Abstract
Maize centromeres are composed of CentC tandem repeat arrays, centromeric retrotransposons (CRs), and a variety of other repeats. One particularly well-conserved CR element, CRM, occurs primarily as complete and uninterrupted elements and is interspersed thoroughly with CentC at the light microscopic level. To determine if these major centromeric DNAs are part of the functional centromere/kinetochore complex, we generated antiserum to maize centromeric histone H3 (CENH3). CENH3, a highly conserved protein that replaces histone H3 in centromeres, is thought to recruit many of the proteins required for chromosome movement. CENH3 is present throughout the cell cycle and colocalizes with the kinetochore protein CENPC in meiotic cells. Chromatin immunoprecipitation demonstrates that CentC and CRM interact specifically with CENH3, whereas knob repeats and Tekay retroelements do not. Approximately 38 and 33% of CentC and CRM are precipitated in the chromatin immunoprecipitation assay, consistent with data showing that much, but not all, of CENH3 colocalizes with CentC.
INTRODUCTION
Although a large fraction of the protein-coding sequences in higher eukaryotes are broadly conserved, centromeric DNA sequences are not (Choo, 2001). Not only is there extreme size variation in eukaryotic centromeres, but there is remarkable structural variability as well. In human, the centromeres are estimated to be 3000 to 4000 kb (Schueler et al., 2001) and are composed primarily of simple ∼171-bp repeats known as α-satellites. In Drosophila, the ∼300-kb centromeres are composed of small repeats of only 5, 7, or 10 bp, interspersed with transposable elements (Sun et al., 1997). In Schizosaccharomyces pombe, the 40-kb centromeres contain central cores surrounded by long tandem repeats (Baum et al., 1994), and in Saccharomyces cerevisiae, the centromere is composed of three single-copy DNA elements that cover only 125 bp (Cottarel et al., 1989). Little or no homology has been found among these well-studied centromeres, and many of the sequences can be removed without consequence (Baum et al., 1994; Sun et al., 1997). The weak constraints on centromere size and sequence have led to extensive speculation that centromeres are activated and maintained primarily by epigenetic mechanisms (Karpen and Allshire, 1997; Henikoff et al., 2001). Noncentromeric sequences can acquire centromere activity de novo under some conditions, adding support to the epigenetic model (Lo et al., 2001a; Maggert and Karpen, 2001).
In higher plants, centromeres vary in size from 3000 to 9000 kb and appear to be composed primarily of tandemly arrayed repeats (satellites) of ∼150 to 180 bp (Kaszas and Birchler, 1996; Ananiev et al., 1998; Copenhaver et al., 1999; Kumekawa et al., 2000; Cheng et al., 2002). These centromeres can be reduced to as little as 5% of their original size and retain much of their capacity to segregate chromosomes (Kaszas and Birchler, 1998). Some homology can be detected among the tandem repeats from closely related species, such as sorghum and sugarcane (Miller et al., 1998a; Nagaki et al., 1998; Zwick et al., 2000) and maize and rice (Ananiev et al., 1998; Cheng et al., 2002). Larger repetitive DNA elements (>400 bp) also have been isolated from the centromeres of maize B chromosome and the maize chromosome 4 (Alfenito and Birchler, 1993; Page et al., 2001). In addition, plant centromeres contain abundant retrotransposons. This centromeric retrotransposon (CR) fraction is particularly noteworthy in cereal centromeres, in which many of the CR elements fall within a highly conserved phylogenetic clade of Ty3/gypsy elements (Miller et al., 1998b; Presting et al., 1998; Langdon et al., 2000). The DNA homology is sufficient that CR probes from sorghum or Brachypodium sylvaticum identify the centromeres in most or all of the chromosomes in agronomically significant cereals (rice, maize, wheat, sorghum, barley, and rye) (Aragon-Alcaide et al., 1996; Jiang et al., 1996; Miller et al., 1998b). This ancient origin (>60 million years) and slow mutation rate sets CR elements apart from the majority of retrotransposons in maize (SanMiguel et al., 1998; Langdon et al., 2000) and suggests that particular sequence elements, not only epigenetic mechanisms, may be responsible for organizing the centromere-kinetochore complex (Dong et al., 1998; Hudakova et al., 2001; Grimes et al., 2002).
Although much of the centromeric DNA is poorly conserved and genetically redundant, many of the kinetochore proteins that bind to centromeres and orchestrate chromosome movement are well conserved (Yu et al., 2000). Perhaps the most conserved of all known kinetochore proteins is centromere-specific histone H3, or CENH3 (Choo, 2001; Henikoff et al., 2001). The CENH3s are known variously as CENP-A (human, mouse, Xenopus), Cse4 (S. cerevisiae), Cnp1 (S. pombe), Cid (Drosophila), HCP-3 (Caenorhabditis elegans), and HTR12 (Arabidopsis) (Choo, 2001; Talbert et al., 2002). Data from several organisms indicate that CENH3 replaces histone H3 on active centromeric DNA (Yoda et al., 2000; Ahmad and Henikoff, 2001; Lo et al., 2001a; Blower et al., 2002) and is required to recruit other key kinetochore proteins such as CENP-C (Hooser et al., 2001; Ando et al., 2002). Each of the known CENH3s shares a common histone H3 core sequence, but they diverge in the N-terminal tail and an internal region known as loop 1 (Malik et al., 2002; Talbert et al., 2002). Both diverged regions interact with DNA in the nucleosome and show evidence of adaptive evolution, suggesting that CENH3 serves as a linker molecule between the rapidly evolving centromeric DNA and the conserved kinetochore machinery (Malik et al., 2002).
Because of its close association with DNA in the context of the centromeric nucleosomes, CENH3 has been used as a tool to identify the centromere sequences that interact with the kinetochore. In situ hybridization (Haaf and Ward, 1994; du Sart et al., 1997) and chromatin immunoprecipitation (ChIP) have been used to show that human CENP-A interacts with the α-satellite as well as novel sequences in human neocentromeres (Vafa and Sullivan, 1997; Lo et al., 2001a). Similarly, antibodies to yeast Cse4 immunoprecipitate yeast core centromere DNA (Meluh et al., 1998). In an effort to make similar advances in maize, we have identified and characterized the maize CENH3 homolog. We show that, as in Arabidopsis (Talbert et al., 2002), maize CENH3 is present in kinetochores throughout the cell cycle. ChIP analysis indicates that maize CENH3 interacts strongly with the centromeric satellite CentC but does not interact with noncentromeric DNA sequences. The ChIP assays also demonstrate that a recently isolated centromeric retrotransposon in maize (CRM) interacts with CENH3 throughout its length. These data provide strong support for the idea that specific sequences confer centromere identity in maize and that a conserved retrotransposable element is part of the functional centromere.
