-CGTCTTGCTGACTGTGCC-ATTGGGTTTGGAAA-3′ and 5′-TGTAATTGTTAGAAATAGTGATGG-CAGTAGA-3′; for Ze572 (TED3-like), 5′-AGTTTGAGTAGCCATTCC-CATTGTTGGAAT-3′ and 5′-CAATAACATCCTCCTCGCCATCGTTAGCAC-3′; for Ze374 (TED4-like), 5′-ATGAGGTCAATTACATAC-ACATCACTGTATG-3′ and 5′-GCATTTCGGTATCGGAACCCCACA-AGCATTGG-3′; for Ze68 (TED2-like), 5′-ATGGTCAAAGCAATT-CGAATTCATGAATTTG-3′ and 5′-TGCATGTTGTGACTTCAGGACCAACTGCTATT-3′; for Ze452 (CESA-like), 5′-CCATTGTCTACC-CGTTTACTTCAATCCCT-3′ and 5′-ACCTTGCAAGCATGTGGTTCC-TTGCTCTT-3′; for Ze94 (LACCASE-like), 5′-CCCTGCCAAGGTTCC-ATTGACTATTGATCA-3′ and 5′-AACACTAATTGAACGGTTGAG-TTGTACTTTA-3′; for Ze126, 5′-CAGATATTTGTGGCTTCGTGC-CGCCTTTTCTG-3′ and 5′-GATGATACTTTTGCTTCTCAAATGCAA-AGATTG-3′; for Ze469, 5′-ACCAAGCTTTATTTCCATTATACAAACGAAAG-3′ and 5′-GGCTTGGCTTATTTTCTCCGGTGATCAGGTTA-3′; for Ze86, 5′-CACCATATGTTATGAAGCTCAAGCTAGGCTTCG-3′ and 5′-GAAGATCCTGTGTTCTGCCTCTGTAGAAGCT-3′; for Ze191 (CLPC-like), 5′-CGGCTTATGGCTTTGACTGCTTCATCTTGA-3′ and 5′-CACAAACTCCGCTACACCGATGATGCACTGGT-3′; for Ze186 (SNF2-like), 5′-AGTAGTACTCGACGCACACAAAGCATGCGAAT-3′ and 5′-AAGGTGAGATGACCATGTCTGGAGATTCACAT-3′, for Ze144 (Inositol 1,3,4-Trisphosphate 5/6 kinase-like), 5′-TCTGTCAGCACAGACTCATAACCCGGCATCT-3′ and 5′-AGTTTACATGGTTCGTTG-TCATTTTCGCAGGT-3′; for Ze396 (COMT-like), 5′-TGGCTCCTG-TGTGCAAGTTTTTGATCAAGAAT-3′ and 5′-GTATCCACTTCATAA-AAATAGCATCTCCT-3′; for Ze91 (SYBL1-like), 5′-GCAAGGTTC-TCTCCTCTATCAATGGCCTT-3′ and 5′-GGCAACTTTCCTGCAATT-GCAACTCAGTGTCTT-3′; and for Ze10 (CELA2-like), 5′-CATCATCGGCTGCTTTTGACGTGACAGTGA-3′ and 5′-ACCTTGCAA-GCATGTGGTTCCTTGCTCTT-3′.
For the analysis of differential expression, two rounds of RT-PCR were conducted with two independently isolated total RNA samples. RT-PCR was performed for 15, 20, 24, 28, 32, and 35 cycles to determine the linearity of the PCR. The thermal cycling parameters used for RT-PCR for all genes were as follows: 94°C for 15 s, 62°C for 30 s, and 72°C for 25 s for a total of 25 cycles. Ze203 was amplified under the same conditions for a total of 33 cycles. As a positive control, a 200-bp 26S rRNA fragment was amplified under the same RT-PCR conditions for a total of 18 cycles using the primer pair 5′-GCACCGATGAAGAAGAATCCATTGAAGA-3′ and 5′-AAAGGATTCTACCAGTCGCTTGATGGGA-3′. The gene specificity of RT-PCR products was confirmed by sequencing.
RNA in Situ Hybridization
Fragments from the Ze469, Ze126, and Ze86 cDNAs (0.5, 0.57, and 0.6 kb, respectively) were cloned into the pGEM-T vector. These fragments were amplified by PCR using the M13-40 and M13 reverse primers. Digoxigenin-labeled sense and antisense RNA probes for Ze469, Ze126, and Ze86 clones were synthesized using SP6 and T7 RNA polymerases (Roche Diagnostics). Tissue preparation and in situ hybridization were performed as described by Shu et al. (2000).
