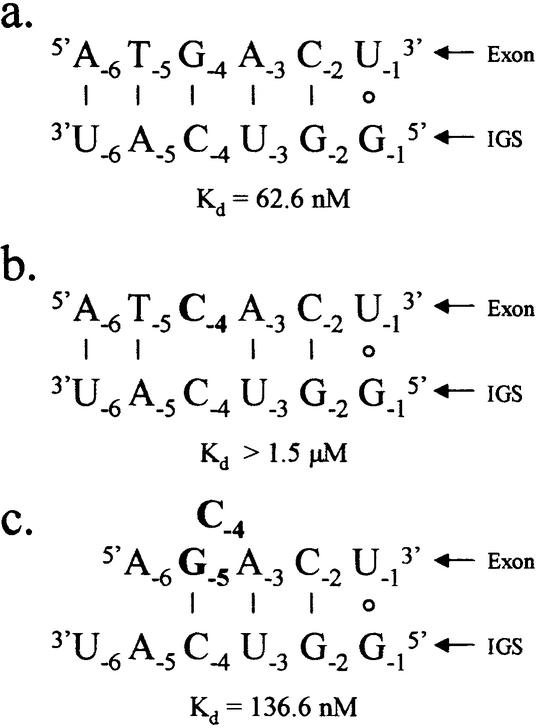Abstract
We report the development, analysis and use of a new combinatorial approach to analyze the substrate sequence dependence of the suicide inhibition, cyclization, and reverse cyclization reactions catalyzed by a group I intron from the opportunistic pathogen Pneumocystis carinii. We demonstrate that the sequence specificity of these Internal Guide Sequence (IGS)-mediated reactions is not high. In addition, the sequence specificity of suicide inhibition decreases with increasing MgCl2 concentration, reverse cyclization is substantially more sequence specific than suicide inhibition, and multiple reverse cyclization products occur, in part due to the formation of multiple cyclization intermediates. Thermodynamic analysis reveals that a base pair at position –4 of the resultant 5′ exon–IGS (P1) helix is crucial for tertiary docking of the P1 helix into the catalytic core of the ribozyme in the suicide inhibition reaction. In contrast to results reported with a Tetrahymena ribozyme, altering the sequence of the IGS of the P.carinii ribozyme can result in a marked reduction in tertiary stability of docking the resultant P1 helix into the catalytic core of the ribozyme. Finally, results indicate that RNA targeting strategies which exploit tertiary interactions could have low specificity due to the tolerance of mismatched base pairs.
INTRODUCTION
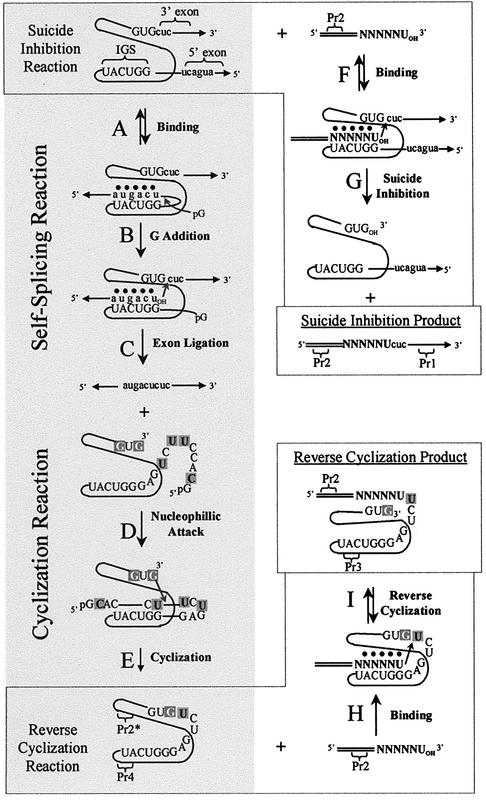
Self-splicing group I introns are catalytic RNAs that splice out of cellular RNA transcripts (Fig. 1A–C), often without the apparent requirement for ancillary proteins (1). The excised introns can then undergo cyclization (Fig. 1D and E) (2–4). These complex catalytic activities are self-contained within RNAs of relatively moderate size, often <400 nt. Therefore, group I introns are frequently exploited as model systems in the continual pursuit of a more thorough understanding of RNA structure and function. As the self-splicing reaction is single-turnover, such studies often utilize group I intron-derived ribozymes, which are introns that lack the endogenous 5′ and 3′ exons (5,6). The use of ribozymes and one or more exogenous exon mimics allow for the isolation of individual binding and reaction steps for thermodynamic and kinetic analyses.
Figure 1.
Schematic diagram of self-splicing and cyclization (steps A→E), suicide inhibition (steps F→G) and reverse cyclization (steps H→I). Addition of an exogenous exon substrate (N represents positional randomization) leads to both suicide inhibition and reverse cyclization products. Note that steps C and G are in competition, as they occur concurrently. Uppercase lettering and black lines represent the intron. Lowercase lettering and their associated lines with arrows represent the exons. The elongated region of the exon mimics is double lined. Tertiary interactions are represented by dots. RT–PCR primers Pr1, Pr2, Pr3 and Pr4 bind to the designated regions. Pr2* indicates that Pr2 and Pr2-d(ATGAC)rU can both act as RT primers even though neither are complementary to the target region for the cyclization. Note that, at least for the P.carinii intron, step B is also achieved through ribozyme-mediated hydrolysis in the absence of pG (as is the case throughout this study) (6). After step C, the intron sequence is shown in more detail. Positions that can act as cyclization nucleophiles are shown with white lettering on a dark gray background and the intron sites of cyclization are shown with black lettering on a dark gray background. The IGS is 5′-GGUCAU-3′.
Group I introns must tightly bind their 5′ exons during the course of the self-splicing reaction. Using a group I intron-derived ribozyme, it was found that this binding event occurs in two well-defined steps (7,8). The 5′ exon first base pairs with a predominantly complementary sequence, the Internal Guide Sequence (IGS) (9,10). The resultant exon–IGS helix, called the P1 helix, then docks into the catalytic core of the intron, stabilized by tertiary interactions (6,11–13). It has been shown, using ribozyme constructs in vitro and in vivo, that altering the IGS of these ribozymes results in a corresponding change in what substrates the ribozyme can bind, such that complementary base pairing is maintained within the P1 helix (10,14–16). This characteristic has been exploited for engineering group I intron-derived ribozymes that target and react with specific sequences, including sequences of medical importance. Examples include RNA repair ribozymes, which involve either replacing mutant regions of defective RNA transcripts with ‘corrected’ regions (trans-splicing; 15,17–21) or selectively excising mutant regions from within transcripts (trans excision-splicing; 22). Alternatively, exogenous 5′ exon mimics can bind and react with transcripts containing introns, at least in vitro, thereby acting as suicide inhibitors of the self-splicing reaction (Fig. 1F and G) (23–25). Exogenous 5′ exon mimics also bind and react with cyclized spliced introns, which result in linearized reverse cyclization products (Fig. 1H and I) (10,26,27). All of these catalytic reactions rely upon the molecular interaction of the IGS with an exon mimic.
Although the sequence dependence of the 5′ exon–IGS interaction has been studied using Tetrahymena ribozymes (in trans-splicing and reverse splicing reactions), the sequence dependence of this interaction has not been analyzed for any Pneumocystis carinii ribozyme reaction, or for the suicide inhibition reaction with any ribozyme. This is of interest because previous results have shown that the P.carinii and Tetrahymena ribozymes utilize a different array of tertiary interactions to dock the P1 helix into the catalytic core (6,12,28). Furthermore, the P.carinii ribozyme catalyzes a unique trans excision-splicing reaction, which along with the suicide inhibition reaction has potential applications. It was of interest then to analyze the sequence dependence (and increase our understanding of this key molecular recognition interaction) for the P.carinii suicide inhibition reaction. In addition to suicide inhibition, cyclization and reverse cyclization occur concurrently and utilize the IGS for substrate binding. Therefore, these reactions were also analyzed.
The results show that certain 5′ exon mimic chimeras, other than those complementary to the IGS, can be substrates in the suicide inhibition and the reverse cyclization reactions. The distribution of substrate sequences is not random, as only certain mismatches are permissible at each of the five randomized 5′ exon positions. Exon sequences that maintain Watson–Crick or wobble base pairs with the IGS, especially within the four 3′ nucleobases of the 5′ exon, are the most effective substrates in these reactions. The sequence specificity of the suicide inhibition reaction is MgCl2 dependent, with a more diverse array of acceptable substrate sequences at higher MgCl2 concentrations. Our results also indicate that the reverse cyclization reaction is substantially more sequence specific than the suicide inhibition reaction. Furthermore, we report that the reverse cyclization products stem from multiple cyclized intron intermediates, which consequently lead to multiple reverse cyclization products. Using a ribozyme derived from the P.carinii intron (4,20), the binding properties of a representative group of synthetic 5′ exon mimic chimeras were quantified. In these studies, we demonstrate the importance of a base pair (position –4 of the P1 helix) for tertiary docking of the P1 helix into the catalytic core of the intron. Finally, the data show that the IGS itself can be altered to target a diverse variety of substrate sequences. Such alterations, however, even though resulting in fully complementary P1 helices, can significantly affect the thermodynamics of P1 helix base pairing and tertiary docking. This combinatorial approach therefore has proven useful for gaining a deeper understanding of the fundamental molecular recognition properties of IGS-mediated reactions.
