Abstract
Seed-type vacuolar processing enzyme (VPE) activity is predicted to be essential for post-translational proteolysis of seed storage proteins in the protein storage vacuole of developing seeds. To test this hypothesis, we examined the protein profiles of developing and germinating seeds from Arabidopsis plants containing transposon-insertional knockout mutations in the genes that encode the two seed-type VPEs in Arabidopsis, βVPE, which was identified previously, and δVPE, which is described here. The effects of these mutations were studied individually in single mutants and together in a double mutant. Surprisingly, we found that most of the seed protein still was processed proteolytically in seed-type VPE mutants. The minor differences observed in polypeptide accumulation between wild-type and βVPE mutant seeds were characterized using a two-dimensional gel/mass spectrometric analysis approach. The results showed increased amounts of propolypeptide forms of legumin-type globulins accumulating in mutant seeds. However, the majority of protein (>80%) still was processed to mature α- and β-chains, as observed in wild-type seeds. Furthermore, we identified several legumin-type globulin polypeptides, not corresponding to pro or mature forms, that increased in accumulation in βVPE mutant seeds compared with wild-type seeds. Together, these results indicate the existence of both redundant and alternative processing activities in seeds. The latter was substantiated by N-terminal sequencing of a napin-type albumin protein, indicating cleavage consistent with previous in vitro studies using purified aspartic protease. Analysis of genome-wide transcript profiling data sets identified six protease genes (including an aspartic protease gene and βVPE) that shared spatial and temporal expression patterns with seed storage proteins. From these results, we conclude that seed-type VPEs constitute merely one pathway for processing seed storage protein and that other proteolytic enzymes also can process storage proteins into chains capable of stable accumulation in mature seeds.
INTRODUCTION
Seed germination is a heterotrophic stage in the life cycle of plants during which the emerging seedling relies on stored materials for continued growth and development. One of these essential materials is reduced nitrogen, which is accumulated predominantly in the form of seed storage proteins. The process of storage protein deposition in maturing seeds and mobilization in germinating seeds appears to be highly specialized, involving dedicated compartments termed protein storage vacuoles (PSVs). Only a few proteins have evolved to survive the lytic environment of the PSV (which probably is the case with proteins that accumulate in any type of plant vacuole). This imposes restrictions on the use of vacuoles for the deposition of recombinant protein. Foreign (nonstorage) proteins and genetically modified storage proteins tend to be proteolytically unstable in vacuoles and consequently often fail to accumulate or are fragmented when expressed in plants (Hoffman et al., 1988; Jung et al., 1993; Kermode et al., 1995; Pueyo et al., 1995; Jung et al., 1998; Frigerio et al., 2000).
Arabidopsis seeds contain two predominant classes of seed storage protein: legumin-type globulins (also referred to as 12S globulin or cruciferin in Arabidopsis) (Sjodahl et al., 1991), and napin-type albumins (also referred to as 2S albumins or arabidin in Arabidopsis) (Krebbers et al., 1988; van der Klei et al., 1993). Characterization of these protein types in various dicotyledonous plants has shown that propolypeptides are targeted from the endoplasmic reticulum lumen to the PSV, where they are processed by vacuolar proteases into specific chains. Prolegumin-type globulins are cleaved at a conserved Asn-Gly peptide bond, converting the pro-form into two disulfide-linked mature polypeptides referred to as α- and β-chain (Muntz, 1998). Pronapin-type albumin proteolytic processing appears to be more complex, requiring removal of three propeptide regions—N-terminal processed fragment, internal processed fragment, and C-terminal processed fragment—to obtain the two di-sulfide-linked mature polypeptides referred to as large and small chains (Krebbers et al., 1988). Several of the napin-type albumin processing steps involve polypeptide cleavage at conserved Asn residues in the P1 position. Although the functional role of processing in the accumulation of napin-type albumin in mature seeds is not understood, studies of Vicia faba bean legumin have demonstrated post–endoplasmic reticulum processing to be essential for trimers of prolegumin to obtain the final higher order molecular forms (hexamers) found in mature seeds (Jung et al., 1998).
Work to isolate an asparaginyl-specific endopeptidase responsible for processing storage proteins from maturing seeds of dicots (Hara-Nishimura et al., 1991, 1993) resulted in the identification of a novel class of Cys proteases in plants that are related to a hemoglobinase from Schistosoma mansoni (C13; EC 3.4.22.34). Because of its specificity for Asn in the P1 position of the proteolytic cleavage site and the apparent property of processing prolegumin in vitro, it was called asparaginyl endopeptidase and legumain (Ishii, 1994; Hara-Nishimura, 1998), respectively, or more commonly vacuolar processing enzyme (VPE) (Hara-Nishimura et al., 1993). Further characterization of members of the VPE family has shown VPE to be localized to the vacuolar matrix (Hara-Nishimura et al., 1993), self-catalytically activated (Kuroyanagi et al., 2002), and capable of cleaving both legumin-type and napin-type storage proteins in vitro (Hara-Nishimura et al., 1991; Shimada et al., 1994; Hiraiwa et al., 1997; Yamada et al., 1999), all of which are hallmarks of a seed protein maturase. Because this was a clearly defined subfamily of proteases and because storage protein accumulation in dicot seed PSVs is highly specialized, it was hypothesized and widely accepted that VPE most likely was responsible for the specific polypeptide processing events of seed storage protein in the PSV. However, members of the VPE family also have been identified in a variety of other tissues throughout the plant, including leaves, roots, nucellar cell walls, and cotyledons of germinating seedlings (Becker et al., 1995; Kinoshita et al., 1995b; Linnestad et al., 1998; Fischer et al., 2000; Hayashi et al., 2001). These VPE family members have been associated with functions other than seed storage protein processing, including tissue senescence and storage protein breakdown during germination, although these functions have not been demonstrated in vivo.
Phylogenetic examination of the VPE family identified two distinct subfamilies of VPEs (Kinoshita et al., 1995a). VPEs associated with seed protein maturation constitute one subfamily (referred to as seed-type VPEs), and VPEs associated with processes other than seed protein maturation constitute the second subfamily (referred to as vegetative-type VPEs). In Arabidopsis, the VPE family has been described as having three members (αVPE, βVPE, and γVPE) (Kinoshita et al., 1995a). Using promoter β-glucuronidase fusion constructs, βVPE was demonstrated to be expressed in seeds, whereas the expression of αVPE and γVPE appeared to be limited to vegetative tissues—root and leaf, respectively. These expression patterns are in accord with the phylogenetic grouping of these genes, identifying βVPE as a member of the seed-type VPE subfamily and αVPE and γVPE as members of the vegetative-type VPE subfamily (Kinoshita et al., 1999).
Confirmation in planta of seed- or vegetative-type VPE function has not been established. Muntz and Shutov (2002) attempted to suppress seed-type VPEs by expressing specific antisense DNA in transgenic tobacco seeds, but they detected no clear processing phenotype. However, the complexity of the VPE family in the tobacco genome is unknown; therefore, undetected, functionally redundant, seed-type VPEs could account for the result. An example of apparent redundancy is provided in this report, in which our examination of the Arabidopsis genome identified a fourth, seed-expressed, VPE family member. Another explanation might involve functionally redundant proteolytic enzymes other than VPE homologs. Although disputed in the literature, support for functionally redundant proteolytic enzymes (other than VPE homologs) has been shown in soybean, from which an activity capable of processing legumin was isolated from seeds. This proteolytic activity was associated with a protein of a markedly different molecular mass than VPEs (Scott et al., 1992). Additionally, aspartic protease activity purified from developing Brassica seeds is capable of cleaving Arabidopsis napin-type albumin propeptide fragments in vitro (D'Hondt et al., 1993), and also has been implicated in the processing of prolectin in barley seeds (Runeberg-Roos et al., 1994). Others have argued that aspartic protease activity may play a role only in trimming C-terminal propeptides (Hiraiwa et al., 1997).
Using the Arabidopsis model, for which a complete genomic sequence and several gene knockout populations are available, we attempted to confirm the specific role of seed-type VPEs in vivo. Here, we provide evidence that seed-type VPEs do not constitute an exclusive proteolytic activity for seed storage protein maturation. Furthermore, we identify other proteases potentially involved in the processing of storage protein into chains competent for stable accumulation in mature seeds.
RESULTS
Identification of δVPE, a Novel Seed-Type VPE Family Member of Arabidopsis
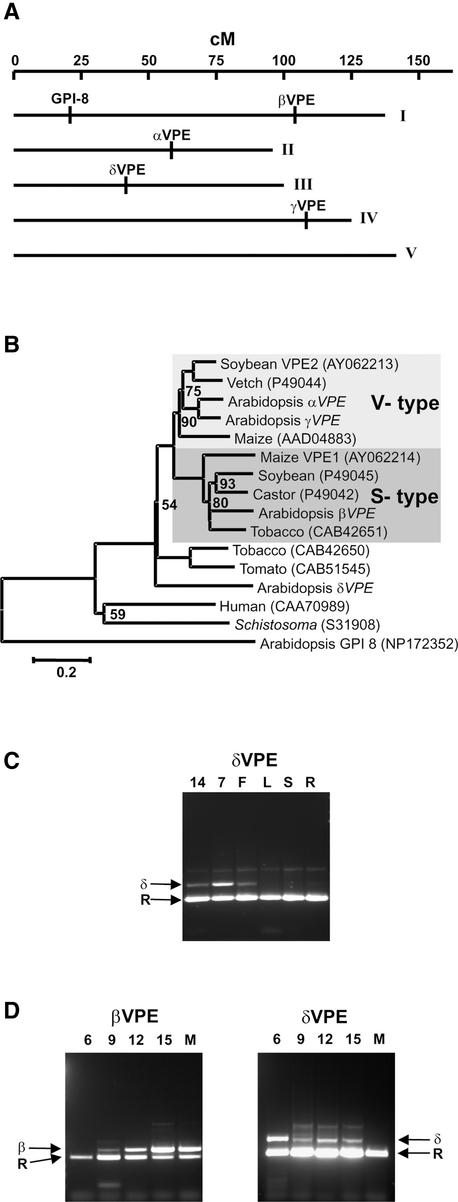
Sequences of seed-type and vegetative-type members of the VPE gene family were used to query the Arabidopsis genomic and EST databases as well as the DuPont-Pioneer Arabidopsis EST database to identify all members of the VPE gene family in Arabidopsis. In addition to the three previously described VPE genes (Kinoshita et al., 1995a), this search located a fourth VPE family member on chromosome III, to which we assigned the name δVPE (Figure 1A). Several δVPE cDNAs, identified in EST libraries derived from green silique tissue, were sequenced. The deduced polypeptide contains a putative N-terminal signal peptide and active site residues characteristic of VPE. δVPE is 48% identical to βVPE and 50% identical to γVPE. It is 22% identical to putative Arabidopsis gpi-8, a member of the GPI-anchor transamidase family, a protein family that appears to be evolutionarily related to VPEs (Benghezal et al., 1996). Examination of the phylogenetic relationship of the VPE protein family indicated that δVPE could not be assigned to either the seed- or the vegetative-type VPE subfamily but represents a novel branch of the VPE family in plants (Figure 1B).
Figure 1.
Chromosomal Location, Phylogenetic Relationship, and Expression of Arabidopsis Seed-Type VPE Genes.
(A) Chromosomal location of each VPE gene and GPI-8. cM, centimorgan.
(B) Dendrogram of VPE protein sequences. The lightly shaded box highlights the vegetative-type (V-type) VPE gene branch, and the darkly shaded box highlights the seed-type (S-type) VPE gene branch. Bootstrapping values of all internal branches exceeded 95% unless indicated otherwise. Accession numbers for non-Arabidopsis proteins are given.
(C) Semiquantitative multiplexed RT-PCR showing the expression of δVPE (δ) and cytosolic ribosomal protein S11 (R) as an internal standard in seeds at 14 DAA (14), seeds at 7 DAA (7), flower (F), leaf (L), stem (S), and root (R) of wild-type plants.
(D) Semiquantitative multiplexed RT-PCR showing the expression of βVPE (β), δVPE (δ), and cytosolic ribosomal protein S11 (R) in developing seeds at 6 DAA (6), 9 DAA (9), 12 DAA (12), and 15 DAA (15) and mature seeds (M).
To determine if δVPE is a seed-type VPE gene, expression was examined using semiquantitative reverse transcriptase PCR (RT-PCR). As shown in Figure 1C, δVPE expression was detected primarily in developing seeds at 7 days after anthesis (DAA). A weaker signal was detected in flowers but not in the vegetative tissues examined. A more detailed examination of expression during seed development showed overlapping expression of δVPE (Figure 1D, right) with βVPE (Figure 1D, left), although peak steady state expression levels of δVPE occur earlier, during the cell division phase of seed development (Meinke, 1994). Notwithstanding the phylogenetic relationship, δVPE expression, occurring primarily in developing seeds, allows for the conclusion that δVPE is a seed-type VPE. Furthermore, because its expression overlaps with that of βVPE and because the half-life of δVPE is unknown, we concluded that it has the potential to act as a βVPE redundant seed protein–processing enzyme.