RESULTS
Maize CENH3 Localizes to the Inner Kinetochore throughout the Cell Cycle
Centromere-specific histones are noteworthy for their high homology with histone H3 in the histone fold domain but striking divergence in the N-terminal and loop-1 domains (Malik et al., 2002). Using BLAST searches, we identified a cDNA clone (as an EST) that shared these characteristics (GenBank AW244191). However, sequence comparisons between AW244191 and other putative CenH3s suggested the presence of an unspliced intron. This suspicion was confirmed when we identified a new cDNA by reverse transcriptase–mediated PCR from young embryo tissue (GenBank AF519807). AF519807 is the only complete and uninterrupted cDNA in the public sequence databases to date, although partial cDNA sequences have been identified in libraries from young tassels and ears as well. As shown in Figure 1, the full-length cDNA encodes a protein of 157 amino acids that shows 56% identity with maize histone H3. Consistent with data from other organisms (Henikoff et al., 2000; Malik et al., 2002; Talbert et al., 2002), the divergent N-terminal and loop-1 domains of CENH3 are longer than the comparable regions of histone H3 (Figure 1). Genomic DNA gel blot analysis produced a single band with a variety of different restriction enzymes, suggesting that CenH3 is a single-copy gene in maize (data not shown).
Figure 1.
Alignment of Maize Histone H3 and CENH3 Protein Sequences.
The underlined sequences are N-terminal and loop-1 domains, respectively. The boldface sequence from positions 48 to 58 was used for CENH3 antibody production. The asterisk indicates the beginning of the sequence cloned into pBAD for bacterial expression.
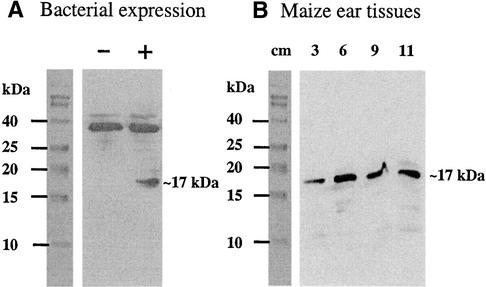
Polyclonal antibodies were raised in rabbits against an 11–amino acid peptide unique to CENH3 (Figure 1). To verify that the antibodies specifically recognized maize CENH3, protein gel blots were prepared from bacteria expressing a partial CENH3 cDNA (Figure 1) and from young maize ears at various stages. These data are shown in Figure 2. Both the bacterially expressed and maize endogenous CENH3s are predicted to encode proteins of 17 kD. As expected, the 17-kD protein was observed in bacteria only when expression was induced with l-arabinose (a higher molecular mass band was observed in both the induced and uninduced cultures; Figure 2A). A single 17-kD band also was readily apparent in young ears, indicating that the antibodies recognized the full-length CENH3 protein with high specificity (Figure 2B).
Figure 2.
Protein Gel Blot Analysis of Maize CENH3 Expression in E. coli and in Maize Young Ear Tissues.
(A) The expression of a partial cDNA of maize CENH3 (see Methods) was analyzed in E. coli before (−) and after (+) l-arabinose induction.
(B) Maize CENH3 expression in inbred W23 line young ears 3 to 11 cm in length.
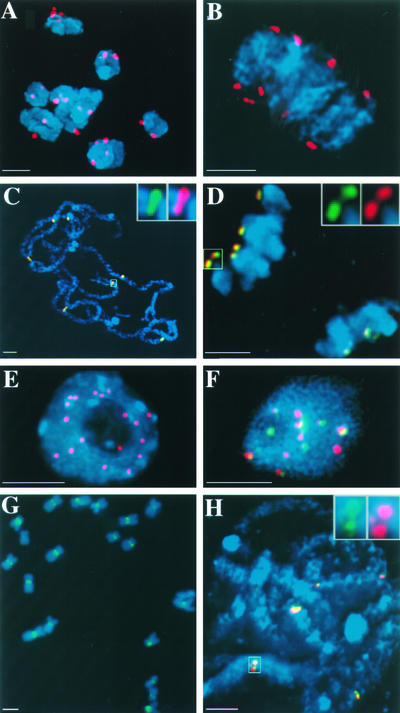
We conducted a variety of experiments to determine the localization of CENH3 with respect to the maize kinetochore protein CENPC (Centromere Protein C) as well as the centromeric DNA sequence CentC. CENPC, a conserved structural component of eukaryotic kinetochores, is localized to the inner kinetochore plate in maize throughout the cell cycle (Dawe et al., 1999; Yu et al., 1999). As illustrated in Figure 3, the CENH3 antiserum identified all 20 kinetochores in meiotic cells (Figures 3A and 3B) and colocalized almost perfectly with CENPC (Figures 3C and 3D). Anti-CENH3 staining was apparent at other prophase stages in meiosis but was relatively difficult to detect in late metaphase and early anaphase (data not shown). CENH3 also was detected throughout mitosis and in interphase, during which ∼20 different spots were apparent (Figure 3E). When interphase nuclei were double labeled with CENH3 and the CentC repeat array (Figure 3F), the two signals colocalized only approximately. Although the majority of CENH3 staining overlapped with the CentC staining, a large portion of the CentC signal did not overlap with the CENH3 staining. The size of the CENH3 spots was consistent among kinetochores (Figure 3E), but the size of the CentC repeat varied considerably from centromere to centromere (Figure 3F), consistent with the original description by Ananiev et al. (1998).
Figure 3.
Localization of CENH3, CENPC, and Centromeric Repeats in Meiosis and Mitosis.
Images are single optical sections unless noted otherwise. Chromosomes are shown in blue.