MATERIALS AND METHODS
Synthesis and purification of nucleic acids
The oligonucleotide d(GCTCGTCGTCGACAACGGCTCATGAC)rU [named Pr2-d(ATGAC)rU] was synthesized on an Applied Biosystems 380B DNA/RNA synthesizer and purified by trityl-on reverse phase column chromatography (Sep-Pak, Milford, MA) as described (6). The oligonucleotides d(GCTCACTCATTAGGCACC) (named Pr1), d(GCTCGTCGTCGACAACGGCTC) (named Pr2), d(GCTCGTCGTCGACAA CGGCTCNNNNN)rU [named Pr2-d(NNNNN)rU], d(CAACATTGTTACTATACCCAGGGCT) (named Pr3), d(GACAGTGTCGCAATCTGAAT) (named Pr4) and all the chimeric hexamers were synthesized, deblocked and desalted by Integrated DNA Technologies (Coralville, IA). N represents machine randomization of all four bases at the designated positions. The all RNA oligonucleotides were synthesized by Dharmacon Research, Inc. (Lafayette, CO). All hexamers (RNAs and chimeras) were purified by thin-layer chromatography as described (6). Designated oligonucleotides were 5′ end radiolabeled and purified by polyacrylamide gel electrophoresis as described previously (6). Oligonucleotide concentrations were calculated based on UV-absorption measurements using a Beckman DU 650 UV-Spectrophotometer (Beckman Coulter, Inc., Fullerton, CA).
The ribozyme rP-8/4x, which was used in the binding assays, was synthesized by run-off transcription of XbaI linearized P-8/4x plasmid (6). The group I intron transcript rP-h, which was used for the reverse cyclization, cyclization, initial suicide inhibition and the kinetic assays was synthesized by run-off transcription of HindIII linearized P-h plasmid (6). This produces an RNA transcript with an 82 base 5′ exon, 344 base intron and 27 base 3′ exon. The group I intron transcript rP-V, used in the combinatorial suicide inhibition assay, was synthesized by run-off transcription of VspI linearized P-h plasmid. This produces an RNA transcript with an 82 base 5′ exon, 344 base intron and 233 base 3′ exon. The rP-V transcript is an elongated version of rP-h with the 3′ exon extended such that it can serve as a primer-binding region for RT–PCR in the combinatorial suicide inhibition assay. Note that the 206 base extended region is derived entirely from the P-h plasmid and is not native to the P.carinii intron or flanking exons.
For designated studies the IGS of the rP-8/4x ribozyme was modified from r(GGUCAU) to either r(GGUGAU) or r(GUGACG). These mutant ribozymes were named rP-8/4x-CtoG and rP-8/4x-oppo, respectively. Site-directed mutagenesis was performed on the plasmid precursor, P-8/4x, using the following mutagenic primer pairs: for rP-8/4x-CtoG, CGACTCACTATAGAGGGTGATGAAAGCGGC and GCC GCTTTCATCACCCTCTATAGTGAGTCG; and for rP-8/4x-oppo, CGACTCACTATAGAGGTGACGGAAAGCGGC and GCCGCTTTCCGTCACCTCTATAGTGAGTCG. Each modified plasmid was generated in a 50 µl reaction consisting of 25 ng P-8/4x plasmid, 2.5 U Pfu DNA polymerase (Stratagene, La Jolla, CA), and 0.5 µM dNTPs in a buffer of 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris–HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100 and 0.1 mg/ml BSA. The mixtures were initially denatured at 95°C for 30 s, then underwent 15 cycles of 95 (30 s), 50 (2 min) and 68°C (6 min) in a PCRExpress Thermal Cycler (Hybaid, Ashford, Middlesex, UK). The parental plasmids were digested with 20 U of DpnI (Gibco BRL, Rockville, MD) in 5.7 μl of the manufacturer’s buffer at 37°C for 2 h. A 3 µl aliquot was used to transform Escherichia coli DH5α competent cells (Gibco BRL). Individual isolates were grown and the modified plasmids were purified using a QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). The plasmids were sequenced for confirmation (ACGT, Inc., Northbrook, IL). The two mutant ribozymes were transcribed as described for rP-8/4x (6).
MgCl2 concentration dependence of group I intron reactions
Initially, as a test of reactivity, the suicide inhibition and reverse cyclization reactions were conducted as a function of MgCl2 concentration using the radiolabeled 5′ exon mimic chimeras d(ATGAC)rU and Pr2-d(ATGAC)rU. In the experiments with d(ATGAC)rU, 3 µl of 667 nM rP-h was pre-annealed at 55°C in HXMg buffer [50 mM HEPES (25 mM Na+), 135 mM KCl and X mM MgCl2 (from 0 to 15) at pH 7.5] for 5 min and slow cooled to 37°C. A 1 µl solution of 1 nM 5′ end radiolabeled d(ATGAC)rU at 37°C was added. The final concentration of rP-h in the d(ATGAC)rU assays was 500 nM because the expected dissociation constant for d(ATGAC)rU was 60 to 100 nM (in 15 mM MgCl2 at 37°C). The reaction was allowed to proceed at 37°C for 2 h. The experiments utilizing Pr2-d(ATGAC)rU were conducted as above, except for using a final concentration of 5 µM rP-h in H4Mg or H15Mg buffer. The concentration of rP-h for the Pr2-d(ATGAC)rU reactions was 10-fold higher than that with d(ATGAC)rU, which was required due to less efficient reactivity with the former. The reactions were quenched by the addition of 4 µl stop buffer (0.1× TBE, 10 M urea and 3 mM EDTA) and the reactants and products were separated on an 8% polyacrylamide, 7.5 M urea gel. The gel was dried under vacuum and the bands were visualized and quantified on a Molecular Dynamics Storm 860 PhosphorImager. Note that this reaction produces visible suicide inhibition and reverse cyclization products, whose ratio depends on the solution concentration of MgCl2. Also note that these reactions were done in the absence of free guanosine monophosphate to enhance suicide inhibition relative to self-splicing (23). The high rate of intron-mediated hydrolysis at the 5′ exon– intron junction, however, still allows a significant amount of self-splicing, and hence formation of the circularization intermediate, to occur (6,23).
Combinatorial assays
For the suicide inhibition reactions, 1 µM of rP-V precursor was pre-annealed at 55°C in 5 µl of either H4Mg or H15Mg buffer for 5 min and slow cooled to 37°C. A 5 µl solution of either 400 nM Pr2-d(ATGAC)rU or Pr2-d(NNNNN)rU in the appropriate buffer at 37°C was added and the reaction was allowed to proceed for 1 h. The reverse cyclization and cyclization reactions were conducted essentially the same as the suicide inhibition reactions, except that the rP-h intron transcript was used and the assay was performed only in H15Mg buffer. In these combinatorial assays, the final concentration of rP-V or rP-h was 500 nM and the final concentration of Pr2-d(ATGAC)rU or Pr2-d(NNNNN)rU was 200 nM. A concentration of 200 nM of the randomized oligonucleotides gives ∼0.2 nM of each particular sequence.
Product isolation and identification
The reaction products were first reverse transcribed using primer Pr1 for suicide inhibition, primer Pr3 for reverse cyclization, and primer Pr4 for the cyclization reaction. The binding sites for these primers are shown in Figure 1. In these reactions, 2 µl of the final non-radiolabeled reaction mixture was added to 3.2 µl sterilized H2O, 0.5 µl of a 10 mM stock of all four dNTPs, as well as 0.3 µl of a 3.3 µM stock of the respective RT primer. This was incubated at 65°C for 5 min and then quick cooled on ice. At this point, 2 µl of 5× reverse transcription buffer [250 mM Tris–HCl (pH 8.3), 375 mM KCl, and 15 mM MgCl2], 1 µl of 0.1 M DTT and 0.5 µl (20 U) RNAseOut (Gibco BRL) was added. This was incubated at 42°C for 1 min, after which 0.5 µl (100 U) of Superscript II (Gibco BRL) was added and the reaction incubated at 42°C for 50 min. The reaction was terminated by incubation at 70°C for 15 min and then was quick cooled on ice. The reverse transcription products were then PCR amplified. Briefly, reactions contained PCR buffer [50 mM KCl, 10 mM Tris–HCl (pH 9.0), 0.1% Triton X-100], 0.1 µl of the reverse transcription reaction from above, 200 nM primer Pr2, 200 nM of the respective reverse transcription primer, 1.2 mM MgCl2, 20 µM dNTP mix and 1 µl of Taq Polymerase (5 U; Gibco BRL). An initial incubation of 2 min at 94°C was followed by 30 cycles of 94 (45 s), 55 (1 min) and 72°C (1.5 min). The resulting PCR products were separated on a 2% agarose gel. The distinct bands at 150 (suicide inhibition reaction), 175 (cyclization reaction) and 200 bp (reverse cyclization reaction) were extracted and purified using QIAquick Gel Extraction Kits (Qiagen).
We exploited the natural 3′ A overhangs of the purified PCR products to ligate them into pGEM-T vectors (Promega, Madison, WI). The 4°C overnight ligation protocol recommended by the manufacturer was followed. The resultant plasmids were transformed into E.coli DH5α cells and plated. Colonies were picked from the plates and grown overnight in LB-ampicillin media. The resultant amplified plasmids were purified using QIAprep Spin Miniprep Kits (Qiagen).
All plasmids were sequenced off the T7 promoter indigenous to the plasmid. Roughly half of the resultant plasmids were sequenced by Davis Sequencing (Davis, CA). Otherwise, sequencing reactions (total volume of 6 µl) consisted of 250 ng plasmid template, 1× sequencing buffer [50 mM Tris–HCl (pH 9.0) and 2 mM MgCl2], 0.10 fmol 5′ end radiolabeled T7 sequencing primer, 0.25 µl Taq Polymerase (1.25 U; Gibco BRL), and one of the following nucleotide mixtures: 300 µM ddATP and 13.3 µM of each of the four dNTPs; 400 µM ddCTP and 13.3 µM of each of the four dNTPs; 33.3 µM ddGTP and 13.3 µM of each of the four dNTPs; or 200 µM ddTTP and 6.6 µM of each of the four dNTPs. The sequencing reactions were run in a PCRExpress thermal cycler. An initial incubation of 2 min at 95°C was followed by 30 cycles of 95 (30 s), 46 (30 s) and 70°C (1 min). The reaction was quenched by adding 3 µl of sequencing stop buffer (10 mM EDTA in de-ionized formamide). The samples were heated for 2 min at 70°C and immediately loaded onto a 6% denaturing polyacrylamide gel. The gels were transferred onto chromatography paper (Whatman 3MM CHR), dried under vacuum at 65°C, and visualized on a Molecular Dynamics Storm 860 PhosphorImager.