Identification of dSpm Transposon Insertion Events in βVPE and δVPE
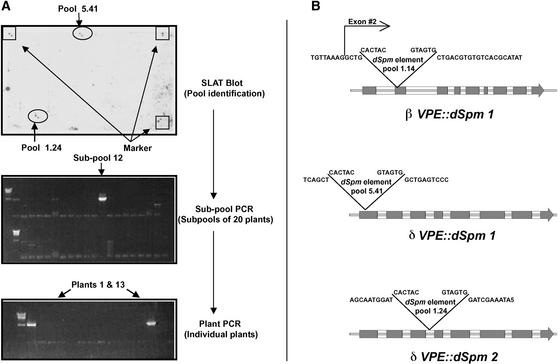
Arabidopsis plants devoid of functional βVPE and δVPE were isolated by identifying plants from the Sainsbury Laboratory dSpm mutant collection containing dSpm transposon insertions in the coding sequences of the corresponding genes (Tissier et al., 1999). A putative insertion allele of βVPE (βVPE::dSpm1) was identified by querying the SINS database of the Sainsbury Laboratory (http://www.jic.bbsrc.ac.uk/Sainsbury-lab/jonathan-jones/SINS-database/database.html). The allele was predicted to be within mutant plant pool 1.14, which we confirmed by PCR screening and sequencing of DNA derived from pool 1.14. We found the dSpm element to be within the beginning of the second exon of βVPE. This insertion disrupts the coding sequence near the N terminus of βVPE, which was expected to cause the complete inactivation of gene function. Plants homozygous for the βVPE::dSpm1 allele were identified using allele-specific PCR by the presence of the βVPE::dSpm1 allele and the absence of the wild-type βVPE allele. DNA gel blot analysis using sequence of the dSpm element as a probe indicated that βVPE::dSpm1 plants have a single dSpm insertion at one locus within their genome (data not shown).
Two independent insertion alleles of δVPE (δVPE::dSpm1 and δVPE::dSpm2) were identified in DNA isolated from mutant plant pools 5.41 and 1.24, respectively, by reverse screening using SLAT (see Methods) blots probed with δVPE (Figure 2A). DNA sequences flanking the δVPE insertion sites were cloned and sequenced to determine the location of the dSpm elements within δVPE (Figure 2B). The allele δVPE::dSpm1 (pool 5.41) contains a dSpm element in the first exon, whereas the allele δVPE::dSpm2 (pool 1.24) contains a dSpm element in the third intron. The δVPE:: dSpm1 allele was selected for functional studies because of the higher probability for gene inactivation by exon transposon insertion. Plants homozygous for the δVPE::dSpm1 allele were isolated as described for βVPE::dSpm1.
Figure 2.
Identification, Isolation, and Location of dSpm Transposon Insertions in δVPE and βVPE.
(A) The top gel shows a chemiluminescence image resulting from hybridizing a SLAT blot with a δVPE probe to identify parental pools containing dSpm insertions (pools 5.41 and 1.24) in or near the δVPE gene. The middle gel shows PCR identification (see Methods) of a subpool of progeny containing an insertion within δVPE (pool 5.41), and the bottom gel shows PCR identification of two individual plants carrying this insertion.
(B) Illustrations showing the locations of dSpm insertions within the δVPE and βVPE genes with respect to DNA sequence and intron/exon regions. Gray boxes indicate exon (coding) regions of the genes.
To obtain double-mutant plants devoid of seed-type VPE activity, plants homozygous for βVPE::dSpm1 and plants homozygous for δVPE::dSpm1 were crossed. Homozygous double-mutant (δVPE/βVPE) plants were identified by PCR in F2 progeny after F1 self-pollination.
Homozygous mutants of δVPE and βVPE as well as double-homozygous δVPE/βVPE mutant plants were examined for visible phenotypes under normal growth conditions. In all cases, no effects were observed on germination rate, vegetative growth, seed set, or macroscopic seed morphology (data not shown).
Confirmation of Gene Knockout by RT-PCR and Protein Immunoblot Analysis
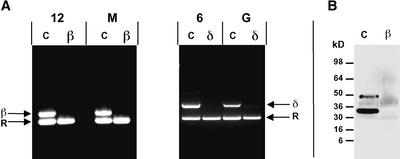
Multiplexed RT-PCR was performed using βVPE or δVPE gene-specific primers downstream of the dSpm insertion site in combination with primers specific for a control transcript (cytosolic ribosomal protein S11). Primers were designed to flank intron segments such that genomic DNA contamination would be recognized as a significant shift in size of the PCR product. As shown in Figure 3A, fragments specific for the cytosolic ribosomal protein S11 transcript were produced in all multiplexed RT-PCR procedures independent of the genotype. By contrast, βVPE- or δVPE-specific RT-PCR fragments of the expected sizes were amplified from developing wild-type seeds but not from extracts of developing seeds from βVPE::dSpm1 or δVPE:: dSpm1 homozygous plants. The lack of detectable VPE-specific fragments even after 45 cycles of amplification was a strong indicator of the absence of functional VPE transcripts.
Figure 3.
Plants Homozygous for the βVPE::dSpm1 or the δVPE::dSpm1 Allele Show No Corresponding Wild-Type Gene Expression.
(A) Multiplexed RT-PCR of 6 DAA (6), 12 DAA (12), mature (M), or germinating (G) seeds of wild-type (C), homozygous βVPE::dSpm1 (β), or homozygous δVPE::dSpm1 (δ) plants to detect cytosolic ribosomal protein S11 (R) and either βVPE (β) or δVPE (δ).
(B) Immunoblot of protein extracted from seeds of wild-type (C) or homozygous βVPE::dSpm1 (β) probed with a βVPE monospecific polyclonal antibody.
Figure 3B shows an immunoblot analysis of seed extracts from mature seeds of homozygous βVPE::dSpm1 mutants and wild-type siblings probed with a monospecific βVPE antibody. Two immunoreactive bands, one minor, with an apparent molecular mass of ∼50 kD, and one major, with an apparent molecular mass of 34 kD, were observed only in wild-type controls and not in homozygous βVPE::dSpm1 plants. These bands are consistent with the reported sizes of VPE propolypeptides and mature VPE, respectively, from other plant species (Hara-Nishimura et al., 1991; Shimada et al., 1994). Together with the RT-PCR data, the immunoblot results strongly support the conclusion that the dSpm insertions in the VPE genes result in an effective knockout of the expression of these genes in seeds of the corresponding homozygous mutant plants.
One-Dimensional Gel Analysis of Polypeptide Content in Seeds during Development and Germination
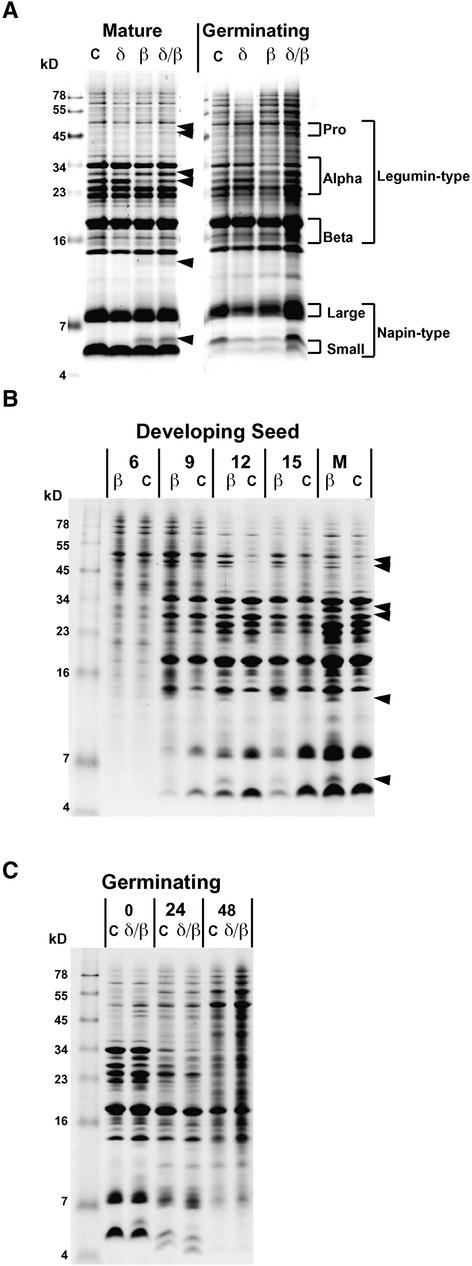
The protein content of mature and germinating seeds was examined using Tris-Tricine SDS-PAGE. Profiles of seed proteins extracted from βVPE::dSpm1 and the wild type are presented in Figure 4. Compared with the wild type, distinct (but minor) alterations in the protein profile were detected in βVPE::dSpm1 seeds, and these changes were observed throughout storage protein accumulation (Figure 4B). The most obvious change related to polypeptides of approximate apparent molecular masses of 5, 13, 33, and 47 to 50 kD, which appear slightly increased, and a polypeptide of approximate apparent molecular mass of 32 kD, which appears slightly decreased (Figures 4A and 4B). This subtle protein phenotype segregated 1:3 in F2 progeny after F1 self-pollination of a cross of βVPE::dSpm1 homozygous plants to the wild type, consistent with a recessive single-gene mutation (data not shown).
Figure 4.
One-Dimensional SDS-PAGE Analysis of Seed Protein Content during Development and Germination.
(A) Mature or germinating seed (24 h) protein from the wild type (C) or homozygous βVPE::dSpm1 (β), homozygous δVPE::dSpm1 (δ), or homozygous βVPE/δVPE double mutants (δ/β). Arrowheads indicate observed differences. The approximate gel locations of legumin-type proglobulins (pro), α- and β-chains, and napin-type large and small chains are shown with brackets.
(B) Seed protein extracted from the wild type (C) or homozygous βVPE::dSpm1 (β) at 6, 9, 12, and 15 DAA and maturity (M). Arrowheads indicate observed differences.
(C) Protein extracted from germinating wild-type (C) or homozygous βVPE/δVPE double-mutant (δ/β) seeds. After cold treatment at 4°C for 48 h, seeds were incubated at 25°C for 0, 24, and 48 h.
Protein profiles of developing and mature seeds from δVPE::dSpm1 and the wild type also were compared. However, in contrast to βVPE::dSpm1 seeds, no differences between mutant and wild-type seeds were detected (only mature seeds are shown; Figure 4A). This result suggested no unique role for δVPE in seed protein accumulation.
Given the overlapping gene expression patterns of βVPE and δVPE, it appeared possible that the genes could compensate partially or completely for each other's function during seed development. However, seed protein profiles of the βVPE/δVPE double mutant appeared identical to profiles of the βVPE::dSpm1 mutant alone (Figure 4A). The identical protein profiles suggest that δVPE processing activity, if any, and βVPE processing activity are not redundant (or additive) to each other.
One seed-type VPE, VsPB2, which is stored in protein bodies of embryonic axes and cotyledons of Vicia sativa, has been implicated in the mobilization of protein reserves during germination (Schlereth et al., 2001). Therefore, we compared seed protein profiles of germinating βVPE/δVPE double-mutant seeds with those of the wild type (Figure 4C). We found that protein degradation appeared to progress similarly in germinating wild-type and mutant seeds. Hence, our data do not support a unique role for βVPE or δVPE in protein degradation during germination.
Comparative Two-Dimensional Gel/Mass Spectrometric Proteome Analysis of βVPE::dSpm1 and Wild-Type Seed Protein
One-dimensional gel analysis of seed proteins indicated that removal of seed-type VPE activity did not result in major alterations in the accumulation of the seed protein. At least two distinct hypotheses could account for this result. The first hypothesis is that βVPE processes only a small, specific subset of seed protein species. The second hypothesis is that βVPE normally processes most seed storage protein species; however, other enzyme activities also are capable of compensating nearly entirely for this function. To address this issue, a comparative two-dimensional gel/mass spectrometric proteomic analysis, capable of specifically quantifying and identifying individual polypeptides, was performed using total protein extracts from mature βVPE::dSpm1 seeds and wild-type seeds. Seeds from δVPE::dSpm1 plants were not analyzed because no differences in protein pattern were observed in one-dimensional gel analysis of these seeds.