RNA Gel Blot Analysis
Total RNA was prepared from Zinnia cells at different stages of in vitro tracheary element formation, as described above. The RNAs (10 μg per slot) were electrophoresed on 1.4% denaturing formaldehyde-agarose gels and transferred onto Hybond N+ nylon membranes (Amersham). After immobilization by UV cross-linking, the blots were hybridized with the digoxigenin-labeled AFLP fragments Ze121, Ze198, Ze48, Ze138, and Ze266. Probe labeling was performed as described above. Hybridization was performed according to the protocol of the DIG Northern Starter Kit (Roche Diagnostics).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession numbers for the genes mentioned in this article are as follows: MONOPTEROS (At1g19850), PIN1 (At1g73590), REVOLUTA/IFL1 (At5g60690), CESA (At5g05170), CSL (At5g22740), TED2 (D30800), TED3 (D30801), TED4 (D30801), ZePEL1 (Y09541), ZeEXP1 (AAF35900), ZCAD1 (BAA19487), LACCASE-like (Y13769), PG1 (At1g10640), ripening-related protein–like gene from Arabidopsis (At5g62350), Arabidopsis putative endo-β-1,4-glucanase (At1g64390) and putative peroxidase (At5g05340), cotton CELA2 (U58284), poplar CEL1 (AF081534), Ze203 (MONOPTEROS-like; AJ316129), Ze519 (ZCAD1-like; AJ316135), Ze414 (CSL-like; AJ409100), Ze349 (PIN1-like; AJ316127), Ze622 (REVOLUTA/IFL1-like; AJ316133), Ze572 (TED3-like; AJ316128), Ze370 (PEL1-like; AJ316131), Ze293 (EXP1-like; AJ316132), Ze452 (CESA-like; AJ409101), Ze374 (TED4-like; AJ316139), Ze68 (TED2-like; AJ316136), Ze94 (LACCASE-like; AJ488929), Ze469 (AJ316118), Ze126 (AJ316137), and Ze86 (AJ316138).
Supplementary Material
Acknowledgments
Many thanks to Sue Bunnewell for photographic assistance, Ronald Sederoff for helpful discussions, Nicholas Carpita and Robert Sablowski for helpful discussions and for review of the manuscript, and Jonathan Jones and Wendy Durrant for assistance and advice with the cDNA-AFLP method. M.C.M., N.J.S., and K.R. gratefully acknowledge the financial support of the Biotechnology and Biological Science Research Council (BBSRC). D.M. was funded by a European Community Marie Curie Fellowship, P.-E.S. was funded by a BBSRC special studentship, and M.C.M. was funded by a Royal Society University Research Fellowship. We are grateful for grants from the Royal Society and the Leverhulme Trust in support of this work.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005231.
Footnotes
Online version contains Web-only data.
References
- Allona, I., Quinn, M., Shoop, E., Swope, K., St. Cyr, S., and Carlis, J. (1998). Analysis of xylem formation in pine by cDNA sequencing. Proc. Natl. Acad. Sci. USA 95, 9693–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Azmi, A., Dewitte, W., Van Onckelen, H., and Chriqui, D. (2001). In situ localization of endogenous cytokinins during shooty tumor development on Eucalyptus globulus Labill. Planta 213, 29–36. [DOI] [PubMed] [Google Scholar]
- Bachem, C.W.B., van der Hoeven, R.S., de Bruijn, S.M., Vreugdenhil, D., Zabeau, M., and Visser, R.G.F. (1996). Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: Analysis of gene expression during potato tuber development. Plant J. 9, 745–753. [DOI] [PubMed] [Google Scholar]
- Berleth, T., Mattsson, J., and Hardtke, C.S. (2000). Vascular continuity and auxin signals. Trends Plant Sci. 5, 387–393. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Sachs, T. (2001). Plant morphogenesis: Long-distance coordination and local patterning. Curr. Opin. Plant Biol. 4, 57–62. [DOI] [PubMed] [Google Scholar]
- Blee, K.A., Wheatley, E.R., Bonham, V.A., Mitchell, G.P., Robertson, D., Slabas, A.R., Burrell, M.M., Wojtaszek, P., and Bolwell, G.P. (2001). Proteomic analysis reveals a novel set of cell wall proteins in a transformed tobacco cell culture that synthesizes secondary walls as determined by biochemical and morphological parameters. Planta 212, 404–415. [DOI] [PubMed] [Google Scholar]
- Boucheron, E., Guivarc'h, A., Azmi, A., Dewitte, W., Van Onckelen, H., and Chriqui, D. (2002). Competency of Nicotiana tabacum L. stem tissues to dedifferentiate is associated with differential levels of cell cycle gene expression and endogenous cytokinins. Planta 215, 267–278. [DOI] [PubMed] [Google Scholar]
- Church, D.L., and Galston, A.W. (1989). Hormonal induction of vascular differentiation in cultured Zinnia leaf discs. Plant Cell Physiol. 30, 73–78. [Google Scholar]