Determination of binding constants
The dissociation constant, Kd, of representative 5′ exon mimics binding to the rP-h intron was approximated using the rP-8/4x ribozyme and the radiolabeled native 5′ exon mimic, r(AUGACU), in competition band-shift gel electrophoresis assays (6,12,29). rP-8/4x (45 nM) was pre-annealed at 55°C for 5 min in 5 µl of a solution of H15Mg buffer containing 4.5% glycerol (v/v). The solution was then slow cooled to 37°C, at which time ∼1 nM radiolabeled r(AUGACU) and various concentrations of representative 5′ exon mimic competitors in a total volume of 2.5 µl (in H15Mg buffer at 37°C) was added. The final concentration of ribozyme was 30 nM and the competitor concentrations varied from 5 to 1500 nM. A ribozyme concentration of 30 nM was used because this will bind essentially all of the r(AUGACU), which has a Kd of 5.2 nM for this ribozyme under these conditions (6). The solutions equilibrated for 90 min. The fraction of 5′ exon mimic bound was partitioned from unbound on a 37°C, 10% native polyacrylamide gel made with H15Mg buffer. The gel was dried under vacuum and the bands quantified with a Molecular Dynamics Storm 860 PhosphorImager. Dissociation constants were calculated as described previously (6) and the data reported are the average of at least two independent assays. Quantification of binding in H4Mg buffer using this assay was not accurate because binding was below detection limits (Kd >1.5 µM).
Optical melting curves
The strength of base pairing between the representative 5′ exon mimics and an oligonucleotide mimic of the intron’s native IGS, r(GGUCAU), was analyzed through thermal denaturation experiments on a Beckman DU 650 UV-Spectrophotometer. Melts were conducted using an incident UV wavelength of 280 nM at 100 µM total strand concentration in H15Mg buffer. Thermodynamic parameters were quantified as described previously (6). The data reported are the average of at least two independent assays.
Kinetics
The observed rate constants for the suicide inhibition and reverse cyclization reactions using the representative 5′ exon mimics were obtained under single-turnover ‘intron excess’ conditions. In these experiments, 15 µl of an 833 nM solution of rP-h was pre-annealed at 55°C in either H4Mg or H15Mg buffer for 5 min and slow cooled to 37°C. A 10 µl solution of ∼1 nM 5′ end radiolabeled 5′ exon mimic in the same buffer at 37°C was added, bringing the final rP-h concentration in the sample to 500 nM. An rP-h concentration of 500 nM was used to permit hexamers with relatively high Kds to bind the intron. A 2 µl aliquot was periodically removed and added to 2 µl stop buffer over typically 120 min. The reactants and products were separated on an 8% polyacrylamide, 7.5 M urea gel. The gel was dried under vacuum and the bands were visualized and quantified on a Molecular Dynamics Storm 860 PhosphorImager. The observed rate constants for the suicide inhibition and reverse cyclization reactions were quantified essentially as described (6) and the data reported are the average of at least two independent assays.
Modified IGS studies
The dissociation constants for IGS-modified ribozymes (rP-8/4x-CtoG and rP-8/4x-oppo) binding various 5′ exon mimics were determined by direct band-shift electrophoresis assays. These assays were conducted using various radiolabeled 5′ exon mimics; rP-8/4x, rP-8/4x-CtoG or rP-8/4-oppo ribozymes at various concentrations (from 50 to 1500 nM); and H15Mg as the binding and electrophoresis buffers. The ribozyme mixtures were pre-annealed at 55°C for 5 min in 5 µl solutions of H15Mg buffer and 4.5% glycerol (v/v). The solutions were then slow cooled to 37°C, at which time 2.5 µl of ∼1 nM radiolabeled 5′ exon mimic (in H15Mg buffer at 37°C) was added. The assay was completed as described for the competitive binding assays and quantified as described (6). Thermal denaturing experiments were conducted as described above, except modified 5′ IGS sequences [r(GGUGAU) and r(GUGACG)] with their complementary 5′ exon mimics [d(ATCAC)rU and d(CGTCA)rU] were analyzed.
RESULTS
Combinatorial assay rationale
It was demonstrated previously that the N3′→P5′ phosphoramidate 5′ exon mimic d(AnTnGnAnCn)rU, where n represents a phosphoramidate linkage (23,28–31), is a suicide inhibitor of the self-splicing reaction in vitro of a group I intron from the opportunistic pathogen P.carinii (Fig. 1F and G). In the suicide inhibition reaction, the exogenous 5′ exon mimic becomes ligated to the endogenous 3′ exon. This property makes the suicide inhibition reaction ideal for developing a combinatorial assay to systematically analyze the sequence dependence of the 5′ exon–IGS molecular interaction. In this combinatorial assay, a 5′ elongated version of the 5′ exon mimic (as seen in Fig. 1) acts as a suicide inhibition substrate and the elongated base region is exploited as a primer-binding site for RT–PCR amplification of the suicide inhibitor sequence. The 3′ bases of the suicide inhibitor, which act as the 5′ exon mimic, are randomized, permitting the simultaneous assessment of all possible sequences to be substrates in the suicide inhibition reaction. In addition to the suicide inhibition product, a reverse cyclization product [described previously as the 350 product in Testa et al. (23)] is concurrently generated in this reaction (23,26,27,32). Reverse cyclization substrates are also 5′ exon mimics in that they first base pair, at least in part, with the IGS of the intron (Fig. 1H). Therefore, this combinatorial assay can also analyze the sequence dependence of the reverse cyclization reaction.
Suicide inhibition and reverse cyclization assay development
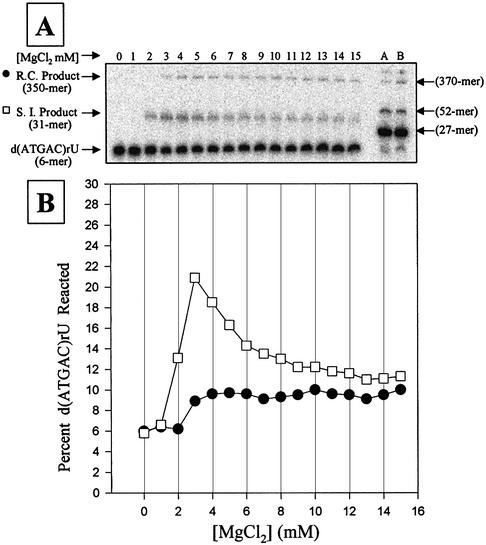
The 5′ exon mimic, d(ATGAC)rU [Kd = 61 nM at 37°C in 15 mM MgCl2], binds to the P.carinii rP-8/4x ribozyme nearly as tightly as the N3′→P5′ phosphoramidate equivalent, d(AnTnGnAnCn)rU [Kd = 16 nM at 37°C in 15 mM MgCl2] (29). The non-modified DNA–RNA chimeras were tested to assess whether they could serve as active substrates in the suicide inhibition and the reverse cyclization reactions. The reactions were run as a function of MgCl2 concentration using the 5′ exon mimic d(ATGAC)rU, as well as the elongated 5′ exon mimic, Pr2-d(ATGAC)rU (Fig. 2). Using d(ATGAC)rU, products of ∼31 nt in length (the suicide inhibition product) and ∼350 nt in length (the reverse cyclization product) were produced, as expected (20). In addition, the MgCl2 dependence of these reactions is similar to that obtained with the phosphoramidate-modified 5′ exon mimics (23). When the elongated 5′ exon mimic, Pr2-d(ATGAC)rU, was used in the reaction, the product of ∼31 nt was replaced by a product of ∼52 nt (Fig. 2, lanes A and B), as expected. For confirmation, the products of the reactions with the elongated 5′ exon mimic Pr2-d(ATGAC)rU (conducted at 4 mM MgCl2 for suicide inhibition and 15 mM MgCl2 for reverse cyclization) were RT–PCR amplified, cloned and sequenced. The sequences obtained were those expected for the two reactions and are diagrammed in Figure 1 [with the randomized regions equal to d(ATGAC)]. These results demonstrate that the elongated, non-modified chimeric 5′ exon mimic is a substrate in these reactions.
Figure 2.
Magnesium dependence of the suicide inhibition (S.I.) and reverse cyclization (R.C.) reactions. (A) A typical polyacrylamide gel of the reactions. Lanes 0–15 show reactions using 1 nM radiolabeled 5′ exon mimic d(ATGAC)rU and 500 nM of rP-h in HXMg buffer [50 mM HEPES (25 mM Na+), 135 mM KCl and X mM MgCl2 (from 0 to 15) at pH 7.5]. Lanes A and B show reactions using 1 nM radiolabeled 5′ exon mimic Pr2-d(ATGAC)rU (27mer) and 500 nM of rP-h in H4Mg (lane A) or H15Mg (lane B) buffer. All reactions were run for 2 h, which allows maximum product formation. The sizes of the 27, 52, 350 and 370mer are approximate. (B) Graph of the MgCl2 concentration dependence for the suicide inhibition (open squares) and the reverse cyclization (solid circles) reactions shown above. Note that both reactions occur simultaneously and that the graph is an average of two reactions.
