Abstract
The Drosophila pre-mRNA splicing factor B52 (SRp55) is essential for fly development, but splicing of RNAs of specific genes tested previously is normal in B52-null animals, presumably due to partial functional redundancy with other SR proteins. To identify B52-dependent splicing substrates in vivo, we selected genomic sequence fragments whose transcripts bind B52. Almost all of the corresponding genes having a known function encode either transcription factors or components of signal transduction pathways, with the B52- binding fragments located to not only exonic but also intronic regions. Some pre-mRNAs from these genes showed splicing defects in the B52-null mutant. These results indicate that B52 has unique functions in the removal of some introns during development, and plays a critical role in cellular regulatory networks.
INTRODUCTION
SR proteins are both essential splicing factors and modulators of alternative splicing that function at early steps of spliceosome assembly (1,2). This family consists of at least six members with sizes ranging from 20 to 75 kDa. SR proteins share a similar structure, with one or two RNA recognition motifs (RRM) at the N-terminus and a region of variable length that is rich in arginine–serine dipeptides (the RS domain) at the C-terminus. Studies have shown that the RRMs mediate the binding of the SR proteins to exonic splicing enhancers (ESEs) that stimulate splicing of the pre-mRNAs containing these elements; inactivation of these ESEs may be the cause of certain genetic diseases (3). While single SR proteins may act in a substrate-specific manner, their functions when studied collectively show partial redundancy, probably due to overlap in binding of multiple different proteins to degenerate sequence motifs (4).
In vitro splicing assays involving complementation of a splicing-deficient cytoplasmic (S100) extract (5) have played a critical role in elucidating the function of SR proteins. Distinct pre-mRNA substrate specificity and different alternative splicing patterns have been found for different mammalian SR proteins in these systems (6). Although assays using synthetic substrates are informative, the identities of genes requiring a specific SR protein for proper processing in vivo have remained elusive. The existence of genes whose splicing depends solely on a single SR protein is suggested from the results of genetic studies. For example, deletion of the Drosophila B52, or SRp55, gene resulted in lethality at the second-instar larval stage (7). Depletion of another SR protein, ASF/SF2, or SRp30a, in the chicken B-cell line DT40 (8) and in Caenorhabdidtis elegans (9) caused cell death. Targeted disruption of SRp20 caused embryonic lethality in mice (10).
Using splicing substrates with multiple splice sites, it had been shown that some SR proteins promote proximal splicing while others shift splicing to distal splice sites (11–13). Native substrates on which SR proteins exhibit unique functions would facilitate mechanistic studies, but their identification is hampered by the family’s functional redundancy. In a previous study, five genes tested, including Hsp83, β1-tubulin, Dopa decarboxylase (Ddc), Sex lethal (Sxl) and Ultrabithorax (Ubx), did not show any splicing deficiency in the developmentally arrested larvae of a B52 deletion mutant (7). We also tested another set of 20 genes known to be transcribed in second instar larvae, but found no splicing defect for their RNAs in the B52-null mutant (S.Kim and J.T.Lis, unpublished results). In most tissues, other SR proteins presumably complement the loss of B52 (14).
In the present study, we used a heuristic scheme that overcomes the difficulties caused by the partial functional redundancy of SR proteins to identify pre-mRNAs that depend on B52 for proper splicing. According to the current view of their function, SR proteins activate splicing by binding to enhancer RNA sequences through the RRM domain and recruiting the splicing machinery through protein–protein interactions via the SR domain (15–17). This led us to search first for the genomic DNA sequence segments whose RNA transcripts bind B52, using the genomic SELEX technology originally developed by Singer et al. (18) and Shtatland et al. (19). In this method, RNA transcripts containing binding sites for the protein of interest are selected by iterative binding, partitioning and amplification steps. The assignment of the isolated segments to genes was facilitated by the availability of the Drosophila genomic sequence (20,21). Equipped with the full sequence of candidate B52-target genes, we compared their splicing patterns in the B52-deletion mutant flies with those in wild-type animals to reveal defects in either constitutive or alternative splicing. A group of B52-dependent splicing substrates emerged from this process. In addition, interesting mechanistic implications were revealed by the nature of their splicing defects in the absence of B52.
MATERIALS AND METHODS
Expression and purification of recombinant B52 protein
Full-length B52 protein was expressed in Sf9 cells using the Baculovirus expression system and prepared as described previously (22).
