Abstract
The human mitochondrial genome encodes 22 tRNAs interspersed among the two rRNAs and 11 mRNAs, often without spacers, suggesting that tRNAs must be efficiently excised. Numerous maternally transmitted diseases and syndromes arise from mutations in mitochondrial tRNAs, likely due to defect(s) in tRNA metabolism. We have systematically explored the effect of pathogenic mutations on tRNAIle precursor 3′ end maturation in vitro by 3′-tRNase. Strikingly, four pathogenic tRNAIle mutations reduce 3′-tRNase processing efficiency (Vmax / KM) to ∼10-fold below that of wild-type, principally due to lower Vmax. The structural impact of mutations was sought by secondary structure probing and wild-type tRNAIle precursor was found to fold into a canonical cloverleaf. Among the mutant tRNAIle precursors with the greatest 3′ end processing deficiencies, only G4309A displays a secondary structure substantially different from wild-type, with changes in the T domain proximal to the substitution. Reduced efficiency of tRNAIle precursor 3′ end processing, in one case associated with structural perturbations, could thus contribute to human mitochondrial diseases caused by mutant tRNAs.
INTRODUCTION
The human mitochondrial genome encodes 22 tRNAs, one for each of 18 amino acids, and two each for Leu and Ser with different anticodons (1). Over 150 pathogenic mutations have been found in the mitochondrial genome [see Mitomap (2) for a compilation], most of them transitions in tRNA genes. Since the tRNA genes combined constitute only approximately one-tenth of the mitochondrial genome, defects in tRNA function apparently contribute disproportionately to inherited mitochondrial disease (reviewed in 3–5).
tRNAs are interspersed among the other functional mitochondrial RNAs (two rRNAs and 11 mRNAs, which encode 13 polypeptide subunits of the respiratory chain complexes) on long precursor transcripts, often without intergenic RNA. The 3′ end CCA common to all mature tRNAs is not mitochondrially encoded, and must be added to the discriminator base by tRNA nucleotidyltransferase after tRNA precursor 3′ end processing. Punctuation of long mitochondrial transcripts with tRNAs thus suggests a need for precise, efficient endonucleolytic cleavage at both their 5′ and 3′ ends (6,7).
Punctuation of long, multi-functional human mitochondrial transcripts by tRNAs suggests that end processing defects arising from pathogenesis-linked tRNA mutations could contribute to disease. Early results consistent with this model include detection of a linked 16S rRNA-tRNALeu(UUR)-ND1 transcript (RNA 19) in northern blots from cells affected by pathogenesis-associated substitutions in tRNALeu(UUR) (8,9). The idea was further investigated by in vitro processing with culture cell extracts, using wild-type and pathogenic tRNALeu(UUR) and tRNASer(UCN) transcripts (10–12), and less efficient tRNA end processing with some of the pathogenic substitutions was observed.
Human mitochondrial RNase P (10,13), 3′-tRNase (10–12) and tRNA nucleotidyltransferase activities (14,15) have been characterized and, in the latter case, cloned and expressed. 3′-tRNase may be important for maturation of both cytoplasmic and mitochondrial tRNAs (reviewed in 16). CC of CCA at the 3′ end of mature tRNAs is a 3′-tRNase anti-determinant (17), and tRNASer(UCN) with the T7445C {note that two numbering schemes are used when referring to mitochondrial tRNAs: based on position in the mitochondrial genome [the Cambridge system (1)], in which tRNAIle runs from 4263 to 4331 (Figs 2, 4 and 5, and Table 1), and the system for numbering tRNAs from +1 to 76 (18,19) used in Figs 1, 2, 3, 6, S1 and S2} substitution, which causes non-syndromic deafness (20,21), changes the sequence of the precursor from G/UCU to G/CCU following the discriminator base, resembling a 3′-tRNase anti-determinant. 3′-tRNase extracted from human culture cell mitoplasts was shown to endonucleolytically cleave wild-type tRNASer(UCN) in vitro at the expected processing site, and found to be unable to utilize the T7445C mutant tRNASer(UCN) precursor as substrate (12). This study laid the groundwork for a more general investigation of a possible relation between pathology-linked mutations in mitochondrial tRNAs and the ability of their precursors to be 3′ end processed.
Figure 2.
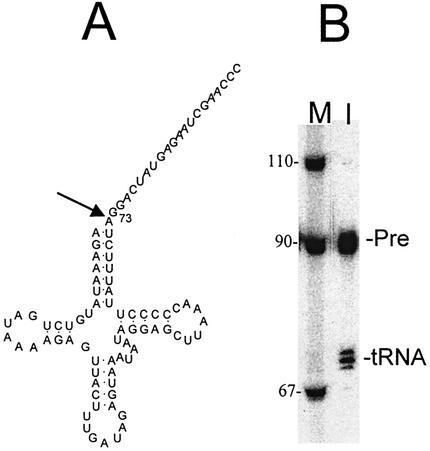
3′ End processing of wild-type tRNAIle precursor. (A) Wild-type tRNAIle precursor with a 20 nt 3′ end trailer used in these processing studies. The arrow following the discriminator base indicates the major 3′-tRNase processing site (after the discriminator base, nucleotide 73). (B) 3′ End processing. Lane M, labeled DNA marker. Lane 1, typical processing reaction using 5′ end labeled precursor. Designations on the right correspond to the tRNA precursor (Pre) and product (tRNA).
Figure 4.