Sequence specificity of suicide inhibition
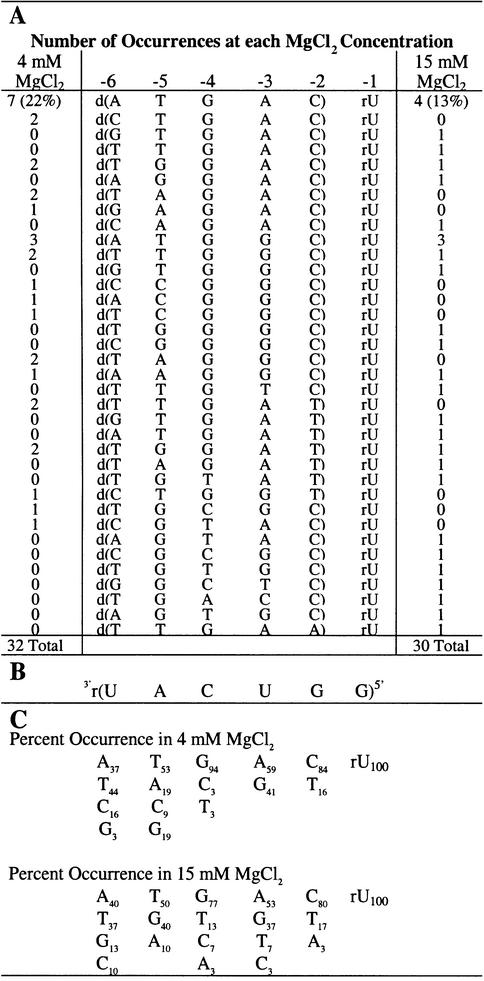
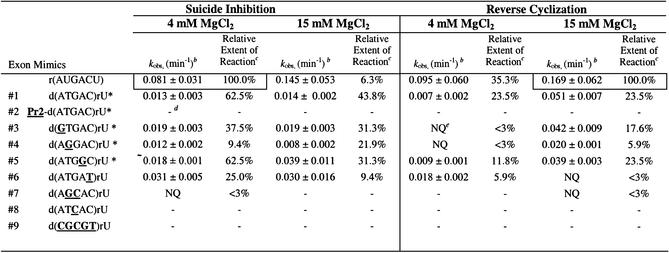
The combinatorial suicide inhibition reaction was initially conducted in H4Mg buffer using the P.carinii rP-V intron and the non-radiolabeled, randomized 5′ exon mimic Pr2-d(NNNNN)rU. Suicide inhibition reactions were run in 4 mM MgCl2 because it is at (23) or near (Fig. 2) the optimum concentration for suicide inhibition reactivity. The 3′ uridine was not randomized, as it is nearly universally conserved. The resulting mixture of products was RT–PCR amplified, cloned and the suicide inhibition junctions were sequenced. The results in Table 1 show that the native 5′ exon sequence, d(ATGAC)rU, occurs most often (22%); however, 56% of the sequences (18 out of 32) obtained are unique. These 5′ exon mimics can effectively compete with the endogenous RNA exon and the exogenous chimeric native exon for the IGS of the intron. Although the sequence specificity is relatively low for this interaction, the distribution of tolerated bases at individual positions within the exon is not random (Table 1C). The exon positions are numbered according to the intron transcript (6) and are shown in Table 1. Position –2 can tolerate a dT, which presumably forms a dT–rG wobble base pair, however, a Watson–Crick base pair is strongly favored. At position –3, dA and dG were found in equal proportions, so a Watson-Crick pair (dA–rU) or a wobble pair (dG–rU) is either strongly favored or required. Position –4 allows a dC and a dT, albeit rarely, thus strongly favoring a Watson–Crick base pair. Positions –5 and –6 are significantly less specific for Watson–Crick or wobble pairs than the other positions, although they are not random and they do prefer Watson–Crick pairing.
Table 1. Suicide inhibitors of the self-splicing reaction.
(A) Suicide inhibitors obtained from the combinatorial assays (written 5′→3′). The number of occurrences for each sequence is indicated in the left column for reactions at 4 mM MgCl2 and in the right column for reactions at 15 mM MgCl2. Note that position –1 is RNA and was not randomized, while positions –2 through to –6 are DNA and were randomized. The numbering system for the inhibitor positions is shown above the sequences. Note that the inhibitors are 5′ exon mimics. (B) Sequence of the IGS of the rP-h intron (and the rP-8/4x ribozyme). (C) The percent occurrence of the four nucleobases at each position. Note that the percent occurrences are different for the two MgCl2 concentrations.
The MgCl2 dependence of suicide inhibition
The combinatorial assay was performed in 15 mM MgCl2 to determine if the concentration of magnesium has an effect on the sequence specificity of the suicide inhibition reaction. The results at 15 mM MgCl2 are similar to results at 4 mM MgCl2 except there is significantly more sequence diversity (83% unique sequences as compared with 56%) at the higher magnesium concentration. A greater number of mismatches are tolerated at positions –2 through to –4, which appear to be the most critical positions for docking of the resultant P1 helices (28; this work). In addition, the native sequence is found almost half as often at the higher MgCl2 concentration. Taken together these results indicate that the sequence specificity of the suicide inhibition reaction decreases at higher MgCl2 concentration.
Sequence specificity of reverse cyclization
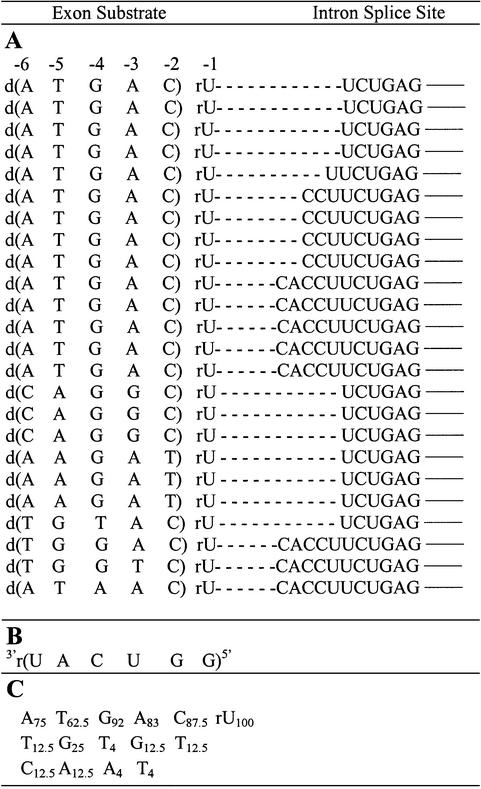
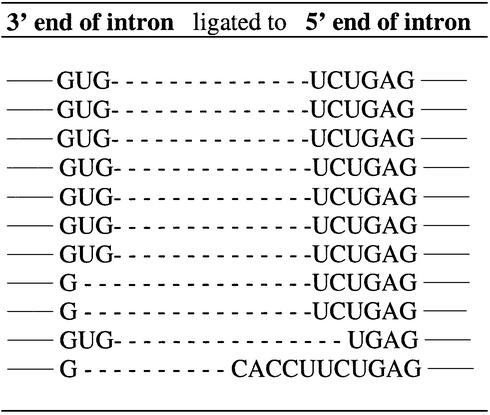
The proportion of substrates that form reverse cyclization products relative to those that form suicide inhibition products increases at higher MgCl2 concentrations (23; see also Fig. 2), perhaps because high salt favors intron cyclization (10,33). Therefore, the sequence specificity of reverse cyclization was analyzed at 15 mM MgCl2. The results show that the native sequence occurs 58% of the time (with 42% unique sequences). Apparently, the reverse cyclization reaction at 15 mM MgCl2 shows a substantially higher preference for the native 5′ exon mimic sequence than the suicide inhibition reaction at either 4 (22%) or 15 mM (13%) MgCl2. The results also show that the 5′ exon mimics ligate to at least four different positions at the 5′ end of the intron (compare results of Table 2 with Fig. 1). Evidently, multiple routes of reverse cyclization occur. It is possible that this stems from the prior formation of multiple cyclized intron intermediates.
Table 2. Reverse cyclization products.
(A) Reverse cyclization products (written 5′→3′) of the combinatorial assay at 15 mM MgCl2 (24 total). The dashed lines represent a single phosphodiester bond. The solid lines represent nucleobase continuation within the intron. The numbering system for exon substrate positions is indicated above the sequences. Note that position –1 is RNA and was not randomized, while positions –2 through to –6 are DNA and were randomized. (B) Sequence of the IGS of the rP-h intron (and the rP-8/4x ribozyme). (C) The percent occurrence of each nucleobase at each exon position.
Identification of cyclization intermediates
Amplification of the reverse cyclization reaction mixture with RT–PCR primers Pr2 and Pr4 resulted in a product whose sequence shows the identity of the intron’s cyclization junction. Table 3 shows the sequences of the cyclization junctions for 11 product sequences. There are two Gs at the 3′ end of the intron (Fig. 1D, shown in white lettering) that can each act as nucleophiles in the cyclization process. In addition, there are at least three different 5′ intron positions (Fig. 1D, shown in black lettering in dark gray background) that are the acceptor sites for the two possible G nucleophiles. Apparently, there are multiple routes to cyclization, which ultimately lead to a mixture of reverse cyclization products. Similar results arise from a Tetrahymena group I intron (4,26,33).
Table 3. Cyclization of the rP-h intron.
Shown are the cyclization junctions, which encompass the 3′ end and the 5′ end of the intron ligated together. The sequences are written in the 5′→3′ direction. The dashed lines represent a single phosphodiester bond. The solid lines represent nucleobase continuation within the intron.
