Abstract
To understand how chloroplast mRNAs are translated into functional proteins, a detailed understanding of all of the components of chloroplast translation is needed. To this end, we performed a proteomic analysis of the plastid ribosomal proteins in the small subunit of the chloroplast ribosome from the green alga Chlamydomonas reinhardtii. Twenty proteins were identified, including orthologs of Escherichia coli S1, S2, S3, S4, S5, S6, S7, S9, S10, S12, S13, S14, S15, S16, S17, S18, S19, S20, and S21 and a homolog of spinach plastid-specific ribosomal protein-3 (PSRP-3). In addition, a novel S1 domain–containing protein, PSRP-7, was identified. Among the identified proteins, S2 (57 kD), S3 (76 kD), and S5 (84 kD) are prominently larger than their E. coli or spinach counterparts, containing N-terminal extensions (S2 and S5) or insertion sequence (S3). Structural predictions based on the crystal structure of the bacterial 30S subunit suggest that the additional domains of S2, S3, and S5 are located adjacent to each other on the solvent side near the binding site of the S1 protein. These additional domains may interact with the S1 protein and PSRP-7 to function in aspects of mRNA recognition and translation initiation that are unique to the Chlamydomonas chloroplast.
INTRODUCTION
Chloroplasts are thought to have evolved by endosymbiosis of a photosynthetic unicellular prokaryote into a eukaryotic host. During chloroplast evolution, a majority of the plastid genes have been translocated into the nuclear genome (reviewed by Subramanian, 1993). During this time, a number of host genes have been used in the plastid. Plastid gene expression is regulated primarily during translation and involves the dynamic interplay between ribosomes, translation factors, and mRNA-protein complexes (Mayfield et al., 1995; Barkan and Goldschmidt-Clermont, 2000). To understand how mRNAs are translated into functional proteins, a detailed understanding is needed of all of the components of chloroplast translation, including the ribosomes, translation factors, and mRNA binding proteins, as well as the RNA elements that interact with them. Proteomic analysis of the spinach plastid ribosome has revealed 6 plastid-specific ribosomal proteins (PSRPs), a stoichiometric amount of plastid ribosome recycling factor (pRRF), and 52 of the 54 Escherichia coli orthologs. Spinach plastids lack only the L25 and L30 orthologs of E. coli ribosomes (Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000). Among the 52 E. coli orthologs in spinach, 12 small subunit proteins and 8 large subunit proteins are plastid encoded, whereas 9 small subunit proteins and 23 large subunit proteins are nucleus encoded. All six PSRPs and the pRRF are nucleus-encoded proteins.
Compared with higher plants (spinach), algal (Chlamydomonas reinhardtii) 30S proteins have been shown to be somewhat different in terms of molecular mass, composition, and plastid gene arrangement (Schmidt et al., 1983; reviewed by Harris et al., 1994). The Chlamydomonas chloroplast ribosomal proteins have been characterized by two-dimensional PAGE (Hanson et al., 1974; Brugger and Boschetti, 1975; Schmidt et al., 1983) as well as immunologically (Schmidt et al., 1984; Randolph-Anderson et al., 1989). Sites of protein synthesis have been studied by pulse labeling of cells in the presence of cytoplasmic or chloroplastic protein synthesis inhibitors. These studies have shown that of the 31 small subunit proteins identified, 14 are synthesized in the chloroplast and 17 are synthesized in the cytosol (Schmidt et al., 1983). It has been reported that the Chlamydomonas chloroplast genome contains an unusual rps2-like gene, orf570 (Leu, 1998), and the rps3-like gene orf712 (Fong and Surzycki, 1992; Liu et al., 1993; Turmel and Otis, 1994). orf570 encodes a protein containing an ∼300–amino acid N-terminal extension (NTE) showing no homology with any known protein and a C-terminal half that shows homology with S2 protein. orf712 encodes a protein that shows significant sequence similarity to bacterial S3 protein, but the similarity is limited to the N and C termini, with an insertion sequence (INS) lacking similarity to other known proteins. Whether the products are actual ribosomal proteins or the NTEs and INSs are processed before the proteins are assembled into ribosomes has not been determined.
Proteomics, which involves gel electrophoresis (one-dimensional or two-dimensional PAGE), liquid chromatography–tandem mass spectrometry (LC-MS/MS), and bioinformatic analysis, has become a powerful tool for the investigation of chloroplast function (reviewed by van Wijk, 2001). We applied proteomics to characterize the small subunit of the Chlamydomonas plastid ribosome. Here, we present the identification of a novel S1 domain–containing protein and three unusually large ribosomal proteins, S2, S3, and S5, from the Chlamydomonas chloroplast small ribosomal subunit. Most of the other expected homologs of E. coli 30S subunit and a plastid-specific ribosomal protein were identified as well.
RESULTS
Biochemical Isolation of the Chlamydomonas Chloroplast 30S Subunit and SDS-PAGE Separation of the 30S Proteins
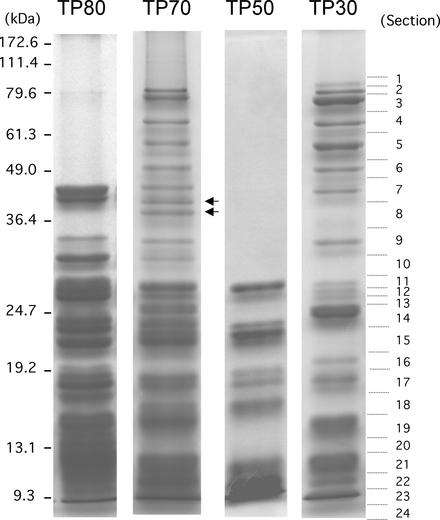
Chloroplast ribosomes used in this study were purified from a postmitochondrial S-40 fraction of total cell extract by successive Suc gradient centrifugation. The chloroplast 70S ribosomes were first separated from the cytoplasmic 80S ribosomes in a 10 to 40% Suc gradient. Purified 70S ribosomes were dissociated into 30S and 50S subunits and separated on a 10 to 30% Suc gradient. The purity of the 70S ribosomes and the 30S and 50S subunits was examined by SDS-PAGE (Figure 1). SDS-PAGE patterns of total proteins in the 80S (TP80) and 70S (TP70) ribosomes are clearly different in both composition and size, indicating that our ribosome preparations were sufficiently pure. Several protein bands of >50 kD are seen in TP70, and several of the proteins from the small subunit (TP30) are much larger than the proteins from the large subunit (TP50). These observations are in agreement with a previous report by Schmidt et al. (1983). Identification of large proteins (>50 kD) in the 30S subunit has not been reported from the higher plant (spinach) chloroplast for which the 30S ribosomal proteins have been characterized fully (ranging from 5 to 40 kD; Yamaguchi and Subramanian, 2000). The TP70 pattern shows a composite profile of the TP30 and TP50 fractions, as expected. However, TP70 also contained two additional protein bands of 41 and 38 kD (Figure 1, arrows) that were not seen in the TP30 or TP50 fractions, suggesting that these two proteins associate only with the 70S ribosome. In spinach, a similar observation has been reported in that 70S ribosomes contain a protein not found in either of the isolated subunits, a homolog of bacterial ribosome recycling factor (Yamaguchi et al., 2000).
Figure 1.
SDS-PAGE Profiles of Chlamydomonas Ribosomal Proteins.
Total proteins extracted from cytosolic ribosomes (80S), chloroplast ribosomes (70S), and chloroplast subunits (50S and 30S) (10 pmol each of TP80, TP70, TP50, and TP30) were subjected to SDS-PAGE. Proteins were stained with Coomassie blue. Two bands (41 and 38 kD) present in TP70 but absent in TP30 and TP50 are indicated by arrows. The TP30 lane was sectioned into 24 pieces for in-gel digestion, as indicated by the dotted lines.
In-Gel Digestion and Identification of Peptides Using LC-MS/MS
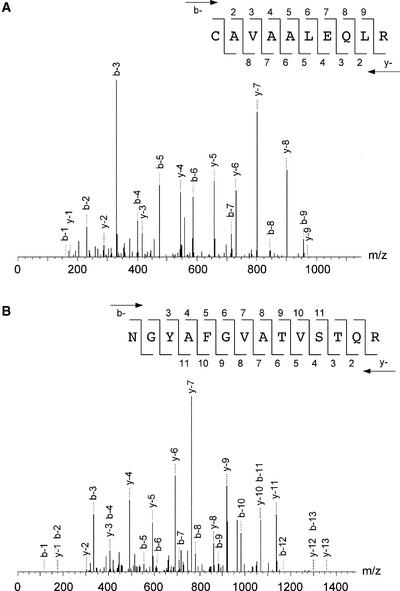
To identify the individual small subunit ribosomal proteins by peptide tandem mass spectra, the 30S proteins were separated by SDS-PAGE and each protein was digested using trypsin and endoproteinase Lys-C. The generated peptide fragments were subjected to LC-MS/MS analyses as described in Methods. Table 1 shows the identified peptide sequences obtained by peptide tandem mass spectra using the SEQUEST algorithm relying on Chlamydomonas EST and chloroplast genome open reading frame (ORF) databases (http://www.biology.duke.edu/chlamy_genome/chloro.txt). Table 1 also shows the corresponding data sets of mass-to-charge ratio (m/z) of parent ions, accession numbers of the identified ESTs or plastid ORFs, protein identification numbers, and SDS-PAGE gel section numbers showing where the peptides originated. All of the peptides listed in Table 1 showed reliable peptide matches with high correlation factors (>2.5). Figure 2 shows examples of collision-induced dissociation spectra derived from the 84- and 57-kD proteins of the small subunit. The collision-induced dissociation spectra show product ions of the tryptic peptide at m/z 565.6 from the 84-kD protein (Figure 2A) and that of a peptide at m/z 735.8 from the 57-kD protein (Figure 2B), identifying those peptide sequences to be CAVAALEQLR and NGYAFGVATVSTQR, respectively. The SEQUEST database search identified these peptides in an EST (AV391351) derived from the nuclear genome and an ORF (CAA76245) of the plastid genome, respectively (Table 1).
Table 1.