(A) CENH3 (red) localization at diakinesis of meiotic prophase I.
(B) CENH3 localization (red) at early meiotic metaphase I.
(C) Colocalization of CENH3 (red) and CENPC (green) in meiotic pachytene.
(D) Colocalization of CENH3 (red) and CENPC (green) in meiotic telophase I.
(E) CENH3 (red) localization at a root tip interphase cell. This image is a full projection showing all of the data in the three-dimensional data set.
(F) Colocalization of CENH3 (red) and CentC (green) in an endosperm nucleus.
(G) CRM (green), identified with the GAG.25 probe, on mitotic metaphase chromosomes. These data were not acquired using three-dimensional light microscopy.
(H) Colocalization of CRM identified with the LTR.50 probe (green) and the CentC probe (red) in meiotic pachytene.
Bars = 3 μm.
CRM, a Centromere-Specific Retrotransposon, Is Highly Conserved and Interspersed with CentC Arrays
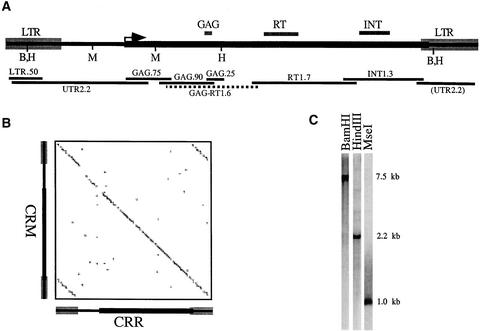
CRM is a recently discovered CR element that shows strong homology with a rice retrotransposable element called CRR (Cheng et al., 2002). One complete CRM element and five partial CRM elements were found in the centromeric BAC clone ZM16H10. To confirm the sequence and organization of CRM, we amplified by PCR and sequenced six different intervals of the retrotransposon from genomic DNA. The locations of these clones are shown in Figure 4A. Two clones from each region were sequenced and found to conform very closely to the CRM sequence of BAC clone ZM16H10 (only clarifying ambiguous bases and changing the overall length slightly, to 7572 bp). To further confirm that the sequenced CRMs are representative of the elements in the genome as a whole, we prepared genomic DNA gel blots of DNA digested with a variety of restriction enzymes. Strong single bands were present on these DNA gel blots when GAG.75 was used as a probe (Figure 4C), and in all cases, the fragment sizes were consistent with the restriction map of the final CRM sequence. These data suggest that most CRMs are intact, full-length elements.
Figure 4.
Structure of CRM, and Its Homology with CRR.
(A) CRM structure. The LTR, GAG, reverse transcriptase (RT), and integrase (INT) domains, and the open reading frame (arrow), are indicated above the line. Below the line are the positions of BamHI (B), HindIII (H), and MseI (M) sites as well as the positions and lengths of the cloned PCR fragments used in this study (GAG-RT1.6 was not used as a probe, but it was sequenced).
(B) Dot plot analysis of CRM and CRR. Areas with a perfect match of at least 11 bp are shown along the diagonal.
(C) Genomic DNA gel blot analysis of CRM. Total genomic DNA was digested with HindIII, BamHI, and MseI and probed with the GAG.75 probe.
CRM contains a single open reading frame that has homology with reverse transcriptase, integrase, and the zinc finger motif (CX2CX4HX4C) typical of the GAG protein (Figure 4A). The sequence homology between CRM and CRR is very high throughout the length of the retrotransposon, with an overall identity of 80% (Figure 4B). Of particular note is the high conservation in the noncoding long terminal repeat and 5′ untranslated region. CRM shows less homology with the previously published maize CR element CentA (Ananiev et al., 1998), with nearly complete divergence in the LTR and only 67% identity in the 5′ untranslated region and coding regions.
In situ localization of CRM confirmed that the retrotransposon is located at centromeres (Figure 3G) and that nearly all of the CRM elements lie within or closely flanking CentC tandem repeat arrays (Figure 3H).
CENH3 Interacts Specifically with CRM and CentC
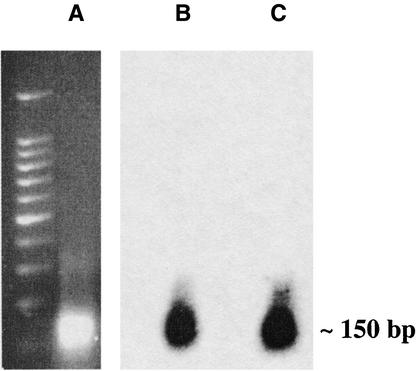
Native ChIP was used to determine if the various centromere repeats interact with CENH3. Young ear tissue was used for the ChIP assay because of its high mitotic index and the fact that CENH3 is abundant in this tissue (Figure 2B). After digestion of nuclei with micrococcal nuclease, nucleosomes were separated on a 2% agarose gel. As shown in Figure 5A, a predominant band (∼150 bp) was visualized after ethidium bromide staining, suggesting the chromatin had been reduced to mononucleosomes. The agarose gel was transferred to a nylon membrane and probed with CentC and the GAG.75 probe from CRM (Figures 5B and 5C). Bands of the same size were detected by both probes, suggesting that the resolution of our ChIP assay in centromeres is ∼150 bp.
Figure 5.
Micrococcal Nuclease Digestion of Chromatin to Produce Mononucleosomes.
(A) shows a 2% ethidium bromide gel. The gel was blotted and probed with CentC (B) and GAG.25 (C).
After incubating the nucleosome preparation with antiserum, immune complexes were precipitated and separated into unbound (S, for supernatant) and bound (P, for pellet) fractions. Equal amounts of the supernatant and pellet factions were blotted on membranes, hybridized with 32P-labeled probes, and quantified using a PhosphorImager. Mock experiments using preimmune rabbit serum served as nonspecific binding controls for each ChIP assay. The percent immunoprecipitation [%IP, defined as P/(P + S)] of the mock experiment was subtracted in each case from the %IP of the anti-CENH3 treatment. As negative controls, we used the 180-bp knob repeat and the Tekay retroelement (Peacock et al., 1981; SanMiguel et al., 1998). The 180-bp repeat serves as an appropriate control for CentC because it is similar in size, tandemly arrayed, and highly abundant (Peacock et al., 1981), it is not found at centromeres (Peacock et al., 1981; Hiatt et al., 2002), and it does not interact with kinetochore proteins (Dawe et al., 1999). Similarly, Tekay serves as an appropriate control for CRM because it is a Ty3/gypsy element closely related to CRM by sequence analysis and is located primarily along chromosome arms (R. Mroczek and R.K. Dawe, unpublished data).