Generation of Drosophila genomic sequence library
The library was constructed essentially according to the procedure described by Singer et al. (18). The initial material was 25 mg DNA from Canton S adult flies prepared using equilibrium density gradients in CsCl (23). Random primed products of ∼200 bp (±50 bp) in length were gel-purified and PCR-amplified for three cycles. RNA was made using the MEGAshortscript in vitro transcription kit (Ambion Inc.). Twenty different primer pairs predicted to anneal to randomly chosen single copy genes were used to carry out PCRs to test the quality of the library. All of them produced PCR products with the predicted sizes (data not shown).
Genomic SELEX
The selection and amplification was performed as previously described (22). The progress was monitored by electrophoretic mobility shift assay using 1% of the labeled RT–PCR transcripts from each generation of selected pools. After six rounds of selection, the pool in the form of DNA was cloned into the vector pGEM-3Z (Promega Inc.). The inserts in individual clones were sequenced.
Electrophoretic mobility shift assay
All RNA–protein binding assays were performed in 20 μl reaction volumes in 1× binding buffer as previously described (22).
RT–PCR
RNA isolation from whole larvae and the RT–PCR assay were performed as described previously (14). An oligo dT primer (5′-TTTTTGTTTTCATTTTTTGACTTT-3′) was used for the RT step and the gene-specific primer pairs listed in Table S2 (Supplementary Material) were used for the PCR. The B52 and β1-tubulin primers were as previously described (7).
Quantitative real-time PCR
RT products made with oligo dT primer were used as templates. The templates and primer sets were mixed with 2× QuantiTect SYBR Green PCR Master Mix, and 20 cycles of PCR were performed using a Rotor-Gene real-time PCR machine (Corbett Research, Inc.). Primers were as above in the gel-based RT–PCR assay. Standards were prepared as described in the Rotor-Gene manual.
Site-directed mutagenesis
Altered sequences were created according to the method described by Carmona et al. (24) with some modifications. Sequencing primers, M13 forward and backward, for the pGEM-3Z vector were used as the flanking primers with the internal mutagenic (site-mutated) primers to introduce mutations into the sequences #1-3 and #1-6. The two contiguous DNA fragments were joined together by fusion PCR (25) instead of blunt-end ligation. The resulting products were gel-purified, further amplified, cloned back into pGEM-3Z vector and sequenced to confirm the introduction of mutations.
RESULTS AND DISCUSSION
B52-binding RNA transcripts of genomic sequences
B52 has two RRM domains that can bind specific RNAs with high affinity as demonstrated by a previous SELEX experiment that started with a random sequence pool (22). The RNA aptamer sequence identified by that study can function as an ESE to stimulate the splicing of a fushi tarazu (ftz)-derived substrate in vitro (26). Based on this observation, we set out to find B52-binding sites in the context of genomic sequences, in the hope of identifying native B52-dependent splicing substrates. Drosophila genomic DNA from adult flies was amplified with primers to generate a sequence library that contains genomic segments of ∼200 bp flanked by a pair of constant regions, one of which contains a promoter for the T7 RNA polymerase (18). An RNA pool was transcribed from this library and used in the selection. The Drosophila genome (<109 bp) should be represented in multiple copies in a pool of 1010 segments of 200 bp, and thus a pool made of several micrograms of DNA is more than enough starting material. Sequences of 200 nt are long enough to allow a homology search and to serve as probes to screen a genomic or cDNA library. To ensure the faithful preservation of genomic sequence through multiple rounds of selection, high-fidelity thermostable DNA polymerases and fewer PCR cycles were used to minimize mutations. In addition, we used fewer rounds of selection than a conventional SELEX to avoid the enrichment of just a few high affinity ‘winners’.