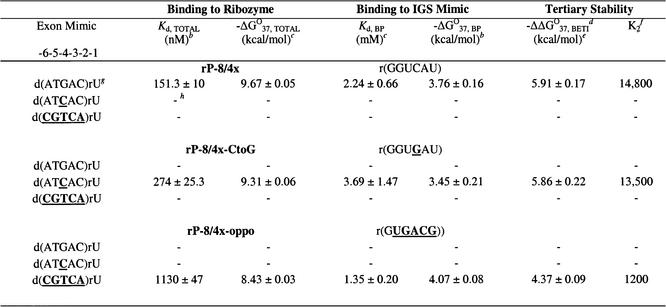
Binding of representative 5′ exon mimics to the rP-8/4x ribozyme
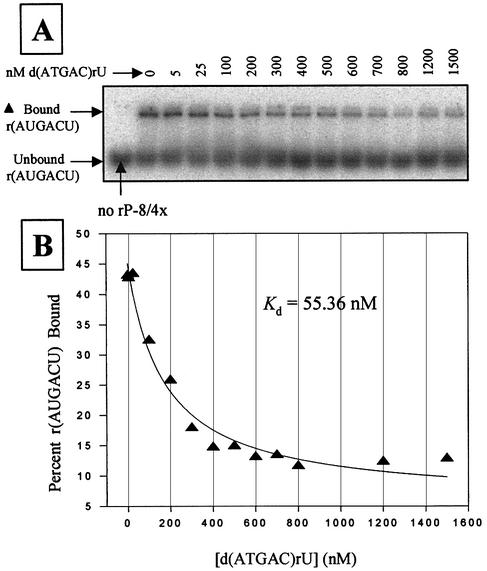
Representative 5′ exon mimics (most sequences were found in the combinatorial assays) were chemically synthesized and their binding strengths to the intron were approximated using competitive band-shift electrophoresis assays in H15Mg buffer with the rP-8/4x ribozyme. Quantification of binding in H4Mg buffer using this assay was not possible because binding was below detection limits (Kd >1.5 µM). The competitive binding assays (Fig. 3 and Table 4) reveal that the hexameric 5′ exon mimics either bind the ribozyme with a Kd <200 nM or they are above the detection limits. Furthermore, addition of the primer-binding region to an exon results in a 2-fold reduction in overall binding. The results also show that a wobble base pair at positions –2, –3 or –6 (exon mimics 6, 5 and 3 in Table 4) changes the Kd at most by ∼2-fold compared with the complementary sequence (exon mimic 1). Therefore, it appears that wobble base pairs, where possible, are well tolerated in terms of overall binding strength.
Figure 3.
Competitive binding assay with d(ATGAC)rU in 50 mM HEPES (25 mM Na+), 15 mM MgCl2 and 135 mM KCl at pH 7.5. (A) A native polyacrylamide gel showing the partitioning of bound and free radiolabeled r(AUGACU) as a function of non-radiolabeled d(ATGAC)rU concentration. Reactions utilized 30 nM rP-8/4x ribozyme, 1 nM radiolabeled 5′ exon mimic r(AUGACU), and 5–1500 nM of the competitor d(ATGAC)rU. (B) Plot and curve-fit of the data in (A). Note that the results in Table 4 are an average of two independent assays.
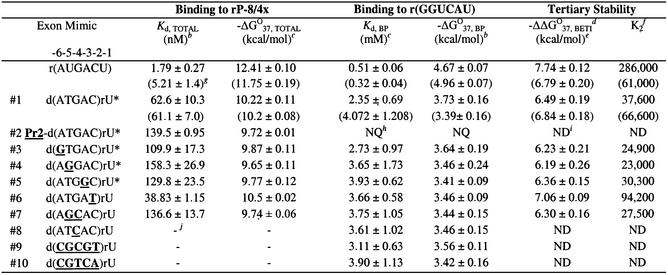
Table 4. hermodynamic parameters for representative hexamers binding to rP-8/4x and to r(GGUCAU) in H15Mg buffera.
aH15Mg buffer consists of 50 mM HEPES (25 mM Na+), 15 mM MgCl2 and 135 mM KCl at pH 7.5. Nucleobases that are bold and underlined represent deviations from the native 5′ exon sequence d(ATGAC)rU. An asterisk (*) indicates oligonucleotides that occur in the combinatorial assay.
bKd, TOTAL was measured by a competition band-shift electrophoresis assay and –ΔG°37, BP was measured by thermal denaturation analysis. The error is the standard deviation of the measurements.
cCalculated from –ΔG°37 = RTln(Kd) where R = 0.001987 kcal mol–1 K–1 and T = 310 K, using more significant digits than listed in this table.
dBETI represents Binding Enhancement by Tertiary Interactions.
eFree energy increment from tertiary interactions calculated from the difference in –ΔG°37 values [(–ΔG°37, TOTAL) – (–ΔG°37, BP)]. The –ΔΔG°37, BETI error was calculated from the square root of the sum of the squares of each individual –ΔG°37 error.
fThe K2 values were calculated by dividing Kd, BP by Kd, TOTAL, using Kd values containing more significant digits than those listed in this table.
gValues in parentheses are from Testa et al. (6).
hValues were not quantifiable (NQ) due to multiphasic transitions.
iND represents values that are not determinable.
jA dash (–) indicates no measurable binding (Kd > 1.5 µM).
A mispair at position –4 of the exon (exon mimic 8) forms a dC–rC pair with the IGS and prohibits binding. From the combinatorial assay, in every exon where a mutation occurs at position –4, there is a T to G mutation at position –5. As seen in Table 4, this double mutant (exon mimic 7) binds the ribozyme reasonably tightly (Kd = 137 nM), which is not due to the single G mutation (exon mimic 4) at position –5 (Kd = 158 nM). Apparently, poor binding due to mismatch formation at position –4 can be rescued by the addition of a specific mutation at position –5.
Base pairing of representative 5′ exon mimics with the IGS, r(GGUCAU)
The stability of the representative 5′ exon mimics base pairing with the IGS of the P.carinii ribozyme was approximated (29) by thermal denaturation analysis with the IGS mimic, r(GGUCAU), in H15Mg buffer. The results of at least two experimental trials (Table 4) indicate that the thermodynamic stabilities (–ΔG°37, BP) of the resultant hybrid helices are very similar, although significantly less than the all RNA helix. The most thermodynamically stable hybrid helix is formed with d(ATGAC)rU, which is the complementary 5′ exon sequence to the IGS. Helices containing one or more mismatches are only at most 0.3 kcal/mol less stable than the native exon hybrid helix, which translates into a 2-fold decrease in base pairing strength. Therefore, none of the mismatched helices studied completely disrupts the ability of the exon mimics to base pair with the IGS. This indicates that exon mimics that do not bind to the ribozyme do so because of an inability to form critical tertiary interactions with the ribozyme.
Kinetics of representative 5′ exon mimics reacting with the rP-h intron
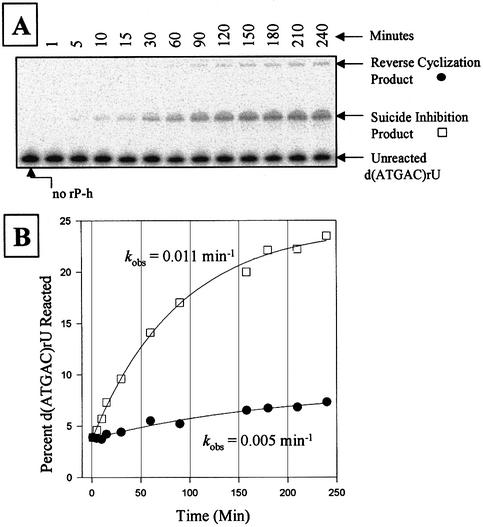
The combinatorial assay developed in this report exploits the ability of each of the 5′ exon mimics to not just bind, but also to react with the intron. Therefore, the observed rate constant, kobs, of each representative oligonucleotide forming suicide inhibition and reverse cyclization products at 4 and 15 mM MgCl2 was measured under single-turnover ‘intron-excess’ conditions. Figure 4 shows a typical gel and graph using the 5′ exon mimic d(ATGAC)rU. Note that each individual assay produces both suicide inhibition and reverse cyclization products. Thus, the extents of reactions for these two products are not altogether independent of each other. The results are shown in Table 5. The observed rate constants are less for the chimeras than the all RNA mimic, indicating that the chimeras are less effective substrates in these reactions. For comparison, the kobs for the suicide inhibition and reverse cyclization reactions using r(AUGACU) are 5–6-fold less than that for the self-splicing reaction under the same conditions (6). For the suicide inhibition reaction using the chimeras, the magnitude of kobs changes <3-fold as a function of either sequence or MgCl2 concentration, at least for those assays that were quantifiable. For the reverse cyclization reaction using the chimeras, the magnitude of kobs changes <3-fold as a function of sequence, but changes up to 7-fold as a function of MgCl2 concentration. Therefore, for the chimeras, increasing the MgCl2 concentration differentially enhances the reverse cyclization reaction relative to the suicide inhibition reaction. Note that the presence of the elongated P2 region disrupts reactivity under the conditions used. This is not problematic in the combinatorial assays, however, as a higher exon mimic concentration was used and the final products were ultimately RT–PCR amplified.
Figure 4.
Kinetic analysis of suicide inhibition and reverse cyclization reactions using 500 nM rP-h and 1 nM radiolabeled d(ATGAC)rU in 50 mM HEPES (25 mM Na+), 4 mM MgCl2 and 135 mM KCl at pH 7.5. (A) A polyacrylamide gel showing a time dependence assay of a typical suicide inhibition and reverse cyclization reaction, using d(ATGAC)rU as the substrate. Note that both reactions occur simultaneously. (B) Graph showing the plot and curve-fit for the suicide inhibition (open square) and reverse cyclization reactions (solid circle) in (A). Note that the results in the Table 5 are an average of two independent assays.
Table 5. Kinetics of representative exons in the suicide inhibition and reverse cyclization reactions as a function of MgCl2 concentrationa.
aBuffers consist of 50 mM HEPES (25 mM Na+), 4 or 15 mM MgCl2 and 135 mM KCl at pH 7.5. Nucleobases that are bold and underlined represent deviations from the native 5′ exon sequence d(ATGAC)rU. An asterisk (*) indicates oligonucleotides that occur in the combinatorial assay.
bThe error is the standard deviation of the measurements.
cThe extent of reaction for the suicide inhibition reactions are reported relative to that obtained with r(AUGACU) at 4 mM MgCl2 (actual extent of reaction = 32%). Similarly, the extent of reaction for the reverse cyclization reactions are reported relative to that obtained with r(AUGACU) at 15 mM MgCl2 (actual extent of reaction = 17%). The relative extent of reaction is calculated by dividing the actual extent of reaction for each oligonucleotide by that for r(AUGACU) and multiplying by 100.
dA dash (–) indicates no measurable activity.
eNQ represents measurements that are not quantifiable due to inconsistent results with low product formation.