Identification of Chlamydomonas 30S PRPs by LC-MS/MS Analyses of Fragments from the Sections of TP30 Gel or Directly from TP30
| Peptide Sequence a | m/z | Protein Header b | Protein ID c | Section No.d or MudPIT e |
|---|---|---|---|---|
| K.RASSTVSNDIQGFK.V | 503.55 | BE024920 | PRP S1 | 6L, 7L |
| R.DPQLVYEK.A | 496.06 | BE024920 | PRP S1 | 6T |
| R.ASSTVSNDIQGFK.V | 677.23 | BE024920 | PRP S1 | 6T |
| R.KRVEQAAQSAAEPQPDTLGAAPEGQPFV.Y f | 964.72 | BE024920 | PRP S1 | 6T |
| R.LNQVSTSLTQNSFK.G | 783.87 | CAA76245 | PRP S2 | 1T, 4T, 5T, 6T, 15T, 16T |
| K.RQELITQSQTLK.S | 722.83 | CAA76245 | PRP S2 | 1TL, 5T, 8T |
| K.LGPNSTGIADLGNNYNNVNK.L | 1038.12 | CAA76245 | PRP S2 | 1TL, 5T, 8T |
| K.AASGLVARAALFSK.K | 681.31 | CAA76245 | PRP S2 | 5L |
| K.NGYAFGVATVSTQR.Lh | 735.81 | CAA76245 | PRP S2 | 5T, 15T |
| K.FASIENGTLTYNVAIAK.I | 906.53 | CAA76245 | PRP S2 | 5TL |
| K.DILSLGSLRVQK.L | 664.79 | Q08365 | PRP S3 | 1L |
| K.AIDNIKVSKDLVTNLQK.T | 633.41 | CAA46980 | PRP S3 | 1L |
| K.NPLVNNDFENAEGLTK.L | 887.96 | Q08365 | PRP S3 | 1L, 2TL, 3TL, 4T, 6L, 7TL, 11T |
| R.TTLVNLFSNLEK.E | 689.80 | CAA46980 | PRP S3 | 1T, 2T, 4T, 11T |
| K.LQTAFLTQIESQRK.M | 831.96 | Q08365 | PRP S3 | 2L, 3L |
| K.ASTVADSIVDALEK.R | 709.79 | Q08365 | PRP S3 | 2L, 3T, 4T, 6L, 7L |
| K.KFANLFLTK.L | 541.17 | S40457 | PRP S3 | 3L, 7L |
| K.LVDDNQAMANESR.K | 731.78 | Q08365 | PRP S3 | 3T |
| R.TENIFYYLATIATAR.K | 874.00 | Q08365 | PRP S3 | 3T |
| K.LQTAFLTQIESQR.K | 767.87 | Q08365 | PRP S3 | 3T, 4T, 7T, 8T |
| K.FANLFLTK.L | 477.08 | S40457 | PRP S3 | 3TL, 4T |
| K.QHANFLFGVNVENAK.E | 562.96 | Q08365 | PRP S3 | 3TL, 7L |
| R.TANTIYGIIGVK.V | 625.24 | Q08365 | PRP S3 | 7T |
| K.TRPYDSSESDYLIRLK.V | 648.05 | AAA81363 | PRP S4 | 13L, 14L |
| K.IKPLGLTSVTAAVELITK.G | 618.42 | AAA81363 | PRP S4 | 14L |
| R.FNYGITER.Q | 500.05 | AAA81363 | PRP S4 | 14T |
| K.TRPYDSSESDYLIR.L | 851.41 | AAA81363 | PRP S4 | 14T |
| R.LDNIVFR.L | 438.52 | AAA81363 | PRP S4 | 14T |
| K.IKPLGLTSVTAAVELITK.G | 927.64 | AAA81363 | PRP S4 | 14T |
| K.KGNEYTAGDVEAALR.F | 531.57 | AV393553 | PRP S5 | 1T, 2T |
| R.SVLELAGVQNVLAK.R | 720.86 | AV391351 | PRP S5 | 1T, 2T, 3T, 4T |
| K.SVVRVPLVGAGTIPHRVEAK.F | 695.50 | AV391351 | PRP S5 | 2L |
| R.VPLVGAGTIPHR.V | 608.73 | AV391351 | PRP S5 | 2T |
| D.VLEEEDGEYVDLEK.E | 833.88 | AV392967 | PRP S5 | 2T |
| K.LEDVDDVEEFR.T | 683.21 | AV392967 | PRP S5 | 2T |
| K.GNEYTAGDVEAALR.F | 733.28 | AV393553 | PRP S5 | 2T |
| R.C*AVAALEQLR.T h | 565.64 | AV391351 | PRP S5 | 2T |
| A.AVAADAAAETVK.Lg | 558.62 | AV388297 | PRP S6 | 16Lj, 23TL |
| A.AAVAADAAAETVK.Lg | 594.16 | AV388297 | PRP S6 | 22T, 23T |
| K.SVAYRIVYNALK.E | 698.83 | CAA37927 | PRP S7 | 12Lj, 18L, 19L |
| K.LKSEILDAYKK.T | 654.28 | CAA37927 | PRP S7 | 19L |
| K.NPVEVFEK.A | 481.05 | CAA37927 | PRP S7 | 19L |
| K.SVAYRIVYNALKEIGDVTQK.N | 756.21 | CAA37927 | PRP S7 | 19L |
| K.LKSEILDAYK.K | 590.20 | CAA37927 | PRP S7 | 19T |
| K.GYLTQDSRVK.E | 583.66 | CAA74006 | PRP S9 | 16L |
| K.DKGYLTQDSRVK.E | 705.29 | CAA74006 | PRP S9 | 17L |
| K.EAVAQVQIK.E | 493.08 | CAA74006 | PRP S9 | 17L |
| K.APFDVLSTSK.K | 532.61 | CAA74006 | PRP S9 | 17L |
| K.QLLEDCVAQIQAVAEATGAVFK.G | 1188.36 | AV623307 | PRP S10 | MudPIT |
| V.PMAESTPLAAAGEK.V g | 686.79 | AV623307 | PRP S10 | MudPIT |
| M.PTIQQLIR.S g | 484.59 | AAA84155 | PRP S12 | 21T |
| K.TYELNEEEINK.L | 691.24 | AV388793 | PRP S13 | 21L, 22TL |
| R.VNNVEIPNSK.R | 557.12 | AV388793 | PRP S13 | 22TL |
| K.KTYELNEEEINK.L | 755.32 | AV388793 | PRP S13 | 22TL |
| K.KTYELNEEEINKLR.S | 890.00 | AV388793 | PRP S13 | 22TL |
| K.TYELNEEEINKLR.S | 825.91 | AV388793 | PRP S13 | 22TL |
| R.EMAHQGLLPGVC*K.S | 720.27 | CAC08471 | PRP S14 | MudPIT |
| R.KLQQLPR.N | 441.66 | CAC08471 | PRP S14 | MudPIT |
| K.RHDADVGSSEVQVAR.L | 542.24 | AV632481 | PRP S15 | 23T |
| R.HDADVGSSEVQVAR.L | 735.27 | AV632481 | PRP S15 | 23T |
| R.RGLEAILSQR.K | 571.67 | AV632481 | PRP S15 | 23T, 24T |
| R.GLEAILSQR.K | 493.57 | AV632481 | PRP S15 | 24T |
| K.GALPSETVENLLR. | 699.80 | AV626226 | PRP S16 | 23T |
| R.DGEPLEYLGWYDPLKK.E | 962.08 | AV626226 | PRP S16 | 23T |
| K.TVVVEAERLATDVTYQK.R | 961.59 | BI531856 | PRP S17 | 15Lj, 16Lj, 17L, 18L, 20L, 21L, 22L, 23L |
| K.RFSVTEVLRKAD.f | 473.88 | BI531856 | PRP S17 | 16Lj |
| V.VVEAERLATDVTYQK.Rg | 861.47 | BI531856 | PRP S17 | 22L |
| K.RFSVTEVLR.K | 553.65 | BI531856 | PRP S17 | 23T |
| R.FSVTEVLR.K | 475.56 | BI531856 | PRP S17 | 23T |
| K.HSGLLQRYIGLGGK.I | 749.88 | CAA76244 | PRP S18 | 19L |
| R.KVLSLSQILSR.L | 622.26 | CAA76244 | PRP S18 | 19T |
| K.VLSLSQILSR.L | 558.18 | CAA76244 | PRP S18 | 19T, 20T |
| K.KGPFVADHLLK.K | 612.74 | pgi | PRP S19 | 22L, 23T |
| K.GPFVADHLLK.K | 548.65 | pgi | PRP S19 | 23TL |
| K.LAETLVAAPSKTEEEVK.N | 908.03 | AV389047 | PRP S20 | 14Lj, 20L, 21TL |
| K.LISEAYTEIDK.A | 641.22 | AV390348 | PRP S20 | 20L, 21L |
| K.TVLM*AAGLYKPAADSPDFARYQK.L | 844.30 | AV390348 | PRP S20 | 20L, 21L |
| K.PAADSPDFARYQK.L | 733.30 | AV390348 | PRP S20 | 21L, 22L |
| K.AGIVDPTYDELYSAELDPK.P | 686.79 | AV388903 | PRP S21 | MudPIT |
| K.PFEDFFQLR.D | 599.68 | AV388903 | PRP S21 | MudPIT |
| K.DSYMVEVEVAEDEPEDVAVR.R | 1141.23 | AV623969 | PRP S21 | MudPIT |
| Y.MVEVEVAEDEPEDVAVR.Rg | 958.56 | AV623969 | PRP S21 | MudPIT |
| K.NLAVAIDQVYSR.G | 674.77 | BG847093 | PSRP-3 | 4Tj, 9T, 10T |
| R.GQVSPLTEYYFWPR.Q | 871.98 | BG847093 | PSRP-3 | 4Tj, 9T, 10T |
| K.LAAGELSEAELSK.V | 659.24 | BE025032 | PSRP-3 | 4TLj, 8L, 9TL, 10T |
| R.LTQLINFWQDEEVK.H | 882.00 | BG847093 | PSRP-3 | 9T, 11T |
| K.TLTGLLAKDEMK.V | 660.30 | AV630899 | PSRP-7 | 4L |
| R.EGADATDDDEDVEVELEDGQVEVR.A | 1317.81 | AV396890 | PSRP-7 | 4T |
| K.VPSSALSAEAQAALR.A | 735.83 | AV630899 | PSRP-7 | 4T, 8T |
C*, S-carbamidomethylated Cys. M*, met sulfoxide.