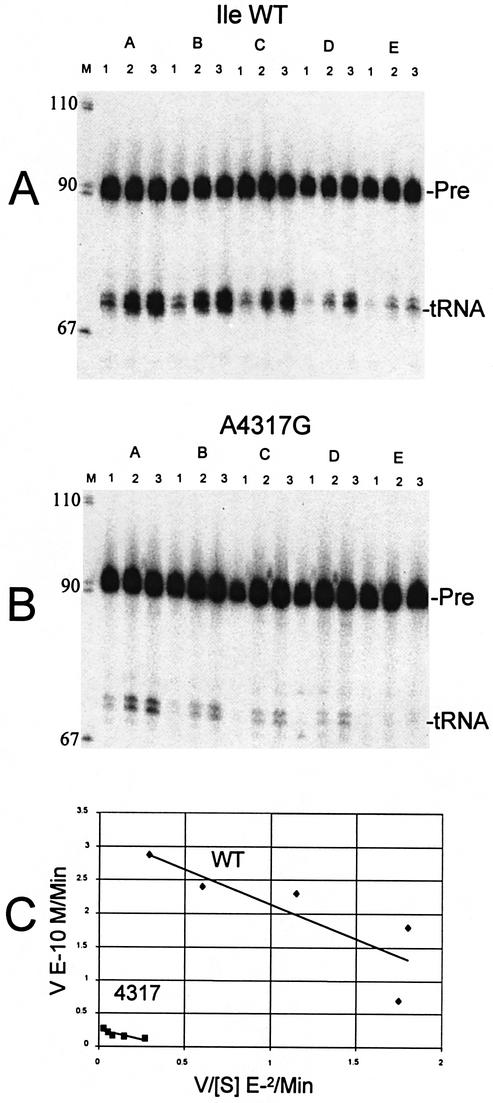
3′-tRNase processing kinetics of wild-type and A4317G precursors. Processing reactions were performed using a series of unlabeled substrate concentrations of 0.4, 1.0, 2.0, 4.0 and 10.0 × 10–8 M in (A) to (E), respectively. Reactions were terminated after 6.6, 13.3 and 20 min of incubation at 37°C in lanes 1–3 at each substrate concentration, respectively. (A) Wild-type; (B) A4317G; (C) Eadie–Hofstee plot of V versus V / [S], in which slope is –KM and V intercept is Vmax. Because input labeled tRNA is constant, the proportion of labeled product obtained per minute of reaction, measured from these gels (A and B), is equivalent to V / [S], which decreases with increasing [S], and V increases as expected with increasing [S]. The reduced processing efficiency of the A4317G mutant relative to wild-type tRNAIle (B) is due principally to reduced Vmax (C). Similar results were obtained with the other mutants (data not shown; Table 1), in which Vmax consistently decreased.
Figure 5.
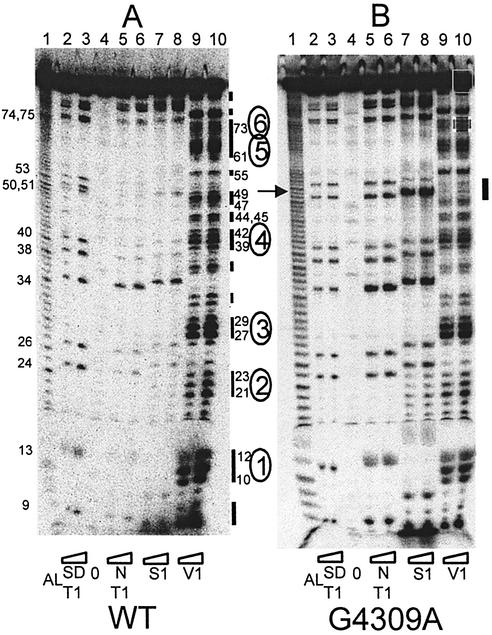
Solution secondary structure probing of wild-type and G4309A tRNAIle. tRNAs [wild-type and G4309A in (A) and (B), respectively] were labeled at their 5′ ends and cleaved with NaHCO3 to produce the alkali ladder (lane 1, designated AL at bottom) and probed with ribonuclease T1 at 1 and 2.5 × 10–3 U/μl under semi-denaturing conditions (lanes 2 and 3, respectively; SD T1). tRNAs re-folded under native conditions (see Materials and Methods) were probed with T1 at 0, 1 and 2.5 × 10–3 U/µl (lanes 4–6; N T1), with ribonuclease S1 at 0.55 and 1.37 U/µl (lanes 7–8) and with ribonuclease V1 at 1 and 2.5 × 10–6 U/µl (lanes 9–10). Numbers on the left designate T1 cleavages at Gs observed in the semi-denaturing T1 lanes. Designations on the right indicate regions and nucleotide numbers corresponding to V1 cleavages, and numbers enclosed in ovals correspond to regions that can be placed on the canonical tRNA structure (see Fig. 6). The arrow on the left and the bar at the right of (B) indicate the position of the G4309A substitution and a corresponding region of altered S1 and V1 sensitivity in this mutant (see Fig. 6).
Table 1. Steady state 3′-tRNase processing kinetics with wild-type and mutant tRNAIle precursors.
| Typea | KM × 10–8 Mb | Vmax × 10–10 M/min | Vmax / KM × 10–2 min–1 | Loss in processing efficiencyc |
|---|---|---|---|---|
| Wild-type | 1.33 ± 0.18 | 2.23 ± 0.23 | 1.68 | 1.0 |
| A4269G (7) | 0.92 ± 0.33 | 0.12 ± 0.004 | 0.13 | 12.5 |
| T4274C (12) | 0.88 ± 0.35 | 0.35 ± 0.05 | 0.40 | 4.2 |
| T4285C (27) | 0.32 ± 0.16 | 0.17 ± 0.03 | 0.53 | 3.2 |
| A4295G (37) | 0.91 ± 0.46 | 0.15 ± 0.03 | 0.16 | 10.3 |
| A4300G (42) | 0.41 ± 0.09 | 0.32 ± 0.04 | 0.78 | 2.1 |
| G4309A (51) | 0.81 ± 0.03 | 0.18 ± 0.01 | 0.22 | 7.7 |
| A4317G (59) | 1.26 ± 0.44 | 0.21 ± 0.03 | 0.17 | 10.0 |
| C4320T (62) | 1.70 ± 0.95 | 0.67 ± 0.20 | 0.39 | 4.2 |
aNumbering of pathogenic mutants is both according to nucleotide position in the mitochondrial genome and in accordance with conventional tRNA numbering (in parentheses).
bStandard error of the mean is indicated. Experiments on mutants were performed at least twice and the wild-type experiment was performed nine times.
cRefers to the factor by which mutant precursor processing efficiency is reduced relative to wild-type.
Figure 1.