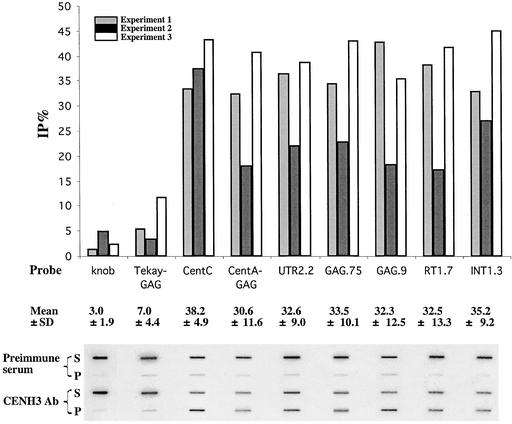
The experiment was repeated three times, and the results from each experiment are reported independently in Figure 6. On average, 38.2% ± 4.9% (mean ± sd) of the signal was found in the pellet when the blots were probed with a CentC probe, whereas only 3.0% ± 1.9% of the signal was detected in the pellet fraction when the same blots were reprobed with the knob repeat. Although there was notable variation among the results of the three experiments, CentC was immunoprecipitated at significantly higher levels than the knob repeat. Five different regions of the CRM element, covering nearly all of the element, also were used as hybridization probes. The %IP for the various CRM fragments ranged from 32.3 to 35.2% and averaged 33.2%. By contrast, the %IP for a 0.9-kb fragment corresponding to the GAG region of Tekay was significantly less, at only 7%. We also probed the blots with a 1.1-kb sequence corresponding to the GAG region of the CR element CentA (Ananiev et al., 1998). An average of 30.6% of the CentA GAG sequence was present in the pellet fraction, consistent with the view that CentA interacts with CENH3 in a manner similar to CRM. These data suggest that CentC and CRM (and probably CentA) interact specifically with CENH3 in the centromere-kinetochore complex. Because we have analyzed only a sample of the sequences present at cereal centromeres, it is likely that other sequences may interact with CENH3 as well.
Figure 6.
Identification of Centromere Repeats That Interact with CENH3 by the ChIP Assay.
Preimmune serum was used in the mock experiment as a background control (see Methods). The same blot was hybridized with the probes shown in Figure 4A. %IP (%IP of anti-CENH3 antibody subtracted from %IP of preimmune serum) from three independent experiments are graphed. A nylon membrane (Hybond N+) was used in experiments 1 and 2, and a nitrocellulose membrane was used in experiment 3. The %IP did not differ significantly between the negative controls (knob and Tekay), nor were there any significant differences in %IP among the CentC, CentA, or CRM sequences. However, the %IP for each of the negative controls differed significantly from the %IP for the CentC, CentA, and CRM sequences (P < 0.05 by t test). P, bound (pellet) fraction; S, unbound (supernatant) fraction.
DISCUSSION
Repetitive sequences such as tandem repeat arrays and retrotransposons make up the majority of DNA in eukaryotic centromeres. Such DNAs generally are regarded as selfish and are known to exploit niches of the genome with reduced recombination (Charlesworth et al., 1994). The idea that selfish elements may function to organize the centromere-kinetochore complex has been viewed with suspicion (Laurent et al., 1997; Langdon et al., 2000). Nevertheless, there are several examples in the literature that suggest that repetitive sequences have been coopted by the cell to perform useful, if not indispensable, functions. The clearest examples involve the HeT-A and TART elements of Drosophila, which function as telomeres (Pardue and Debaryshe, 2000). Instead of the more common telomerase-mediated extension of chromosome ends, HeT-A and TART continually transpose to the ends of chromosomes to maintain chromosome length. Another example involves the α-satellite at human centromeres. The α-satellite has been shown to interact with the kinetochore proteins by high-resolution immunocytochemistry (Haaf and Ward, 1994; du Sart et al., 1997) and by ChIP with antibodies to the conserved kinetochore protein CENP-A (Vafa and Sullivan, 1997; Lo et al., 2001a). In addition, efforts by several independent groups have established that α-satellites can initiate the formation of artificial chromosomes in human cell lines (Grimes et al., 2002).
Here, we provide evidence in maize that two major classes of repetitive centromeric DNA, CentC arrays and CRM elements, have roles in the organization and function of the centromere-kinetochore complex. CentC appears to be the structural homolog of the human α-satellite, and by comparison with the data in humans, might be expected to have a centromere function. The CR elements have received intensive previous study because of their remarkable sequence conservation. At 80% sequence identity (Figure 4B), the CR elements are evolving at rates similar to typical protein-coding genes and are among the most highly conserved retrotransposon families yet identified.
CENH3, a Conserved Kinetochore Component, Colocalizes with Maize CENPC in the Inner Kinetochore
The most intensively studied kinetochore protein is CENH3, which substitutes for histone H3 in active centromeric chromatin. Several lines of evidence from animal studies suggest that CENH3 serves as the core of the centromere-kinetochore complex (Choo, 2001; Henikoff et al., 2001). As the primary histone H3 protein in centromeric nucleosomes (Yoda et al., 2000; Ahmad and Henikoff, 2001; Lo et al., 2001a; Blower et al., 2002), CENH3 is necessary (Howman et al., 2000) and sufficient to recruit other key kinetochore proteins such as CENPC (Hooser et al., 2001; Ando et al., 2002). Using the sequence features of known CENH3s as a guide, we identified a candidate of maize CENH3 as an EST and confirmed its identity using reverse transcriptase–mediated PCR (Figure 1). In addition to conforming well to the structure of other centromeric histones, maize CENH3 localizes to kinetochores, as expected (Figure 3). CENH3 shows striking colocalization with the previously characterized maize homolog of CENPC (Dawe et al., 1999), suggesting that the two proteins interact closely in the centromere-kinetochore complex. CENH3 and CENPC lie in the inner kinetochore, beneath an outer regulatory domain that contains the cell cycle checkpoint proteins MAD2 and the 3F3/2 antigen (Yu et al., 1999).