- Demura, T., and Fukuda, H. (1994). Novel vascular cell-specific genes whose expression is regulated temporally and spatially during vascular system development. Plant Cell 6, 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo, C., Roberts, K., Stacey, N.J., Connerton, I., Ruíz-Teran, F., and McCann, M.C. (1998). A pectate lyase from Zinnia elegans is auxin inducible. Plant J. 13, 17–28. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E., and Jones, J.D.G. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, H. (1996). Xylogenesis: Initiation, progression and cell death. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 299–325. [DOI] [PubMed] [Google Scholar]
- Fukuda, H., and Komamine, A. (1980). Establishment of an experimental system for the tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 52, 57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Haecker, A., and Laux, T. (2001). Cell-cell signaling in the shoot meristem. Curr. Opin. Plant Biol. 4, 441–446. [DOI] [PubMed] [Google Scholar]
- Hertzberg, M., et al. (2001). A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. USA 98, 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp, M., and Kirik, V. (2000). Trichome differentiation and morphogenesis in Arabidopsis. Adv. Bot. Res. 31, 237–260. [Google Scholar]
- Im, K.H., Cosgrove, D.T., and Jones, A.M. (2000). Subcellular localization of expansin mRNA in xylem cells. Plant Physiol. 123, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFayette, P.R., Eriksson, K.E., and Dean, J.F. (1999). Characterization and heterologous expression of laccase cDNAs from xylem tissues of yellow-poplar (Liriodendron tulipifera). Plant Mol. Biol. 40, 23–35. [DOI] [PubMed] [Google Scholar]
- McCann, M.C. (1997). Tracheary element formation: Building up to a dead end. Trends Plant Sci. 2, 333–338. [Google Scholar]
- McCann, M.C., and Roberts, K. (2000). Xylogenesis: The birth of a corpse. Curr. Opin. Plant Biol. 3, 517–522. [DOI] [PubMed] [Google Scholar]
- McCann, M.C., Stacey, N.J., Dahiya, P., Milioni, D., Sado, P.E., and Roberts, K. (2001). Zinnia: Everybody needs good neighbors. Plant Physiol. 127, 1480–1482. [PMC free article] [PubMed] [Google Scholar]
- Milioni, D., Sado, P.E., Stacey, N.J., Domingo, C., Roberts, K., and McCann, M.C. (2001). Differential expression of cell-wall-related genes during the formation of tracheary elements in the Zinnia mesophyll cell system. Plant Mol. Biol. 47, 221–238. [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.S., Cavell, A.C., Dolan, L., Roberts, K., and Grierson, C.S. (2000). Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12, 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Riechmann, J.L., and Zhang, J.Z. (2000). INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA. Plant Cell 12, 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, T. (2000). Higher plant cellulose synthases. Genome Biol. 1, 300.1–300.6. [DOI] [PMC free article] [PubMed]
- Richmond, T.A., and Somerville, C.R. (2001). Integrative approaches to determining Csl function. Plant Mol. Biol. 47, 131–143. [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126, 4405–4419. [DOI] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 24, 463–472. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Watanabe, T., Komamine, A., Hibino, T., Shibata, D., Sugiyama, M., and Fukuda, H. (1997). Changes in the activity and mRNA of cinnamyl alcohol dehydrogenase during tracheary element differentiation in Zinnia. Plant Physiol. 113, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, G.G., Baum, D.A., and Mets, L.J. (2000). Detection of gene expression patterns in various plant tissues using non-radioactive mRNA in situ hybridisation. Biochem. News 1, 37–38. [Google Scholar]
- Sinnott, E.W., and Bloch, R. (1945). The cytoplasmic basis of intercellular patterns in vascular differentiation. Am. J. Bot. 32, 151–156. [Google Scholar]
- Sterky, F., et al. (1998). Gene discovery in the wood-forming tissues of poplar: Analysis of 5,692 expressed sequence tags. Proc. Natl. Acad. Sci. USA 95, 13330–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Adler, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla, C., Moritz, T., Sandberg, G., and Sundberg, B. (1996). Auxin as a positional signal in pattern formation in plants. Proc. Natl. Acad. Sci. USA 93, 9282–9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, R.E., Burn, J.E., and Hocart, C.H. (2001). Cellulose synthesis: Mutational analysis and genomic perspectives using Arabidopsis thaliana. Cell. Mol. Life Sci. 58, 1475–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.