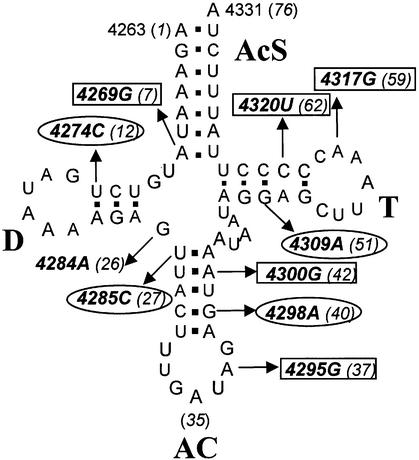
Human mitochondrial tRNAIle secondary structure and pathological substitutions. tRNAIle runs from nucleotides 4263 to 4331 using the Cambridge mitochondrial genome numbering (1). Substitutions are identified in the text by wild-type nucleotide, nucleotide number and mutation, thus A4269G indicates that the wild-type A at position 4269 is replaced by G in the mutant tRNA. Conventional tRNA nucleotide numbering (18,19) is indicated in parentheses. Domain designations (bold) are: AcS, acceptor stem; D, D domain; AC, anticodon domain; T, T domain. Nucleotide 4331 is the discriminator base, the last (unpaired) encoded nucleotide beyond the acceptor stem which is retained after 3′ end maturation, to which CCA is added by tRNA nucleotidyltransferase. Mutations enclosed by rectangles and ovals are those which cause cardiomyopathy and ophthalmoplegeia, respectively. G4284A has a mixed presentation. References for these mutations are listed on the web site www.gen.emory.edu/mitomap.html.
Figure 3.
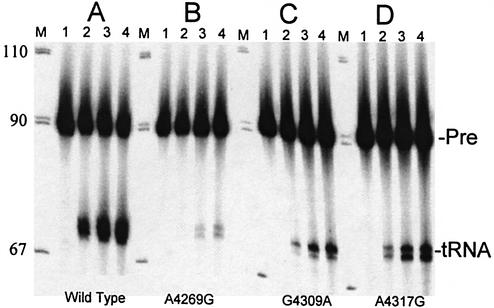
tRNAIle precursor 3′ end processing of wild-type and selected pathological mutants. 5′ End labeled tRNAs were processed without added unlabeled tRNA. (A) Wild-type, (B) A4269G, (C) G4309A, (D) A4317G. Reactions were sampled after 0, 5, 10 and 15 min incubation at 37°C in lanes 1–4 of each set. M is same as in Figure 2.
Figure 6.
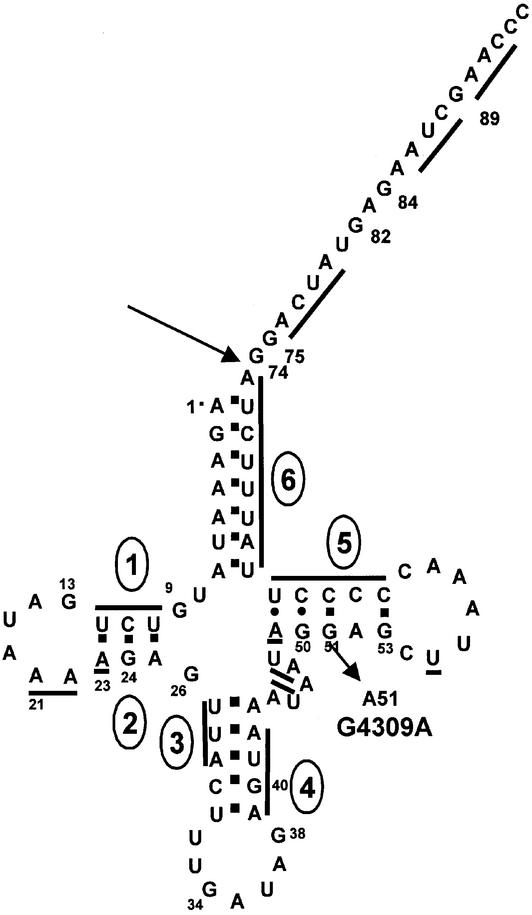
Secondary structure of the tRNAIle precursor. Nucleotides are numbered according to Guan et al. (21) and Degoul et al. (22). Stems designated by lines and numbers enclosed in ovals correspond with the regions of V1 sensitivity designated in Figure 5. The oblique arrow at nucleotide 51 indicates the G4309A substitution.
Among human mitochondrial tRNAs, tRNALeu(UUR), tRNALys and tRNAIle are hot spots, with 21, 10 and 10 known pathogenesis-associated mutations, respectively (2). The effect of mutations in tRNAIle on aminoacylation by mitochondrial isoleucyl-tRNA synthetase has already been analyzed (22–25). Here, we further systematically consider the effects of a number of pathogenesis-linked mutations in tRNAIle on 3′ end maturation by 3′-tRNase and on tRNA structure in solution.
Two pathological presentations are linked to mutations in tRNAIle (Fig. 1): cardiomyopathies (mutations A4269G, A4295G, A4300G, A4317G and C4320T) and ophthalmoplegia (mutations T4274C, T4285C, G4298A and G4309A). For one further mutation, G4284A, a varied familial presentation including both fatal cardiomyopathy and ophthalmoparesis was observed (26).
The positions of the mutations within the tRNA 2D cloverleaf structure (Fig. 1) do not display an obvious pattern with respect to the different tRNA domains, nor is there an evident relationship between the position of each mutation and the disease it causes [the same can generally be said for pathogenesis-associated mutations in human mitochondrial tRNAs (27)]. All mutations are transitions; in stems, most of them convert a Watson–Crick pair into a C·A or A·C mismatch and one mutation (C4320T) converts a mismatch into a Watson–Crick pair.
Here the 3′ end processing of wild-type and variant tRNA-Ile precursors was investigated using a fractionated mitoplast extract, in which interactions could occur between substrate and various factors which may be present. All the pathological substitutions reduce 3′-tRNase processing efficiency in vitro, in several cases by ∼10-fold. Thus, defective tRNA 3′ end processing could contribute to mitochondrial pathology. Interestingly, the G4309A substitution, located close to the A·C mismatch in the wild-type T stem, affects T domain structure, which is generally important for tRNA recognition and processing.