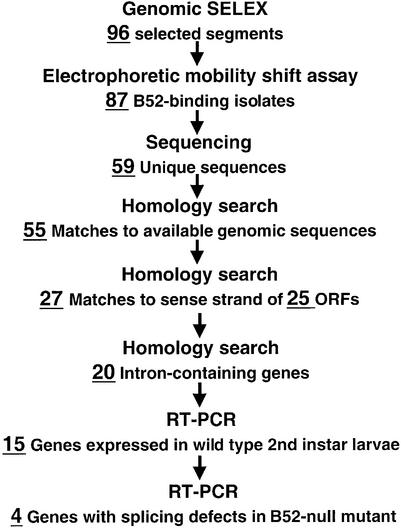
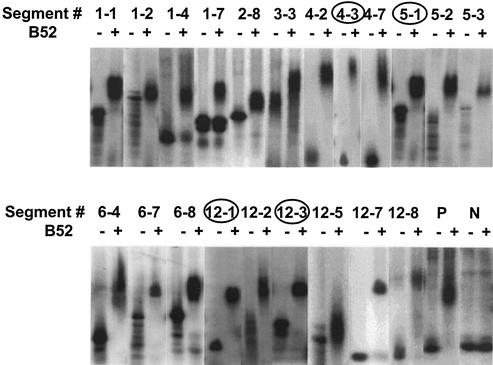
Selection and amplification were carried out against full-length, Baculovirus-expressed B52. The affinity of the RNA pool for B52 increased as the selection progressed (data not shown). After six rounds of selection, we generated and cloned cDNAs from the final pool of selected RNA. As summarized in Figure 1, we transcribed DNA of 96 clones to generate corresponding RNAs for individual electrophoretic mobility shift assays with the B52 protein. Eighty-seven out of the 96 clones tested showed binding to B52. Figure 2 shows results for those clones later identified as parts of intron-containing genes. Using the genomic SELEX process, we narrowed the size of the candidate population to be further investigated in this study from the 13 601 genes predicted in the Drosophila genome (27) to a few dozen genes.
Figure 1.
An overview of the process leading to the isolation of B52-dependent pre-mRNA splicing substrates. The number of candidates in each step is indicated.
Figure 2.
B52-binding activity of the RNA transcripts derived from exonic as well as intronic regions as demonstrated by electrophoretic mobility shift assay. The binding results of in vitro transcripts of 21 genomic sequence segments selected in genomic SELEX and identified as parts of intron- containing genes are shown. Segments in those genes confirmed to have splicing defects in the B52-null mutant are indicated by circles around their names. The positive control (P) is BBS #8, an aptamer isolated in a previous SELEX experiment. The negative control (N) is MGM #1, an RNA that binds to the nitrocellulose filter used as partitioning matrix (22).
Genes identified by the selected sequence segments
We operationally defined a B52-specific splicing substrate as one that fits two critical and independent constraints: the binding of the RNA to B52 and the splicing defects in B52-null flies. We sequenced the 87 individual clones whose RNA transcripts showed binding to B52, and used the resulting 59 different sequences to search FlyBase (http://flybase.harvard. edu/) to pinpoint the genomic regions that contain these sequences. As shown in Figure 1, we applied several additional constraints in the sequence analysis to generate a short-list to test for splicing defects. For example, we required that the B52-binding site be located in the coding region of intron-containing genes and present in the sense orientation, in order to consider a selected sequence as a candidate B52-target gene.
As shown in Table 1, two different types of convergence are evident when the candidates are surveyed collectively. First, some clones were repeated isolates of the same insert. Secondly, in some other cases, different selected sequences were derived from separate regions of a single gene. For example, sequences #6-4 and #6-8 are located respectively in intron 3 and 12 of the arrest gene. Fourteen different sequences from 21 isolated clones are parts of 13 known genes: Ecdysone receptor (EcR), ladybird late (lbl), frizzled (fz), Furin 1 (Fur1), longitudinals lacking (lola), Rx, skiff (skf), Mlx interactor (Mio), arrest (aret), RhoGAP16F, faint sausage (fas), Syndecan (Sdc) and homothorax (htn). According to information in Flybase, all but one of these genes encode transcription factors (EcR, lbl, lola, Rx, Mio and hth) and/or components of signal transduction pathways (EcR, fz, Fur1, skf, RhoGAP16F, fas and Sdc). This suggests an important role played by B52 in cellular regulatory networks, consistent with our previous findings that proper B52 expression levels are critical to survival and normal development of the organism (7,28,29). Another eight different sequences from 12 isolates were located to genes with unidentified functions, including one without an intron (CG5228). Five different sequences from nine isolates matched open reading frames (ORFs) of four transposable elements. All sequence segments that locate to ORFs in their sense orientation are summarized in Table 1. In addition to these 27 matches to 25 ORFs, we identified 28 sequences that matched ‘intergenic regions’ based on the annotation in Flybase. While these regions lack the statistical signature of a normal protein-coding gene, we cannot rule out the possibility that they may be transcribed into RNA in some physiological context.