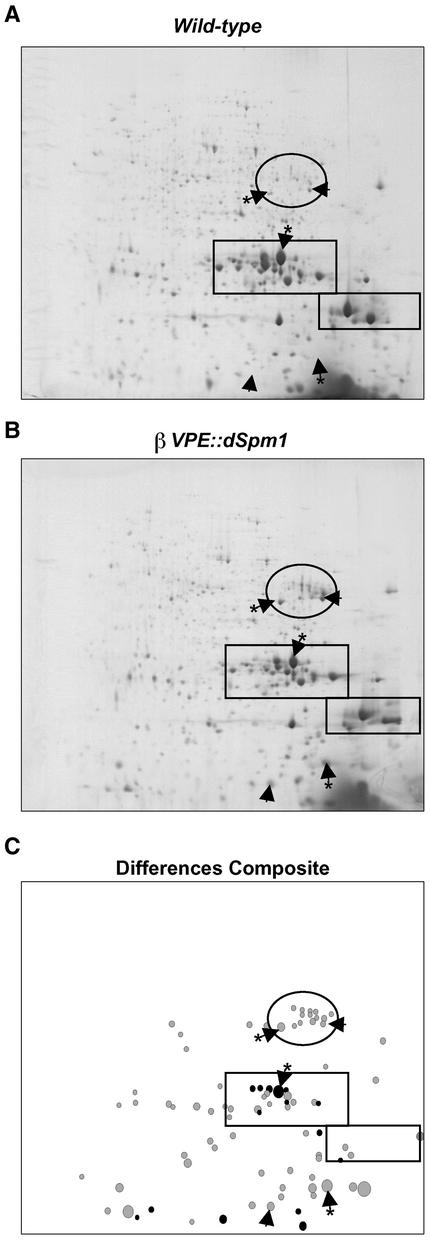
Representative gel images shown in Figure 5 illustrate several differences in seed protein content. The proteins were visualized using a fluorescent dye that binds noncovalently to the SDS moiety attached to each polypeptide (Page et al., 1999). This permitted quantification of protein spots (features) in a wide dynamic range. Based on the cumulative fluorescence intensity of all features, individual feature quantities were calculated as percentage values of the total protein quantity. The gel separation of each sample was replicated three times, enabling quantitative evaluation of digitally captured gel images. This analysis was performed using ROSETTA software (see Methods) and resulted in the detection of 1364 unique reproducible features at a level of >1 ng of protein. Eighty-four of these features were altered consistently between the mutant and wild-type samples by more than twofold (Figure 5C), illustrating the increased sampling depth of the two-dimensional gel analysis compared with the one-dimensional gel analysis.
Figure 5.
Two-Dimensional Gel Analysis of βVPE::dSpm1 and Wild-Type Seed Protein.
(A) Representative gel image of wild-type seed protein.
(B) Representative gel image of homozygous βVPE::dSpm1 seed protein. The region identified as containing prolegumin-type globulin polypeptides is circled. The large rectangle and the small rectangle indicate regions identified as containing α- and β-chains, respectively. Arrowheads indicate a selection of differentially accumulating protein spots (features) to aid in comparing images. Arrowheads with asterisks indicate a selection of differentially accumulating features identified successfully using mass spectrometry (see Figure 6).
(C) A composite image generated using Decision Site software (Spotfire, Somerville, MA) identifying all differentially accumulating features. Lightly shaded spots indicate features with increased accumulation in βVPE::dSpm1 seeds compared with wild-type seeds, and darkly shaded spots indicate features with increased accumulation in wild-type seeds compared with βVPE::dSpm1 seeds.
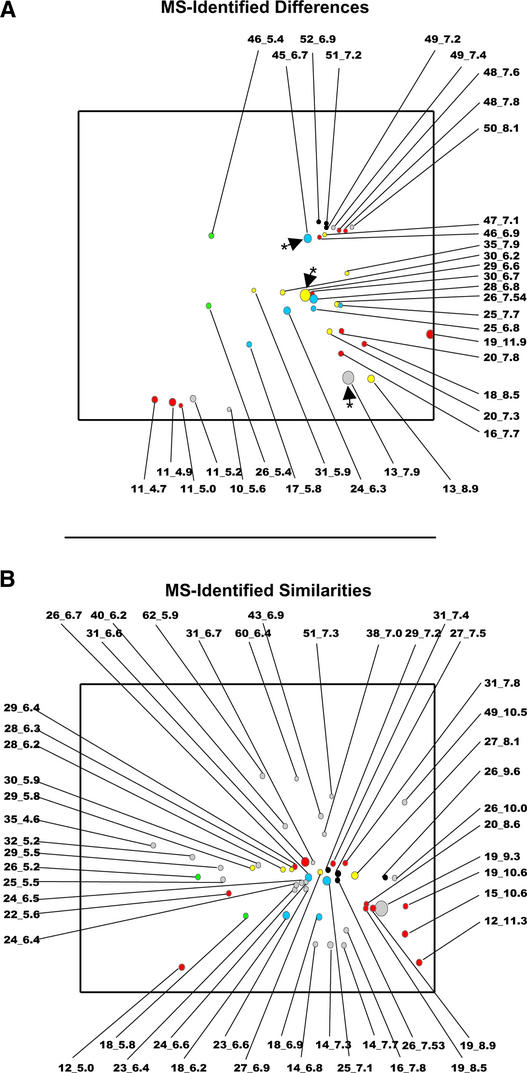
Mass spectrometry (MS) identification of two groups of features was attempted. One group was composed of all differentially accumulating features (84 protein spots) detected (Figures 5C and 6A, Table 1). A second group was composed of a selection of abundant (>0.01% of total detected protein), nondifferentially accumulating (<2-fold change between samples) features (73 protein spots) (Figure 6B, Table 2) designed to identify storage protein accumulation unaffected by βVPE knockout. A total of 85 protein spots were identified by MS, of which 34 were accumulated differentially and 51 were not changed significantly in accumulation between mutant and wild-type controls (Figure 6, Tables 1 and 2).
Figure 6.
Mass Spectrometric Identification of Protein Features.
Composite gel images showing MS-identified features with either a significant change in accumulation (A) or no significant change in accumulation (B). Labels indicate apparent molecular mass and pI of each feature and refer to results detailed in Tables 1 and 2. Arrowheads with asterisks are provided as an aid for comparison with Figure 5. Features identified as legumin-type globulin-related polypeptides are color coded as follows: red (PID 1628583), yellow (PID 9759513), green (PID 4204298), blue (PID 4204299), and black (PID 9279583). All other features are shown in gray.
Table 1.
Mass Spectrometric Identification of Seed Protein Features That Were Changed in Accumulation between Homozygous βVPE::dSpm1 and Wild-Type Seeds
| ID No.a | pI | kD | % WT b | % βVPE b | Change c | t Test d | Annotation Detail | PIDe |
|---|---|---|---|---|---|---|---|---|
| 52_6.9 | 6.88 | 52171 | 0.001 | 0.044 | 33.4 | 0.05 | Vicilin-like precursor f | 9279583 |
| 51_7.2 | 7.15 | 51335 | 0.022 | 0.233 | 10.7 | 0.08 | Vicilin-like precursor f | 9279583 |
| 50_8.1 | 8.07 | 49734 | 0.000 | 0.033 | NAg | 0.03 | Vicilin-like precursor f and legumin-type globulin precursor f |
9279583 1628583 |
| 49_7.4 | 7.39 | 49332 | 0.052 | 0.135 | 2.6 | 0.08 | Vicilin-like precursor f and catalase 2 |
9279583 1705620 |
| 49_7.2 | 7.15 | 49412 | 0.025 | 0.100 | 4.0 | 0.08 | Vicilin-like precursor f | 9279583 |
| 48_7.8 | 7.84 | 47906 | 0.038 | 0.705 | 18.6 | 0.12 | Legumin-type globulin precursor f | 1628583 |
| 48_7.6 | 7.61 | 48268 | 0.011 | 0.115 | 10.3 | 0.15 | Legumin-type globulin precursor f | 1628583 |
| 47_7.1 | 7.09 | 46509 | 0.007 | 0.117 | 16.8 | 0.11 | Legumin-type globulin precursor f | 9759513 |
| 46_6.9 | 6.92 | 45515 | 0.003 | 0.023 | 6.7 | 0.03 | Legumin-type globulin precursor f | 1628583 |
| 46_5.4 | 5.39 | 46101 | 0.011 | 0.210 | 18.6 | 0.11 | Legumin-type globulin precursor f | 4204298 |
| 45_6.7 | 6.69 | 45347 | 0.076 | 0.724 | 9.5 | 0.01 | Legumin-type globulin precursor f | 4204299 |
| 35_7.9 | 7.88 | 34774 | 0.040 | 0.095 | 2.4 | 0.23 | Legumin-type globulin precursor f,h | 9759513 |
| 31_5.9 | 5.85 | 30545 | 0.016 | 0.034 | 2.1 | 0.01 | Legumin-type globulin precursor f,h | 9759513 |
| 30_6.7 | 6.74 | 29573 | 0.542 | 0.118 | −4.6 | 0.02 | α-chain of legumin-type globulinf,h | 1628583 |
| 30_6.2 | 6.20 | 29787 | 0.350 | 0.099 | −3.5 | 0.01 | α-chain of legumin-type globulinf,i | 9759513 |
| 29_6.6 | 6.60 | 29117 | 6.057 | 2.154 | −2.8 | 0.50 | α-chain of legumin-type globulinf,i | 9759513 |
| 28_6.8 | 6.76 | 27722 | 0.975 | 2.559 | 2.6 | 0.08 | Legumin-type globulin precursor f,h | 4204299 |
| 26_7.54 | 7.54 | 25955 | 0.000 | 0.614 | NA | 0.10 | α-chain of legumin-type globulinf,h | 9759513 |
| 26_5.4 | 5.36 | 25569 | 0.068 | 0.200 | 2.9 | 0.06 | α-chain of legumin-type globulinf,i | 4204298 |
| 25_7.7 | 7.66 | 25357 | 0.033 | 0.000 | NA | 0.01 | α-chain of legumin-type globulinf,i | 4204299 |
| 25_6.8 | 6.76 | 24819 | 0.148 | 0.387 | 2.6 | 0.10 | α-chain of legumin-type globulinf,i | 4204299 |
| 24_6.3 | 6.26 | 24249 | 0.066 | 0.160 | 2.4 | 0.00 | α-chain of legumin-type globulinf,h | 4204299 |
| 20_7.8 | 7.77 | 19736 | 0.105 | 0.005 | −22.8 | 0.00 | β-chain of legumin-type globulinf,i | 1628583 |
| 20_7.3 | 7.27 | 19614 | 0.006 | 0.223 | 39.8 | 0.07 | β-chain of legumin-type globulin with N-terminalf,h extension |
9759513 |
| 19_11.9 | 11.92 | 19146 | 0.092 | 0.019 | −4.9 | 0.34 | β-chain of legumin-type globulinf,i | 1628583 |
| 18_8.5 | 8.50 | 17673 | 0.152 | 1.342 | 8.8 | 0.12 | β-chain of legumin-type globulin with N-terminalf,h extension |
1628583 |
| 17_5.8 | 5.81 | 17598 | 0.039 | 0.113 | 2.9 | 0.03 | β-chain of legumin-type globulinf,h | 4204299 |
| 16_7.7 | 7.68 | 16299 | 0.006 | 0.117 | 18.5 | 0.06 | β-chain of legumin-type globulinf,h | 1628583 |
| 13_8.9 | 8.87 | 13231 | 0.051 | 0.145 | 2.9 | 0.09 | α-chain of legumin-type globulinf,h | 9759513 |
| 13_7.9 | 7.93 | 13337 | 0.030 | 1.463 | 49.1 | 0.00 | Napin-type albumin 2S 3 precursor f | 166616 |
| 11_5.2 | 5.18 | 11198 | 0.082 | 0.026 | −3.1 | 0.24 | Embryo-specific protein 3 | 7546697 |
| 11_5.0 | 5.00 | 10615 | 0.150 | 0.057 | −2.6 | 0.00 | α-chain of legumin-type globulinf,h | 1628583 |
| 11_4.9 | 4.88 | 11118 | 0.165 | 0.513 | 3.1 | 0.03 | α-chain of legumin-type globulinf,h | 1628583 |
| 11_4.7 | 4.65 | 11111 | 0.051 | 0.118 | 2.3 | 0.03 | α-chain of legumin-type globulinf,h | 1628583 |
Identification number assigned for reference to Figure 6.
Percentage of integrated fluorescence intensity of a specific protein spot in relation to the integrated fluorescence intensity of all detected protein spots.
Fold increase (positive numbers; calculated as the quotient of % βVPE and % WT) or fold decrease (negative numbers; calculated as the quotient of % WT and % βVPE) of the integrated fluorescence intensity of a particular protein spot in βVPE mutant seeds compared with wild-type seeds. This calculation was not applicable (NA) when a specific protein spot was not detected in both samples.
Significance of comparisons between three gels each of wild-type and βVPE mutant seeds.
PID, protein identification number in GenBank.
Mass spectrometry coverage data were used to further suggest the relationship of the individual polypeptide fragment with respect to its corresponding full-length protein form.
NA, not applicable.
Legumin-type globulin polypeptides identified as alternatively processed forms (see Results for details).
Legumin-type globulin polypeptides identified as normally processed or redundantly processed forms (see Results for details).
Table 2.