MATERIALS AND METHODS
Preparation of tRNA precursors
A set of plasmids with tRNAIle inserts (wild-type, A4269G, T4274C, T4285C, A4295G, G4298A, A4300G, A4317G and C4320T; Fig. 1) linked at +1 to a hammerhead ribozyme and T7 promoter was a kind gift of Shana Kelley. In these inserts [originally designed to run off at a BstNI site, producing full-length CCA-terminated transcripts (+76) for aminoacylation studies (23)], the CCA (nucleotides 74–76) was replaced by PCR amplification with the first 20 nt of the 3′ end sequence flanking the human mitochondrial tRNAIle precursor (5′-GGACUAUGAGAAUCGAACCC-3′; Fig. 2A), including a SmaI runoff site. Inserts were recloned into the small high copy vector pHC624 and confirmed by DNA sequencing. The G4309A mutant tRNAIle precursor was constructed from wild-type by amplification using a 63 nt reverse primer (from the subcloning site at the 5′ end of the oligonucleotide through sequence complementary to the 3′ end trailer, including the sequence from nucleotide 73 through the substitution at nucleotide 51, and ending with the 3′ end of the oligonucleotide at nucleotide 41).
Unlabeled transcripts were prepared with T7 RNA polymerase as previously described (28), followed by phenol/chloroform deproteinization and ethanol precipitation before hammerhead self-cleavage of tRNAIle at the 5′ side of +1. tRNA precursors were purified from the hammerhead on denaturing 6% polyacrylamide gels, visualized by UV shadowing, extracted from gel slices by diffusion for 30 min at 37°C and recovered by ethanol precipitation, using oyster glycogen as a carrier. Concentration of recovered tRNA was determined by reading A260, using a conversion factor of 975 000 A260 M–1.
RNA labeling
tRNA precursors were labeled at their 5′ ends with T4 polynucleotide kinase and [γ-32P]ATP for 30 min at 37°C using RNasin as a ribonuclease inhibitor, gel purified, visualized by autoradiography, and recovered from the gel as described above. 3′ End labeling was performed using T4 RNA ligase and [α-32P]Cp for 16 h at 4°C and precursors were recovered as described above.
tRNA refolding
Both labeled and unlabeled tRNAs were re-folded by heating in water for 5 min at 75°C and mixing with an equal volume of heated buffer to a final concentration of 25 mM Tris–HCl pH 7.5, 250 mM KCl, 5 mM MgCl2 and 5% glycerol, and allowed to cool to room temperature for 5 min.
3′ End processing
The 3′-tRNase processing extract was prepared from cultured HeLa cell mitoplasts fractionated by DEAE–Sepharose chromatography as previously described (12). Enrichment by anion exchange removes most contaminating RNAs and some contaminating proteins. Attempts to express human 3′-tRNase based on homology with Arabidopsis RNase Z (29) have so far been unsuccessful (L.Levinger, unpublished results).
For Michaelis–Menten kinetics, re-folded unlabeled tRNA precursor covering a concentration range centered on KM (usually 0.4–10 × 10–8 M) was mixed with a fixed amount of 5′ end labeled precursor in a volume of 5 µl and placed at 4°C. Processing reactions were initiated by adding 20 µl of a reaction mix to make final concentrations of 25 mM Tris–HCl pH 7.5, 50 mM KCl, 1 mM MgCl2, 5% glycerol, 3 mM DTT, 40 U/ml RNasin with the 3′-tRNase at a dilution of 1:60, and placing the tubes at 37°C. Samples (5 µl) were taken after 6.6, 13.3 and 20 min of incubation and terminated by adding to 2.5 µl of formamide containing 0.5× TBE, 0.01% bromophenol blue and 0.01% xylene cyanol at 4°C. Products were separated by electrophoresis on 6% denaturing polyacrylamide gels until the xylene cyanol migrated 10 cm above the bottom of the gels. Images were taken from dried gels using a Fuji BAS 2000 phosphorimager and analyzed with MacBas100 software.
Processing efficiency experiments were performed using matched input radiolabeled substrates without added unlabeled tRNA precursors. [S] is known to be much less than KM under these conditions because the tRNA is labeled to high specific activity and no unlabeled tRNA is added. The increase in percentage of product per minute of reaction is thus a direct measure of reaction efficiency (Vmax / KM). Michaelis–Menten kinetics provide a more reliable estimate of reaction efficiency, however, since it is based on the independent determination of KM and Vmax. Enzyme concentration is also expected to be much less than [S], as required by the steady-state assumption of Michaelis–Menten kinetics. This is difficult to substantiate using mitoplast extract, but it is generally thought that the concentration of mitochondrial housekeeping enzymes such as those involved in tRNA metabolism is limiting [see, for example, the arguments of Puranam and Attardi (30) concerning the intramitochondrial concentration of RNase P].
Structure probing
For alkaline ladders, the end-labeled tRNA was incubated at 90°C for 10 min in 50 mM NaHCO3, pH 9. For semi-denaturing ribonuclease (RNase) T1 reactions, the labeled tRNA was incubated at 55°C for 10 min in 12.5 mM Na citrate, pH 4.5, 3.5 M urea, 0.5 mM EDTA. Unlabeled Escherichia coli tRNA was used as a carrier at a final concentration of 1 µg/10 µl reaction. To achieve native conditions, tRNAs were refolded as described above for processing, and incubated in the processing buffer (but without RNasin) for 5 min at 37°C with the structure probing nucleases. Final concentrations were 1 and 2.5 × 10–3 U/µl (RNase T1), 0.55 and 1.37 U/µl (nuclease S1) or 1 and 2.5 × 10–6 U/µl (RNase V1). Reactions were terminated by placing samples at –20°C. For electrophoresis on 30 × 40 cm, 6, 8 or 10% denaturing polyacrylamide gels, 5 µl of formamide-marker dye mix was added and 7.5 µl samples were loaded directly, without heating. Gels were electrophoresed until the bromophenol blue migrated to 10 cm above the bottom. Imaging and analysis were performed as for processing gels.