Table 1. ORFs identified by selected B52-binding segments.
| Segment name | Number of isolates | ORF identity | Gene function | Location within ORF |
|---|---|---|---|---|
| #1-1 | 4 | Ecdysone receptor (EcR) | TF/ST | Intron 2 |
| #1-2 | 1 | ladybird late (lbl) | TF | Intron 1 |
| #1-4 | 1 | frizzled (fz) | ST | Intron 1 |
| #2-8 | 1 | Furin 1 (Fur1) | ST | Intron 1a |
| #3-3 | 3 | longitudinals lacking (lola) | TF | Last intron |
| #4-3 | 1 | Rx | TF | Intron 4 |
| #4-7 | 1 | skiff (skf) | ST | Exon 1 and intron 1 |
| #5-1 | 1 | Mlx interactor (Mio) | TF | Intron 5a |
| #6-4 | 1 | arrest (aret) | RNA-binding | Intron 3 |
| #6-8 | 1 | arrest (aret) | Intron 12 | |
| #12-1 | 1 | RhoGAP16F | ST | Intron 3 and exon 4 |
| #12-5 | 1 | faint sausage (fas) | ST | Intron 9 |
| #12-7 | 3 | Syndecan (Sdc) | ST | Intron 2 |
| #12-8 | 1 | homothorax (hth) | TF | Intron 4 |
| #1-7 | 1 | CG11760 | Exon 1 | |
| #2-1 | 1 | CG5228 | Intron-less gene | |
| #4-2 | 1 | CG14796 | Exon 3 | |
| #5-2 | 3 | CG5953 | Intron (only one) | |
| #5-3 | 1 | CG15593 | Exon 3 and boundariesb | |
| #6-7 | 1 | CG9080 | Last exon (3′ UTR) | |
| #12-2 | 3 | CG3950 | Intron 3 | |
| #12-3 | 1 | CG14670 | Exon 6 and boundariesb | |
| #1-3 | 3 | F element (F-element) | TE | |
| #1-6 | 1 | Doc element (Doc) | TE | |
| #6-2 | 3 | roo element (roo) | TE | |
| #11-7 | 1 | HeT-A element (HeT-A) | TE | |
| #12-6 | 1 | Doc element (Doc) | TE |
TF, RNA polymerase II transcription factor; ST, component of signal transduction pathways; TE, transposable element.
aA region within the gene that is alternatively spliced.
bBoundaries cover part of the adjacent introns.
Previously, we developed an in vitro splicing assay to test B52 function using a ftz-derived substrate. However, we did not isolate any sequence segment of the ftz gene through genomic SELEX. This may be a consequence of the limited number of B52-binding clones sequenced, or it may also reflect a limitation of our scheme. For example, sequences that require the presence of additional protein for B52 binding would not be selected.
Further inspection of the location of the selected sequence segments within the 13 known genes revealed an interesting pattern. Consistent with previous studies that the RNA sequences that form high-affinity binding sites for individual SR proteins are sufficient to function as ESEs (30,31), approximately one-quarter of the selected sequences in this group match exonic regions. However, intron sequences seem to be more prevalent, with more than half in this group matching the intronic regions of pre-mRNAs. Moreover, many intronic sequences so identified are located in the middle of large introns (>6 kb, for example, in the case of segment #4-3). In principle, binding sites located in either intronic or exonic region could recruit B52 and allow the regulatory function of the SR protein to be exerted in the early steps of splicing. Our method has the advantage of being unbiased and suggests a more prominent role for intronic sites in splicing regulation than has been appreciated; however, the full significance of this finding will require further investigation. Also of note is that one segment that binds B52, segment #6-7, is located in a 3′ untranslated region. The binding of B52 to both exonic and intronic segments of RNA is shown in Figure 2.
B52-specific splicing defects
The B52 gene is essential for Drosophila development. Homozygous B52-null flies develop to late second instar larvae and remain at this stage for several days until they die (7). These developmentally arrested larvae provide unique and convenient material to test the candidates for splicing defects in vivo, even though it limits our assay to genes that are expressed at this stage. To evaluate the effect of B52 binding on splicing in vivo, we compared the splicing products of candidate B52-target genes in B52-null larvae to those in wild-type animals. We examined each of the 20 intron-containing candidates, using primer sets designed to amplify segments with boundaries in the exons flanking the B52-binding region in RT–PCR assays. Fifteen transcripts were detectable by this assay in wild-type flies. The others presumably are either not expressed during this stage or are not amplifiable under the conditions used. Of the 15, several showed different patterns in the B52 mutant relative to wild type. The four showing the most striking differences were chosen for further analysis (Fig. 3). Several repeats of the RT–PCR assay using independent RNA preparations produced the same results. A second primer set that amplifies within one exon was used as a control in the same PCR. Also, an internal standard, β1-tubulin mRNA, was amplified from the same RNA samples. With each RNA preparation, the B52 mRNA was measured by RT–PCR to confirm that it is indeed depleted in the B52-null larvae assayed. To confirm our assignments of the RT–PCR bands, we also sequenced the PCR products.