Identified peptides are shown between dots, indicating neighboring residues based on sequence databases.
Accession numbers for identified EST or ORF with highest similarity score.
Identification and designation was performed based on the results of BLAST search, pairwise comparison, and sequence alignment with other proteins (see Table 2).
TP30 gel section numbers shown in Figure 1 followed by enzymes used for digestion: typsin (T), Lys-C (L) or both (TL). Major peptide products from a PRP are indicated with boldface type.
Peptides were identified using MudPIT (multidimensional protein identification technology) directly from the TP30 digest.
Probable C-terminal peptide.
Probable N-terminal peptide.
Collision-induced dissociation spectra are shown in Figure 2.
An ORF encoded in the plastid genome (J. Maul, J.W. Lilly and D.B. Stern, unpublished data).
Probably generated from a protein dimer (see Results).
Figure 2.
Collision-Induced Dissociation Spectra of Tryptic Peptides Derived from the 84- and 57-kD Proteins of the Small Subunit.
The SEQUEST algorithm assigned b-ion and y-ion series to the observed mass spectra based on sequence databases.
(A) Fragmentation of the doubly charged precursor ion at an m/z of 735.8 yielded detectable singly charged b-ion species (b-2 to b-9) and y-ion species (y-2 to y-8) for which the sequence is indicated. The peptide sequence obtained from the 84-kD protein belonged to a nucleus-encoded protein (PRP S5).
(B) Fragmentation of the doubly charged precursor ion at an m/z of 565.6 yielded detectable singly charged b-ion species (b-3 to b-11) and y-ion species (y-2 to y-11) for which the sequence is indicated. The peptide sequence obtained from the 57-kD protein belonged to a plastid-encoded protein (PRP S2).
In the case of EST hits, we searched EST contigs (assembled cDNAs in silico) using a WU-BLAST search. The obtained EST contig or ORF was examined by similarity search using BLAST, and the identified proteins and genes were designated according to the PRP nomenclature (Yamaguchi et al., 2000). We determined that the 84- and 57-kD proteins were homologs of the bacterial S5 and S2 proteins. The same procedure was performed for every peptide listed in Table 1. Seventeen proteins were identified by this process, including orthologs of E. coli S1, S2, S3, S4, S5, S6, S7, S9, S12, S13, S15, S16, S17, S18, S19, and S20, a homolog of spinach PSRP-3, and a novel S1 domain–containing ribosomal protein. Although no nonribosomal proteins were identified from our 30S preparation, several cytoplasmic ribosomal proteins were identified (data not shown). We used ∼10 pmol of 30S subunit for the mass spectrometric analysis. Because LC-MS/MS analysis is able to detect femtomole levels of peptides, contaminants of ribosomal proteins from Chlamydomonas cytoplasmic subunits might be expected. All of the identified cytoplasmic ribosomal protein peptides were identical to those obtained from cytosolic ribosomal proteins prepared from pure 80S (our unpublished results). Therefore, it is unlikely that these proteins are components of the 30S subunit; rather, they likely arise from contaminating 40S subunits.
Direct Analysis of the Protein Components of the Chlamydomonas Chloroplast Ribosomal 30S Subunit
Multidimensional protein identification technology also was applied to identify 30S ribosomal proteins as a complementing analysis. Multidimensional protein identification, which involves two-dimensional LC-MS/MS, identifies protein components directly from an enzymatic digest of total protein from a large protein complex without resolving proteins on one-dimensional or two-dimensional PAGE gels (Link et al., 1999; Washburn et al., 2001; Wolters et al., 2001). TP30 was digested with endoproteinase Lys-C and trypsin, and the digest was subjected to two-dimensional LC-MS/MS as described in Methods. Three additional orthologs of E. coli S10, S14, and S21, which were not identified from in-gel digests, were identified (Table 1). Other small subunit proteins identified by this method were orthologs of E. coli S1, S2, S3, S4, S5, S6, S7, S9, S12, S15, S16, S18, and S20 plus the homolog of spinach PSRP-3 and the novel S1 domain–containing ribosomal protein (data not shown).
Characteristics of Chlamydomonas 30S Chloroplast Ribosomal Proteins
The complete set of spinach 30S PRPs was identified using a proteomic analysis that included HPLC purification, two-dimensional PAGE, Edman chemistry, and LC-MS and matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (Yamaguchi et al., 2000). Based on the spinach PRP data (N-terminal/internal peptide and some cDNA sequences), a computational search allowed for the identification of a set of 30S PRP homologs from the complete sequences of the Arabidopsis nuclear and chloroplast genomes. In the same way, Synechocystis and E. coli homologs were searched from the complete genome sequences. The Arabidopsis 30S PRP sequences were used to search homologs of Chlamydomonas from the EST/ORF databases. Among the 23 spinach/Arabidopsis PRP homologs (Table 2), our proteomic analyses identified 20 PRPs: S1, S2, S3, S4, S5, S6, S7, S9, S10, S12, S13, S14, S15, S16, S17, S18, S19, S20, S21, and PSRP-3. In addition to the spinach and Arabidopsis PRP homologs, we identified a novel S1 domain–containing protein (see below). Therefore, 21 PRPs were identified positively from the small subunit of Chlamydomonas chloroplast ribosome in this study. The Chlamydomonas 30S PRPs that were identified by LC-MS/MS analyses and by computational analysis of the Chlamydomonas EST/ORF databases using the Arabidopsis 30S PRP homologs were identical and complement each other. The homology search using spinach and Arabidopsis 30S PRP sequences identified S8, PSRP-1, and PSRP-4 genes that were missing from the LC-MS/MS analyses of the Chlamydomonas chloroplast 30S subunit (Table 2). These three proteins might have escaped detection by mass spectrometry for a variety of reasons. (1) Few detectable peptides (m/z 400 to 1400) were generated by trypsin or endoproteinase Lys-C digestion, or the peptides were post-translationally modified, resulting in the lack of identification by the SEQUEST algorithm. (2) Although the genes are present in the genomes, the proteins are not expressed because the genes are silent or pseudogenes. Homologs of S11 protein and PSRP-2 were not identifiable from the Chlamydomonas EST/ORF databases or by proteomic analysis. This may be attributable to either incomplete EST/ORF databases or an absence of these genes and/or proteins in Chlamydomonas.
Table 2.
Percentage Similarities of Chlamydomonas 30S PRP Homologs Found in Arabidopsis, Synechocystis and E. coli
| Chlamydomonas
|
Arabidopsis
|
Synechocystis
|
E. Coli
|
||||
|---|---|---|---|---|---|---|---|
| 30S PRP a | Accession No. | Gene Allocation | Accession No. | Gene Allocation | % Similarity | % Similarity | % Similarity |
| S1 | 20001021.681.1b | Nucleus | AF370232 | Nucleus | 64 | 63 | 50 |
| S2 | orf570 (R)c | Plastid | P56797 | Plastid | 52 | 58 | 58 |
| S3 | orf712 (F)c | Plastid | P56798 | Plastid | 63 | 74 | 63 |
| S4 | rps4 (F)c | Plastid | P56799 | Plastid | 62 | 68 | 55 |
| S5 | AY093615 | Nucleus | U78721 | Nucleus | 73 | 68 | 63 |
| S6 | 20011023.5203.12b | Nucleus | AC009519 | Nucleus | 50 | 56 | 48 |
| S7 | rps7 (R)c | Plastid | P56800 | Plastid | 66 | 67 | 61 |
| (S8) | rps8 (F)c | Plastid | P56801 | Plastid | 69 | 69 | 69 |
| S9 | rps9 (R)c | Plastid | BAA82396 | Nucleus | 46 | 46 | 44 |
| S10 | 20011023.3083.1b | Nucleus | BAB01403 | Nucleus | 61 | 72 | 69 |
| (S11) | NDe | – | P56802 | Plastid | NAd | NAd | NAd |
| S12 | rps12 (R)c | Plastid | BAA84409 | Plastid | 89 | 90 | 83 |
| S13 | 20011023.6351.1b | Nucleus | P42732 | Nucleus | 66 | 71 | 69 |
| S14 | rps14 (R)c | Plastid | P56804 | Plastid | 67 | 81 | 60 |
| S15 | 20011023.5052.1b | Nucleus | P56805 | Plastid | 64 | 61 | 61 |
| S16 | 20011023.5085.1b | Nucleus | P56806 | Plastid | 66 | 69 | 58 |
| S17 | 20011023.676.1b | Nucleus | P16180 | Nucleus | 59 | 62 | 56 |
| S18 | rps18 (R)c | Plastid | P56807 | Plastid | 68 | 75 | 59 |
| S19 | rps19 (F)c | Plastid | P56808 | Plastid | 83 | 87 | 80 |
| S20 | 20011023.538.1b | Nucleus | AF361807 | Nucleus | 62 | 62 | 50 |
| S21 | 20011023.1239.1b | Nucleus | BAB01933 | Nucleus | 49 | 60 | 59 |
| (PSRP-1) | 20011023.2607.2b | Nucleus | BAB11201 | Nucleus | 58 | 55 | NDe |
| (PSRP-2) | NDe | – | AY039568 | Nucleus | NAd | NDe | NDe |
| PSRP-3 | 20011023.3226.1b | Nucleus | CAC01775 | Nucleus | 67 | 74 | NDe |
| (PSRP-4) | 20011023.5064.1b | Nucleus | AF236826 | Nucleus | 54 | NDe | NDe |
| PSRP-7 | 20001021.681.1b | Nucleus | NDe | – | NDe | NDe | NDe |
Unidentified PRPs in this proteomic study were searched using spinach and Arabidopsis PRPs as probes and are indicated in parentheses.
Accession numbers of EST contigs were obtained by WU-BLAST search (http://www.biology.duke.edu/chlamy_genome/blast/blast_form.html).
Gene in the chloroplast genome (http://www.biology.duke.edu/chlamy_genome/chloro_table.html): F, forward strand; R, reverse strand.
Proteins are present but similarity score is not available.
Homologs are not detected in database searches.