RESULTS
Human mitochondrial tRNAIle precursor 3′ end processing
Endonucleolytic removal of the 3′ end trailer is central to the maturation of human mitochondrial tRNA precursors. We designed a tRNAIle precursor with a mature 5′ end [the presence of a 5′ end leader can interfere with 3′ end processing (12,31,32)] and a 20 nt 3′ end trailer (Fig. 2A; arrow indicates the expected 3′-tRNase cleavage site directly following the discriminator base). The 5′ end labeled precursor can be efficiently processed by 3′-tRNase extracted from human culture cell mitochondria (Fig. 2B). Three bands of expected product tRNA size are reproducibly obtained in similar proportions, the most intense arising from the predicted cleavage after the discriminator base (relative intensities of nucleotide 73 > 74 >> 72). Product heterogeneity is not due to 3′ end heterogeneity of T7 transcripts, since the precursor was labeled at its 5′ end. Such heterogeneity of 3′-tRNase products (Fig. 2B) has been observed with other combinations of processing enzymes and substrates (12,32), including the cloned and expressed Arabidopsis thaliana and Methanococcus janeschii 3′-tRNase (29). Quantitation of processing kinetics was performed by combining the intensities in the product region.
Selected pathological substitutions reduce the ability of tRNAIle precursors to be processed by 3′-tRNase
Ten mutations in human mitochondrial tRNAIle have been shown to cause disease (Fig. 1). All the pathogenic tRNAIle mutants tested reduced 3′-tRNase processing efficiency; Figure 3 illustrates the processing effects of the A4269G, G4309A and A4317G substitutions. A4269G causes the greatest decrease in processing efficiency (∼12-fold less than wild-type), and four of the eight mutants tested reduced processing efficiency to ∼10-fold below wild-type (see below). In this experiment, almost 40% of the wild-type precursor was converted to product in 15 min of reaction (Fig. 3A, lane 4), close to the upper limit for 3′-tRNase processing of wild-type substrate produced by in vitro transcription. Some mutant substrates display a lower end point for processing than wild-type, as well as a reduced processing efficiency.
Reduced 3′-tRNase processing efficiency of mutation- carrying tRNAIles is principally due to lower Vmax
Of the 10 tRNAIle mutations known to cause disease (Fig. 1), we performed Michaelis–Menten kinetics on eight (Fig. 4 and Table 1). G4298A was not included in the kinetic analysis because it is located close to A4300G in the anticodon stem (which was included) and its relative processing efficiency was only slightly reduced (data not shown). A4317G was used to illustrate wild-type and mutant processing kinetics (Fig. 4) because it is located in the T loop, which is rich in processing determinants (see Discussion), and displays a typically strong reduction in Vmax while KM is practically unaffected. With every mutant tested, Vmax for 3′-tRNase was lower than with wild-type. KMs for pathogenic tRNAIles were also consistently less than for wild-type, but the decrease in Vmax (up to ∼20-fold, in the case of A4269G) was consistently greater than that for KM, so that processing efficiency (Vmax / KM) was reduced for all the mutants, ∼10-fold in the case of four mutant tRNAIles (A4269G, A4295G, G4309A, A4317G; bold figures in the right-most column of Table 1).
Effect of mutations on tRNAIle precursor structure
We used secondary structure probing nucleases to investigate precursor tRNAIle folding and to search for structural differences between the mutants and wild-type. Ribonuclease T1 is specific for G residues, nuclease S1 for single-stranded regions, and ribonuclease V1 for paired stems and otherwise structured regions [note, however, that V1 cleavage as a literal indicator of helicity can be misleading, missing some helices and cutting to the sides of others (33); reliable comparisons between mutant and wild-type V1 data can be made, however]. Ribonuclease T1 used under semi-denaturing as well as native conditions can reveal differences in exposure of Gs which arise from tRNA folding. The four mutants which gave the lowest processing efficiencies in Michaelis–Menten experiments (A4269G, A4295G, G4309A, A4317G; see Fig. 1 for location) were 5′ end labeled and probed with nucleases alongside wild-type tRNAIle. No differences were observed between wild-type precursor and three of the mutants (A4269G, A4295G, A4317G; results not shown). Structural differences observed between the wild-type and G4309A patterns (Figs 5 and S1) were tightly clustered surrounding the site of the G4309A substitution, as indicated by a vertical bar to the right of Figure 5B.
All of the Gs from nucleotide 9 to the 3′ end of the precursor can be detected with roughly equal intensity in the semi-denaturing T1 lanes (lanes 2 and 3 in both panels of Fig. 5). The absence of G51 from the G4309A pattern (arrow at left of Fig. 5B, lanes 2 and 3) confirms the G4309A substitution. G34 in the anticodon loop is the most fully exposed T1 site in the folded tRNA (native state, lanes 5 and 6, designated N T1), consistent with the canonical tRNA fold. The S1 pattern closely follows the T1 pattern (lanes 7 and 8), displaced up by a single nucleotide (nucleases S1 and V1 leave a 3′-OH, while T1 leaves a 3′-PO42–, so that corresponding T1 bands migrate faster due to greater negative charge). Some secondary structure was also observed in the 3′ end trailer, based on alternating S1 and V1 sensitivity downstream of nucleotide 73.
The distribution of V1 cleavages within tRNAIle precursors is consistent with the cloverleaf structure of tRNA (bars and numbers enclosed in ovals placed on the secondary structure diagram in Fig. 6 correspond with those in Fig. 5). The bottom of the T stem (nucleotides 49–53) displays low V1 sensitivity; only nucleotide 49 is a strong V1 site in wild-type tRNAIle precursor. The variable domain appears to be structured, as suggested by V1 bands at nucleotides 44, 45 and 47, 48. V1 cleavage at nucleotide 55 and in the D loop could indicate tertiary interactions (see Discussion).
Most structural changes in the G4309A precursor relative to wild-type are just upstream from the mutation at nucleotide 51. Nucleotide 50 becomes highly susceptible to S1 (Fig. 5B, lanes 7 and 8, scans of gels in Fig. S1) and nucleotide 49 becomes much less susceptible to V1 (Fig. 5B, lanes 9 and 10; Fig. S1). In addition, V1 sensitivity of G4309A increases at T loop nucleotide 55. Thus the structural effects of the G4309A substitution are local, and only slightly propagated. The V1 sensitivity of the acceptor stem and top of the T stem (72–66 and 65–61, Fig. S1) in G4309A do not appear to be affected. Comparison of native T1 (lanes 5–6) and S1 (lanes 7–8) with semi-denaturing T1 (lanes 2–3) patterns in the G4309A mutant (B) relative to wild-type (A) suggests that, in addition to pronounced local effects noted above, the entire mutant tRNA may be more exposed, consistent with a weakened tertiary structure. Results with 3′ end labeled tRNAIle precursors are essentially the same (Fig. S2).