Figure 3.
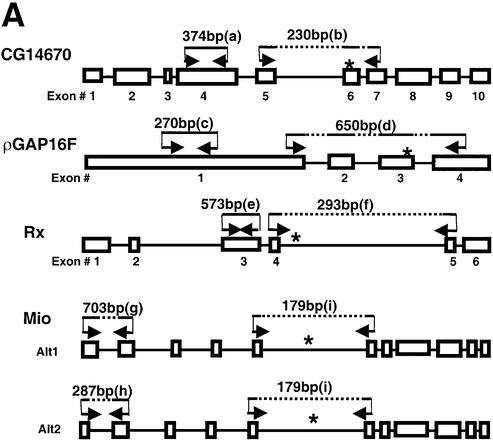
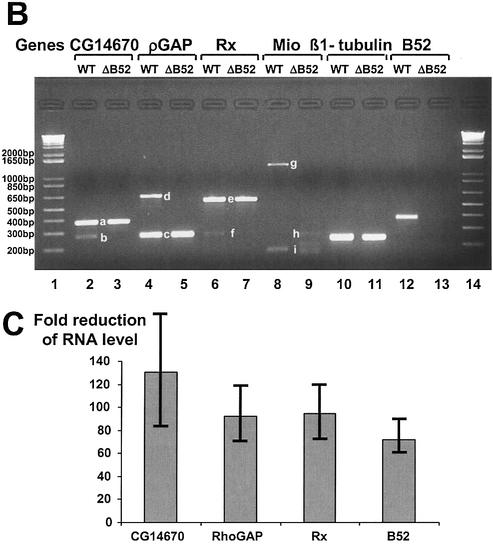
Splicing deficiency of four B52-binding pre-mRNAs in B52 deletion flies revealed by RT–PCR assay. (A) Schematic diagrams (not to scale) of the four pre-mRNAs. Exons are represented by boxes, and introns by lines. Asterisks indicate the location of segments containing B52-binding sites. The location of primer sets used in RT–PCR and the predicted lengths of RT–PCR products from wild type are also indicated. All primers reside completely within exon sequences. (B) RT–PCR products visualized on an agarose gel. The identities of the lettered bands are indicated in (A). (C) Real-time PCR assays of specific RNAs in the B52 mutant and wild-type larvae. The fold reductions in the RNAs’ abundance in B52-null mutant larvae are plotted with standard deviations (N = 3). The reduction represents the ratio of the RNA level in wild-type to the B52-null mutants using β1-tubulin mRNA levels as a standard. β1-Tubulin mRNA levels are unaffected in the B52 mutant (7).
The gene containing segment #12-1 is RhoGAP16F, a member of the Drosophila RhoGAP family, which is involved in regulating axon branch stability (32). The gene containing #12-3, CG14670, apparently encodes a biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase (EC 6.3.4.10). B52 protein binds to exonic regions of these pre-mRNAs, consistent with previous data on ESEs in other genes (30,31). Figure 3B shows that the pre-mRNA of CG14670 is not properly spliced between exon 5 and 7 in B52 mutants (lane 3), but is spliced properly in wild-type flies (lane 2). However, exon 4 of the same gene was detected in both B52 mutant and wild type, indicating that the absence of B52 only affected splicing, but not transcription or stability, of this RNA. Similarly, while the pre-mRNA of RhoGAP16F could not be spliced properly between exons 1 and 4 in the B52 mutant (compare lanes 4 and 5), similar amounts of exon 1 product were observed in both the wild type and B52 mutant.
The gene containing #4-3 is Rx, which encodes a transcription factor with a homeobox domain and an OAR domain. The vertebrate homolog of Rx has a function during brain and eye development. In situ hybridization experiments on Drosophila larvae suggested a role of Rx in the development of larval brain (33). The gene has a large intron where B52 binds, separating exon 4 and 5. We used a primer pair that covers a region across the splice junction of exons 4 and 5 to assay the level of its spliced product. The B52 deletion mutant showed a severe decrease in the exon 4–5 splicing product of the Rx gene, relative to wild type (Fig. 3B, lanes 6 and 7). However, the level of exon 3 RNA is not affected significantly by the B52 deletion. While this result suggests that this intronic B52-binding site plays a role in the removal of this intron, further studies are required to prove the causal relationship. Similar to this case, in the alternative splicing of interferon regulatory factor-3 (IRF-3) pre-mRNA, a subset of SR proteins seem to interact with an intronic position to promote the use of the upstream 5′ splicing site (34).