An overview of the characteristics of the Chlamydomonas 30S subunit is shown in Table 3 comparing Chlamydomonas PRPs with other ribosomal proteins. The accession numbers and gene allocations for Chlamydomonas and Arabidopsis 30S PRPs are summarized in Table 2, as are percentage similarities of 30S PRPs found in Chlamydomonas, Arabidopsis, Synechocystis, and E. coli. The mature protein sizes and pI values were calculated after removal of the predicted transit peptide (for nucleus-encoded proteins) or N-formyl Met (for plastid-encoded proteins) and are summarized in Table 3. Gene allocations for Chlamydomonas and Arabidopsis 30S PRPs also are summarized in Table 3.
Table 3.
Characteristics of Chlamydomonas 30S Plastid Ribosomal Proteins
| Predicted Number of Amino Acid Residues and pI Values
|
Molecular Mass (kD)
|
|||||
|---|---|---|---|---|---|---|
| 30S PRP | Precursor or Proproteina | Transit Peptideb or Met Removalc | Mature Protein | pId | Sequence | SDS-PAGEe |
| S1 | >289 | NA | >289 | 4.6 | >32.0 | 49 |
| S2 | 570 | Met (+) | 570 | 11.0 | 63.2 | 57 |
| S3 | 712 | Met (−) | 711 | 10.7 | 81.7 | 76 |
| S4 | 257 | Met (−) | 256 | 11.3 | 29.9 | 24 to 26 |
| S5 | 673 | 51 | 622 | 4.0 | 66.7 | 84 |
| S6 | 171 | 54 | 117 | 7.0 | 13.3 | 10 to 11 |
| S7 | 168 | Met (−) | 167 | 10.4 | 19.0 | 14 to 16 |
| S8 f | 153 | Met (−) | 152 | 10.1 | 17.1 | ? |
| S9 | 191 | Met (−) | 190 | 9.8 | 20.9 | 18 to 19 |
| S10 | 169 | 37 | 132 | 5.8 | 14.7 | 14 to 16 |
| S12 | 133 | Met (−) | 132 | 11.5 | 14.5 | 11 to 13 |
| S13 | 164 | 29 | 135 | 10.5 | 15.4 | 10 to 11 |
| S14 | 100 | Met (−) | 99 | 11.3 | 11.6 | ? |
| S15 | 141 | 33 | 108 | 9.9 | 12.4 | <10 |
| S16 | 128 | 28 | 100 | 9.9 | 11.5 | <10 |
| S17 | >105 | >25 | 80 | 10.1 | 9.0 | <10 |
| S18 | 137 | Met (+) | 137 | 11.9 | 16.3 | 14 to 16 |
| S19 | 92 | Met (−) | 91 | 10.8 | 10.3 | <10 |
| S20 | 166 | 38 | 128 | 9.9 | 14.2 | 11 to 13 |
| S21 | 184 | 28 | 156 | 4.7 | 17.7 | ? |
| PSRP-1f | 286 | 59 | 227 | 6.1 | 25.4 | ? |
| PSRP-3 | 298 | 36 | 262 | 4.1 | 29.3 | 34 |
| PSRP-4f | >135 | >53 | 127 | 10.5 | 13.5 | ? |
| PSRP-7 | >378 | 28 | >350 | ? | >38.0 | 66 |
Cytosolic precursor and plastid proprotein sequences were deduced from nucleic acid sequences (see Table 2).
Predicted by ChloroP (Emanuelsson et al., 1999).
Predicted by penultimate amino acid residues (Giglione and Meinnel, 2001).
From predicted mature protein sequence.
Estimated from major peptides found in the TP30 gel section by LC-MS/MS (see Table 1).
Unidentified proteins by LC-MS/MS analyses.
Identification of the S1 Protein and a Novel S1 Domain–Containing Protein
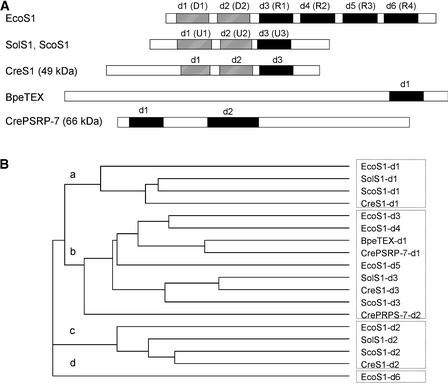
Four peptides from TP30 gel section 6, containing a 49-kD protein, belonged to the translated sequence of an EST (BE024920) that showed significant sequence homology with plastid 30S ribosomal protein S1. This 49-kD protein appears to be a nucleus-encoded homolog of S1 (Table 1). An additional S1 domain–containing protein also was identified. Peptides obtained from section 4, containing a 66-kD protein, belonged to two ESTs (AV396890 and AV630899). The two ESTs were identified as being from the same contig (20,001,021.681.1) by WU-BLAST search. The EST contig encodes an S1 domain–containing protein having significant sequence homology, in the S1 domain, with the TEX protein, a transcription accessory protein from Bordetella pertussis (Fuchs et al., 1996). Although available EST contigs did not contain the full-length 49- or 66-kD proteins, the partially assembled cDNA sequences allowed us to confirm the 49-kD protein as a bacterial S1 counterpart and the 66-kD protein as a novel S1 domain–containing protein. The S1 domains in the 49- and 66-kD proteins could be aligned with those of other S1 proteins and the TEX protein (Figure 3A). The E. coli S1 protein has six S1 domains, each composed of ∼70 amino acids (Subramanian, 1983; Gribskov, 1992). The first two domains (designated D1 and D2) are involved in ribosome binding, and the next three domains (R1 to R3) are involved in mRNA binding (Subramanian, 1983). The last domain (R4) is involved in the autoregulation of its own mRNA (Boni et al., 2000). In spinach chloroplasts and cyanobacteria, S1 proteins have truncated structures with only three S1 domains, designated U1 to U3 (Franzetti et al., 1992; Sugita et al., 1995). Spinach plastid ribosomal protein S1 has RNA binding activity at the C-terminal half but not at the N-terminal half (Shteiman-Kotler and Schuster, 2000). This finding indicates that the plastid and cyanobacterial S1 proteins have functions similar to the N-terminal half of E. coli S1.
Figure 3.
Two S1 Domain–Containing Proteins Are Present in the Chlamydomonas Chloroplast 30S Ribosomal Subunit: An Ortholog of E. coli S1 and a Novel S1 Domain–Containing Protein (PSRP-7).
(A) Schemes of the E. coli S1 protein (EcoS1), spinach PRP S1 (SolS1), Synechococcus S1 (ScoS1), Chlamydomonas PRP S1 (CreS1), Bordetella pertussis TEX protein (BpeTEX), and Chlamydomonas PSRP-7 (CrePSRP-7). The ribosome binding domains of the S1 proteins are indicated by striped boxes. The RNA binding domains of the S1 proteins are indicated by closed boxes.
(B) Phylogenetic tree of the relationships among S1 domains from EcoS1, SolS1, ScoS1, CreS1, BpeTEX, and CrePSRP-7. The tree was created using CLUSTAL W (Thompson et al., 1994). The S1 domains indicated by closed boxes in (A) were grouped in branch b, except d6 of E. coli. These domains contain conserved aromatic residues believed to be involved in RNA binding, based on the NMR structure of the S1 domain of E. coli polynucleotide phosphorylase (Bycroft et al., 1997). The first two domains (d1 and d2) of the S1 proteins, indicated by striped boxes in (A), were grouped in branches a and c. Domains grouped in branches a, c, and d (d6 of E. coli S1) lack the aromatic residues required for mRNA binding. The first two domains in E. coli S1 do not interact with mRNA, nor does the last domain (d6) of E. coli S1, which is required for the autoregulation of its own mRNA but not for overall mRNA recognition (Boni et al., 2000).
We designated the S1 domains in Chlamydomonas proteins d1 to d3 from the N to the C terminus in order of their domain arrangement. The 49-kD protein has three S1 domains (d1 to d3) showing significant homology with d1 to d3 of the spinach and cyanobacterial S1 proteins. The 66-kD protein has at least two S1 domains. The first domain (d1) has strong sequence homology (55.7% identity) with the S1 domain of TEX. The second domain (d2) follows d1 after an 80-residue spacer (Figure 3A), has weak sequence similarity (∼20% identity) to other S1 domains, and contains a 30–amino acid insertion. Although d1 of the 66-kD protein has strong sequence similarity to the S1 domain of TEX, the remainder of the 66-kD protein does not share homology with the TEX protein (Figure 3A). The S1 domain arrangement in the 66-kD protein has not been reported for any other characterized protein. We tentatively designated the 66-kD protein PSRP-7. The designation followed the discoveries of spinach PSRP-1 to PSRP-6 (Yamaguchi et al., 2000; Yamaguchi and Subramanian, 2000). The protein bands of 49 and 66 kD shown in Figure 1 appear to accumulate to similar amounts as other 30S subunit proteins, suggesting that Chlamydomonas plastid ribosomal protein S1 and PSRP-7 are present in the 30S subunit in stoichiometric amounts with other ribosomal proteins.
A phylogenetic tree was created to align the two S1 domains from the 49- and 66-kD proteins with other S1 protein domains (Figure 3B). This phylogenetic analysis suggests that the two S1 domains of PSRP-7 evolved from an ancestral S1 domain that had RNA binding ability and that they may be required for mRNA interaction on the 30S subunit.