Maize CENH3 Occurs in a Complex with CentC and CRM
A recent survey of the sequences in one centromeric BAC from maize provides an indication of the type and abundance of sequences that are likely to be present in maize centromeres. The primary sequence appears to be the tandem repeat array CentC, and a secondary, but very significant, fraction of the centromere consists of retro-transposons. Some of the retrotransposons are spread throughout the genome, whereas others (CR elements) are found exclusively at centromeres. Among the CR elements, the most highly conserved is CRM (Figure 4), which is present primarily as complete uninterrupted elements (Figure 4C) interspersed with CentC (Figure 3H). Retrotransposons have been reported at the centromeres of other organisms as well (Sun et al., 1997; Cambareri et al., 1998; Copenhaver et al., 1999), but none appears to be as well conserved as those in the cereal grains.
As a first step toward determining whether CentC and CRM have functional roles in the centromere, we used anti-CENH3 antiserum to immunoprecipitate the DNA in the centromere-kinetochore complex. We found that the antiserum specifically precipitated CentC and CRM but not the 180-bp knob repeat or the Tekay retroelement (Figure 6). In our experiments, 38% of the CentC and 33% of the CRM elements in young ear nuclei were precipitated selectively by CENH3 antiserum. Although these numbers are significantly high relative to those of negative controls, large fractions of the CentC and CRM (62 and 67%, respectively) were not immunoprecipitated by anti-CENH3 antiserum. To some extent, the incomplete recovery probably is a result of inefficiencies in the protocol. However, it is likely that not all of the CentC and CRM are components of the centromere-kinetochore complex. Indeed, our immunolocalization and in situ localization data suggest that a significant fraction of the CentC in interphase cells is not associated with CENH3 (Figure 3F). Based on the results from a detailed analysis of human centromeres (Schueler et al., 2001), it has been suggested that only the most recently evolved satellites have the capacity to bind to CENH3 (Henikoff, 2002). According to this view, newer satellites are evolving in central regions of the centromeres and older sequences are displaced into the pericentromeric regions. On the flanks of the centromere, the satellites are likely to degenerate, lose their capacity to interact with CENH3, and become riddled with transposons (Henikoff, 2002). Our data are consistent with this notion.
Potential Roles for CentC and CR Elements
Broad sequence comparisons suggest that tandem repeat arrays are the major structural elements of most multicellular eukaryotic centromeres (Csink and Henikoff, 1998). Aside from the neocentromeres in human cancer cell lines, there are no examples of centromeres from complex eukaryotes that lack extensive arrays of satellite DNA. However, the near absence of primary sequence identity among centromeric satellites (du Sart et al., 1997), the awareness that tandem repeat arrays also are found in noncentromeric regions of the genome (Sun et al., 1997), and the prevalent view that tandem repeats are a form of selfish DNA all have served as barriers to a clearer understanding of how satellites function in centromeres. Malik and Henikoff (2001) recently proposed an intriguing model to explain how satellites might evolve and function in centromeres. The key to this model is meiotic drive, a process in which chromosomes compete for access to the next generation by skewing Mendelian segregation in their favor (Novitski, 1967). Under this model, centromeric DNAs that are most efficient at binding to kinetochore proteins are likely to arrive at spindle poles first and be segregated to the functional megaspore (plants) or pronucleus (animals). The N-terminal and loop-1 regions of CENH3 show clear evidence of adaptive evolution (Henikoff et al., 2001; Talbert et al., 2002), suggesting that CENH3 may provide a molecular link between the kinetochore and the rapidly evolving centromere. It is possible that the CR elements have sequence features that allow them to insert into the centromere without disrupting the interaction of CENH3 with satellite arrays and that they, too, are being propagated either directly or indirectly by meiotic drive.
At odds with the proposal that CENH3 is a sequence-specific linker molecule is the lack of evidence that CENH3 specifically recognizes satellite DNA. Also difficult to reconcile with the model is the recent observation that Arabidopsis thaliana CENH3 interacts well with the centromeres of a different species (A. arenosa) when the two are introduced together in A. suecica polyploids (Talbert et al., 2002). Although these data do not exclude the idea that CENH3 and satellite DNAs are evolving as a part of a complex, they suggest that other factors may mediate the CENH3–satellite interaction. The interspersion of CRM elements with tandem repeat arrays may allow the centromere to adopt a chromatin configuration that is better suited to attracting kinetochore proteins. Alternatively, the CRM RNA molecule could bind to centromeric chromatin in a manner similar to the interaction of Xist (human) and roX (Drosophila) RNAs with the sex chromosomes during X inactivation (Clemson et al., 1996; Meller et al., 1997; Cohen and Lee, 2002). In these cases, the noncoding RNAs participate in effecting epigenetic changes that result in dramatic alterations in histone methylation and acetylation. Finally, the proteins encoded in CRM may interact with satellite DNA and/or CENH3 to facilitate the formation of the centromere-kinetochore complex. The most likely candidates are the integrase and GAG proteins, which have been shown to associate with specific regions of the chromosomes in yeast and Drosophila, respectively (Xie et al., 2001; Rashkova et al., 2002). Additional localization experiments with the predicted products of CRM will help to narrow the range of possibilities.
METHODS
Identification of a Full-Length CenH3 cDNA
We used a maize (Zea mays) histone H3 sequence as a query to search a maize EST database (http://www.zmdb.iastate.edu/), with the understanding that CENH3s are highly conserved in the histone fold but have variable N-terminal and loop-1 regions. Of 151 H3 homologs retrieved at the time of the BLAST search, one clone (AW244191) was identified as a maize CenH3 candidate. AW244191 was derived from developing embryos of Illinois High Oil Maize Strain Cycle 90. Comparison of this sequence with a putative rice homolog (identified by Steve Henikoff, Fred Hutchinson Cancer Research Center, Seattle, WA) led us to suspect that AW244191 contains an unspliced intron. Therefore, we identified another cDNA by reverse transcriptase–mediated PCR using primers homologous with the 5′ and 3′ regions of the AW244191 sequence (we thank Suneng Fu [Department of Plant Biology, University of Georgia, Athens, GA] for supplying the B73 young embryo cDNA [16 days after pollination] for this experiment). The primer sequences were 5′-ATGGETCGAACCAAGCACCA-3′ (forward) and 5′-TGCCCAACGCCTTCCTCCGAT-ACGCCTTGCAAGTT-3′ (reverse). Sequencing of the new cDNA (AF519807) and a partial genomic clone covering the 5′ region (derived by PCR) indicated that AW244191 does contain an unspliced intron at position 35 in the protein sequence (Figure 1).