The gene containing #5-1 encodes Mlx interactor (Mio), also known as dMondo, a basic helix–loop–helix–leucine zipper transcription factor involved in a Max-like transcriptional regulatory network (35). It has two alternative splicing products, Alt1 and Alt2 (Fig. 3A). One primer set, which can reveal the different sizes of Alt1 (predicted to be 703 bp) and Alt2 (predicted to be 287 bp), produced only Alt2 product (lane 9) in the B52-null mutant. In contrast, Alt1 product was produced by this primer pair in wild-type flies (lane 8). (This Alt1 product, the band g, is longer than predicted, and we have confirmed its sequence as being part of this gene.) The observed B52 binding far from this affected region (Fig. 3A) may exert its effect at a distance, kilobases away, or alternatively, another binding site may exist near the affected region. Using another primer set that covers the exons flanking B52-binding sites, we detected PCR products both in B52-null mutant and wild type, and here too, the patterns of small products differ slightly in the mutant and wild-type larvae. Further tests of these sequences and characterization of splicing defects require more comprehensive analyses that go beyond this screen and initial identification of targets.
The splicing defects in the B52-null flies were also quantified using a real-time PCR assay. We used five sets of primers with QuantiTect SYBR Green PCR Mix to amplify β1-tubulin mRNA, CG14670 (from exons 5 to 7), RhoGAP16F (from exons 3 to 4), Rx (from exons 4 to 5) and B52 mRNA, from the RT products. As summarized in Figure 3C, this assay revealed a severe decrease in the level of the spliced RNAs containing the corresponding segments in the B52 mutant.
Together, the results in Figure 3 indicate that the B52 protein binds exonic or intronic regions and regulates constitutive or alternative splicing of a specific subset of pre-mRNAs in vivo. Of the 15 candidates with detectable transcripts in late second instar larvae, at least four genes showed clear splicing defects. Screening by genomic SELEX followed by testing in B52-null mutants is a significantly more effective search scheme for B52 targets than randomly testing transcripts for splicing defects (P = 0.015 by Fisher’s exact test). Many candidates with B52-binding capability were, nevertheless, properly spliced in the B52-null mutant larvae. This is likely due to functional redundancy among SR proteins in splicing of many introns. Consistent with this model, depletion of ASF/SF2 only affected the splicing of a few pre-mRNAs (8). The heuristic nature of our approach, guided by the available knowledge of B52’s function in splicing, while efficient, could also limit the scope of identified target genes. First, the set of B52 targets is defined operationally by RNA-binding and splicing; other modes of SR protein interaction and SR protein function are excluded. Secondly, our splicing assays surveyed splicing efficiency of exons residing in or near the B52-binding segments and could have missed more long-range effects on splicing. Thirdly, our whole-larva assay would have missed defects in certain particular tissues, since different tissues have different SR protein levels that may compensate for the absence of B52 to different degrees (14).
The RNA-binding site in one sequence segment resembles the B52 aptamer
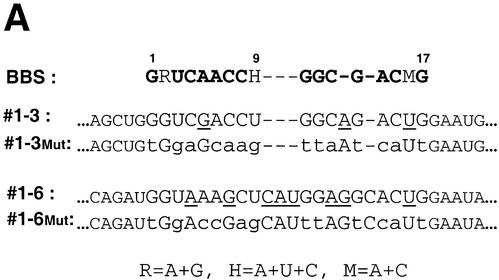
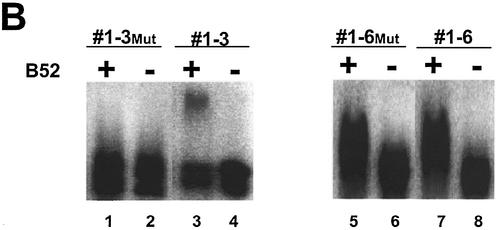
While the search for the target genes was facilitated by using longer inserts (200 nt) in the genomic SELEX than normally used in conventional in vitro selection, this method made it more difficult to precisely locate the B52-binding sites, since those are usually <20 nt in length. The B52 protein previously selected a family of aptamers, a group of 17mers termed B52-binding sequences (BBS), from a large (1013) unbiased RNA sequence pool (22). But attempts to identify native B52-dependent genes by searching the Drosophila genome for genes with the BBS sequence were not productive. After obtaining the sequence data of the genomic SELEX, we searched for BBS-related sequences in the clones that showed B52 binding. Using the multiple sequence alignment program ClustalW (ClustalW www Service at the European Bioinformatics Institute http://www.ebi.ac.uk/clustalw) (36) we found sequences similar to BBS in segments #1-3 and #1-6 (Fig. 4A), with the one in #1-3 appearing to have a higher similarity to BBS. Compared to the BBS consensus, the site in #1-3 has a G-U combination at the co-variant positions #2-16. It also has an A→G transition at position #5 and an A insertion between positions 12 and 13. Introduction of multiple G↔T and A↔C transversions to this homologous region abolished the ability of the segment to bind B52 (Fig. 4B, lane 1). A database search revealed that segment #1-3 is part of the F element, a transposable element (Table 1). The other segment, #1-6, contains a stretch that has less similarity to the BBS (Fig. 4A), and the same mutation strategy did not affect its binding to B52 (Fig. 4B, lane 5). Other selected sequences listed in Table 1 did not have extensive homology with BBS, indicating that some B52-binding sites specified by genomic sequences must be different from the highly-selected aptamers from large random sequence pools, whose affinity for B52 is stronger than any isolated from the genomic SELEX.