S2 Homolog
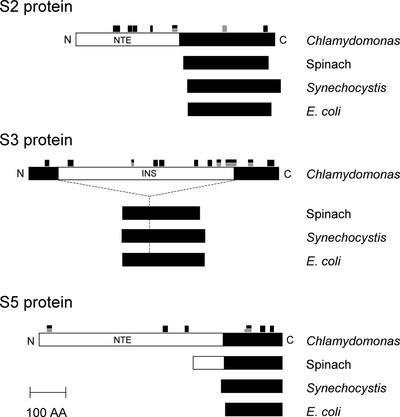
Two open reading frames (ORF208 and ORF570), which have sequence similarity to the bacterial S2 protein, were identified in the Chlamydomonas plastid genome (http://www.biology.duke.edu/chlamy_genome/chloro.txt). The N-terminal half of ORF208 has no sequence similarity to any known protein, whereas the C-terminal half has significant homology with plant PRP S2 and the S2 proteins of cyanobacteria. orf570 was reported to be a rps2-like gene encoding an NTE with no homology with known proteins and a C-terminal half with S2 homology (Leu, 1998). Compared with ORF570, ORF208 is closer in size and shares greater homology with other S2 proteins. Our proteomic analysis, however, identified only the ORF570 product. A trans-splicing event would not be expected to connect the N-terminal region of ORF570 and the C-terminal region of ORF208, as is the case for the S12 gene prps12 in higher plants (Koller et al., 1987), because we identified peptides from all regions of ORF570 (Figure 4), showing that the gene was not spliced. Thus, in Chlamydomonas, the orf570 gene appears to be the functional prps2 gene, whereas the orf208 gene may be a pseudogene. Major peptides belonging to PRP S2 (ORF570 product) were identified from gel section 5, which contains a 57-kD protein (Figure 1, Table 1). The apparent mass (57 kD) is in agreement with the predicted mass for ORF570 (63.2 kD). Figure 2B shows an example of the collision-induced dissociation spectrum as well as the peptide sequence from the NTE of PRP S2. Some of the peptides identified belong to portions of the NTE, demonstrating that the NTE constitutes part of the mature protein (Figure 4).
Figure 4.
Chlamydomonas 30S PRPs S2, S3, and S5 Are Two to Four Times Larger Than Their Counterparts in Spinach, Synechocystis, and E. coli as a Result of the Existence of Additional Domains in the Chlamydomonas Proteins.
Mature polypeptides (N to C terminus) are represented schematically and aligned. Closed boxes represent regions homologous with E. coli proteins. Open boxes represent nonhomologous sequences. The locations of tryptic fragments (black boxes) and Lys-C fragments (gray boxes), which were identified by MS/MS, are indicated. These NTE (PRP S2 and PRP S5) and INS (PRP S3) sequences have no similarity to those of characterized proteins from other organisms. AA, amino acids.
S3 Homolog
The Chlamydomonas chloroplast orf712 gene has been reported to encode an unusual S3-like protein (Fong and Surzycki, 1992; Liu et al., 1993; Turmel and Otis, 1994). The predicted amino acid sequence of ORF712 shows significant similarity to that of bacterial and chloroplast ribosomal protein S3, but the similarity is limited to the N and C termini of the ORF. An INS of 472 amino acids, lacking similarity to other known proteins, is found in ORF712. The orf712 gene is reported to be essential for cell growth and is conserved in the Chlamydomonas lineage (Liu et al., 1993). Thirteen peptides obtained from several gel sections, mainly from section 3 containing a 76-kD protein, matched the orf712 protein product, identifying this gene as prps3. Some of the peptides identified by MS/MS spectra belong to portions of the INS, demonstrating that the INS constitutes part of the mature protein (Figure 4).
S5 Homolog
Eight peptides obtained from gel section 2, which contains an 84-kD protein band, identified an EST (AV391531) encoding an S5 homologous sequence and two ESTs (AV392967 and AV393533) encoding unknown proteins (Figure 1, Table 1). In eubacteria, S5 proteins have molecular masses of ∼17 kD, whereas spinach PRP S5 (28 kD) harbors an NTE (Yamaguchi and Subramanian, 2000). Because a portion of the S5 homolog was identified in the 84-kD band, we speculated that the unknown EST clones could encode portions of a large NTE. Because EST contigs covering a full-length S5 protein could not be found in a WU-BLAST search, the longest EST clone (AV396809, clone CL69a03) was obtained from the Kazusa DNA Research Institute (Chiba, Japan) and sequenced. The nucleotide sequence identified a protein of 673 amino acids. Two of the identified ESTs (AV392967 and AV393533) belong to the N-terminal region of this protein. This protein contains a chloroplast transit peptide, as predicted by the ChloroP program (Emanuelsson et al., 1999). The transit peptide (51 amino acids) shows the common characteristic amino acid composition of chloroplast transit peptides: abundance of Ser and Thr, primarily a net positive charge, absence of Trp and Tyr, and a general amphiphilic nature (reviewed by Bruce, 2000). Thus, the mature protein was considered to consist of the remaining 622 amino acids. Alignment of the Chlamydomonas PRP S5 with corresponding proteins from spinach, Synechocystis, and E. coli clearly shows that the mature Chlamydomonas PRP S5 protein contains a 452-residue NTE followed by a 170-residue C-terminal domain homologous with S5 proteins (Figure 4). The NTE of PRP S5 (amino acids 52 to 503 in the precursor sequence) showed no significant matches to proteins characterized in BLAST searches. The C-terminal S5 homologous sequence shows ∼70% similarity and 50% identity to S5 proteins from Arabidopsis, spinach, and Synechocystis (Table 2).
An unusual charge distribution was identified in the Chlamydomonas PRP S5, with a strong negatively charged NTE (pI of 3.7) and a positively charged C-terminal S5 homologous region (pI of 9.9), generating an acidic net charge (pI of 4.0). With a strongly acidic NTE, the Chlamydomonas PRP S5 is one of the most acidic plastid ribosomal proteins identified to date. The NTE in spinach PRP S5 (87 residues, pI of 4.1) also is strongly acidic, but the sequence shows no similarity to the NTE of Chlamydomonas PRP S5. The apparent size of PRP S5 on SDS-PAGE (84 kD) is 17 kD larger than the predicted size (67 kD), suggesting that the unusual charge distribution of this protein might affect migration in SDS-PAGE. However, post-translational modifications cannot be excluded.
S6 Homolog
Two peptides from gel section 23 belonged to the nucleus-encoded PRP S6. The observed size (gel section 23 corresponds to 10 to 11 kD) is in agreement with the mass as predicted from the gene (13.3 kD). Another peptide was identified from gel section 16, which corresponds to ∼22 kD (Table 1). It is twice as large as the estimated size of PRP S6. Thus, part of the protein may exist as a dimer, even on SDS-PAGE gels. Similar observations were seen in PRPs S7, S17, S20, and PSRP-3 (Table 1). It has been reported that some spinach 30S PRPs separated by two-dimensional PAGE exist in both monomer and dimer forms (Yamaguchi et al., 2000). Two peptides (AAAVAADAAAETVK and AAVAADAAAETVK) from gel section 23 are likely the N-terminal peptides of PRP S6, because trypsin or endoproteinase Lys-C usually do not cleave peptide bonds of Val-Ala or Ala-Ala, and the peptide locations are immediately after the predicted transit peptide. This observation indicates that PRP S6 exists in at least two forms, different by one amino acid, as a result of cleavages at different positions of the transit peptides or N-terminal trimming after the removal of the transit peptides. In spinach, PRP S6 exists in five forms (α to ɛ), each with a different N-terminal length (Yamaguchi and Subramanian, 2000).
S12 Homolog
A single tryptic peptide obtained from gel section 21 identified PRP S12 (Figure 1, Table 1). This peptide sequence corresponds to the deduced N-terminal sequence (MPTIQQLIR) of the prps12 gene, except for the N-terminal Met. This finding indicates that the N-terminal Met of the protein is removed post-translationally. N-Formyl Met of many organelle proproteins is removed by Met aminopeptidase after removal of the N-formyl group by peptide deformylase (reviewed by Giglione and Meinnel, 2001). A rule for Met excision by Met aminopeptidase has been postulated (Giglione and Meinnel, 2001) in which the N-terminal Met is removed if the penultimate amino acid is a small residue (G, A, P, S, T, or C), whereas the Met is not removed if it is a bulky residue (N, D, L, I, H, Q, E, F, M, K, Y, W, or R). The rule appears to be applicable for plastid-encoded PRPs in spinach (Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000) and those in Chlamydomonas as well. This rule also is supported by the presence of an E. coli Met aminopeptidase homolog in the Chlamydomonas EST database (EST contig number 20,011,023.2219.1). Using the postulated rule, we predicted N-terminal Met removal for other plastid-encoded PRPs (Table 3).
S15 and S16 Homologs
Plastid ribosomal proteins S15 and S16 are interesting in Chlamydomonas in terms of gene allocation. In land plants, the prps15 gene is present in the plastid genome, whereas these genes are absent from the plastid genomes of Euglena gracilis and Porphyra purpurea and from the cyanelle genome. In various angiosperms, PRP S16 is encoded by a plastid gene, but the prps16 gene is absent from the Marchantia plastid genome (reviewed by Harris et al., 1994). Both the prps15 and prps16 genes are missing from the Chlamydomonas plastid genome (http://www.biology.duke.edu/chlamy_genome/chloro.txt). It is unclear whether these proteins are missing from the ribosome or present as nucleus-encoded proteins. PRP S15 and PRP S16 were identified from Chlamydomonas ESTs (Table 3), showing that functional PRP S15 and PRP S16 are encoded in the nucleus in Chlamydomonas.
S18 Homolog
Two peptides from gel section 19 identified plastid-encoded PRP S18. One of the two peptides belonged to an NTE of 59 amino acids, demonstrating that the NTE is present in the functional PRP S18 protein, as is the NTE of PRP S2. In land plants, PRP S18 contains a heptapeptide repeat in the NTE (sevenfold SKQPFRK in monocots, twofold similar heptapeptide in dicots; Weglöhner et al., 1995). No heptapeptide repeat was identified in the NTE of the Chlamydomonas PRP S18. The NTE of the Chlamydomonas PRP S18 shows weak sequence similarity to that of the Chlorella PRP S18 (data not shown). Functions for the NTEs of land plant or algal PRP S18 have not been identified.
PSRP-3 Homolog
A nucleus-encoded protein was identified as a homolog of the spinach PSRP-3. PSRP-3 is one of the plastid-specific ribosomal proteins whose counterparts are not identifiable in the E. coli ribosome (Yamaguchi et al., 2000; Yamaguchi and Subramanian, 2000). The Chlamydomonas PSRP-3 is approximately twice as large as its spinach counterpart as a result of the presence of an N-terminal extension (Figure 5). In Chlorella and Euglena, PSRP-3 homologs are identifiable in those plastid genomes as ycf65. Other PSRP homologs were not identified in our proteomic analysis, but PSRP-1 and PSRP-4 homologs were identified in the Chlamydomonas EST database (Table 2). PSRP-1 has been identified in Chlamydomonas plastid ribosome by immunological cross-reactivity using anti-spinach PSRP-1 antiserum (Bubunenko and Subramanian, 1994). The function or localization of PSRP-3 on the 30S subunit is unclear, but PSRP-3 might be involved in a plastid-specific function (e.g., light-dependent translation regulation), as was suggested previously (Yamaguchi et al., 2000).