Production of Antibodies
Anti-CENH3 antibodies were raised against a peptide (GDSVKKTKP-RH, which is unique to maize CENH3; Figure 1) conjugated to keyhole limpet hemocyanin. The preparation and affinity purification of antiserum was performed by BioSource International, Inc. (Camarillo, CA). We also generated chicken anti-CENPC antibodies, because the rabbit anti-CENPC serum described previously (Dawe et al., 1999) are difficult to use in double-labeling experiments with other rabbit antiserum. The same CENPC peptide used previously (KVK-SFVPEQYSDLVAKSARY) was injected into chickens at Aves Laboratories (Tigard, OR). The egg IgY fraction was column affinity purified against the CENPC peptide using the UltraLink Immobilization Kit (Pierce, Rockford, IL).
Protein Gel Blot Analysis
To verify the specificity of the CENH3 antibodies, the partial CenH3 coding region from AW244191, including 125 amino acids with a predicted molecular mass of 14 kD, was amplified by PCR and cloned into the pBAD expression vector (Invitrogen, Carlsbad, CA); the expressed protein has a His tag of ∼3 kD. Expression was induced in Escherichia coli with 0.0002% l-arabinose for 2 h. The bacteria were sonicated and boiled with loading buffer (Yu et al., 1997) for 20 min before loading. Total protein from young ear nuclei (see below for preparation) was prepared by boiling the nuclei in loading buffer for 15 min. All samples were separated by 15% SDS-PAGE and transferred to nitrocellulose. CENH3 was detected by enhanced chemiluminescence (Amersham Pharmacia Biotech) after incubating the blots with CENH3 antiserum at a dilution of 1:5000 (0.46 μg/mL).
Immunocytochemistry
Cells were prepared from the inbred W23 line as described by Yu et al. (1999) except as noted. Endosperm nuclei were fixed directly on the slides using 4% paraformaldehyde in PHEM (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, and 0.3 mM Sorbitol; pH 6.8) for 20 min. Rabbit anti-CENH3 antibody (1:2000) and chicken anti-CENPC antibody (1:75) were applied to the fixed samples overnight. Rhodamine-conjugated goat anti-rabbit antibodies (1:50) (Jackson Immunoresearch, West Grove, PA) were used to detect CENH3, and fluorescein isothiocyanate–conjugated goat anti-chicken antibodies (1:30) (Jackson Immunoresearch) were used to detect CENPC. For colocalization of CENH3 and CentC, the primary and secondary antisera were applied first, and then the slides were hybridized with fluorescein isothiocyanate–labeled CentC oligonucleotides (Hiatt et al., 2002). Procedures for the necessary washing steps, mounting, and 4′,6-diamidino-2-phenylindole staining have been described previously (Yu et al., 1997). The cells were analyzed using DeltaVision three-dimensional light microscopy (Applied Precision, Issaquah, WA; Yu et al., 1997).
In Situ Localization of CRM
Anthers from the inbred W23 line were fixed for 2 h as described previously (Hiatt et al., 2002). Meiocytes were spun down onto poly-l-Lys–coated cover slips and washed with the following solutions for 10 min each (1 × buffer A [80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, and 15 mM PIPES; pH 7.0] , 1 × SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate], and 20% deionized formamide) and twice with 2 × SSC and 30% formamide. Centromeres were localized with a probe (Rhod-C) consisting of three nonoverlapping, rhodamine-labeled oligonucleotides that are homologous with CentC (Ananiev et al., 1998). The oligonucleotides are as follows: 1, 5′-ACTCGTGCTTTCTATGCA-3′; 2, 5′-AAATTGCGCGAAACCACC-3′; and 3, 5′-AAGTAGTGGATTGGGCAT-3′. For each cover slip, the probe mixture contained 0.85 ng/mL of each oligonucleotide, giving a total concentration of 2.65 ng/mL. The LTR.50 PCR product was labeled with fluorescein using a random priming labeling kit (Yu et al., 1997; Hiatt et al., 2002), mixed with the Rhod-C oligonucleotides, and hybridized to the meiocytes overnight at 28°C in a solution of 2 × SSC and 30% deionized formamide. After hybridization, cover slips were washed with the following solutions for 10 min each: 2 × SSC, 10% deionized formamide, and 1.0% Tween 20; 2 × Tris-buffered saline (TBS) and 2 × SSC; 2 × TBS; and 1 × TBS and 0.1 mg/mL 4′,6-diamidino-2-phenylindole. The cells were analyzed using Delta-Vision three-dimensional light microscopy.
Mitotic chromosomes (Figure 3G) were prepared from the inbred B73 line by the nitrous oxide–air drying method (Kato, 1999). A CRM sequence (GAG.25; Figure 4A), identified initially from randomly amplified PCR products, was used as a probe. The PCR product was labeled with biotin-14-dATP (Invitrogen) by nick translation. Tyramide-amplified fluorescence in situ hybridization signal detection and image processing were performed as described previously (Kaszas et al., 2002).