Figure 4.
Identification of a B52-binding site on a sequence segment selected by the genomic SELEX. (A) BBS-like elements found in two segments by the multiple alignment program ClustalW (36) (http://www. ebi.ac.uk/clustalw) are shown as are their mutated derivatives. Underlined letters in the sequence of #1-3 and #1-6 indicate deviations from the BBS consensus. Lower case letters in the ‘Mut’ constructs indicate sequence differences from the original isolates. (B) A mobility shift assay with B52 shows the BBS-like element of segment #1-3 is critical for B52 binding.
To identify RNA sequences recognized by SR proteins, several SELEX experiments using short, randomized pools have been performed. These experiments yielded several tight-binding RNA sequences for the RRM domain(s) of several SR proteins (22,37,38). However, the identification of native genes whose splicing is regulated by single SR proteins has not been effectively facilitated by the availability of these sequences. One reason is that the sequences were usually short, which made it difficult to search the genome database to identify native genes. Another reason is that the SELEX procedure selects the tightest binders from a vast sequence space, but native sequences may not necessarily have, and the functioning of the SR protein may not necessarily require, a very high affinity. As a result, the native binding sites may exhibit considerable variability. Also, it has been noted that the sequences identified by SELEX using different schemes may be different, adding another level of complication (39).
Three important points emerge from this work. First, the alteration or disruption of splicing of specific pre-mRNAs by the B52 deletion might explain the developmental arrest and lethality of the B52-null animals. Secondly, as first noted by Singer et al. (18) and Shtatland et al. (19), the genomic SELEX approach provides an efficient means of ‘ribonomic’ search for natural candidate RNAs that bind specific proteins. Thirdly, the B52-specific splicing substrates identified here will facilitate mechanistic studies of this SR protein on natural substrates.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Martha Hamblin for advice on statistical analysis. We also thank Drs Volker Vogt and Mariana Wolfner for their thoughtful comments on the manuscript. This project was funded by NIH.
REFERENCES
- 1.Fu X.-D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- 2.Valcarcel J. and Green,M.R. (1996) The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci., 21, 296–301. [PubMed] [Google Scholar]
- 3.Blencowe B.J. (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci., 25, 106–110. [DOI] [PubMed] [Google Scholar]
- 4.Hastings M.L. and Krainer,A.R. (2001) Pre-mRNA splicing in the new millennium. Curr. Opin. Cell. Biol., 13, 302–309. [DOI] [PubMed] [Google Scholar]
- 5.Fu X.D. and Maniatis,T. (1990) Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature, 343, 437–441. [DOI] [PubMed] [Google Scholar]
- 6.Fu X.-D. (1993) Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature, 365, 82–85. [DOI] [PubMed] [Google Scholar]
- 7.Ring H.Z. and Lis,J.T. (1994) The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol., 14, 7499–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., Takagaki,Y. and Manley,J.L. (1996) Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev., 10, 2588–2599. [DOI] [PubMed] [Google Scholar]
- 9.Longman D., Johnstone,I.L. and Caceres,J.F. (2000) Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J., 19, 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jumaa H., Wei,G. and Nielsen,P.J. (1999) Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol., 9, 899–902. [DOI] [PubMed] [Google Scholar]
- 11.Ge H. and Manley,J.L. (1990) A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell, 62, 25–34. [DOI] [PubMed] [Google Scholar]