Figure 5.
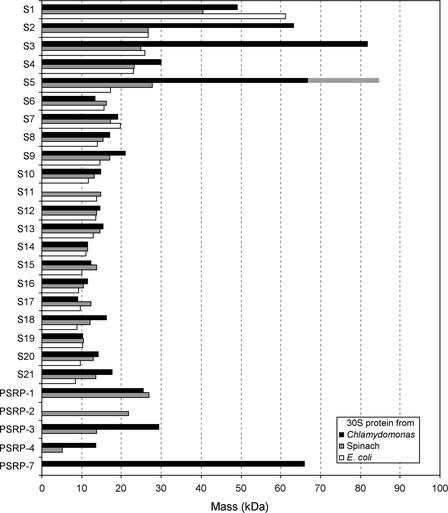
Comparison of the Molecular Masses of the 30S Proteins from Chlamydomonas Chloroplast, Spinach Chloroplast, and E. coli.
Molecular masses of spinach 30S PRPs (Yamaguchi et al., 2000) and E. coli 30S r-proteins (Arnold and Reilly, 1999) were compared with those in Chlamydomonas (see Table 3). The apparent size of PRP S5 is 17 kD larger (striped bar) than that of the predicted mature protein from the sequence (see text).
DISCUSSION
We performed a proteomic analysis of ribosomal proteins from the small subunit of the chloroplast 70S ribosome from the green alga Chlamydomonas. Although 31 small subunit protein spots have been identified by two-dimensional PAGE (Schmidt et al., 1983), this number does not likely correspond to the total number of proteins in the 30S subunit. In spinach, proteomic analyses of the 30S subunit identified 25 proteins, whereas 30 protein spots were identified on two-dimensional PAGE gels (Yamaguchi et al., 2000). Because proteins exist in multiple forms, a single protein can be found in different spots. This also would be expected for the Chlamydomonas 30S subunit proteins as well. The Chlamydomonas chloroplast 16S rRNA (1474 nucleotides; http://www.biology.duke.edu/chlam_genome/chloro.txt) and spinach chloroplast 16S rRNA (1491 nucleotides) are conserved at both the nucleotide level (74% identity) and in terms of secondary structure, closely resembling those of prokaryotes (http://www.rnaicmb.utexas.edu). The 30S ribosomal subunit of Chlamydomonas has a majority of proteins in common with those of higher plants, cyanobacteria, and bacteria. However, as shown in Figure 5, some of the Chlamydomonas 30S PRPs are prominently larger than their counterparts in spinach chloroplasts or in E. coli, as a result of the presence of NTEs, INSs, or C-terminal extensions. The prokaryotic homologous regions of Chlamydomonas 30S PRPs are conserved (60 to 90% similarity) along with their counterparts in other organisms (Table 2). The total mass of Chlamydomonas 30S PRPs is at least 590 kD (the sum of 21 predicted mature proteins that were identified by LC-MS/MS analyses), which exceeds the total mass of spinach plastid 30S PRPs (430 kD) and that of E. coli 30S ribosomal proteins (350 kD). The total mass of the Chlamydomonas 30S PRPs could reach 640 kD if we include the three proteins missing from the proteomic analysis (S8, PSRP-1, and PSRP-4) that were identified in the EST/ORF database searches. The sedimentation value of the Chlamydomonas chloroplast ribosomal small subunit has been identified as 41S by Bourque et al. (1971) and as 33S by Chua et al. (1973). These values are greater than those of the spinach chloroplast or E. coli 30S subunits.
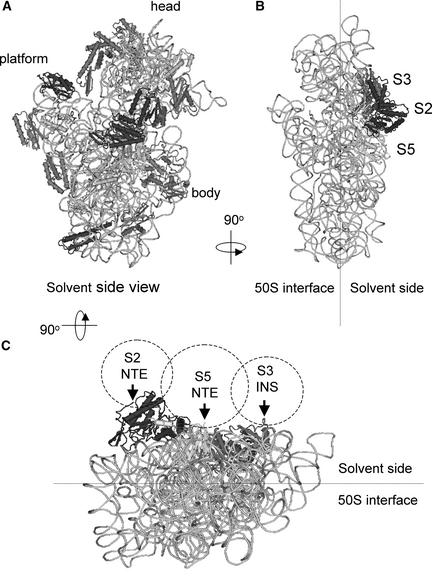
Four unusually large ribosomal proteins were identified: PRPs S2 (63 kD), S3 (82 kD), and S5 (67 to 84 kD) and PSRP-7 (66 kD). PRPs S2, S3, and S5 are clear orthologs of E. coli ribosomal proteins, whereas PSRP-7 appears to share function with prokaryotic S1 proteins. Such large ribosomal proteins have not been identified by analyses of other organellar ribosomes (chloroplast [Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000] and mitochondria [Koc et al., 2001; Suzuki et al., 2001]), eukaryotic cytosolic ribosomes (Louie et al., 1996), or bacterial ribosomes (Arnold and Reilly 1999). As shown in Figure 4, the Chlamydomonas PRPs S2, S3, and S5 contain additional domains that appear to localize on the solvent side of the small subunit (Figure 6), as predicted using the crystal structure of the Thermus thermophilus 30S subunit (Wimberly et al., 2000). The S2, S3, and S5 proteins all are located adjacent to each other and around the neck of the 30S subunit. The N termini of T. thermophilus S2 and S5 proteins, and a loop region of the S3 protein (where the INS is connected in the Chlamydomonas counterpart), are exposed to the solvent side. The additional domains in Chlamydomonas are likely anchored to the 16S rRNA by the prokaryotic homologous region. The positions of these domains suggest that orthologs of these proteins may not be ribosomal proteins in other systems but may be ribosomal associated proteins that have not yet been identified.
Figure 6.
Localization of the Chlamydomonas PRPs S2, S3, and S5 and Predicted Locations of Their Extra Domains on the 30S Subunit, Based on T. thermophilus 30S Subunit Structure.
The three-dimensional structure of the T. thermophilus 30S subunit (Wimberly et al., 2000) was represented using the Cn3D program provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml).
(A) Front view showing all of the 30S proteins and 16S rRNA.
(B) Side view showing only the S2, S3, and S5 proteins and 16S rRNA.
(C) Bottom view showing only the S2, S3, and S5 proteins and 16S rRNA. Probable localizations of the S2 NTE, S3 INS, and S5 NTE in the Chlamydomonas 30S PRPs are indicated by dotted circles. The N termini of T. thermophilus S2 and S5 proteins, and a loop region of the S3 protein (where the Chlamydomonas S2 NTE, S3 INS, and S5 NTE are connected in the Chlamydomonas counterpart), are exposed on the solvent side of the subunit (arrows).
PSRP-7 was identified as a novel S1 domain–containing protein. A direct counterpart for this protein was not reported from the spinach chloroplast 30S proteome. In spinach, PSRP-2, a homolog of cpRNP (chloroplast RNA binding protein), has been identified as a plastid-specific ribosomal protein. It has been proposed that PSRP-2 might be involved in mRNA interaction, complementing mRNA binding by the truncated S1 protein (Yamaguchi and Subramanian, 2000). We have not identified a PSRP-2 counterpart in the Chlamydomonas 30S subunit by either LC-MS/MS analysis or database searches. Because the spinach 30S ribosome does not contain a PSRP-7 homolog (Yamaguchi and Subramanian, 2000), Chlamydomonas might use PSRP-7 instead of PSRP-2 as a functional counterpart. The S1 domains of PSRP-7 may complement mRNA recognition of the plastid S1 protein that was identified as a truncated ortholog of E. coli S1. The S1 protein has been visualized at the junction of the head, platform, and main body of the E. coli 30S subunit in the cryoelectron microscopic map (Sengupta et al., 2001). The N-terminal loop region of S5 is oriented toward S1, and the solvent side of S1 faces the large concave surface of S2. These data suggest that each of the extra domains (NTE/INS) identified in PRPs S2, S3, and S5 might function in conjunction with PSRP-7 near the S1 protein binding site.
Immunological cross-reactivity has shown that the Chlamydomonas chloroplast 30S subunit has diverged from those of E. coli and spinach, whereas the 50S subunit is relatively conserved (Schmidt et al., 1984; Randolph-Anderson et al., 1989). This observation is consistent with our proteomic analysis of the 30S subunit (this study) and the 50S subunit (our unpublished data). In general, the 30S subunit is responsible for decoding mRNA and for translation initiation, interacting directly with initiation factors and mRNAs, whereas the 50S subunit is responsible for peptide synthesis (reviewed by Ramakrishnan, 2002). The spinach chloroplast 30S subunit contains a complete set of orthologs of the E. coli 30S subunit (S1 to S21) and four additional plastid-specific ribosomal proteins. The Chlamydomonas chloroplast 30S subunit also contains a majority of the E. coli 30S subunit protein orthologs and a few additional plastid-specific ribosomal proteins. The mammalian mitochondrial ribosomal 30S subunit lacks orthologs of E. coli S1, S3, S4, S8, S13, S19, and S20, whereas it contains 15 mitochondria-specific ribosomal proteins (Koc et al., 2001). Although the mammalian mitochondrial ribosomal 30S subunit has been largely reformed, the chloroplast 30S ribosomal subunit maintains a bacterial 30S-type core structure. Additional plastid-specific ribosomal proteins have been added, perhaps to regulate the 30S subunit in response to specific demands on plastid gene expression (e.g., light-dependent, redox-regulated translation) (reviewed by Somanchi and Mayfield, 2001).