Chromatin Immunoprecipitation
A native chromatin immunoprecipitation (ChIP) assay was performed as described previously (Lo et al., 2001b) with minor modifications. Young kernels (inbred W23 line ears 6 to 11 cm long) were ground to a fine powder with liquid N2 and resuspended in TBS containing 0.5% Tween 40 (2v/3v), and the nuclei were separated in a Suc gradient. We did not chemically cross-link the chromatin, as is common in other ChIP protocols. Nuclei were digested with micrococcal nuclease (Amersham Pharmacia Biotech) to liberate nucleosomes. The nucleosome sample was incubated with preimmune rabbit serum (1:1000 dilution for 4 h) and 2% protein A–Sepharose (Amersham Pharmacia Biotech) for 2 h and then centrifuged. This preclearing step reduces the amount of nonspecific binding in the ChIP assay. The precleared supernatant was incubated with anti-CENH3 antibodies (1:2000 dilution) overnight and with 12.5% protein A–Sepharose for 4 h. After centrifugation, the sample was separated into supernatant (unbound) and pellet (bound) fractions. The bound fraction was subjected to a series of washes and SDS treatment to dissociate the immune complexes. Nucleic acids were extracted from both the supernatant and washed-dissociated bound fractions with phenol/chloroform, resuspended in the same volume (30 μL), and slot blotted onto nitrocellulose (one experiment; Protran; Schleicher and Schuell) or nylon (two experiments; Hybond N+; Amersham Pharmacia Biotech) membranes. Each sample was blotted multiple times to account for loading errors, and the results were averaged. The experiment was repeated in its entirety three times over 3 weeks (beginning with fresh ear tissue and preceding through the blotting step). The membranes were probed with cloned DNA sequences (generated using PCR; see below for primers), and the results were quantified with a PhosphorImager (Storm 860; Molecular Dynamics, Sunnyvale, CA). The 32P signal in each slot was quantified using software supplied by the manufacturer.
Primers Used to Isolate Fragments for Sequencing and Probes
CRM clones were amplified from maize genomic DNA using the following primers, where F indicates forward primer and R indicates reverse primer: LTR.50 (F) 5′-TCGTCAACTCAACCATCAGGTGAT-3′ and (R) 5′-GCAAGTAGCGAGAGCTAAACTTGA-3′; UTR2.2 (F) 5′-GACCCGTGCACTCTTGTTTTGCTTAGGAA-3′ and (R) 5′-GGTGAA-AACCACCCATACCTCTACGGTTA-3′; GAG.75 (F) 5′-TCTGTTCAT-CTAACCATGGCAGGAT-3′ and (R) 5′-ACCCTGTCTCAATTGTGT-GCAACCT-3′; GAG.90 (F) 5′-CTGTTTGGTGGATAGAACATGGTA-AGA-3′ and (R) 5′-GATTCGGCAAAGATGCACCAGGAA-3′; GAG-RT1.6 (sequencing only) (F) 5′-CTGTTTGGTGGATAGAACATGGTA-AGA-3′ and (R) 5′-GATTCGGCAAAGATGCACCAGGAA-3′; RT1.7 (F) 5′-ATGCAGCATTCTTTGCCTCCTGTTA-3′ and (R) 5′-CAACGAGCA-ACCAATCTCACCACAT-3′; and INT1.3 (F) 5′-CTTTTGTTGTTACAGGAGGCGCATGGAGG-3′ and (R) 5′-TCCTAAGCAAAACAAGAGTGC-ACGGGTCT-3′. The CentA GAG primers were (F) 5′-GACCACAAAGATGGAGCAGGA-3′ and (R) 5′-GCAGCTGCCACTATCGATGAT-3′, amplifying bases 6430 to 7536 of BAC ZM16H10. The Tekay GAG primers were (F) 5′-GTGAAGGCTGTCTTCGGTACA-3′ and (R) 5′-GTGCCTGAAGATGCCGACATA-3′, amplifying bases 54167 to 55053 of GenBank AF448416. All PCR fragments were cloned into pCR4-TOPO (Invitrogen) and verified by sequencing.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: maize CENH3 cDNA (AF519807), CRM (AY129008), CRR (AC022352), and BAC clone ZM16H10 (AC116034).
Acknowledgments
We thank Evelyn Hiatt and Carolyn Lawrence for critically reading the manuscript, K.H. Choo for providing the detailed protocol for ChIP, and Wayne Parrott for his support and encouragement. This work was supported by Grant 9975827 from the National Science Foundation to R.K.D., J.J., and J.A.B.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006106.
References
- Ahmad, K., and Henikoff, S. (2001). Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, S., Yang, H., Nozaki, N., Okazaki, T, and Yoda, K. (2002). CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22, 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S., and Moore, G. (1996). A cereal centromeric sequence. Chromosoma 105, 261–268. [DOI] [PubMed] [Google Scholar]
- Baum, M., Ngan, V., and Clarke, L. (1994). The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol. Biol. Cell 5, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M., Sullivan, B., and Karpen, G. (2002). Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri, E.B., Aisner, R., and Carbon, J. (1998). Structure of the chromosome VII centromere region in Neurospora crassa: Degenerate transposons and simple repeats. Mol. Cell. Biol. 18, 5465–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., Sneglowski, P., and Stephan, W. (1994). The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., Dong, F., Langdon, T., Ouyang, S., Buell, C.R., Blattner, F.R., and Jiang, J. (2002). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H. (2001). Domain organization at the centromere and neocentromere. Dev. Cell 1, 165–177. [DOI] [PubMed] [Google Scholar]
- Clemson, C.M., McNeil, J.A., Willard, H.F., and Lawrence, J.B. (1996). XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, D., and Lee, J. (2002). X-chromosome inactivation and the search for chromosome-wide silencers. Curr. Opin. Genet. Dev. 12, 219–224. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286, 2468–2474. [DOI] [PubMed] [Google Scholar]
- Cottarel, G., Shero, J., Hieter, P., and Hegemann, J. (1989). A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 3342–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink, A.K., and Henikoff, S. (1998). Something from nothing: The evolution and utility of satellite repeats. Trends Genet. 14, 200–204. [DOI] [PubMed] [Google Scholar]
- Dawe, R.K., Reed, L., Yu, H.-G., Muszynski, M.G., and Hiatt, E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., Miller, J.T., Jackson, S.A., Wang, G.-L., and Ronald, P.C. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95, 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart, D., Cancilla, M.R., Earle, E., Mao, J.I., Saffery, R., Tainton, K.M., Kalitsis, P., Martyn, J., Barry, A.E., and Choo, K.H.A. (1997). A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 16, 144–153. [DOI] [PubMed] [Google Scholar]
- Grimes, B., Rhoades, A., and Willard, H. (2002). α-Satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol. Ther. 5, 798–805. [DOI] [PubMed] [Google Scholar]