Mass Spectrometric Identification of Select Seed Protein Features That Did Not Change in Accumulation Between Homozygous βVPE::dSpm1 and Wild-Type Seeds
| ID No.a | pI | kD | % WT b | % βVPE b | Changec | t Test d | Annotation Detail | PIDe |
|---|---|---|---|---|---|---|---|---|
| 62_5.9 | 5.93 | 61525 | 0.186 | 0.305 | 1.6 | 0.13 | Putative seed maturation protein | 4559335 |
| 60_6.4 | 6.42 | 60215 | 0.038 | 0.046 | 1.2 | 0.42 | β-Glucosidase | 9294684 |
| 51_7.3 | 7.31 | 51336 | 0.021 | 0.024 | 1.1 | 0.77 | Glycine hydroxymethyltransferase and vicilin-like precursor f |
7433539 9279583 |
| 49_10.5 | 10.53 | 48745 | 0.284 | 0.282 | −1.0 | 0.96 | Putative elongation factor 1-a | 8778823 |
| 43_6.9 | 6.90 | 43080 | 0.036 | 0.046 | 1.3 | 0.58 | Alcohol dehydrogenase class III | 1498024 |
| 40_6.2 | 6.22 | 40128 | 0.073 | 0.133 | 1.8 | 0.03 | 11-β-Hydroxysteroid dehydrogenase-like | 8777393 |
| 38_7.0 | 7.04 | 38045 | 0.122 | 0.095 | −1.3 | 0.41 | Putative glyceraldehyde-3-phosphate dehydrogenase | 9958054 |
| 35_4.6 | 4.61 | 35056 | 0.014 | 0.017 | 1.2 | 0.74 | Embryonic protein LEA.D34 homolog type 1 | 2129632 |
| 32_5.2 | 5.15 | 31872 | 0.190 | 0.211 | 1.1 | 0.83 | LEA-like protein | 4140257 |
| 31_7.8 | 7.79 | 30753 | 0.136 | 0.097 | −1.4 | 0.74 | α-chain of legumin-type globulinf,g | 1628583 |
| 31_7.4 | 7.37 | 30586 | 0.458 | 0.488 | 1.1 | 0.90 | α-chain of legumin-type globulinf,g | 1628583 |
| 31_6.7 | 6.72 | 30624 | 0.103 | 0.076 | −1.3 | 0.23 | α-chain of legumin-type globulinf and vicilin-like precursor C-terminal portionf |
1628583 9279583 |
| 31_6.6 | 6.59 | 30740 | 4.909 | 3.000 | −1.6 | 0.42 | α-chain of legumin-type globulinf,g | 1628583 |
| 30_5.9 | 5.88 | 29800 | 0.108 | 0.149 | 1.4 | 0.20 | Strong similarity to Brassica Asp protease and alpha chain of legumin-type globulinf |
2160151 1628583 |
| 29_7.2 | 7.16 | 28511 | 0.130 | 0.206 | 1.6 | 0.24 | C-terminal chain of vicilin-like storage proteinf | 9279583 |
| 29_6.4 | 6.38 | 29379 | 0.152 | 0.114 | −1.3 | 0.82 | α-chain of legumin-type globulinf,h | 1628583 |
| 29_5.8 | 5.83 | 28853 | 0.089 | 0.054 | −1.7 | 0.01 | α-chain of legumin-type globulinf | 9759513 |
| 29_5.5 | 5.48 | 29033 | 0.096 | 0.098 | 1.0 | 0.95 | Hypothetical protein desiccation-related | 7486973 |
| 28_6.3 | 6.32 | 28436 | 0.080 | 0.060 | −1.3 | 0.85 | α-chain of legumin-type globulinf,h | 9759513 |
| 28_6.2 | 6.19 | 27756 | 0.528 | 0.281 | −1.9 | 0.05 | α-chain of legumin-type globulinf,h | 9759513 |
| 27_8.1 | 8.13 | 26535 | 2.649 | 4.043 | 1.5 | 0.19 | α-chain of legumin-type globulinf,h | 9759513 |
| 27_7.5 | 7.53 | 27023 | 1.416 | 0.825 | −1.7 | 0.12 | N-terminal chain of vicilin-like storage proteinf | 9279583 |
| 27_6.9 | 6.89 | 27390 | 1.163 | 1.124 | −1.0 | 0.85 | α-chain of legumin-type globulinf,h | 9759513 |
| 26_9.6 | 9.56 | 25779 | 0.084 | 0.121 | 1.4 | 0.48 | N-terminal chain of vicilin-like storage proteinf | 9279583 |
| 26_7.53 | 7.53 | 25725 | 0.254 | 0.313 | 1.2 | 0.66 | N-terminal chain of vicilin-like storage proteinf | 9279583 |
| 26_6.7 | 6.65 | 25851 | 0.100 | 0.071 | −1.4 | 0.05 | α-chain of legumin-type globulinf,g | 4204299 |
| 26_5.2 | 5.22 | 26207 | 0.410 | 0.300 | −1.4 | 0.52 | α-chain of legumin-type globulinf,g | 4204298 |
| 26_10.0 | 10.02 | 25679 | 0.918 | 0.980 | 1.1 | 0.67 | LEA76 homolog type2 | 1592677 |
| 25_7.1 | 7.13 | 25135 | 4.609 | 3.047 | −1.5 | 0.10 | α-chain of legumin-type globulinf,g | 4204299 |
| 25_5.5 | 5.50 | 25237 | 0.079 | 0.113 | 1.4 | 0.15 | Cytosolic triose phosphate isomerase | 414550 |
| 24_6.6 | 6.59 | 24474 | 0.301 | 0.239 | −1.3 | 0.12 | Similar to rehydrin homolog | 8778528 |
| 24_6.5 | 6.51 | 24410 | 0.207 | 0.197 | −1.1 | 0.82 | Unknown protein | 8778719 |
| 24_6.4 | 6.37 | 24277 | 0.035 | 0.012 | −2.9 | 0.23 | C-terminal chain of putative seed storage protein (vicilin-like)f | 4510397 |
| 23_6.6 | 6.60 | 22616 | 0.090 | 0.077 | −1.2 | 0.34 | Manganese superoxide dismutase-like protein | 7572933 |
| 23_6.4 | 6.42 | 22700 | 0.169 | 0.147 | −1.2 | 0.32 | C-terminal chain of putative seed storage protein (vicilin-like)f | 4510397 |
| 22_5.6 | 5.56 | 21758 | 0.231 | 0.150 | −1.5 | 0.16 | α-chain of legumin-type globulinf,h | 1628583 |
| 20_8.6 | 8.55 | 19627 | 0.089 | 0.024 | −3.7 | 0.11 | β-chain of legumin-type globulinf,g | 1628583 |
| 19_9.3 | 9.32 | 18813 | 13.671 | 13.831 | 1.0 | 0.93 | β-chain of legumin-type globulinf,g | 1628583 |
| β-chain of legumin-type globulinf,g | 9759513 | |||||||
| 19_8.9 | 8.90 | 18955 | 0.907 | 0.484 | −1.9 | 0.40 | β-chain of legumin-type globulinf,g | 1628583 |
| 19_8.5 | 8.53 | 18913 | 0.317 | 0.182 | −1.7 | 0.12 | β-chain of legumin-type globulinf,g | 1628583 |
| 19_10.6 | 10.59 | 19392 | 0.123 | 0.185 | 1.5 | 0.60 | Legumin-type globulin precursor f,h | 1628583 |
| 18_6.9 | 6.85 | 17614 | 4.090 | 3.765 | −1.1 | 0.68 | β-chain of legumin-type globulinf,g | 4204299 |
| 18_6.2 | 6.22 | 17849 | 0.111 | 0.128 | 1.2 | 0.46 | β-chain of legumin-type globulinf,g | 4204299 |
| 18_5.8 | 5.76 | 17736 | 0.434 | 0.409 | −1.1 | 0.73 | β-chain of legumin-type globulinf,g | 4204298 |
| 16_7.8 | 7.81 | 15698 | 0.150 | 0.153 | 1.0 | 0.73 | Peptidylprolyl isomerase ROC1 | 1076367 |
| 15_10.6 | 10.58 | 15289 | 0.092 | 0.090 | −1.0 | 0.90 | α-chain of legumin-type globulinf,h | 1628583 |
| 14_7.7 | 7.74 | 13868 | 0.040 | 0.017 | −2.3 | 0.13 | Albumin 2S 3 precursor f | 166616 |
| 14_7.3 | 7.25 | 13972 | 0.234 | 0.194 | −1.2 | 0.38 | Major latex protein type 3 | 2129642 |
| 14_6.8 | 6.76 | 14045 | 0.243 | 0.251 | 1.0 | 0.82 | Major latex protein type 1 | 2129641 |
| 12_5.0 | 4.98 | 11548 | 0.085 | 0.081 | −1.0 | 0.77 | α-chain of legumin-type globulinf,h | 1628583 |
| 12_11.3 | 11.27 | 12135 | 0.160 | 0.113 | −1.4 | 0.62 | Legumin-type globulin precursor f,h | 1628583 |
Identification number assigned for reference to Figure 6.
Percentage of integrated fluorescence intensity of a specific protein spot in relation to the integrated fluorescence intensity of all detected protein spots.
Fold increase (positive numbers; calculated as the quotient of % βVPE and % WT) or fold decrease (negative numbers; calculated as the quotient of % WT and % βVPE) of the integrated fluorescence intensity of a particular protein spot in βVPE mutant seeds compared with wild-type seeds.
Significance of comparisons between three gels each of wild-type and βVPE mutant seeds.
PID, protein identification number in GenBank.
Mass spectrometry coverage data were used to further suggest the relationship of the individual polypeptide fragment with respect to its corresponding full-length protein form.
Legumin-type globulin polypeptides identified as normally processed or redundantly processed forms (see Results for details).
Legumin-type globulin polypeptides identified as alternatively processed forms (see Results for details).
The data shown in Tables 1 and 2 support the concept of βVPE being involved specifically with storage protein maturation in seeds. Almost all of the differences (33 of 34) were identified as seed storage proteins (legumin type, napin type, or vicilin type), whereas despite the identification of several nonstorage proteins (17 of 85 in the project), only one nonstorage protein was changed significantly in quantity (relative to total protein) in the mutant (Table1, feature 11_5.2). The list of identified differences (Table 1) also supports βVPE involvement in the deposition of several distinct seed storage proteins. Differentially deposited polypeptide forms of four Arabidopsis legumin-type globulin proteins, a vicilin-type protein, and one napin-type albumin protein were identified. Note that this analysis did not capture polypeptides of approximate apparent molecular mass of <10 kD, complicating the interpretation of changes of low molecular mass storage proteins (e.g., napin-type albumins). To assess net changes in seed storage protein accumulation between βVPE mutants and wild-type controls, we considered mostly legumin-type globulins.
Predicted apparent molecular mass and pI (supported by direct MS identification data) were used to identify gel regions corresponding to pro and processed forms of legumin-type globulins. The circles in Figure 5 indicate a gel region of apparent molecular mass and pI predicted to correspond to propolypeptide forms of legumin-type globulin storage proteins. Indeed, all of the differentially accumulating polypeptides in this region were identified by MS as either legumin-type globulins (all four proteins) or a vicilin-like protein (Table 1, features 52_6.9 to 45_6.7, Figure 6A), indicating that knockout of βVPE results in the accumulation of storage protein precursors. The sum of the protein detected in this region amounted to 3.7% ± 1.2% and 0.7% ± 0.05% of the total protein quantity detected in the mature seeds of βVPE mutants and wild-type controls, respectively. Therefore, this area of the gel shows a threefold to sevenfold increase in detected prolegumin-type globulin protein in βVPE mutants compared with the wild type. Similarly, the rectangles in Figure 5 indicate regions of the gel corresponding to the predicted apparent molecular mass and pI of the α- and β-chains of legumin-type globulin proteins. As expected, MS identified many of the predominant proteins in these gel regions as α- and β-chains (Tables 1 and 2, Figure 6). The sum of the protein quantities detected in the α-chain region averaged 36.6% ± 3.8% and 42.8% ± 3.4% for βVPE mutant and wild-type controls, respectively. Therefore, our results showed a trend for the reduction of α-chain accumulation in βVPE mutant seeds compared with wild-type seeds. The sum of the amount of protein detected in the β-chain region averaged 29.3% ± 3.6% and 29.5% ± 0.6% for the βVPE mutant and the wild-type control, respectively, indicating no significant change in β-chain accumulation as defined by this type of analysis.
In addition to the propolypeptide forms of legumin-type globulin, we detected several polypeptides of apparent molecular mass and pI not consistent with either pro-forms or mature α- and β-chains (Figure 6, Tables 1 and 2). This point is illustrated by examination of all of the polypeptides derived from a single legumin-type globulin protein (GenBank protein identification number [PID] 1628583). Twenty-seven gel features have been identified as containing derivatives of this protein (Tables 1 and 2). Twenty-three of these 27 gel features were identified as containing exclusively derivatives of this specific protein (the remaining 4 were identified as mixed features or spots—i.e., containing more than one polypeptide type). Each of these 23 gel features was detected in both βVPE::dSpm1 and wild-type seeds, indicating that their presence was not unique to either background. One feature was identified by apparent molecular mass and abundance (>2% of total protein detected) as a mature α-chain (Table 2, feature 31_6.6), a second feature was identified as containing mature β-chain (Table 2, feature 19_9.3), and three features could be accounted for by apparent molecular mass as corresponding to propolypeptide forms (Table 1, features 48_7.8, 48_7.6, and 46_6.9). Of the remaining 18 features, 8 were altered significantly in accumulation (4 decreasing and 4 increasing in response to βVPE knockout; Table 1) and 10 were not changed significantly in accumulation (Table 2). Assuming equal distribution of the polypeptide quantity of the β-chain gel feature (Table 2, feature 19_9.3), these 18 features accounted for approximately one-fourth of the total quantity of protein identified as PID 1628583 in wild-type seeds. Therefore, most of the protein (75%) could be assigned to the expected pro- and mature forms; however, the remainder appeared to be processed alternatively.