The additional domains in the Chlamydomonas PRPs S2, S3, and S5 and PSRP-7 are unique to the chloroplast 30S subunit of Chlamydomonas. These additional domains and PSRP-7 may be related to unique attributes of Chlamydomonas chloroplast translational regulation, or they may be orthologs of nonribosomal proteins in other systems. It is possible that these domains do not directly affect ribosome assembly or general translation; thus, they could be dispensable for Chlamydomonas translation. Evolutionarily, these domains may represent fusions between 30S subunit proteins and loosely associated nonribosomal proteins and have not yet been identified in 30S subunits from other organisms. Future experiments should demonstrate whether other (bacterial) ribosomes could tolerate such large modifications. Likewise, it will be interesting to determine if these domains are dispensable in Chlamydomonas and to identify how these domains function in chloroplast translation.
METHODS
Preparation of Chlamydomonas reinhardtii Chloroplast Ribosomes and Subunits
The chloroplast ribosomes and subunits were prepared using a modification of a previously reported procedure (Chua et al., 1973). The Chlamydomonas reinhardtii strain Arg7/cw15 was grown at 25°C under constant light in 2 L of liquid TAP medium (Harris, 1989) with 50 μg/mL l-Arg to a density of 5 to 8 × 106 cells/mL. The cells were harvested by centrifugation at 4°C for 5 min at 2800g. All subsequent operations were performed at 4°C or on ice. The cell pellet was suspended in 40 mL of T25K25M25D5 buffer (25 mM Tris-HCl, pH 8.0, 25 mM KCl, 25 mM MgCl2, and 5 mM DTT), and the cells were harvested by centrifugation for 5 min at 4000g. The cell pellet was resuspended in 15 mL of T25K25M25D5 buffer and disrupted in a chilled N2 bomb (Parr Instrument Co., Moline, IL) under 600 p.s.i. of N2 for 6 min. The cell debris was centrifuged in a JA-17 rotor (Beckman, Fullerton, CA) at 8500 rpm (10,000g) for 10 min. The supernatant was transferred to a 13- × 51-mm polyallomer centrifuge tube (Beckman) for centrifugation in a TLA-100.3 fixed-angle rotor (Beckman) at 27,000 rpm (40,000g) for 30 min. The resulting postmitochondrial supernatant (designated the S-40 fraction; 2.0 to 2.5 mL of ∼10 mL) was layered on 36 mL of 10 to 40% Suc gradient in T25K25M10D5 buffer in a 25- × 89-mm polycarbonate centrifuge tube. After centrifugation at 22,500 rpm (91,000g) for 12 h in a SW28 rotor, ribosomes were fractionated with Auto Densi-Flow monitoring at 254 nm (Buchler Instruments, Fort Lee, NJ).
Fractions containing the chloroplast 70S ribosomes and cytoplasmic 80S ribosomes were collected separately. The ribosomes were recovered from the pooled fractions by centrifugation in a Ti 70.1 rotor at 60,000 rpm (250,000g) for 14 h in a 16- × 76-mm polycarbonate thick-wall centrifuge tube. The resulting ribosome pellet was resolved in T25K25M10D5 buffer, and 70S and 80S ribosomes were purified by the second Suc gradient as described above. Chloroplast ribosomal subunits were obtained from the 70S pellet. The 70S pellet was dissolved in T25K100M5D5 buffer and layered on 36 mL of 10 to 30% Suc gradient in the same buffer in a 25- × 89-mm polycarbonate centrifuge tube. After centrifugation at 22,500 rpm for 20 h in a SW28 rotor, 30S and 50S subunits were fractionated. The subunits were recovered from the pooled fractions by centrifugation in a Ti 70.1 rotor at 60,000 rpm for 16 h in a 16- × 76-mm polycarbonate thick-wall centrifuge tube. Ribosomes and subunits were kept at −70°C until use.
Protein Extraction and Electrophoresis
Ribosomal proteins were extracted from ribosomes or subunits by the addition of one-third volume of 0.05 M magnesium acetate in acetic acid as described previously (Yamaguchi and Subramanian, 2000). SDS-PAGE was performed by the method of Laemmli (1970) using a 12% acrylamide gel. Molecular mass markers used were the BENCHMARK prestained protein ladder (Gibco BRL). For liquid chromatography–tandem mass spectrometry analysis, 10 pmol of total protein from the 30S subunit (TP30) was separated by SDS-PAGE, stained with 0.25% Coomassie Brilliant Blue R 250 in 50% methanol and 10% acetic acid for 2 h, and destained in 50% methanol and 10% acetic acid for 2 h and then in 12.5% methanol and 2.5% acetic acid for 16 h. The gel was washed in distilled water for 30 min before in-gel digestion.
In-Gel Digestion
The TP30 gel was sectioned into 24 pieces (Figure 1), and each gel piece was fragmented further into 1-mm2 pieces and transferred into 1 well of a 96-well plate. The plates were transferred to a Massprep digestion robot (Micromass, Beverly, MA) for destaining (Gharahdaghi et al., 1999), reduction/alkylation (iodoacetamide), and in-gel digestion with trypsin or endoproteinase Lys-C (Shevchenko et al., 1996). After digestion, tryptic peptides were extracted from the gel pieces with 5% formic acid and 5% acetonitrile on the Massprep robot. The extracted peptides were diluted to 100 μL per well with 0.1% formic acid.
HPLC Tandem Mass Spectrometry
A microbore HPLC system (Surveyor; ThermoFinnigan, San Jose, CA) was modified to operate at capillary flow rates using a T-piece flow splitter. Columns (10 cm × 75 mm i.d.) were prepared by packing 100-Å, 5-μm Zorbax C18 resin at 500 p.s.i. pressure into New Objectives Pico Frits (New Objectives, Woburn, MA) columns with integral spray needles. Peptides were eluted in a gradient using buffer A (5% [v/v] acetonitrile and 0.1% formic acid) and buffer B (90% [v/v] acetonitrile and 0.1% formic acid) at a flow rate of 300 nL/min. After an initial wash with buffer A for 10 min, peptides were eluted with a linear gradient from 0 to 100% buffer B over a 30-min interval. Samples were introduced onto the analytical column using a Surveyor autosampler that first transferred the 100-μL peptide extract onto a C18 (300 × 5 mm) cartridge (LC Packings, San Francisco, CA) and then used a switching valve to transfer the eluted peptides onto the analytical column. The HPLC column eluent was eluted directly into the electrospray ionization source of a ThermoFinnigan LCQ-Deca ion trap mass spectrometer. Spectra were scanned over the range 400 to 1400 mass units. Automated peak recognition, dynamic exclusion, and product ion scanning of the top two most intense ions were performed using Xcalibur software, as described previously (Haynes et al., 1998).
Direct Analysis of Total Small Subunit Proteins
TP30 (10 μg) was resolved in 40 μL of 8 M urea and 100 mM Tris-HCl, pH 8.5. TP30 was reduced by adding 0.12 μL of 1 M Tris(2-carboxyethyl) phosphine and then incubated at room temperature for 20 min. The reduced TP30 was alkylated by adding 0.88 μL of 500 mM iodoacetamide at room temperature for 15 min. Endoproteinase Lys-C (Boehringer Mannheim) was added to a final substrate-to-enzyme ratio of 100:1, and the reaction was incubated at 37°C for 4 h. The Lys-C digest was diluted fourfold with 100 mM Tris-HCl, and then 100 mM CaCl2 was added to the solution and the final concentration was adjusted to 1 mM. The diluted Lys-C digest was digested further with 10 μL of Porozyme Trypsin Beads (Applied Biosystems, Foster City, CA) at 37°C for 16 h. The tryptic peptide mixture was analyzed by multidimensional protein identification technology, as described previously (Link et al., 1999; Washburn et al., 2001; Wolters et al., 2001).
Database Searching and Data Interpretation
Tandem mass spectrometry data were analyzed using SEQUEST, a computer program that allows the correlation of experimental data with theoretical spectra generated from known protein sequences (Eng et al., 1994; Yates et al., 1995). In this work, the general criteria for a preliminary positive peptide identification for a doubly charged peptide were a correlation factor of >2.5, a delta cross-correlation factor of >0.1 (indicating a significant difference between the best match reported and the next best match), a minimum of one tryptic peptide terminus, and a high preliminary score. For triply charged peptides, the correlation factor threshold was set at 3.5. All matched peptides were confirmed by visual examination of the spectra. All spectra were searched against the FASTA format database generated from Chlamydomonas ESTs and reported and unpublished open reading frames in the Chlamydomonas plastid genome (SWISS-PROT; J. Maul, J.W. Lilly, and D.B. Stern, unpublished data).
Computational Analyses
The program BLAST (National Center for Biotechnology Information) was used for sequence searches. Assembled EST sequence (EST contig) was obtained by WU-BLAST search (http://www.biology.duke.edu/chlamy_genome/blast/blast_form.html). Homology comparison was performed using BLAST 2 sequences. Multiple sequence alignment and phylogenetic tree representation were performed by CLUSTAL W (Thompson et al., 1994). pI values and sequence masses were calculated by the ProtParam tool (http://www.expasy.ch/tools/protparam.html). Predictions of cleavage sites for chloroplast transit peptides were obtained using the ChloroP program (Emanuelsson et al., 1999).
Nomenclature for Ribosomes, Subunits, Ribosomal Proteins, and Genes
We describe Chlamydomonas chloroplast ribosomes, the large subunits, and the small subunits as 70S, 50S, and 30S, respectively, although the sedimentation values of ribosomes and subunits from this algal chloroplast have been reported to be somewhat higher (Bourque et al., 1971; Margulies and Tiffany, 1979). Similarly, we use 80S for the cytoplasmic ribosomes in Chlamydomonas. Previously, Chlamydomonas chloroplast ribosomal proteins were characterized by two-dimensional PAGE and designated S-1 to S-31 in order of decreasing molecular mass (Schmidt et al., 1983; Randolph-Anderson et al., 1989). However, we have adopted the PRP nomenclature system used for plant plastid ribosomal proteins, based on sequence similarity to bacterial r-proteins (Yamaguchi et al., 2000). The PRP nomenclature was used for easier comparison with other bacterial ribosomal proteins. That is, chloroplast/plastid orthologs of E. coli S1 to S21 are designated PRP S1 to S21 (plastid ribosomal proteins S1 to S21). In accordance with the approved Commission on Plant Gene Nomenclature designation for plant genes, gene names are written in italics, with nuclear genes having uppercase first letters and organelle genes having lowercase first letters (Price and Reardon, 2001). For example, the gene for nucleus-encoded PRP S1 is Prps1, whereas the gene for plastid-encoded PRP S2 is prps2. The prefix PRP may be omitted when it is obvious that one is referring to plastid ribosomal proteins.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
Accession numbers for sequences mentioned in this article that are not given in Table 2 are as follows: Prps5 (AY093615), prps12 (AAA84155), spinach chloroplast 16S rRNA (AJ400848), and T. thermophilus 30S subunit (1FJF).