- 12.Mayeda A., Zahler,A.M., Krainer,A.R. and Roth,M.B. (1992) Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 89, 1301–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahler A.M. and Roth,M.B. (1995) Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc. Natl Acad. Sci. USA, 92, 2642–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman B.E. and Lis,J.T. (2000) Pre-mRNA splicing by the essential Drosophila protein B52: tissue and target specificity. Mol. Cell. Biol., 20, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J.Y. and Maniatis,T. (1993) Specific interactions beween proteins implicated in splice site selection and regulated alternative splicing. Cell, 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- 16.Kohtz J.D., Jamison,S.F., Will,C.L., Zuo,P., Luhrmann,R., Garcia-Blanco,M.A. and Manley,J.L. (1994) Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature, 368, 119–124. [DOI] [PubMed] [Google Scholar]
- 17.Xiao S.H. and Manley,J.L. (1997) Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev., 11, 334–344. [DOI] [PubMed] [Google Scholar]
- 18.Singer B.S., Shtatland,T., Brown,D. and Gold,L. (1997) Libraries for genomic SELEX. Nucleic Acids Res., 25, 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shtatland T., Gill,S.C., Javornik,B.E., Johansson,H.E., Singer,B.S., Uhlenbeck,O.C., Zichi,D.A. and Gold,L. (2000) Interactions of Escherichia coli RNA with bacteriophage MS2 coat protein: genomic SELEX. Nucleic Acids Res., 28, E93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 21.Rubin G.M., Hong,L., Brokstein,P., Evans-Holm,M., Frise,E., Stapleton,M. and Harvey,D.A. (2000) A Drosophila complementary DNA resource. Science, 287, 2222–2224. [DOI] [PubMed] [Google Scholar]
- 22.Shi H., Hoffman,B.E. and Lis,J.T. (1997) A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol., 17, 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippsen P., Stotz,A. and Scherf,C. (1991) DNA of Saccharomyces cerevisiae. Methods Enzymol., 194, 169–182. [DOI] [PubMed] [Google Scholar]
- 24.Carmona S., Passman,M., Kew,M. and Arbuthnot,P. (1999) Site-directed mutagenesis by fusion of contiguous DNA fragments. Biotechniques, 26, 382–384, 386. [DOI] [PubMed] [Google Scholar]
- 25.Mullinax R.L., Gross,E.A., Hay,B.N., Amberg,J.R., Kubitz,M.M. and Sorge,J.A. (1992) Expression of a heterodimeric Fab antibody protein in one cloning step. Biotechniques, 12, 864–869. [PubMed] [Google Scholar]
- 26.Shi H. (1997) Perturbing protein function with RNA aptamers. PhD Dissertation, Cornell University, Ithaca, NY.
- 27.Rubin G.M., Yandell,M.D., Wortman,J.R., Gabor Miklos,G.L., Nelson,C.R., Hariharan,I.K., Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W., Cherry,J.M. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraus M.E. and Lis,J.T. (1994) The concentration of B52, an essential splicing factor and regulator of splice site choice, is critical for Drosophila development. Mol. Cell. Biol., 14, 5360–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H., Hoffman,B.E. and Lis,J.T. (1999) RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA, 96, 10033–10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watakabe A., Tanaka,K. and Shimura,Y. (1993) The role of exon sequences in splice site selection. Genes Dev., 7, 407–418. [DOI] [PubMed] [Google Scholar]
- 31.Sun Q., Mayeda,A., Hampson,R.K., Krainer,A.R. and Rottman,F.M. (1993) General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev., 7, 2598–2608. [DOI] [PubMed] [Google Scholar]
- 32.Billuart P., Winter,C.G., Maresh,A., Zhao,X. and Luo,L. (2001) Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell, 107, 195–207. [DOI] [PubMed] [Google Scholar]
- 33.Eggert T., Hauck,B., Hildebrandt,N., Gehring,W.J. and Walldorf,U. (1998) Isolation of a Drosophila homolog of the vertebrate homeobox gene Rx and its possible role in brain and eye development. Proc. Natl Acad. Sci. USA, 95, 2343–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karpova A.Y., Howley,P.M. and Ronco,L.V. (2000) Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes Dev., 14, 2813–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Billin A.N., Eilers,A.L., Coulter,K.L., Logan,J.S. and Ayer,D.E. (2000) MondoA, a novel basic helix–loop–helix–leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol. Cell. Biol., 20, 8845–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tacke R. and Manley,J.L. (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J., 14, 3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinrichs V. and Baker,B.S. (1995) The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J., 14, 3987–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H.X., Zhang,M. and Krainer,A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.