Although the specific post-translational modifications resulting in the observed shifts of apparent molecular mass and pI were not determined, it was observed that several of these polypeptides were reduced in apparent molecular mass compared with the highly abundant mature α- and β-chains. We interpreted this information as indicative of post-translational proteolysis and sought to compare wild-type and βVPE mutant seeds. MS coverage data (location of mass matches of the individual polypeptide with respect to the conserved P1 Asn residue of the full-length protein), apparent molecular mass, and polypeptide quantity were used to define legumin-type globulin polypeptides as pro-forms, wild-type mature α- or β-forms, or alternatively processed forms. Alternatively processed forms were defined for each legumin-type globulin protein as having MS coverage data on both sides of the conserved P1 Asn but smaller in apparent molecular mass (>5-kD difference) than the precursor pro-form or having MS coverage data identifying a polypeptide as corresponding to either the α- or β-form but smaller in apparent molecular mass (≥0.5-kD difference) than the respective highly abundant mature form (>2% of total protein detected). Using the entire data set, we found that the MS-identified alternative polypeptide processing derivatives constituted 11.8% ± 1.4% and 7.4% ± 0.8% of the total detected protein in βVPE::dSpm1 seeds and wild-type seeds, respectively. The quantities of the remaining mature polypeptide forms of the legumin-type globulins identified by MS were 28.6% ± 2.1% and 35.0% ± 0.8% of the total protein detected in βVPE::dSpm1 seeds and wild-type seeds, respectively. Therefore, a significant shift from wild-type processed forms of legumin-type globulins to alternatively processed forms occurred in response to the removal of βVPE activity in seeds.
N-Terminal Amino Acid Sequence Analysis
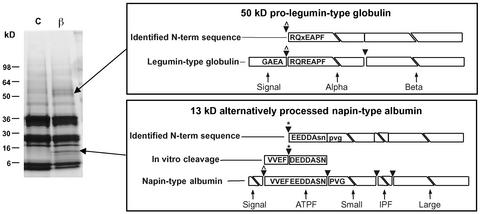
Although it is a powerful technology for protein identification, MS is less suited for the determination of N-terminal amino acid sequences. Therefore, Edman degradation (Matsudaira et al., 1993) was performed with two prominent polypeptides accumulated in βVPE::dSpm1 seeds (Figure 7). The larger polypeptide (∼50 kD) was identified as a legumin-type globulin (PID 4204298) with an N terminus corresponding to amino acids immediately downstream of the predicted signal sequence (Figure 7). Sequence and molecular mass identify this polypeptide as an unprocessed legumin-type proglobulin precursor. It appears to be identical to the two-dimensional gel feature 46_5.4 (Table 1), identified as the same specific legumin-type globulin protein.
Figure 7.
N-Terminal Sequence Analysis of Polypeptides Accumulated in βVPE::dSpm1 Seeds.
At left is a SDS-PAGE 4 to 20% mini-gel comparing mature seed protein of homozygous βVPE::dSpm1 (β) and the wild type (C) to indicate the polypeptide bands sequenced. The boxes at right show the alignment of the identified N-terminal sequence for each polypeptide band with respect to the corresponding full-length seed storage protein. Arrowheads illustrate cleavage sites resulting in mature seed storage protein chains. Arrowheads with darts indicate N-termini identified at sites consistent with signal peptidase cleavage, and arrowheads with asterisks indicate N termini identified at sites consistent with processing using purified Asp protease in vitro.
The smaller polypeptide (∼13 kD) was identified as a product of a napin-type albumin (PID 166616) that is not yet processed within the internal processed fragment region to produce the typical large and small albumin subunits found in the wild type. It appears to be identical to two-dimensional gel feature 13_7.9 (Table 1). The N-terminal residues of this polypeptide did not correspond to the residues immediately downstream of the signal peptide sequence, as would be predicted for a propolypeptide precursor; instead, it corresponded to the central portion of the N-terminal processed fragment sequence (Figure 7).
Analysis of Massively Parallel Signature Sequencing Transcript Profiling Data
Results of the analysis of seed-type VPE knockout mutants suggested the existence of both alternative and redundant processing pathways for storage proteins in maturing seeds. Additionally, our results demonstrated that a protease gene (βVPE) with peak levels of transcription occurring during mid seed development (14 DAA) was involved in storage protein processing. The mid seed developmental window also was the stage at which large amounts of storage protein accumulated (Figure 4B). Therefore, to identify other protease genes with expression patterns that correlated with storage protein accumulation, we queried a database of precomputed gene expression clusters (GECs) derived from several Arabidopsis Lynx Therapeutics massively parallel signature sequencing (MPSS) high-resolution gene expression data sets (D.A. Selinger, F. Gruis, and R. Jung, unpublished data). A GEC is a group of genes that share a significantly similar spatial and temporal expression level relationship. Lynx Therapeutics MPSS gene expression data sets are essentially very deep EST sequencing experiments, each of which consists of 1 to 2 million sequences obtained from a single tissue source (Brenner et al., 2000). This depth of EST sequencing provides quantitative and comprehensive gene expression data. The GEC database used here included Lynx Therapeutics MPSS data sets from seeds during early development (cell division phase; 7 DAA), seeds during mid- development (storage protein accumulation phase; 14 DAA), germinating seeds (radicle protrusion phase), whole-plant seedlings (stage 1), leaves, whole roots, and inflorescences.
Queries of the GEC database (which contains 128 unique GECs) using conceptual Lynx Therapeutics MPSS ESTs (see Methods) for Arabidopsis napin-type albumins, legumin-type globulins, and vicilin-like gene sequences resulted in the identification of a single GEC (GEC98) that contained ESTs for 10 of the most abundant seed storage protein genes. Table 3 lists the locus names and accession numbers for the seed storage protein genes used to identify GEC98 as the relevant seed storage protein–specific GEC. Figure 8A illustrates the relative expression of the storage protein genes identified in GEC98 across all tissue types examined. In addition to the 10 unique ESTs corresponding to storage protein gene transcripts, GEC98 contained 230 other MPSS ESTs. To determine if any protease gene transcripts also were identified with GEC98, conceptual MPSS ESTs were determined for all 127 annotated proteases (classified as members of the Cys, Asp, Ser and metallo-protease gene families) identified in the Arabidopsis genome (NCBI, February 5, 2002) and compared with the 230 MPSS ESTs in GEC98. As expected, the EST corresponding to the βVPE transcript (Table 3) was present in GEC98. Also as expected, we observed no MPSS EST corresponding to the δVPE transcript in GEC98. This finding is in accord with those of RT-PCR experiments (Figure 1) indicating that δVPE had an expression pattern unlike that of βVPE (maximum δVPE expression before maximum βVPE expression in developing seeds). In addition to βVPE, we identified five gene transcripts annotated as proteases (Table 3).
Table 3.
Locus Names and Accession Numbers for Genes That Encode Seed Proteins or Seed-Specific Proteases
| Locus | Accession No. | Expression Profile in GEC98 Cluster |
Expression Detected in Seeds |
Description |
|---|---|---|---|---|
| At4g27140 | NP_194444 | NDa | +a | Napin-type albumin 1 (NWMU1), 2S albumin 1 precursor |
| At4g27150 | NP_194445 | + | + | Napin-type albumin 2 (NWMU2), 2S albumin 2 precursor |
| At4g27160 | NP_194446 | + | + | Napin-type albumin 3 (NWMU3), 2S albumin 3 precursor |
| At4g27170 | NP_194447 | + | + | Napin-type albumin 4 (NWMU4), 2S albumin 4 precursor |
| At5g54740 | NP_200285 | + | + | Napin-type albumin 5, 2S storage protein-like |
| At4g28520 | NP_194581 | + | + | Legumin-type globulin Cruciferin 1, 12S seed storage protein |
| At1g03890 | NP_171885 | + | + | Legumin-type globulin Cruciferin 2, 12S seed storage protein |
| At1g03880 | NP_171884 | + | + | Legumin-type globulin Cruciferin 3, 12S seed storage protein |
| At5g44120 | NP_199225 | + | + | Legumin-type globulin Cruciferin 4, 12S seed storage protein |
| At2g28490 | NP_180416 | + | + | Vicilin-like seed storage protein (globulin) |
| At4g36700 | NP_195388 | + | + | Vicilin-like seed storage protein (globulin) |
| At2g18540 | NP_179444 | NDb | −b | Vicilin-like seed storage protein (globulin) |
| At3g22640 | NP_566714 | − | + | Vicilin-like seed storage protein (globulin) |
| At1g62710 | NP_176458 | + | + | βVPE |
| At1g62290 | NP_176419 | + | + | Asp protease |
| At3g54940 | NP_567010 | + | + | Cys proteinase 2 |
| At4g11310 | NP_567376 | + | + | Cys proteinase 1 |
| At5g09640 | NP_568215 | + | + | Carboxypeptidase-like protein |
| At4g16640 | NP_193397 | + | + | Similar to soybean metalloendoproteinase 1 |
Not determined; this gene has no valid Lynx Therapeutics MPSS tag, but it is expressed in seeds based on EST clones in GenBank.
Not determined; this gene has no valid Lynx Therapeutics MPSS tag and no corresponding EST clones.
Figure 8.
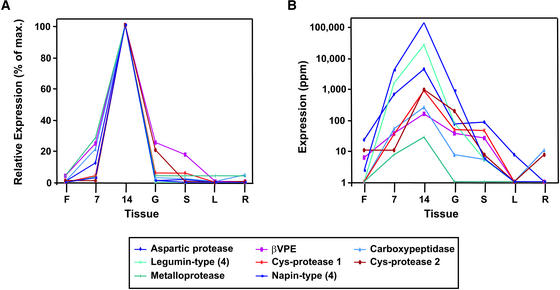
Expression Profiles of the Seed Storage Protein and Protease Genes Identified in the Seed Protein–Specific Expression Cluster (GEC98).
(A) The expression profile of each gene or of a storage protein gene group (average of four genes; see key) is plotted as a percentage of expression relative to maximum observed expression across all tissues sampled. Tissues sampled were inflorescences (F), seeds at 7 DAA (7), seeds at 14 DAA (14), germinating seeds (G), seedlings (S), leaves (L), and roots (R).
(B) Expression profiles plotted as a log scale of transcript levels in parts per million. Tissues and genes are the same as in (A).
Figure 8B shows MPSS EST levels in parts per million for the seed storage and protease genes identified in the seed storage protein–specific cluster (GEC98). Three of these protease genes were highly expressed: two belong to the papain-type subfamily of Cys proteases, and the other belongs to the aspartic protease family. A Ser carboxypeptidase homolog and the homolog of soybean metalloendoproteinase I also were found in GEC98, but they were expressed at lower levels based on MPSS EST counts (parts per million). The aspartic protease family member shares 80% identity with an aspartic protease cloned previously from Arabidopsis seeds (D'Hondt et al., 1997). The two Cys proteases are not related closely to each other, sharing only 30% identity. One (Cys protease 1) appears to be related more closely to RD21A (43% identity), whereas the other (Cys protease 2) appears to be related more closely to RD19A (51% identity) (Koizumi et al., 1993). The closest homolog of Cys protease 2 that we identified was a soybean protein annotated as the “40-kD seed maturation protein” (Nong et al., 1995), with which it shares 62% identity.
DISCUSSION
The understanding and control of the cellular mechanisms responsible for protein accumulation and turnover in seeds are of importance to biotechnological efforts to produce either foreign proteins or engineered endogenous proteins in this tissue. It is believed that seed storage protein deposition in seed vacuoles follows a specific path involving restricted proteolytic processing by specialized proteases (Muntz, 1998). It has been suggested that proteases involved in seed storage protein processing also may determine the stability of recombinant proteins targeted to PSV in seeds (Jung et al., 1993, 1998). Members of the VPE family of proteases were isolated from seeds and identified as capable of processing legumin-type globulin and napin-type albumin seed storage proteins in dicots on the basis of proteolytic processing assays in vitro (Hara-Nishimura et al., 1993). Additionally, seed-type VPEs have been localized to the PSV and shown to be capable of self-catalytic activation in acidic vacuoles (Hara-Nishimura et al., 1993; Kuroyanagi et al., 2002). Together, this evidence has led to the paradigm that seed-type VPEs are the enzymes responsible for processing seed storage proteins (Hara-Nishimura, 1998; Muntz and Shutov, 2002). However, to our knowledge, VPE function with regard to seed storage protein processing has never been demonstrated successfully or disproved directly in planta. This may be attributable to functional redundancy provided either from within the VPE gene family or by alternative proteases. To address these challenges, we used a plant model system with a fully sequenced genome (Arabidopsis) to test the validity of the model of VPE function in seeds.