Acknowledgments
We thank Emma Brown, Scott Franklin, Aravind Somanchi, and David Hambly for helpful advice and critical review. This work was supported by funds from the Department of Energy (ER15313) and the National Institutes of Health (GM54659) to S.P.M. K.Y. was supported by a Skaggs Postdoctoral Fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004341.
References
- Arnold, R.J., and Reilly, J.P. (1999). Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem. 269, 105–112. [DOI] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Boni, I.V., Artamonova, V.S., and Dreyfus, M. (2000). The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control. J. Bacteriol. 182, 5872–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, D.P., Boynton, J.E., and Gillham, N.W. (1971). Studies on the structure and cellular location of various ribosome and ribosomal RNA species in the green alga Chlamydomonas reinhardtii. J. Cell Sci. 8, 153–183. [DOI] [PubMed] [Google Scholar]
- Bruce, B.D. (2000). Chloroplast transit peptides: Structure, function and evolution. Trends Cell Biol. 10, 440–447. [DOI] [PubMed] [Google Scholar]
- Brugger, M., and Boschetti, A. (1975). Two-dimensional gel electrophoresis of ribosomal proteins from streptomycin-sensitive and streptomycin-resistant mutants of Chlamydomonas reinhardtii. Eur. J. Biochem. 58, 603–610. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M.G., and Subramanian, A.R. (1994). Recognition of novel and divergent higher plant chloroplast ribosomal proteins by Escherichia coli ribosome during in vivo assembly. J. Biol. Chem. 269, 18223–18231. [PubMed] [Google Scholar]
- Bycroft, M., Hubbard, T.J., Proctor, M., Freund, S.M., and Murzin, A.G. (1997). The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell 88, 235–242. [DOI] [PubMed] [Google Scholar]
- Chua, N.H., Blobel, G., and Siekevitz, P. (1973). Isolation of cytoplasmic and chloroplast ribosomes and their dissociation into active subunits from Chlamydomonas reinhardtii. J. Cell Biol. 57, 798–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, J., McCormack, A.L., and Yates, J.R., III (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Fong, S.E., and Surzycki, S.J. (1992). Organization and structure of plastome psbF, psbL, petG and ORF712 genes in Chlamydomonas reinhardtii. Curr. Genet. 21, 527–530. [DOI] [PubMed] [Google Scholar]
- Fuchs, T.M., Deppisch, H., Scarlato, V., and Gross, R. (1996). A new gene locus of Bordetella pertussis defines a novel family of prokaryotic transcriptional accessory proteins. J. Bacteriol. 178, 4445–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzetti, B., Carol, P., and Mache, R. (1992). Characterization and RNA-binding properties of a chloroplast S1-like ribosomal protein. J. Biol. Chem. 267, 19075–19081. [PubMed] [Google Scholar]
- Gharahdaghi, F., Weinberg, C.R., Meagher, D.A., Imai, B.S., and Mische, S.M. (1999). Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605. [DOI] [PubMed] [Google Scholar]
- Giglione, C., and Meinnel, T. (2001). Organellar peptide deformylases: Universality of the N-terminal methionine cleavage mechanism. Trends Plant Sci. 6, 566–572. [DOI] [PubMed] [Google Scholar]
- Gribskov, M. (1992). Translational initiation factors IF-1 and eIF-2 alpha share an RNA-binding motif with prokaryotic ribosomal protein S1 and polynucleotide phosphorylase. Gene 119, 107–111. [DOI] [PubMed] [Google Scholar]
- Hanson, M.R., Davidson, J.N., Mets, L.J., and Bogorad, L. (1974). Characterization of chloroplast and cytoplasmic ribosomal proteins of Chlamydomonas reinhardtii by two-dimensional gel electrophoresis. Mol. Gen. Genet. 132, 105–118. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook. (San Diego, CA: Academic Press).
- Harris, E.H., Boynton, J.E., and Gillham, N.W. (1994). Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 58, 700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, P.A., Fripp, N., and Aebersold, R. (1998). Identification of gel-separated proteins by liquid chromatography-electrospray tandem mass spectrometry: Comparison of methods and their limitations. Electrophoresis 19, 939–945. [DOI] [PubMed] [Google Scholar]
- Koc, E.C., Burkhart, W., Blackburn, K., Moseley, A., and Spremulli, L.L. (2001). The small subunit of the mammalian mitochondrial ribosome: Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 276, 19363–19374. [DOI] [PubMed] [Google Scholar]
- Koller, B., Fromm, H., Galun, E., and Edelman, M. (1987). Evidence for in vivo trans splicing of pre-mRNAs in tobacco chloroplasts. Cell 48, 111–119. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 259, 680–685. [DOI] [PubMed] [Google Scholar]
- Leu, S. (1998). Extraordinary features in the Chlamydomonas reinhardtii chloroplast genome. (1) rps2 as part of a large open reading frame. (2) A C. reinhardtii specific repeat sequence. Biochim. Biophys. Acta 1365, 541–544. [DOI] [PubMed] [Google Scholar]
- Link, A.J., Eng, J., Schieltz, D.M., Carmack, E., Mize, G.J., Morris, D.R., Garvik, B.M., and Yates, J.R., III (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682. [DOI] [PubMed] [Google Scholar]
- Liu, X.Q., Huang, C., and Xu, H. (1993). The unusual rps3-like orf712 is functionally essential and structurally conserved in Chlamydomonas. FEBS Lett. 336, 225–230. [DOI] [PubMed] [Google Scholar]
- Louie, D.F., Resing, K.A., Lewis, T.S., and Ahn, N.G. (1996). Mass spectrometric analysis of 40 S ribosomal proteins from Rat-1 fibroblasts. J. Biol. Chem. 271, 28189–28198. [DOI] [PubMed] [Google Scholar]
- Margulies, M.M., and Tiffany, H.L. (1979). Sedimentation behavior of chloroplast ribosomes from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 563, 171–180. [DOI] [PubMed] [Google Scholar]
- Mayfield, S.P., Yohn, C.B., Choen, A., and Danon, A. (1995). Regulation of chloroplast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 147–166. [Google Scholar]
- Price, C.A., and Reardon, E.M. (2001). Mendel, a database of nomenclature for sequenced plant genes. Nucleic Acids Res. 29, 118–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, V. (2002). Ribosome structure and the mechanism of translation. Cell 108, 557–572. [DOI] [PubMed] [Google Scholar]
- Randolph-Anderson, B.L., Gillham, N.W., and Boynton, J.E. (1989). Electrophoretic and immunological comparisons of chloroplast and prokaryotic ribosomal proteins reveal that certain families of large subunit proteins are evolutionarily conserved. J. Mol. Evol. 29, 68–88. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., Myers, A.M., Gillham, N.W., and Boynton, J.E. (1984). Immunological similarities between specific chloroplast ribosomal proteins from Chlamydomonas reinhardtii and ribosomal proteins from Escherichia coli. Mol. Biol. Evol. 1, 317–334. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., Richardson, C.B., Gillham, N.W., and Boynton, J.E. (1983). Sites of synthesis of chloroplast ribosomal proteins in Chlamydomonas. J. Cell Biol. 96, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, J., Agrawal, R.K., and Frank, J. (2001). Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. USA 98, 11991–11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shteiman-Kotler, A., and Schuster, G. (2000). RNA-binding characteristics of the chloroplast S1-like ribosomal protein CS1. Nucleic Acids Res. 28, 3310–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanchi, A., and Mayfield, S.P. (2001). Regulation of chloroplast translation. In Advances in Photosynthesis and Respiration, Vol. 11, Regulation of Photosynthesis, E.-M. Aro and B. Andersson, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 137–151.
- Subramanian, A.R. (1983). Structure and functions of ribosomal protein S1. Prog. Nucleic Acids Res. Mol. Biol. 28, 101–142. [DOI] [PubMed] [Google Scholar]
- Subramanian, A.R. (1993). Molecular genetics of chloroplast ribosomal proteins. Trends Biochem. Sci. 18, 177–181. [DOI] [PubMed] [Google Scholar]
- Sugita, M., Sugita, C., and Sugiura, M. (1995). Structure and expression of the gene encoding ribosomal protein S1 from the cyanobacterium Synechococcus sp. strain PCC 6301: Striking sequence similarity to the chloroplast ribosomal protein CS1. Mol. Gen. Genet. 246, 142–147. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Terasaki, M., Takemoto-Hori, C., Hanada, T., Ueda, T., Wada, A., and Watanabe, K. (2001). Proteomic analysis of the mammalian mitochondrial ribosome: Identification of protein components in the 28 S small subunit. J. Biol. Chem. 276, 33181–33195. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel, M., and Otis, C. (1994). The chloroplast gene cluster containing psbF, psbL, petG and rps3 is conserved in Chlamydomonas. Curr. Genet. 27, 54–61. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J. (2001). Challenges and prospects of plant proteomics. Plant Physiol. 126, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates, J.R., III (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Weglöhner, W., Kauschmann, A., and Subramanian, A.R. (1995). Evolution of the NH2- and COOH-terminal extensions of chloroplast ribosomal protein S18: Nucleotide sequence of pea and rye chloroplast rps 18 genes. Biochem. Mol. Biol. Int. 36, 265–273. [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons, W.M., Jr., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. (2000). Structure of the 30S ribosomal subunit. Nature 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Wolters, D.A., Washburn, M.P., and Yates, J.R., III (2001). An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., and Subramanian, A.R. (2000). The plastid ribosomal proteins: Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275, 28466–28482. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., von Knoblauch, K., and Subramanian, A.R. (2000). The plastid ribosomal proteins: Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275, 28455–28465. [DOI] [PubMed] [Google Scholar]
- Yates, J.R., III, Eng, J.K., McCormack, A.L., and Schieltz, D. (1995). Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436. [DOI] [PubMed] [Google Scholar]