Analysis of the sequence requirements at the –4 position of the 5′ exon
Our results suggest that there is a critical requirement for a base pair at position –4. To directly test whether a G(exon)–C(IGS) pair at exon position –4 is required [in contrast to a C(exon)–G(IGS) pair], a ribozyme (rP-8/4x-CtoG) was synthesized with a G in position –4 of the IGS (instead of a C) and its binding to the exon mimics d(ATGAC)rU and d(ATCAC)rU was analyzed (Table 6). Apparently, a C(exon)–G(IGS) base pair at position –4 does permit substrate binding, although binding strength decreases 2-fold relative to the native G(exon)–C(IGS) base pair (274 versus 151 nM). This decrease is entirely at the level of base pairing, and thus does not result from a loss of tertiary interactions. Therefore, although the IGS can be changed to alter the sequence specificity of the ribozyme, certain types of base pairs at certain positions are thermodynamically favored over other base pairs. In addition, the 5′ exon mimic d(ATCAC)rU does not bind to the ribozyme with the native IGS sequence (which forms a dC–rC mismatch at position –4) and the native 5′ exon mimic d(ATGAC)rU does not bind the rP-8/4x-CtoG ribozyme (which forms a dG-rG mismatch at position –4). Apparently in these two later cases the P1 helix loses the ability to dock into the catalytic core of the ribozyme, most likely through disruption of base pair specific tertiary interactions.
Table 6. Thermodynamic parameters for representative exon mimics binding to ribozymes and to IGS mimics in H15Mg buffera.
aH15Mg buffer consists of 50 mM HEPES (25 mM Na+), 15 mM MgCl2 and 135 mM KCl at pH 7.5. Nucleobases that are bold and underlined represent deviations from the native 5′ exon sequence d(ATGAC)rU.
bKd, TOTAL was measured by a direct band-shift electrophoresis assay with r(AUGACU) and –ΔG°37, BP was measured by thermal denaturation analysis. The error is the standard deviation of the measurements.
cCalculated from –ΔG°37 = RTln(Kd) where R = 0.001987 kcal mol–1 K–1 and T = 310 K, using more significant digits than listed in this table.
dBETI represents Binding Enhancement by Tertiary Interactions.
eFree energy increment from tertiary interactions calculated from the difference in –ΔG°37 values [(–ΔG°37, TOTAL) – (–ΔG°37, BP)]. The –ΔΔG°37, BETI error was calculated from the square root of the sum of the squares of each individual –ΔG°37 error.
fThe K2 values were calculated by dividing Kd, BP by Kd, TOTAL, using Kd values containing more significant digits than those listed in this table.
gDissociation constants (Kds) obtained through direct band-shift assays are typically 2.5 times larger than those obtained through the competitive band-shift assay (6,53).
hA dash (–) indicates no measurable binding (Kd > 1.5 µM).
As a preliminary test to see how much the IGS can be altered and still retain complementary exon mimic binding, the rP-8/4x-oppo ribozyme was synthesized. In this ribozyme, every purine of a C–G pair in the IGS is replaced by a pyrimidine of a U–A pair, and vice versa (except for the conserved terminal U-G pair). Results in Table 6 show that the exon mimic d(CGTCA)rU does bind to this ribozyme, although the strength of binding is reduced ∼8-fold (1130 versus 151 nM) compared with the native sequences. Interestingly, this decrease is entirely due to a loss of tertiary interactions, as the base pairing stability of the resultant 5′ exon–IGS helix increases (Table 4). This exon mimic does not bind the native 5′ IGS of rP-8/4x, nor did the native 5′ exon mimic bind the modified rP-8/4x-oppo ribozyme. Therefore, although changing the 5′ IGS can result in a change in the sequence that the ribozyme targets, this change can result in a drastic reduction in the binding strength of the exon mimic– catalytic RNA interaction (even when using complementary 5′ exons). Also note that these results show that a dT–rA base pair at position –4 is tolerated.
DISCUSSION
In this report we show that 5′ exon mimic chimeras can contain normal phosphodiester backbones and still be substrates in the suicide inhibition and reverse cyclization reactions. These non-modified chimeras, however, are less effective substrates in the suicide inhibition and reverse cyclization reaction, as compared with the N3′→P5′ phosphoramidates used previously (23,28). This is not unexpected, as the phosphoramidate-containing duplexes are significantly more thermodynamically stable than the unmodified chimera-containing duplexes (31). In addition, the modified duplexes are expected to more closely resemble A-form RNA duplexes as compared with the unmodified duplexes (31), thus maintaining helix geometry more amenable to tertiary interactions.
The combinatorial approach developed in this work identifies substrate sequences that are able to effectively compete with a pool of all available sequences for being a substrate in the suicide inhibition and reverse cyclization reactions. The resultant sequences, therefore, are not a comprehensive list of all possible substrates, but rather those most able to compete for the 5′ IGS binding site for subsequent reactivity. Note, however, that this assay can detect very poor substrates as well, albeit rarely, as long as the rate of substrate dissociation is not substantially faster than the rate of chemistry.
For those exon mimics that were obtained in the combinatorial assay, we could find no correlation between the number of times the exon mimics occur and their observed rate constants. This indicates that the rate of chemistry is likely not the limiting factor in determining how often the 5′ exon mimics occur (for those that are obtained in the assay). The number of occurrences of each exon mimic appears to correlate somewhat with the binding strength of the mimics with the ribozyme. That the correlation is weak perhaps stems from limitations of the binding assay itself. Analyzing exon mimic binding using the ribozyme approximates the actual binding that occurs when the exon mimic has to compete with the endogenous 5′ exon in the combinatorial assay. Note that we do have one example of a representative exon mimic, d(AGCAC)rU, that binds the ribozyme but does not react in the assay. Therefore, it is expected that either the rate of chemistry or the strength of substrate binding can be a limiting factor in determining whether a particular exon mimic is detected in the combinatorial assay.
Sequence specificity of suicide inhibition
As expected, the native 5′ exon sequence, d(ATGAC)rU, occurred most often in the suicide inhibition reaction at 4 and 15 mM MgCl2, indicating that the native sequence is the most effective suicide inhibitor and that the reactions were run under reasonably stringent conditions. However, a number of other sequences are also permitted. In fact, all five randomized exon positions tolerate nucleobases not present in the native 5′ exon. In general, the requirement for even moderate sequence specificity decreases as you get further away from the reaction site (position –1 of the 5′ exon), with the specificity decreasing considerably at positions –5 and –6. In addition, it appears that a wobble pair is an acceptable substitute for Watson–Crick base pairs at positions –2 and –3. The binding data (Table 4) and reactivity data (Table 5) using representative hexamers show that the 5′ exon sequences obtained in the combinatorial assay are indeed specific substrates in the suicide inhibition reaction. Therefore, similar to the specificity of trans-splicing and reverse splicing reactions for Tetrahymena ribozymes (21,34–37), the sequence specificity of the suicide inhibition reaction for the P.carinii intron is not high.
The physiological environment within which the group I intron-catalyzed self-splicing reaction takes place is largely unknown. Reports indicate that the amount of free magnesium in a cell is at or below 1 mM (38,39). We have shown previously that this is not enough magnesium for the intron to fold and catalyze the self-splicing reaction in vitro (6,23). There is evidence, however, that proteins can aid in the folding of introns (40), which diminishes the need for a high salt environment in the cell. In our assays, a greater diversity of sequences (and of positional mismatches) is permitted at higher salt. One possible explanation for this is that the differential stability of the various 5′ exon mimic–IGS helices decreases at increased MgCl2 concentration. Extrapolating these results to physiological concentrations of MgCl2 suggests that a low salt environment (perhaps containing RNA folding chaperones) would naturally facilitate IGS-mediated reactions with higher sequence specificity than reported here.
It has been noted that one possible way to enhance specificity is by weakening the strength of the 5′exon–IGS interaction, for example by mismatch incorporation, such that the discrimination threshold increases between similar sequences (32,41). As we see here, mismatch incorporation does not always lead to weaker binding [compare d(ATGAT)rU and d(ATGAC)rU in Table 4]. Nevertheless, the MgCl2 concentration-dependent studies do appear to confirm that weaker binding (at 4 mM relative to 15 mM MgCl2) results in increased specificity.
Sequence specificity of reverse cyclization
Using the combinatorial approach, we have shown that the reverse cyclization product is actually a combination of products. The sequence dependence of the reverse cyclization reaction was of interest because it was shown previously with a Tetrahymena intron system that reverse cyclization substrates, which are also 5′ exon mimics, can be as short as a dinucleotide, indicating that the 5′ exon mimics might base pair with only relatively short regions of the IGS (11,26,27). The results (Table 2) for the P.carinii intron show there is a functional advantage to having all or most of each reverse cyclization substrate complementary to the corresponding IGS region. In addition, the sequence specificity for the suicide inhibition and the reverse cyclization reactions are substantially dissimilar. For example, 58% of the substrates in the reverse cyclization reaction are the native 5′ exon sequence, while in the suicide inhibition reaction just 13% of the substrates are the native sequence (each at 15 mM MgCl2). In addition, unlike suicide inhibition substrates, reverse cyclization substrates do not require a dG at either position –4 or –5 of the exon. One possible explanation for this is that complementary base pairing at position –4 (and other positions) is beneficial in the reverse cyclization reaction because it enhances base pairing stability, while in the suicide inhibition reaction this position is additionally involved in critical tertiary interactions. Apparently, factors that govern the molecular recognition of the IGS for the 5′ exon in the reverse cyclization and suicide inhibition reactions are not identical, indicating a dynamic use of the IGS between the two reactions.