Redundant Proteolytic Mechanisms Compensate Incompletely for the Loss of βVPE Expression in Developing Seeds
Arabidopsis was an especially attractive model system because the VPE family in this species is small, with only one seed-type VPE member (βVPE) described previously. Surprisingly, knockout of βVPE expression did not abolish seed storage protein processing of either the legumin-type globulins or the napin-type albumins. However, in mutant seeds, we observed a consistent accumulation of small amounts of novel storage protein polypeptide derivatives that apparently were absent or minor components in wild-type seeds. During germination, these novel polypeptides as well as the mature storage protein polypeptide chains appeared to be mobilized with kinetics similar to those of wild-type seeds. These observations support the conclusion that βVPE is involved in storage protein accumulation but not in storage protein mobilization. This finding is in agreement with the idea that the enzymes that process storage proteins during seed development are not the same enzymes that degrade them during germination (Muntz, 1996). Furthermore, these observations support the conclusion that the same cellular machinery responsible for mobilizing mature wild-type storage protein polypeptides in germinating seeds also is capable of mobilizing the accumulated novel polypeptides in βVPE knockout seeds.
A Second Seed-Expressed VPE Gene Identified in Arabidopsis (δVPE) Does Not Contribute Significantly to Seed Storage Protein Processing
We predicted that the redundancy of the VPE family might be the explanation for the normal processing of the majority of storage protein in βVPE::dSpm1 seeds. Examination of the genome led to the identification of a second seed-expressed VPE gene (δVPE). Further searches did not identify any additional ESTs or genomic sequence beyond those corresponding to the four VPE genes described, even at homology levels low enough to identify the more distantly related putative gpi-8 gene. Therefore, it is highly unlikely that other expressed VPE genes remain undiscovered in the Arabidopsis genome. Although our results showed that the δVPE transcript is expressed highly in developing seeds, its expression pattern differed from that of βVPE in that the highest levels of δVPE expression are found in seeds before βVPE expression. Nonetheless, δVPE expression clearly is seed preferred and partially overlaps that of βVPE, making it a good candidate for functional redundancy to βVPE. However, examination of mature seed protein profiles of δVPE knockout mutants and βVPE/δVPE double mutants failed to identify any role for δVPE in seed storage protein processing. It remains to be determined whether the increased sampling depth of two-dimensional gel analysis could reveal polypeptide changes in seeds of δVPE::dSpm1 that were not detected by one-dimensional gel analysis.
Recently, two closely related fruit/seed-expressed VPE family members, one from tomato and one from tobacco, were described (Fischer et al., 2000; Muntz and Shutov, 2002). As with δVPE, it has not been possible to clearly assign these family members to either the seed-type or the vegetative-type phylogenetic group, which suggests the possibility of a third functional VPE subclass. To date, no direct function has been identified for this group of VPE genes, yet it has been proposed that the tobacco VPE may have a functional role in early embryogenesis (Fischer et al., 2000; Muntz and Shutov, 2002). Here, we present no evidence that δVPE knockout results in any embryogenic abnormalities, nor do we present evidence of disadvantageous consequences when δVPE knockout plants are propagated under typical laboratory conditions. Therefore, the function of this VPE family member remains unclear.
A second alternative functional role for VPE family members (VsPB2 and proteinase B from V. sativa) relates to storage protein mobilization in germinating seeds (Becker et al., 1995; Schlereth et al., 2001). However, such a function for δVPE appears unlikely, because we did not find a reduction of storage protein mobilization in the βVPE/δVPE double null mutant during germination.
Specific and Direct Involvement of βVPE in Storage Protein Processing
The unexpected result that deleting βVPE in Arabidopsis resulted in only a minor change in seed protein accumulation led us to hypothesize that βVPE may act on a specific subset of storage proteins or is active in only a specific vacuolar subcompartment within the cells (Jiang et al., 2001). A comparative two-dimensional gel/MS analysis to determine all protein accumulation changes between wild-type seeds and βVPE::dSpm1 seeds demonstrated that a wide variety of storage proteins are altered in accumulation (both legumin-type globulins and napin-type albumins), whereas only one nonstorage seed protein was determined to be altered in accumulation. These results support the hypothesis of the specific and direct involvement of βVPE in storage protein processing. Additionally, we determined that the absence of βVPE significantly increased the accumulation of pro-forms of legumin-type globulins and a napin-type albumin in mature seeds. The increase in pro-forms of legumin-type globulins was accompanied by a trend toward reduced accumulation of mature α-chains in the βVPE mutant. It is important to note that the amount of protein accumulated as a prolegumin-type globulin is only a fraction (∼5 to 10%) of the total amount of legumin-type globulin in the seed; therefore, it is difficult to detect the corresponding reduction in mature α- and β-chains in the βVPE mutant with a high degree of confidence. Nevertheless, these observations are consistent with the conclusion that βVPE does perform a function in processing seed storage protein from pro-forms to mature chains, as predicted previously (Kinoshita et al., 1999). However, in contrast to previous predictions, we found that other redundant proteolytic mechanisms are capable of compensating for this function almost completely.
Alternative Proteolytic Processing of Arabidopsis Seed Proteins
In addition to detecting pro-forms of legumin-type globulins and mature legumin-type globulin α- and β-chains, our comparative proteomic analysis also detected a number of novel polypeptide-processing derivatives of legumin-type globulins. For each legumin-type globulin gene, several polypeptides accumulated in seeds, many of them differing in apparent molecular mass and pI from the predominantly accumulating α- and β-chains. The decrease in molecular mass of the derivative polypeptides compared with their respective mature α- and β-chains is evidence that the post-translational modifications involved most likely are the result of alternative or additional proteolytic mechanisms. We observed these legumin-type globulin polypeptide derivatives in both wild-type and βVPE::dSpm1 seeds. Individually, these polypeptide derivatives do not appear to account for a large amount of the legumin-type globulin protein from any single gene; however, together, they can constitute as much as one-fourth of the protein from a single gene. We also found that the amounts of a subset of these derivatives, but not all of them, were altered in response to βVPE knockout. Although the accumulated quantity of some derivatives decreased and others increased in βVPE::dSpm1 seeds, a significant overall increase in this pool of legumin-type globulin was observed in βVPE::dSpm1 seeds. We interpret this observation as support for the conclusion that alternative proteolytic processing pathways exist in developing seeds. We hypothesize that more legumin-type globulin protein is processed by these alternative pathways in response to an increase in the amount of pro-forms of storage protein in the PSVs of seeds.
There are few benchmarks in the literature for comparison of our comparative two-dimensional gel/MS analysis of mature Arabidopsis seeds. However, a recent proteomic analysis of Arabidopsis also described alternative forms of legumin-type globulins accumulating in mature wild-type seeds (Gallardo et al., 2001). Although the molecular mass and pI associated with protein features (spots) in the two-dimensional separations in that analysis are technically difficult to compare with the features observed in our analysis, a similar origin of these features is likely. Gallardo's group suggested that these polypeptides were products of proteolysis by proteases that are active predominantly during germination. We find this interpretation tenuous for the following reasons. First, the majority of legumin-type globulin polypeptides identified by Gallardo et al. (2001) (18 features) remained constant in relative quantity and did not increase or decrease during germination, as might be expected if they were transient products of legumin-type globulin mobilization. Second, although Gallardo and colleagues observed an increase in quantity of six legumin-type globulin polypeptides during germination, they also observed a decrease in four legumin-type globulin polypeptides corresponding in molecular mass to pro-forms of the protein. Therefore, the increase in processed polypeptide forms might be accounted for by processing of the pro-forms remaining in mature seeds. Finally, in our experiments using one-dimensional gels, we observed no transient accumulation in wild-type germinating seeds of polypeptides similar in molecular mass to those that accumulated in developing βVPE::dSpm1 seeds (Figure 4). To the contrary, polypeptides found to accumulate in βVPE::dSpm1 seeds diminished during germination at rates similar to mature α- and β-chains.
Aspartic Protease Activity Likely Is Involved in Napin-Type Albumin Processing
In addition to the comparative two-dimensional gel/MS analysis, Edman degradation sequencing was used to determine the N-terminal amino acids of two polypeptides that accumulated in βVPE::dSpm1 seeds. One polypeptide (50 kD) was identified by sequence and apparent molecular mass as a prolegumin-type globulin and provided further supporting evidence that pro-forms can and do accumulate in mature seeds in response to βVPE knockout. A second polypeptide (13 kD) was identified by its sequence and apparent molecular mass as a napin-type albumin that was not cleaved in the internal processed fragment region; cleavage at this site produces the typical mature large and small chains. Interestingly, the N-terminal sequence of this napin-type polypeptide was determined to start immediately after a Phe residue in the middle of the N-terminal processed fragment region. This site is downstream of the predicted signal sequence cleavage site and is consistent in sequence context with cleavage by a member of the aspartic protease gene family (D'Hondt et al., 1993). D'Hondt et al. (1993) dem-onstrated that aspartic protease activity purified from developing canola seed cleaved a short synthetic polypeptide derived from the middle of the N-terminal processed fragment region in vitro. Here, we report that the cleavage site we observed in planta is the same cleavage site observed previously in the in vitro processing assays, providing evidence of a functional role for aspartic protease activity in seed storage protein processing in planta.
The in planta identification of processed storage proteins with cleavage sites consistent with aspartic protease activity in plants that lack βVPE implies a different conclusion than that derived from in vitro processing assays by Hiraiwa et al. (1997). In that report, the authors discounted a possible primary role of aspartic proteases in the processing of seed storage protein and proposed that VPE activity was the primary or initial activity involved in the processing of storage protein. It was further concluded that aspartic protease activity was capable of performing only a secondary function of trimming the C-terminal propeptides after cleavage by VPE into chains. However, contrary to these conclusions, our results suggest a discrete and independent in vivo role (with no primary VPE activity needed) for aspartic proteases in the processing of napin-type albumins. Additionally, we observed aspartic protease–cleaved napin-type albumin at all investigated stages during storage protein accumulation (Figure 4B), suggesting the colocalization of aspartic protease and napin-type albumins in planta throughout seed development.
Candidate Genes for Redundant and Alternative Seed Protein Processing Enzymes in Arabidopsis
Protease genes that might constitute redundant or alternative proteolytic pathways for seed storage proteins were identified by the fact that they shared a spatial and temporal expression relationship with seed storage protein genes. Queries of a gene expression cluster database derived from several Arabidopsis Lynx Therapeutics MPSS expression profiling data sets (D.A. Selinger, F. Gruis, and R. Jung, unpublished data) identified a single cluster of 240 genes that shared a similar expression profile. In addition to containing transcripts from 10 of the most abundantly expressed storage protein genes, the cluster also contained 6 annotated protease genes. The discovery that one of the six genes was βVPE and that a second gene was a highly expressed putative aspartic protease gene supports the validity of this approach. This information enables testing of the hypothesis that the specific aspartic protease gene identified encodes the protease involved in the alternative cleavage of napin-type albumins by virtue of gene suppression in a βVPE null background. Of the remaining four genes identified in the analysis, two are highly expressed. One is a papain-type Cys proteinase that has homology with a soybean gene identified in developing seeds during the storage protein deposition stage (Nong et al., 1995). The second also is a Cys proteinase with greatest homology with RD21A, a drought-inducible Cys proteinase of Arabidopsis (Koizumi et al., 1993). Interestingly, a recent report indicated that RD21A and γVPE both were accumulated specifically in “endoplasmic reticulum bodies” in leaf epidermal cells of Arabidopsis (Hayashi et al., 2001). Although it is unclear whether these two proteases can act synergistically, it is attractive to hypothesize that their colocalization and coexpression are of functional consequence. The RD21A homolog we identified is coexpressed in a transient pattern similar to βVPE; therefore, we predict that it also may have a potential functional synergy. The remaining two genes expressed at a much lower level include a putative metalloprotease and a putative Ser protease that has homology with carboxypeptidases. Activities related to similar genes have been described in developing buckwheat seeds. Although the precise function of these genes is not known, some indicators suggest a role during germination (Dunaevsky et al., 1989; Belozersky et al., 1990).
In conclusion, our results show that seed-type VPEs are not the only proteases involved in seed storage protein maturation. Furthermore, we provide evidence to support the existence of both redundant and alternative proteolytic pathways in developing Arabidopsis seeds. Finally, we provide a list of potential protease genes that might be involved, with the caveat that the analysis described here may not identify protease genes upregulated specifically in response to βVPE knockout. Using the current model system, it is expected that we will be able to delineate the entire proteolytic process of seed storage protein maturation, commencing with the suppression or knockout of these seed-expressed protease genes.