DISCUSSION
3′-tRNase processing of a human mitochondrial tRNAIle precursor
Mitochondrial tRNAs are embedded in long primary transcripts, punctuating the two ribosomal RNAs and 11 mRNAs (1). Production of mature, functional mitochondrial RNAs requires endonucleolytic cleavage of the precursors at the 5′ and/or 3′ ends of the tRNAs (6,7). tRNAIle overlaps by 1 nt on its 5′ end with the 3′ end of ND1 mRNA, which therefore requires that 1 A be added after processing to complete the ND1 translation termination codon. On the 3′ side, tRNAIle is flanked by a 70 nt spacer followed by tRNAMet, making the reductions in 3′-tRNase processing efficiency that arise from pathogenesis-associated mutations still more remarkable. The same is true for tRNASer(UCN) (12), which is followed by a 2.5 kb spacer on the 3′ side.
Human mitochondrial tRNAIle undergoes post-transcriptional modification at five positions in vivo [to produce m1G9, m22G26, Ψ27, Ψ28 and t6A37 (25)]. The in vitro 3′-tRNase reaction using unmodified wild-type transcript is efficient (Figs 2B and 3A), and excision of the tRNAs from precursor transcripts is expected to occur early in tRNA metabolism; the stage of tRNA maturation at which modification occurs and in most cases, the significance of the modifications, are unknown. Nonetheless, tRNA modification could alter the relative effects of pathogenesis-linked mutations on tRNA precursor 3′ end processing. Interestingly, three of the five tRNAIle modifications occur at positions that coincide with pathogenesis-associated mutations (m22G26–G4284A; Ψ27–T4285C; t6A37–A4295G).
Several pathogenic tRNAIle precursors substantially reduce 3′-tRNase processing efficiency, principally due to lower Vmax
To date, 10 substitutions in tRNAIle have been linked to mitochondrial diseases. Of these, we initially investigated the effect of nine pathogenic tRNAIle substitutions on 3′-tRNase processing [we became aware of the G4284A mutation (26) toward the end of this investigation]. All these substitutions reduce the efficiency with which tRNAIle can be processed by 3′-tRNase. The greatest reductions were observed with mutations A4269G, A4295G, G4309A and A4317G.
On the basis of these reduced processing efficiencies, we performed Michaelis–Menten kinetics on wild-type and eight mutant tRNAIles (Fig. 4 and Table 1). A tRNAIle precursor with the pathogenic A4317G mutation displays substantially reduced Vmax compared with wild-type, with a lesser reduction in KM. Most of the tRNAIle mutants display a similar pattern of larger reductions in Vmax, reflecting the chemical step of catalysis, accompanied by smaller reductions in KM (Table 1). Reduced KM usually indicates tighter substrate binding, which would improve catalytic efficiency (Vmax / KM), but the consistently larger reductions in Vmax that arise from pathogenic mutations cause catalytic efficiency to be reduced. KM would be expected to increase in the event of structure changes that affect substrate recognition and binding, but in most cases, no such structure changes were observed (see below). Processing efficiencies with A4317G and three other mutant precursors are all reduced ∼10-fold relative to wild-type. 3′ End maturation defects could thus contribute to pathogenicity of some of the mutations.
Wild-type tRNAIle precursor is in the cloverleaf structure, and G4309A displays local structural differences from wild-type
Mitochondrial tRNAs display many differences in sequence and secondary structure from cytoplasmic tRNAs; tRNAIle is one of the most highly conserved mammalian mitochondrial tRNAs (reviewed in 19). Despite an unusually small D domain and an A·C apposition in the T stem (Fig. 1), wild-type tRNAIle displays a secondary structure consistent with canonical tRNA. The bottom of the T stem (nucleotides 49–53) is weakly cleaved by V1 except for nucleotide 49, consistent with the suggestion that the 52·62 A·C apposition weakens the T stem (23,24). G18,19 in the D loop and TΨC in the T loop, which are universally conserved in cytoplasmic tRNAs, are replaced in human mitochondrial tRNAIle by AA and CUU, respectively. Interestingly, V1 cleavages at D loop nucleotides 19, 20 and at T loop nucleotide 55 could reflect tertiary A-U pairs which replace the canonical D/T loop base pairs.
Structure modeling predicts alternate foldings of mitochondrial tRNAIle (D.Friede and P.Schuster, personal communication), but the structure probing results (Fig. 6) are generally more consistent with the canonical fold. The 3′ end trailer is evidently structured, but without obvious pairing partners. The sequence of the 3′ end trailer used in vitro (Fig. 2A), characteristic of the tRNAIle precursor in vivo, is also present on the mutants, thus the 3′ end trailer is unlikely to interfere differentially with 3′ end processing.
Effect of a pathogenesis-linked substitution on secondary structure
Mutant and wild-type secondary structures were compared in search of a structural basis for reduced processing efficiency. Apart from G4309A, the mutants do not display different structures. G4309A is of particular interest because the substitution is 1 nt upstream from a 52·62 A·C mismatch in wild-type tRNAIle, and because it shows strongly reduced 3′-tRNase processing efficiency. Differences between wild-type and mutant G4309A precursors are observed with enzymatic probes. S1 sensitivity of G4309A at nucleotide 50 increases, V1 sensitivity at nucleotides 49, 50 decreases just upstream from the pathogenic substitution at nucleotide 51, and nucleotide 55 increases in susceptibility to V1 nuclease in the G4309A mutant. These local and slightly propagated structure changes are located in a region which could be important for 3′-tRNase recognition and catalysis. End processing reactions by RNase P, 3′-tRNase and NTase require an intact, coaxially stacked acceptor stem and T domain (33). The location of the G4309A mutation in the T stem and its structural, functional and physiological effects are consistent with the idea that the structural changes could interfere with its ability to be cleaved by 3′-tRNase, contributing to pathology.
Mutations A4269G and A4317G are also in the acceptor stem and T domain of the tRNA. While these mutations do not lead to detectable changes in secondary structure, the wild-type nucleotides at these positions could be directly recognized by 3′-tRNase. For mutant A4295G, the strong reduction in 3′-tRNase processing efficiency is less interpretable, since a mutation in the anticodon loop would not be expected to interfere with 3′ end processing.