Molecular recognition
In all assays conducted, the permissibility of non-pairing mismatches increases the farther away from the reactivity center (exon position –1) that the mismatch occurs, with a marked decrease after position –4. This supports previous experimental data that show nearly all of the tertiary stability of this molecular interaction occurs within the four 3′ bases of the 5′ exon–IGS interaction (28). Thus, the occurrence of base pairs at positions –5 and –6 may serve to simply increase the stability of the P1 helix in preparation for subsequent tertiary interactions during the docking stage(s) of folding. In addition, replacing Watson–Crick base pairs with wobble base pairs at positions –2, –3 and –6 does not significantly interfere with the overall binding of the mimic to the catalytic core of the intron. This was not anticipated because wobble pairs have some different accessible functional groups for tertiary interactions than Watson–Crick pairs (42). Apparently, these structural differences at these positions do not alter the ability of the resultant P1 helices to form tertiary interactions with the catalytic core of this intron.
The decrease in occurrence of dG at exon position –4 in the suicide inhibition assay as the MgCl2 concentration increases directly correlates with an increase in the occurrence of dG at position –5. The binding assays, where a dG at position –5 rescues lost binding that occurs in the absence of dG at position –4, shows that this is not a random trend. In addition, P1 helices with dC–rC or dG–rG mismatches at position –4 are not able to stably dock into the catalytic core of the ribozyme. This suggests that there is an absolute requirement for a base pair at the –4 exon–IGS position. In cases where positional rescue of binding occurs, it is likely that the dG at position –5 base pairs with the corresponding position –4 of the IGS, leaving the nucleobase at exon position –4 as a single nucleotide bulge in the P1 helix (Fig. 5). The overall stability that results from having a base pair at position –4 appears to be strong enough to dominate the disruption of base pairs at exon positions –3 and –5. It is interesting to note that the corresponding position in an Azoarcus intron likely participates in a structurally important A-minor tertiary motif (43). Perhaps in support of this, A-minor motifs appear to thermodynamically favor canonical base pairs over base pair mismatches (44), which is the same trend we see at the –4 position.
Figure 5.
Schematics of 5′ exon mimics base pairing with the IGS of the P.carinii ribozyme. (a) The native 5′ exon, (b) the 5′ exon with a single G to C mutation at exon position –4 and (c) the 5′ exon with a G to C mutation at exon position –4 and a T to G mutation at exon position –5. The Kd values given are for the respective 5′ exon mimics binding to the IGS through base pairing and tertiary interactions (in 15 mM MgCl2). Exon positions –2 through to –6 are deoxyribonucleotides. Note that the double mutant (c) rescues binding lost with the single mutant (b).
Interestingly, the exon mimic d(AGCAC)rU does not react in the suicide inhibition assays (exon mimic 7 in Table 5), but does bind the ribozyme (exon mimic 7 in Table 4). Apparently, binding alone is not enough to specify reactivity, as local and global structure of the P1 helix also plays a role in determining reactivity (45). This is further supported with the 5′ exon mimic d(ATGAT)rU (exon mimic 6), which binds very tightly to the ribozyme, but its extent of reaction is relatively low.
We show that IGS-modified P.carinii ribozymes do favor binding to 5′ exon mimics that are complementary to their particular IGS. However, the strength of the overall binding of the exon to the ribozyme can change considerably even though the 5′ exon–IGS helices are complementary at exon positions –2 through to –6. In the case of the rP-8/4x-oppo ribozyme, this change in overall binding stability is primarily due to a reduction in tertiary stability (∼1.5 kcal/mol). This indicates that the ribozyme likely forms tertiary interactions with only specific base pairs at certain P1 helix positions, in contrast to that reported with the Tetrahymena ribozyme (45,46). One possible source of base pair specific tertiary interactions is Type I A-minor motifs (47), although there is no direct evidence that such a motif occurs here.
It does not appear that the large sequence diversity of 5′ exons obtained in the combinatorial suicide inhibition and reverse cyclization assays can be explained by the 5′ exons misaligning (or slipping) on the IGS (48,49), although in a few cases it could be occurring. The different cyclization products obtained (and hence different sites of reverse cyclization), however, can be explained by different regions of the 5′ end of the spliced intron base pairing with the IGS (Fig. 1). Note that the cyclization reaction is similar to the self-splicing reaction in that the reaction substrates bind to the IGS (50). The forward and reverse cyclization data reveal that in three cases (Fig. 6) the cyclization sites in the exons contain three contiguous pyrimidines that possibly pair with the G-rich end of the IGS. In these cases the cyclization junction is likely to be situated along the IGS at the same position as the self-splicing junction. The positioning of the exon cyclization junction along the IGS is not obvious for the fourth case, and so may not utilize the same positioning as the self-splicing junction. The data also reveal that substrate slipping occurs in the P9.0 helix, as either of the last two guanosines can act as cyclization nucleophiles. This is possible because either of these guanosines can bind the G-binding site, while upstream bases are able to form a P9.0 helix. Implicit then is the possibility that either guanosine can act as the terminal guanosine of the intron during the second step of self-splicing under the reaction conditions used here.
Figure 6.
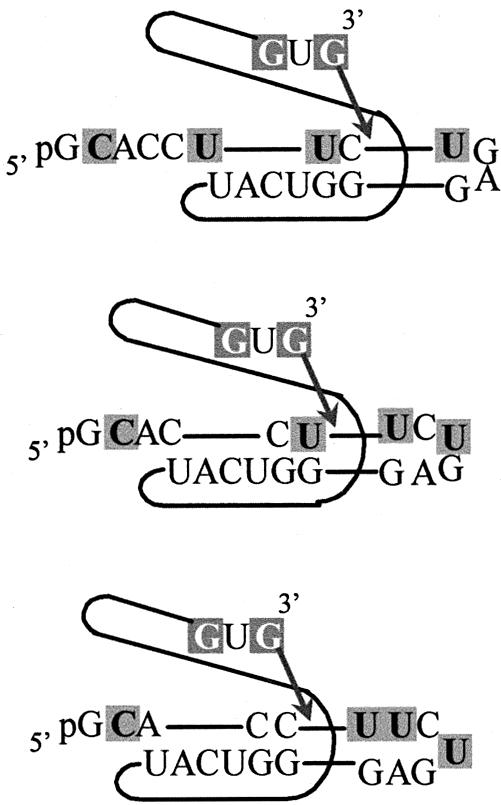
Schematics of the 5′ end of the intron base pairing with the IGS before cyclization. These are three possible routes for the 5′ end of the intron binding to the G-rich region of the IGS that are consistent with the data. The cyclization reaction is similar to the self-splicing reaction with the substrate binding to the IGS, allowing the free 3′ OH on the 3′ G to attack at the –1 exon position.
All of the above results show that the combinatorial assay developed in this report has proven useful for gaining a more fundamental understanding of the molecular recognition properties of IGS-mediated reactions.
Implications
Previous reports have shown that oligonucleotides that mimic the 5′ exon of the P.carinii rRNA group I intron are suicide inhibitors of the self-splicing reaction in vitro (23). Such inhibitors are novel in that they specifically exploit the tertiary interactions of the structured RNA. This was coined Binding Enhancement by Tertiary Interactions (BETI) (25,35). This strategy was developed to potentially alleviate the problem of non-specific binding, which appears to be a factor in using conventional antisense approaches (41,51,52). As a general rule, these results show that BETI (and more specifically suicide inhibition) likely will suffer from the same problem, although perhaps to a lesser degree: mismatches can be tolerated when short helices are stabilized by tertiary interactions (unless the mismatches severely disrupt tertiary interactions).
RNA targeting strategies such as BETI (25,35) and mRNA repair (15,22) rely on tertiary interaction formation between the catalytic RNA enzyme and its substrate (after base pairing). This report shows that the strength of these tertiary interactions can depend on the presence of certain base pairs at certain positions along the catalytic RNA–substrate helix. This could limit the choice of effective targets that the catalytic RNA can act on. It could also, however, increase the specificity of the catalytic RNA, as the strength of the catalytic RNAs binding to similar, but undesired targets could be decreased.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Kylie Sutter for technical assistance. This work was supported by The Kentucky Research Challenge Trust Fund, an award from The Research Corporation, and the Muscular Dystrophy Association.