METHODS
Identification and Phylogenetic Analysis of δVPE
BLAST (Basic Local Alignment Search Tool; Altschul et al., 1990) was used to query all Arabidopsis genomic and EST sequence databases (GenBank and DuPont). Queries were performed using all previously identified Arabidopsis vacuolar processing enzyme (VPE) genomic and cDNA sequences. Sequences with scores equal to or greater than the score obtained for gpi-8 were examined further using CLUSTAL W (Thompson et al., 1994). δVPE ESTs were identified from a library created from mRNA of Arabidopsis thaliana fertilized carpels with developing seeds at 6 to 7 days after fertilization. Three δVPE cDNA clones were sequenced in their entirety, and a genomic (GenBank) sequence annotation corresponding to these cDNAs was identified later. Phylogenetic analysis of the VPE gene family, including bootstrap tests of the resulting dendrograms (500 iterations of neighbor joining), was accomplished using MEGA 2b3 software (Kumar et al., 2001).
Reverse Transcriptase–PCR
Poly(A) RNA was extracted from ∼2 mg of fresh frozen tissue (roots, leaves, inflorescences, stems, seeds dissected from pods days after anthesis, and mature seeds) using Dynabeads (Dynal, Oslo, Norway), and the entire contents of the isolation were used to produce first-strand cDNA using Superscript II (Stratagene, La Jolla, CA) according to the manufacturers' protocols. A total of 2 μL of the cDNA reaction was used for PCR using oligonucleotide primers 5′-GTC-GTATCTTAGCTGGTACTTGCCACAGTGCGAA-3′; and 5′-CAGCAG-CAGCTTACATTCCAGTGAATGCCTTCTT-3′; to identify cytosolic ribosomal protein S11 transcript multiplexed with either 5′-GGA-AGATGGGTCAAGGAAGAAGGATGACACAT-3′; or 5′-AGATTGATGGATGCACCGTGTAGCGAGC-3′; to identify βVPE transcript or 5′-CCAGAGACTTCTCATGTATGCCGTTTCGGAA-3′; and 5′-CCG-TTGCACCGCAGTGATTCTTGAAGCTATTAA-3′; to identify δVPE transcript. For semiquantitative measurement of transcript levels, 35 cycles were performed, and for conformation of gene knockout, 45 cycles were performed using Expand High Fidelity enzyme (Roche Molecular Biochemicals, Indianapolis, IN) at an annealing temperature of 65°C after a hot-start protocol (Sambrook and Russell, 2001). Expected product sizes of the spliced transcript amplified were 288 bp for cytosolic ribosomal protein S11, 362 bp for βVPE, and 424 bp for δVPE.
SLAT Filter Hybridization
A SLAT (Sainsbury Laboratory Arabidopsis thaliana dSpm Transposants) filter blot, obtained from the Sainsbury Laboratory (Norwich, UK), displaying flanking DNA of the Sainsbury dSpm transposon insertion population was probed with a genomic DNA probe corresponding to the δVPE gene. A digoxigenin (DIG)-labeled probe was produced by PCR with the oligonucleotides 5′-CTGAGTCCC-GCAAAACCCAATTGCTGAA-3′; and 5′-CCGTTGCACCGCAGTGAT-TCTTGAAGCTATTAA-3′; using the PCR DIG Probe Synthesis kit (Roche Molecular Biochemicals) according to the manufacturer's protocol. The SLAT blot membrane orientation marker was detected with a DIG 1-kb ladder marker probe generated using the High Prime kit (Roche Molecular Biochemicals). SLAT blot membranes were hybridized with these probes using the DIG Easy Hyb kit (Roche Molecular Biochemicals) by incubation overnight at 42°C. After hybridization, SLAT filters were washed for 1 h at 65°C with 1 × SSC (150 mM NaCl and 15 mM sodium citrate, pH 7.2) containing 0.1% SDS. DIG-labeled probes hybridizing to the blot were detected with an anti-DIG antibody conjugated with alkaline phosphatase (Roche Molecular Biochemicals) and the disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2-(5-chloro) tricyclo [3.3.1.13,7] decan}-4-yl) phenyl phosphate (Roche Molecular Biochemicals).
SINS Database
Searches of the SINS (Sequenced Insertion Sites) database (http://www.jic.bbsrc.ac.uk/Sainsbury-lab/jonathan-jones/SINS-database/database.html) were performed using BLAST (Altschul et al., 1990).
Cloning and Sequencing of Genomic DNA Flanking dSpm Transposon Alleles
Pooled genomic DNA, isolated from parents giving rise to each seed pool (obtained from the Sainsbury Laboratory), was subjected to several rounds of PCR using transposon border primers in combination with gene-specific primers. Primers used include the following: 5′-GTGTCGGCTTATTTCAGTAAGAGTGTGGGGTTTTG-3′ and 5′-GTGTGGGGTTTTGGCCGACACTCCTTAC-3′ corresponding to the 3′ end of the dSpm element, 5′-CGGCCCCGACACTCTTTAATT-AACTGACACTCCTT-3′ and 5′-CGGTGCAGCAAAACCCACACTTTT-ACTTCGAT-3′ corresponding to the 5′ end of the dSpm element, 5′-TCTGTTAGTTCTTTTGGTTCATGCCGAGTCA-3′ corresponding to the 5′ end of βVPE, 5′-AGATTGATGGATGCACCGTGTAGCGAGC-3′ corresponding to the 3′ end of βVPE, 5′-GAACGCTAT-CCTCTATATCTCGTCCTGGTCCAAATGT-3′; corresponding to the 5′ end of δVPE, and 5′-CCGTTGCACCGCAGTGATTCTTGAAGCTATTAA-3′; corresponding to the 3′ end of δVPE. DNA products from these PCRs were separated on Tris-acetate EDTA agarose gels, isolated using the QIAquick gel extraction kit (Qiagen, Valencia, CA), and cloned into the pCR TOPO 2.1 plasmid vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturers' protocols. Sequences of the cloned DNA fragments were analyzed using CLUSTAL W (Thompson et al., 1994) to confirm the location of the dSpm insertion sites within VPE genes.
PCR Identification of dSpm Alleles
Seeds (obtained from the Sainsbury Laboratory) from pools 5.41 and 1.14 (50 progenitors per pool) were grown (∼500 plants). Before flowering, genomic DNA from individual plants was isolated using the DNeasy Plant Mini kit (Qiagen) according to the manufacturer's protocol. Plants containing the dSpm insertion allele(s) of interest were identified by PCR using the primers mentioned above. Plants homozygous for the βVPE::dSpm1 and/or δVPE::dSpm1 allele(s) were defined by the absence of amplification of wild-type alleles and the presence of amplification of mutant alleles. Homozygosity was confirmed by the lack of detectable wild-type alleles in the progeny resulting from self-pollination.
βVPE Monospecific Polyclonal Antibody and Protein Immunoblotting
A keyhole limpet hemocyanin–conjugated synthetic polypeptide (5′-SSVTAANFYAVLLGDQKA-3′) corresponding to amino acids 128 to 145 of βVPE was synthesized (Research Genetics, Huntsville, AL) and used as an antigen to generate antisera in rabbits at Strategic Biosolutions (Newark, DE). Antiserum from rabbit 99090 was affinity purified against the immobilized peptide antigen at Research Genetics. The purified antibody was used at a dilution of 1:500 in an immunoblot analysis of protein extracted from mature βVPE::dSpm1 seeds and wild-type seeds. Total protein was extracted from whole seeds in a 20-fold (v/w) excess of 2% SDS, 160 mM DTT, and 50 mM Tris-HCl, pH 6.8. After the addition of buffer, the samples were incubated immediately for 5 min at 100°C, vortexed, and centrifuged for 10 min at 20,800g. Supernatants were collected (avoiding the low-density lipid layer), and protein samples were adjusted to ∼1 mg/mL in SDS-PAGE sample buffer (Laemmli, 1970). Proteins (10 μg/sample) were separated electrophoretically by SDS-PAGE using a 4 to 20% gradient mini-gel (Bio-Rad, Hercules, CA) and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon P; Millipore, Bedford, MA) using a semidry electroblotter (SemiPhor TE70; Hoefer, San Francisco, CA), as described previously (Matsudaira et al., 1993). Prestained molecular mas protein standards (SeeBlue; Novex, San Diego, CA) were used to determine the apparent molecular masses of proteins. The detection of antigens on PVDF blots was performed according to standard procedures (Harlow and Lane, 1988) using a 1:10,000 dilution of horseradish peroxidase–conjugated goat anti-rabbit antibody (Bio-Rad) and the enhanced chemiluminescence kit (Pharmacia, Piscataway, NJ) according to the manufacturers' protocols.
Tris-Tricine SDS-PAGE of Developing and Germinating Seeds
Developing seeds were obtained from plants grown at 23°C with a 16-h-light/8-h-dark cycle. Flowers (pods) were marked at anthesis. At various times after anthesis, marked pods were removed from the plant and developing seeds were dissected and frozen immediately in liquid nitrogen. For germinating seed samples, mature seeds were allowed to imbibe in water at 4°C for 48 h before being placed on sterile filter paper and incubated at 25°C. Radicle protrusion typically occurred at ∼24 h after the start of the 25°C incubation period. After 0, 24, or 48 h of incubation at 25°C, seed samples were collected and frozen immediately in liquid nitrogen. Protein was extracted from all samples, including mature seeds, as described above. Approximately 30 μg/sample was separated electrophoretically by SDS-PAGE on Tris-Tricine gels (8% spacer and 15% separating) (Schagger and von Jagow, 1987) using a SE400 vertical slab gel unit (Hoefer) and visualized by Coomassie Brilliant Blue R250 staining (Sambrook and Russell, 2001).
Comparative Two-Dimensional Gel/Mass Spectrometry Proteome Analysis
Pooled mature seeds (50 mg) from plants homozygous for βVPE::dSpm1 and wild-type siblings (five plants each) were ground in and defatted by three hexane extractions at room temperature. After vacuum desiccation, protein was extracted using denaturing two-dimensional lysis buffer (8 M urea, 2 M thiourea, 4% [w/v] 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, and 65 mM DTT). After brief vortexing, samples were incubated on ice for 15 min and subjected to centrifugation at 20,800g to remove insoluble debris. The supernatants were aspirated, and protein was adjusted to a concentration of 1 mg/mL in lysis buffer.
Two-dimensional electrophoresis was conducted at Oxford GlycoSciences (Oxfordshire, UK), with the second-dimension gels bound to one of the glass plates according to procedures detailed by Page et al. (1999). The gels were fixed and stained with a fluorescent dye (OGT MP17), and images were obtained exactly as described (Page et al., 1999) using Escherichia coli proteins as landmark features to determine the pI and apparent molecular mass for each protein spot (feature). Three replica gels were run for each sample, and primary gel images were processed using proprietary software at Oxford GlycoSciences. Individually resolved protein features were enumerated and quantified on the basis of fluorescence signal intensity. A total of 1364 unique reproducible features were identified in wild-type and βVPE::dSpm1 seed samples and compiled into a database table and into composite images. The data obtained from the three replica gels of each protein sample were analyzed statistically (variance) to remove outliers. In addition, the composite images of the gels were examined for artifacts to eliminate perturbed features from the analysis. Analysis to identify differentially accumulated proteins then was undertaken using proprietary software (ROSETTA) provided by Oxford GlycoSciences and confirmed by visual examination of all gels.
Protein features of interest were identified as described by Page et al. (1999). A mass list of peptides from tryptic peptide pools of each feature was compiled using matrix-assisted laser desorption ionization time-of-flight spectrometry. Fragmentation spectra from one-dimensional mass windows were recorded using a nanospray ionization source (Z-spray) on a Quadrupole Time-of-Flight Mass Spectrometry instrument (Micromass, Manchester, UK). Protein identification was accomplished using SEQUEST software (Thermo Finnigan, San Jose, CA) to search the SWISS-PROT and DuPont-Pioneer genome databases and was confirmed when an ion series consistent with y-type fragmentation was observed for the complete peptide sequence. Insufficient spectral coverage of a polypeptide was the predominant reason for identification failures.
N-Terminal Amino Acid Sequence Analysis
Proteins were extracted, separated electrophoretically by SDS-PAGE using a 4 to 20% gradient mini-gel (Bio-Rad), and transferred to PVDF membranes (Immobilon Psq; Millipore) as described above. Proteins on PVDF membranes were visualized with Coomassie Brilliant Blue as described previously (Matsudaira et al., 1993). Protein bands of interest were excised and air-dried, and samples were Edman sequenced using a Procise 492 Sequencer (Applied Biosystems, Foster City, CA) at the University of Iowa Molecular Analysis Facility (Iowa City). Sequence identity was determined using FASTA (Pearson and Lipman, 1988) to query the GenBank and SWISS-PROT protein databases.
Massively Parallel Signature Sequencing Transcript Profiling
The mRNA from a variety of tissue samples was isolated previously (F. Gruis, unpublished data), and massively parallel signature sequencing (MPSS) was performed by Lynx Therapeutics (Hayward, CA) as described (Brenner et al., 2000). The resulting MPSS ESTs, here defined as the first 17 bp including and downstream of the most 3′ Sau3A site (GATC) of a transcript, are quantified and reported as parts per million (1 to 2 million sequencing reactions performed per sample).