- Haaf, T., and Ward, D.C. (1994). Structural analysis of α-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet. 3, 697–709. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. (2002). Near the edge of a chromosome's ‘black hole’. Trends Genet. 18, 165–167. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., Platero, J.S., and Steensel, B.V. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt, E., Kentner, E., and Dawe, R. (2002). Independently-regulated neocentromere activity of two classes of satellite sequences in maize. Plant Cell 14, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooser, A.V., Ouspensik, I., Gregson, H., Starr, D., Yen, T., Goldberg, M., Yokomori, K., Earnshaw, W., Sullivan, K., and Brinkley, B. (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529–3542. [DOI] [PubMed] [Google Scholar]
- Howman, E.V., Fowler, K.J., Newson, A.J., Redward, S., MacDonald, A.C., Kalitsis, P., and Choo, K.H.A. (2000). Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudakova, S., Michalek, W., Presting, G., ten Hoopen, R., dos Santos, K., Jasencakova, Z., and Schubert, I. (2001). Sequence organization of barley centromeres. Nucleic Acids Res. 29, 5029–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Nasuda, A., Dong, F., Scherrer, C., Woo, S.-S., Wing, R., Gill, B., and Ward, D. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93, 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G.H., and Allshire, R.C. (1997). The case of epigenetic effects on centromere identity and function. Trends Genet. 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1996). Misdivision analysis of centromere structure in maize. EMBO J. 15, 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1998). Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150, 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., Kato, A., and Birchler, J.A. (2002). Cytological and molecular analysis of centromere misdivision in maize. Genome 43, 759–768. [DOI] [PubMed] [Google Scholar]
- Kato, A. (1999). Air drying method using nitrous oxide for chromosome counting in maize. Biotech. Histochem. 74, 160–166. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. (2000). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res. 7, 315–321. [DOI] [PubMed] [Google Scholar]
- Langdon, T., Seago, C., Mende, M., Leggett, M., Thomas, H., Forster, J., Jones, R., and Jenkins, G. (2000). Retrotransposon evolution in diverse plant genomes. Genetics 156, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent, A., Puechberty, J., and Roizes, G. (1997). Hypothesis: For the worst and for the best, L1Hs retrotransposons actively participate in the evolution of the human centromeric alphoid sequences. Chromosome Res. 7, 305–317. [DOI] [PubMed] [Google Scholar]
- Lo, A., Craig, J., Saffery, R., Kalitsis, P., Irvine, D., Earle, E., Magliano, D., and Choo, K. (2001. a). A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A., Magliano, D., Sibson, M., Kalitsis, P., Craig, J., and Choo, K. (2001. b). A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11, 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert, K., and Karpen, G. (2001). The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158, 1615–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H., Vermaak, D., and Henikoff, S. (2002). Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 99, 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.S., and Henikoff, S. (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, V.H., Wu, K.H., Roman, G., Kuroda, M.I., and Davis, R.L. (1997). roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88, 445–457. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B., Yang, P., Glowczewski, L., Koshland, D., and Smith, M.M. (1998). Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607–613. [DOI] [PubMed] [Google Scholar]
- Miller, J.T., Dong, F., Jackson, S.A., Song, J., and Jiang, J. (1998. a). Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150, 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.T., Jackson, S.A., Nasuda, S., Gill, B.S., Wing, R.A., and Jiang, J. (1998. b). Cloning and characterization of a centromere-specific repetitive DNA element from Sorghum bicolor. Theor. Appl. Genet. 96, 832–839. [Google Scholar]
- Nagaki, K., Tsujimoto, H., and Sasakuma, T. (1998). A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Res. 6, 295–302. [DOI] [PubMed] [Google Scholar]
- Novitski, E. (1967). Nonrandom disjunction in Drosophila. Annu. Rev. Genet. 1, 71–86. [Google Scholar]
- Page, B., Wanous, M., and Birchler, J. (2001). Characterization of a maize chromosome 4 centromeric sequence: Evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue, M., and Debaryshe, P. (2000). Drosophila telomere transposons: Genetically active elements in heterochromatin. Genetica 109, 45–52. [DOI] [PubMed] [Google Scholar]
- Peacock, W.J., Dennis, E.S., Rhoades, M.M., and Pryor, A.J. (1981). Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78, 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presting, G.G., Malysheva, L., Fuchs, J., and Schubert, I. (1998). A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 16, 721–728. [DOI] [PubMed] [Google Scholar]
- Rashkova, A., Karam, S., and Pardue, M. (2002). Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc. Natl. Acad. Sci. USA 99, 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B., Tikhonov, A., Nakajima, A., and Bennetzen, J. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- Schueler, M., Higgins, A., Rudd, M., Gustashaw, K., and Willard, H. (2001). Genomic and genetic definition of a functional human centromere. Science 294, 109–114. [DOI] [PubMed] [Google Scholar]
- Sun, X., Wahlstrom, J., and Karpen, G. (1997). Molecular structure of a functional Drosophila centromere. Cell 91, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P., Masuelli, R., Tyagi, A., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa, O., and Sullivan, K.F. (1997). Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Xie, W., Gai, X., Zhu, Y., Zappulla, D., Sternglanz, R., and Voytas, D.F. (2001). Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21, 6606–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda, K., Ando, S., Morishita, S., Houmura, K., Hashimoto, K., Takeyasu, K., and Okazaki, T. (2000). Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97, 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.-G., Hiatt, E.N., Chan, A., Sweeney, M., and Dawe, R.K. (1997). Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.-G., Hiatt, E.N., and Dawe, R.K. (2000). The plant kinetochore. Trends Plant Sci. 5, 543–547. [DOI] [PubMed] [Google Scholar]
- Yu, H.-G., Muszynski, M.G., and Dawe, R.K. (1999). The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 145, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick, M., Islam-Faridi, M., Zhang, H., Hodnett, G., Gomez, M., Kim, J., Price, H., and Stelly, D. (2000). Distribution and sequence analysis of the centromere-associated repetitive element CEN38 of Sorghum bicolor (Poaceae). Am. J. Bot. 87, 1757–1764. [PubMed] [Google Scholar]