An overview of tRNA maturation and aminoacylation in the context of mitochondrial disease
Pathogenesis-linked mutations in mitochondrial tRNAs cause a wide variety of diseases, syndromes and symptoms. The specific position of a mutation in a particular tRNA does not correlate in a simple, obvious way with the disease, but interestingly, four pathogenic mutations in tRNASer(UCN) cause sensorimotor deafness [see Mitomap (2) and Toompuu et al. (34)], and three mutations in tRNAIle that cause ophthalmoplegia (T4274C, T4285C and G4298A) also severely reduce their ability to be aminoacylated (23).
Aminoacylation is the most-studied aspect of tRNAIle metabolism for which the effects of pathogenesis-linked mutations have been investigated (22–25). Results are summarized in Table 2, along with the present findings on 3′ end maturation. Mutations which most strongly reduce 3′ end processing efficiency (A4269G, A4295G, A4317G; bold in Table 2, column 5) have little or no effect on aminoacylation, and those which affect processing the least (T4274C, T4285C, G4298A; bold in Table 2, column 6) display reduced capacity for aminoacylation, suggesting a reciprocal relationship. Interference of a mutation mainly with processing could reduce the level of mature but fully aminoacylable tRNA, while interference of a mutation with aminoacylation could lead to a normal steady state level of tRNA, but a decreased level of aminacyl-tRNA.
Table 2. Effects of mutations in tRNAIle associated with diseases on structure, precursor 3′-tRNase processing and aminoacylation.
| Location in mitochondrial genome | Location in tRNA (nucleotide position) | Diseasea | Structural effect | Loss in 3′-tRNase processing efficiencyb | Loss in aminoacylation efficiency |
|---|---|---|---|---|---|
| A4269G | Acceptor stem (7) | CM | Stability decreasedc | 12.5 | 1.3d |
| T4274C | D-stem (12) | OP | – | 4.2 | 25d |
| G4284A | Connector (26) | Mixed | – | – | – |
| T4285C | Anticodon stem (27) | OP | – | 3.2 | 50d |
| A4295G | Anticodon loop (37) | CM | No changeb | 10.3 | 0.77e |
| G4298A | Anticodon stem (40) | OP | – | –f | >1000d |
| A4300G | Anticodon stem (42) | CM | – | 2.1 | 5e |
| G4309A | T-stem (51) | OP | Local changeb | 7.7 | – |
| A4317G | T-loop (59) | CM | No changeb | 10.0 | 3.75d |
| C4320T | T-stem (62) | CM | – | 4.2 | 0.6d |
aCM, cardiomyopathy; OP, ophtalmoplegia; Mixed, both cardiomyopathy and ophtalmoplegia.
bThis work.
cYasukawa et al. (25); (–) not determined.
eS. Kelley, personal communication.
fNot determined, but cleavage efficiency comparable to mutant A4300G.
Bold text is used to indicate those mutants which display the strongest reduction in either 3′-tRNase or aminoacylation efficiency, but not both (as discussed in text).
A defect in any aspect of tRNA metabolism could lead to a decrease in the steady-state level of functional tRNA. If severe, multiple molecular defects arise from the same mutation, the total decrease in steady-state level of functional tRNA might be lethal. On the other hand, two mutations (A4300G, C4320T) barely affect either 3′ end processing or aminoacylation; in these instances, the molecular defect could lie elsewhere.
Kelley et al. (23,24) suggested that the 52·62 A·C mismatch in wild-type tRNAIle is destabilizing and that mutations elsewhere throughout the tRNA compound its effect. Of the mutant tRNAIle structures we investigated, only G4309A (51A; see Figs 1 and 6) clearly affects secondary structure, reduces the ability of the precursor to be 3′ end processed, and causes disease. The G4309A effect on secondary structure is local, affecting the two nucleotides upstream from the mutation in mid-T stem, and nucleotide 55 in the T loop. Yasukawa et al. (25) showed A4269G to be unstable relative to wild-type tRNAIle by using thermal melting and susceptibility to nucleases present in mitochondrial extract (Table 2), but we did not detect a difference in secondary structure between this mutant and wild-type. Subtle problems with structure, folding and stability of pathogenic tRNAs might lead to their reduced ability to be processed and contribute, along with other deficiencies, to mitochondrial disease. Detection of the postulated mild or tertiary structural defects which arise from pathogenesis-linked mutations would require more sensitive structure probing methods.
The generally reduced ability of pathogenesis-linked mutant tRNAIle precurors to be processed at their 3′ ends observed here (∼10-fold, in the case of four of the mutants investigated) is consistent with the suggestion that inefficient end processing of mutant tRNAs could contribute to pathology.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the initial technical assistance of V. Greene and L. Toussaint, structure prediction by D. Friede and P. Schuster (U Wien) and helpful conversations with A. Mohan (YC), M. Sissler (IBMC) and D. Thurlow (Clark U). The tRNAIle plasmid set was a gift of S. Kelley (Boston College). This work was supported by the Centre National de la Recherche Scientifique (CNRS) and by NIH grants S06GM08153 and T34GM08498. Sabbatical support for L.L. was provided by NIH fellowship F33-GM64266 and by Univerité Louis Pasteur, Strasbourg.
REFERENCES
- 1.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J., Staden,R. and Young,I.G. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 2.Center for Molecular Medicine (2001) MITOMAP: A Human Mitochondrial Genome Database. Center for Molecular Medicine, Emory University, Atlanta, GA, USA (http://www.gen.emory.edu/mitomap.html).