The quantity (parts per million) of each unique MPSS EST across all seven MPSS experiments was computed previously such that each unique MPSS EST was assigned to a specific gene expression cluster (GEC) (D.A. Selinger, F. Gruis, and R. Jung, unpublished data). Therefore, each GEC in the GEC database constitutes a unique collection of MPSS ESTs sharing a similar temporal and spatial expression relationship across these seven tissue samples.
Identification of seed storage protein–specific GEC98 was accomplished by identifying conceptual MPSS ESTs from the respective storage protein genes (the first 17 bp including and downstream of the most 3′ Sau3A site of a conceptually spliced transcript) and searching the GEC database of MPSS sequence tags for an exact string match to experimentally derived MPSS ESTs. Identification of proteases contained in GEC98 was accomplished in a similar manner except that exact string matches of conceptual annotated protease transcript MPSS ESTs were identified from only the 230 unique experimentally derived MPSS ESTs found in GEC98.
Novel materials described in this article are available for noncommercial research purposes upon acceptance and signing of a material transfer agreement. In some cases, such materials may contain or be derived from materials obtained from a third party. In such cases, distribution of material will be subject to the requisite permission from any third-party owners, licensors, or controllers of all or parts of the material. Obtaining permission will be the sole responsibility of the requestor.
Accession Numbers
Protein accession numbers for the polypeptides identified by MS are given in Tables 1 and 2. Accession numbers for proteases identified in GEC98 are provided in Table 3. Other accession numbers for sequences discussed in this article are AF521661 (δVPE), NP_172352 (GPI-8), L07877 (cytosolic ribosomal protein S11), AAD46920 and CAA83673 (40-kD seed maturation protein), D61393 (αVPE), D61395 (γVPE), NP_564497 (RD21A), and NP_568052 (RD19A). Accession number for βVPE is NP_176458.
Acknowledgments
We are grateful to Vincent Sewalt, Enno Krebbers, Odd-Arne Olsen, Keith Roesler, Jan Schulze, Rebecca Boston, and Con Moothart for critically reading the manuscript and providing helpful comments. We thank Jan Schulze, Ryan Dove, Mark Mucha, Jason Campbell, Jason Boehme, Eva Wojcik, Jeremy Hayter (Oxford GlycoSciences), Jim Lawrence, Bill Van Zante, Henry Mirsky, and Brian Zeka for technical support. We also appreciate the efforts of Matthew Smoker, Yvette Reader, and other members of the Sainsbury Laboratory for graciously providing dSpm library materials. Finally, we are indebted to members of the DuPont-Pioneer genomics and bioinformatics groups for expert support, and we thank Paul Anderson and Larry Beach, who encouraged this work.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005009.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Becker, C., Shutov, A.D., Nong, V.H., Senyuk, V.I., Jung, R., Horstmann, C., Fischer, J., Nielsen, N.C., and Muntz, K. (1995). Purification, cDNA cloning and characterization of proteinase B, an asparagine-specific endopeptidase from germinating vetch (Vicia sativa L.) seeds. Eur. J. Biochem. 228, 456–462. [PubMed] [Google Scholar]
- Belozersky, M.A., Dunaevsky, Y.E., and Voskoboynikova, N.E. (1990). Isolation and properties of a metalloproteinase from buckwheat (Fagopyrum esculentum) seeds. Biochem. J. 272, 677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal, M., Benachour, A., Rusconi, S., Aebi, M., and Conzelmann, A. (1996). Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 15, 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., et al. (2000). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol 18, 630–634. [DOI] [PubMed] [Google Scholar]
- D'Hondt, K., Bosch, D., Van Damme, J., Goethals, M., Vandekerckhove, J., and Krebbers, E. (1993). An aspartic proteinase present in seeds cleaves Arabidopsis 2 S albumin precursors in vitro. J. Biol. Chem. 268, 20884–20891. [PubMed] [Google Scholar]
- D'Hondt, K., Stack, S., Gutteridge, S., Vandekerckhove, J., Krebbers, E., and Gal, S. (1997). Aspartic proteinase genes in the Brassicaceae Arabidopsis thaliana and Brassica napus. Plant Mol. Biol. 33, 187–192. [DOI] [PubMed] [Google Scholar]
- Dunaevsky, Y.E., Sarbakanova, S.T., and Belozersky, M.A. (1989). Wheat seed carboxypeptidase and joint action on gliadin of proteases from dry and germinating seeds. J. Exp. Bot. 40, 1323–1329. [Google Scholar]
- Fischer, J., Becker, C., Hillmer, S., Horstmann, C., Neubohn, B., Schlereth, A., Senyuk, V., Shutov, A., and Muntz, K. (2000). The families of papain- and legumain-like cysteine proteinases from embryonic axes and cotyledons of Vicia seeds: Developmental patterns, intracellular localization and functions in globulin proteolysis. Plant Mol. Biol. 43, 83–101. [DOI] [PubMed] [Google Scholar]
- Frigerio, L., Vine, N.D., Pedrazzini, E., Hein, M.B., Wang, F., Ma, J.K., and Vitale, A. (2000). Assembly, secretion, and vacuolar delivery of a hybrid immunoglobulin in plants. Plant Physiol. 123, 1483–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo, K., Job, C., Groot, S.P., Puype, M., Demol, H., Vandekerckhove, J., and Job, D. (2001). Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura, I. (1998). Asparaginyl endopeptidase. In Handbook of Proteolytic Enzymes, A. Barrett, N. Rawlings, and J. Woessner, eds (London: Academic Press), pp. 746–749.
- Hara-Nishimura, I., Inoue, K., and Nishimura, M. (1991). A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett. 294, 89–93. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Takeuchi, Y., and Nishimura, M. (1993). Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell 5, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hayashi, Y., Yamada, K., Shimada, T., Matsushima, R., Nishizawa, N.K., Nishimura, M., and Hara-Nishimura, I. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42, 894–899. [DOI] [PubMed] [Google Scholar]
- Hiraiwa, N., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (1997). An aspartic endopeptidase is involved in the breakdown of propeptides of storage proteins in protein-storage vacuoles of plants. Eur. J. Biochem. 246, 133–141. [DOI] [PubMed] [Google Scholar]
- Hoffman, L.M., Donaldson, D.D., and Herman, E.M. (1988). A modified storage protein is synthesized, processed, and degraded in the seeds of transgenic plants. Plant Mol. Biol. 11, 717–729. [DOI] [PubMed] [Google Scholar]
- Ishii, S. (1994). Legumain: Asparaginyl endopeptidase. Methods Enzymol. 244, 604–615. [DOI] [PubMed] [Google Scholar]
- Jiang, L., Phillips, T.E., Hamm, C.A., Drozdowicz, Y.M., Rea, P.A., Maeshima, M., Rogers, S.W., and Rogers, J.C. (2001). The protein storage vacuole: A unique compound organelle. J. Cell Biol. 155, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, R., Saalbach, G., Nielsen, N.C., and Muntz, K. (1993). Site-specific limited proteolysis of legumin chloramphenicol acetyl transferase fusions in vitro and in transgenic tobacco seeds. J. Exp. Bot. 44, 343–349. [Google Scholar]
- Jung, R., Scott, M.P., Nam, Y.W., Beaman, T.W., Bassuner, R., Saalbach, I., Muntz, K., and Nielsen, N.C. (1998). The role of proteolysis in the processing and assembly of 11S seed globulins. Plant Cell 10, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermode, A.R., Fisher, S.A., Polishchuk, E., Wandelt, C., Spencer, D., and Higgins, T.J. (1995). Accumulation and proteolytic processing of vicilin deletion-mutant proteins in the leaf and seed of transgenic tobacco. Planta 197, 501–513. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Nishimura, M., and Hara-Nishimura, I. (1995. a). The sequence and expression of the gamma-VPE gene, one member of a family of three genes for vacuolar processing enzymes in Arabidopsis thaliana. Plant Cell Physiol. 36, 1555–1562. [PubMed] [Google Scholar]
- Kinoshita, T., Nishimura, M., and Hara-Nishimura, I. (1995. b). Homologues of a vacuolar processing enzyme that are expressed in different organs in Arabidopsis thaliana. Plant Mol. Biol. 29, 81–89. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yamada, K., Hiraiwa, N., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (1999). Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 19, 43–53. [DOI] [PubMed] [Google Scholar]
- Koizumi, M., Yamaguchi-Shinozaki, K., Tsuji, H., and Shinozaki, K. (1993). Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129, 175–182. [DOI] [PubMed] [Google Scholar]
- Krebbers, I., Herdies, L., De Clercq, A., Seurinck, J., Leemans, J., Van Damme, J., Sequra, M., Gheysen, G., Van Montagu, M., and Vandekerckhove, J. (1988). Determination of the processing sites of an Arabidopsis 2S albumin and characterization of the complete gene family. Plant Physiol. 87, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi, M., Nishimura, M., and Hara-Nishimura, I. (2002). Activation of Arabidopsis vacuolar processing enzyme by self-catalytic removal of an auto-inhibitory domain of the C-terminal propeptide. Plant Cell Physiol. 43, 143–151. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Linnestad, C., Doan, D.N., Brown, R.C., Lemmon, B.E., Meyer, D.J., Jung, R., and Olsen, O.A. (1998). Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiol. 118, 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira, P., Aebersold, R., Charbonneau, H., LeGendre, N., Mansfield, M., Martin, S.A., Scoble, H.A., Stone, K.L., Vath, J.E., Weiss, A., Williams, K.R., and Yu, W. (1993). A Practical Guide to Protein and Peptide Purification for Microsequencing. (San Diego, CA: Academic Press).
- Meinke, D. (1994). Seed development in Arabidopsis thaliana. In Arabidopsis, E. Meyerowitz and C. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 253–295.
- Muntz, K. (1996). Proteases and proteolytic cleavage of storage proteins in developing and germinating dicotyledonous seeds. J. Exp. Bot. 47, 605–622. [Google Scholar]
- Muntz, K. (1998). Deposition of storage proteins. Plant Mol. Biol. 38, 77–99. [PubMed] [Google Scholar]
- Muntz, K., and Shutov, A. (2002). Legumains and their functions in plants. Trends Plant Sci. 7, 340–344. [DOI] [PubMed] [Google Scholar]
- Nong, V.H., Becker, C., and Muntz, K. (1995). cDNA cloning for a putative cysteine proteinase from developing seeds of soybean. Biochim. Biophys. Acta 1261, 435–438. [DOI] [PubMed] [Google Scholar]
- Page, M.J., Amess, B., Townsend, R.R., Parekh, R., Herath, A., Brusten, L., Zvelebil, M.J., Stein, R.C., Waterfield, M.D., Davies, S.C., and O'Hare, M.J. (1999). Proteomic definition of normal human luminal and myoepithelial breast cells purified from reduction mammoplasties. Proc. Natl. Acad. Sci. USA 96, 12589–12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W.R., and Lipman, D.J. (1988). Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueyo, J.J., Chrispeels, M.J., and Herman, E.M. (1995). Degradation of transport-competent destabilized phaseolin with a signal for retention in the endoplasmic reticulum occurs in the vacuole. Planta 196, 586–596. [DOI] [PubMed] [Google Scholar]
- Runeberg-Roos, P., Kervinen, J., Kovaleva, V., Raikhel, N.V., and Gal, S. (1994). The aspartic proteinase of barley is a vacuolar enzyme that processes probarley lectin in vitro. Plant Physiol. 105, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schagger, H., and von Jagow, G. (1987). Tricine-sodium dodecylsulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schlereth, A., Standhardt, D., Mock, H.P., and Muntz, K. (2001). Stored cysteine proteinases start globulin mobilization in protein bodies of embryonic axes and cotyledons during vetch (Vicia sativa L.) seed germination. Planta 212, 718–727. [DOI] [PubMed] [Google Scholar]
- Scott, M.P., Jung, R., Muntz, K., and Nielsen, N.C. (1992). A protease responsible for post-translational cleavage of a conserved Asn-Gly linkage in glycinin, the major seed storage protein of soybean. Proc. Natl. Acad. Sci. USA 89, 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, T., Hiraiwa, N., Nishimura, M., and Hara-Nishimura, I. (1994). Vacuolar processing enzyme of soybean that converts proproteins to the corresponding mature forms. Plant Cell Physiol. 35, 713–718. [DOI] [PubMed] [Google Scholar]
- Sjodahl, S., Rodin, J., and Rask, L. (1991). Characterization of the 12S globulin complex of Brassica napus: Evolutionary relationship to other 11-12S storage globulins. Eur. J. Biochem. 196, 617–621. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D. (1999). Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei, H., Van Damme, J., Casteels, P., and Krebbers, E. (1993). A fifth 2S albumin isoform is present in Arabidopsis thaliana. Plant Physiol. 101, 1415–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., Shimada, T., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (1999). Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme. J. Biol. Chem. 274, 2563–2570. [DOI] [PubMed] [Google Scholar]