REFERENCES
- 1.Kruger K., Grabowski,P.J., Zaug,A.J., Sands,J., Gottschling,D.E. and Cech,T.R. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell, 31, 147–157. [DOI] [PubMed] [Google Scholar]
- 2.Zaug A.J., Grabowski,P.J. and Cech,T.R. (1983) Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature, 301, 578–583. [DOI] [PubMed] [Google Scholar]
- 3.Zaug A.J., Kent,J.R. and Cech,T.R. (1984) A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science, 224, 574–578. [DOI] [PubMed] [Google Scholar]
- 4.Zaug A.J., Been,M.D. and Cech,T.R. (1986) The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature, 324, 429–433. [DOI] [PubMed] [Google Scholar]
- 5.Zaug A.J. and Cech,T.R. (1986) The intervening sequence RNA of Tetrahymena is an enzyme. Science, 231, 470–475. [DOI] [PubMed] [Google Scholar]
- 6.Testa S.M., Haidaris,C.G., Gigliotti,F. and Turner,D.H. (1997) A Pneumocystis carinii group I intron ribozyme that does not require 2′ OH groups on its 5′ exon mimic for binding to the catalytic core. Biochemistry, 36, 15303–15314. [DOI] [PubMed] [Google Scholar]
- 7.Bevilacqua P.C., Kierzek,R., Johnson,K.A. and Turner,D.H. (1992) Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science, 258, 1355–1358. [DOI] [PubMed] [Google Scholar]
- 8.Herschlag D. (1992) Evidence for processivity and two-step binding of the RNA substrate from studies of J1/2 mutants of the Tetrahymena ribozyme. Biochemistry, 31, 1386–1399. [DOI] [PubMed] [Google Scholar]
- 9.Waring R.B., Scazzocchio,C., Brown,T.A. and Davies,R.W. (1983) Close relationship between certain nuclear and mitochondrial introns. Implications for the mechanism of RNA splicing. J. Mol. Biol., 167, 595–605. [DOI] [PubMed] [Google Scholar]
- 10.Been M.D. and Cech,T.R. (1986) One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell, 47, 207–216. [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto N., Tomka,M., Kierzek,R., Bevilacqua,P.C. and Turner,D.H. (1989) Effects of substrate structure on the kinetics of circle opening reactions of the self-splicing intervening sequence from Tetrahymena thermophila: evidence for substrate and Mg2+ binding interactions. Nucleic Acids Res., 17, 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pyle A.M. and Cech,T.R. (1991) Ribozyme recognition of RNA by tertiary interactions with specific ribose 2′-OH groups. Nature, 350, 628–631. [DOI] [PubMed] [Google Scholar]
- 13.Bevilacqua P.C. and Turner,D.H. (1991) Comparison of binding of mixed ribose-deoxyribose analogues of CUCU to a ribozyme and to GGAGAA by equilibrium dialysis: evidence for ribozyme specific interactions with 2′ OH groups. Biochemistry, 30, 10632–10640. [DOI] [PubMed] [Google Scholar]
- 14.Murphy F.L. and Cech,T.R. (1989) Alteration of substrate specificity for the endoribonucleolytic cleavage of RNA by the Tetrahymena ribozyme. Proc. Natl Acad. Sci. USA, 86, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullenger B.A. and Cech,T.R. (1994) Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing. Nature, 371, 619–622. [DOI] [PubMed] [Google Scholar]
- 16.Sargueil B. and Tanner,N.K. (1993) A shortened form of the Tetrahymena thermophila group I intron can catalyze the complete splicing reaction in trans. J. Mol. Biol., 233, 629–643. [DOI] [PubMed] [Google Scholar]
- 17.Lan N., Howrey,R.P., Lee,S.W., Smith,C.A. and Sullenger,B.A. (1998) Ribozyme-mediated repair of sickle beta-globin mRNAs in erythrocyte precursors. Science, 280, 1593–1596. [DOI] [PubMed] [Google Scholar]
- 18.Phylactou L.A., Darrah,C. and Wood,M.J. (1998) Ribozyme-mediated trans-splicing of a trinucleotide repeat. Nature Genet., 18, 378–381. [DOI] [PubMed] [Google Scholar]
- 19.Kohler U., Ayre,B.G., Goodman,H.M. and Haseloff,J. (1999) Trans-splicing ribozymes for targeted gene delivery. J. Mol. Biol., 285, 1935–1950. [DOI] [PubMed] [Google Scholar]
- 20.Zarrinkar P.P. and Sullenger,B.A. (1999) Optimizing the substrate specificity of a group I intron ribozyme. Biochemistry, 38, 3426–3432. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe T. and Sullenger,B.A. (2000) Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc. Natl Acad. Sci. USA, 97, 8490–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell M.A., Johnson,A.K. and Testa,S.M. (2002) Ribozyme-catalyzed excision of targeted sequences from within RNAs. Biochemistry, 41, 15327–15333. [DOI] [PubMed] [Google Scholar]
- 23.Testa S.M., Gryaznov,S.M. and Turner,D.H. (1999) In vitro suicide inhibition of self-splicing of a group I intron from Pneumocystis carinii by an N3′→P5′ phosphoramidate hexanucleotide. Proc. Natl Acad. Sci. USA, 96, 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Disney M.D., Testa,S.M. and Turner,D.H. (2000) Targeting a Pneumocystis carinii group I intron with methylphosphonate oligonucleotides: backbone charge is not required for binding or reactivity. Biochemistry, 39, 6991–7000. [DOI] [PubMed] [Google Scholar]
- 25.Disney M.D., Matray,T., Gryaznov,S.M. and Turner,D.H. (2001) Binding enhancement by tertiary interaction and suicide inhibition of a Candida albicans group I intron by phosphoramidate and 2′-O-methyl hexanucleotides. Biochemistry, 40, 6520–6526. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan F.X. and Cech,T.R. (1985) Reversibility of cyclization of the Tetrahymena rRNA intervening sequence: implication for the mechanism of splice site choice. Cell, 42, 639–648. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto N., Kierzek,R. and Turner,D.H. (1988) Kinetics for reaction of a circularized intervening sequence with CU, UCU, CUCU, and CUCUCU: mechanistic implications from the dependence on temperature and on oligomer Mg2+ concentrations. Biochemistry, 27, 6384–6392. [DOI] [PubMed] [Google Scholar]
- 28.Disney M.D., Gryaznov,S.M. and Turner,D.H. (2000) Contributions of individual nucleotides to tertiary binding of substrate by a Pneumocystis carinii group I intron. Biochemistry, 39, 14269–14278. [DOI] [PubMed] [Google Scholar]
- 29.Testa S.M., Gryaznov,S.M. and Turner,D.H. (1998) Antisense binding enhanced by tertiary interactions: binding of phosphorothioate and N3′→P5′ phosphoramidate hexanucleotides to the catalytic core of a group I ribozyme from the mammalian pathogen Pneumocystis carinii. Biochemistry, 37, 9379–9385. [DOI] [PubMed] [Google Scholar]
- 30.Gryaznov S.M. and Letsinger,R.L. (1992) Synthesis and properties of oligonucleotides containing aminodeoxythymidine units. Nucleic Acids Res., 20, 3403–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gryaznov S.M., Lloyd,D.H., Chen,J.K., Schultz,R.G., DeDionisio,L.A., Ratmeyer,L. and Wilson,W.D. (1995) Oligonucleotide N3′→P5′ phosphoramidates. Proc. Natl Acad. Sci. USA, 92, 5798–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaug A.J., Grosshan,C.A. and Cech,T.R. (1988) Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry, 27, 8924–8931. [DOI] [PubMed] [Google Scholar]
- 33.Been M.D. and Cech,T.R. (1987) Selection of circularization sites in a group I IVS RNA requires multiple alignments of an internal template-like sequence. Cell, 50, 951–961. [DOI] [PubMed] [Google Scholar]
- 34.Roman J. and Woodson,S.A. (1995) Reverse splicing of the Tetrahymena IVS: evidence for multiple reaction sites in the 23S rRNA. RNA, 1, 478–490. [PMC free article] [PubMed] [Google Scholar]
- 35.Jones J.T., Lee,S.W. and Sullenger,B.A. (1996) Tagging ribozyme reaction sites to follow trans-splicing in mammalian cells. Nature Med., 2, 643–648. [DOI] [PubMed] [Google Scholar]
- 36.Roman J. and Woodson,S.A. (1998) Integration of the Tetrahymena group I intron into bacterial rRNA by reverse splicing in vivo. Proc. Natl Acad. Sci. USA, 95, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roman J., Rubin,M.N. and Woodson,S.A. (1999) Sequence specificity of in vivo reverse splicing of the Tetrahymena group I intron. RNA, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison P.M. and Hoare,R.J. (1980) Metals in Biochemistry. Chapman & Hall, New York, pp. 8–9.
- 39.Macquire M.E. (1990) In Sigel,H. and Sigel,A (eds), Metals in Biological Systems. Dekker, New York, Vol. 26, pp. 135–153.
- 40.Weeks K.M. and Cech,T.R. (1995) Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell, 82, 221–230. [DOI] [PubMed] [Google Scholar]
- 41.Herschlag D. (1991) Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: more isn’t always better. Proc. Natl Acad. Sci. USA, 88, 6921–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strobel S.A. and Cech,T.R. (1995) Minor groove recognition of the conserved G.U pair at the Tetrahymena ribozyme reaction site. Science, 267, 675–679. [DOI] [PubMed] [Google Scholar]
- 43.Soukup J.K., Minakawa,N., Matsuda,A. and Strobel,S.A. (2002) Identification of A-minor interactions within a bacterial group I intron active site by 3-deazaadenosine interference mapping. Biochemistry, 41, 10426–10438. [DOI] [PubMed] [Google Scholar]
- 44.Battle D.J. and Doudna,J.A. (2002) Specificity of RNA-RNA helix recognition. Proc. Natl Acad. Sci. USA, 99, 11676–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doudna J.A., Cormack,B.P. and Szostak,J.W. (1989) RNA structure, not sequence, determines the 5′ splice-site specificity of a group I intron. Proc. Natl Acad. Sci. USA, 86, 7402–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell T.B. and Cech,T.R. (1996) Mutations in the Tetrahymena ribozyme internal guide sequence: effects on docking of the P1 helix into the catalytic core and correlation with catalytic activity. Biochemistry, 35, 11493–11502. [DOI] [PubMed] [Google Scholar]
- 47.Doherty E.A., Batey,R.T., Masquida,B. and Doudna,J.A. (2001) A universal mode of helix packing in RNA. Nature Struct. Biol., 8, 339–343. [DOI] [PubMed] [Google Scholar]
- 48.Been M.D. and Cech,T.R. (1987) Selection of circularization sites in a group I IVS RNA requires multiple alignments of an internal template-like sequence. Cell, 50, 951–961. [DOI] [PubMed] [Google Scholar]
- 49.Strobel S.A. and Cech,T.R. (1994) Translocation of an RNA duplex on a ribozyme. Nature Struct. Biol., 1, 13–17. [DOI] [PubMed] [Google Scholar]
- 50.Cech T.R. (1990) Self-splicing of group I introns. Annu. Rev. Biochem., 59, 543–568. [DOI] [PubMed] [Google Scholar]
- 51.Roberts R.W. and Crothers,D.M. (1991) Specificity and stringency in DNA triplex formation. Proc. Natl Acad. Sci. USA, 88, 9397–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woolf T.M., Melton,D.A. and Jennings,C.G.B. (1992) Specificity of antisense oligonucleotides in vivo. Proc. Natl Acad. Sci. USA, 89, 7305–7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pyle A.M., McSwiggen,J.A. and Cech,T.R. (1990) Direct measurement of oligonucleotide substrate binding to wild-type and mutant ribozymes from Tetrahymena. Proc. Natl Acad. Sci. USA, 87, 8187–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]