- 3.Larsson N.G. and Clayton,D.A. (1995) Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet., 29, 151–178. [DOI] [PubMed] [Google Scholar]
- 4.Schon E.A., Bonilla,E. and DiMauro,S. (1997) Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomembr., 29, 131–149. [DOI] [PubMed] [Google Scholar]
- 5.Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 482–488. [DOI] [PubMed] [Google Scholar]
- 6.Montoya J., Ojala,D. and Attardi,G. (1981) Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature, 290, 465–470. [DOI] [PubMed] [Google Scholar]
- 7.Ojala D., Montoya,J. and Attardi,G. (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature, 290, 470–474. [DOI] [PubMed] [Google Scholar]
- 8.King M.P., Koga,Y., Davidson,M. and Schon,E.A. (1992) Defects in mitochondrial protein synthesis and respiratory chain activity segregate with tRNALeu(UUR) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and strokelike episodes. Mol. Cell. Biol., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bindoff L.A., Howell,N., Poulton,J., McCullough,D.A., Morten,K.J., Lightowlers,R.N., Turnbull,D.M. and Weber,K. (1993) Abnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA. J. Biol. Chem., 268, 19559–19564. [PubMed] [Google Scholar]
- 10.Rossmanith W., Tullo,A., Potushak,T., Karwan,R. and Sbisa,E. (1995) Human mitochondrial tRNA processing. J. Biol. Chem., 270, 12885–12891. [DOI] [PubMed] [Google Scholar]
- 11.Rossmanith W. and Karwan,R.M. (1998) Impairment of tRNA processing by point mutations in mitochondrial tRNALeu(UUR) associated with mitochondrial diseases. FEBS Lett., 433, 269–274. [DOI] [PubMed] [Google Scholar]
- 12.Levinger L., Jacobs,O. and James,M. (2001) In vitro 3′-end endonucleolytic processing defect in a human mitochondrial tRNASer(UCN) precursor with the U7445C substitution, which causes non-syndromic deafness. Nucleic Acids Res., 29, 4334–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doersen C.J., Guerrier-Takada,C., Altman,S. and Attardi,G. (1985) Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J. Biol. Chem., 260, 5942–5949. [PubMed] [Google Scholar]
- 14.Nagaike T., Suzuki,T., Tomari,Y., Takemoto-Hori,C., Negayama,F., Watanabe,K. and Ueda,T. (2001) Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J. Biol. Chem., 276, 40041–40049. [DOI] [PubMed] [Google Scholar]
- 15.Reichert A.S., Thurlow,D.L. and Morl,M. (2001) A eubacterial origin for the human tRNA nucleotidyltransferase? Biol. Chem., 382, 1431–1438. [DOI] [PubMed] [Google Scholar]
- 16.Morl M. and Marchfelder,A. (2001) The final cut. The importance of tRNA 3′-processing. EMBO Rep., 2, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan A., Whyte,S., Wang,X., Nashimoto,M. and Levinger,L. (1999) The 3′ end CCA of mature tRNA is an antideterminant for eukaryotic 3′-tRNase. RNA, 5, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helm M., Brulé,H., Friede,D., Giegé,R., Pütz,J. and Florentz,C. (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA, 6, 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid F.M., Rovio,A., Holt,I.J. and Jacobs,H.T. (1997) Molecular phenotype of a human lymphoblastoid cell-line homoplasmic for the np 7445 deafness-associated mitochondrial mutation. Hum. Mol. Genet., 6, 443–449. [DOI] [PubMed] [Google Scholar]
- 21.Guan M.-X., Enriquez,J.A., Fischel-Ghodsian,N., Puranam,R.S., Lin,C.P., Law,M.A. and Attardi,G. (1998) The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol. Cell. Biol., 18, 5868–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degoul F., Brulé,H., Cepenac,C., Helm,M., Marsac,C., Leroux,J., Giegé,R. and Florentz,C. (1998) Isoleucylation properties of native human mitochondrial tRNAIle and tRNAIle transcripts. Implications for cardiomyopathy-related point mutations (4269, 4317) in the tRNAIle gene. Hum. Mol. Genet., 7, 347–354. [DOI] [PubMed] [Google Scholar]
- 23.Kelley S.O., Steinberg,S.V. and Schimmel,P. (2000) Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nature Struct. Biol., 7, 862–865. [DOI] [PubMed] [Google Scholar]
- 24.Kelley S.O., Steinberg,S.V. and Schimmel,P. (2001) Fragile T-stem in disease-associated human mitochondrial tRNA sensitizes structure to local and distant mutations. J. Biol. Chem., 276, 10607–10611. [DOI] [PubMed] [Google Scholar]
- 25.Yasukawa T., Hino,N., Suzuki,T., Watanabe,K., Ueda,T. and Ohta,S. (2000) A pathogenic point mutation reduces stability of mitochondrial mutant tRNA(Ile). Nucleic Acids Res., 28, 3779–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corona P., Lamantea,E., Greco,M., Carrara,F., Agostino,A., Guidetti,D., Dotti,M.T., Mariotti,C. and Zeviani,M. (2002) A pathogenic point mutation reduces stability of mitochondrial mutant tRNA(Ile). Ann. Neurol., 51, 118–122. [DOI] [PubMed] [Google Scholar]
- 27.Florentz C. and Sissler,M. (2001) Disease-related versus polymorphic mutations in human mitochondrial tRNAs. Where is the difference? EMBO Rep., 2, 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fechter P.J., Rudinger,R., Giegé,A. and Theobald-Dietrich,A (1998) Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett., 436, 99–103. [DOI] [PubMed] [Google Scholar]
- 29.Schiffer S., Rösch,S. and Marchfelder,A. (2002) Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J., 21, 2769–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puranam R.S. and Attardi,G. (2001) The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol. Cell. Biol., 21, 548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunzmann A., Brennicke,A. and Marchfelder,A. (1998) 5′ end maturation and RNA editing have to precede tRNA 3′ processing in plant mitochondria. Proc. Natl Acad. Sci. USA, 95, 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nashimoto M., Weisemann,D.R., Geary,S., Tamura,M. and Kaspar,R.L. (1999) Long 5′ leaders inhibit removal of a 3′ trailer from a precursor tRNA by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res., 27, 2770–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maizels N. and Weiner,A.M. (1999) The genomic tag hypothesis: what molecular fossils tell us about the evolution of tRNA. In Gesteland,R., Cech,T. and Atkins,J. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, NY, pp. 79–111.
- 34.Toompuu M., Yasukawa,T., Suzuki,T., Haakinen,T., Spelbrink,J.N., Watanabe,K. and Jakobs,H.T. (2002) The 7472insC mitochondrial DNA mutation impairs the synthesis and extent of aminoacylation of tRNASer(UCN) but not its structure or rate of turnover. J. Biol. Chem., 277, 22240–22250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.