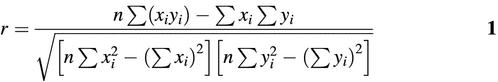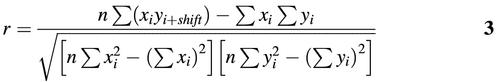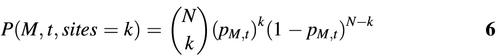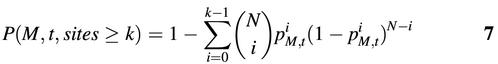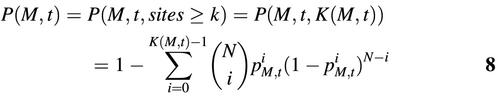Abstract
Computational identification of transcription factor binding sites is an important research area of computational biology. Positional weight matrix (PWM) is a model to describe the sequence pattern of binding sites. Usually, transcription factor binding sites prediction methods based on PWMs require user-defined thresholds. The arbitrary threshold and also the relatively low specificity of the algorithm prevent the result of such an analysis from being properly interpreted. In this study, a method was developed to identify over-represented cis-elements with PWM-based similarity scores. Three sets of closely related promoters were analyzed, and only over- represented motifs with high PWM similarity scores were reported. The thresholds to evaluate the similarity scores to the PWMs of putative transcription factors binding sites can also be automatically determined during the analysis, which can also be used in further research with the same PWMs. The online program is available on the website: http://www.bioinfo.tsinghua.edu.cn/∼zhengjsh/OTFBS/.
INTRODUCTION
A complex network of regulatory controls governs the patterns of gene expression (1). The regulation occurs in several steps of gene expression, including transcriptional regulation, mRNA splicing and modification, and translational regulation. Among the various regulation steps, transcription regulation determine the proper time for a gene to be transcribed into RNA molecules, so the energy can be utilized more efficiently because only the RNA sequences necessary for further translation are produced. Tightly orchestrated spatial and temporal regulation of gene transcription is critical to the proper development of all metazoans (2). Part of the blueprint of transcriptional regulation is stored in a large number of cis-regulating elements and enhancers surrounding the coding regions.
A cis-regulating element is a segment of DNA sequence which can interact with specific transcription factors to recruit basal transcription apparatuses at the transcription start site. The binding affinity of transcription factors and their corresponding cis-elements is largely affected by the sequence pattern of the cis-elements (3). After more and more transcription factors and their binding sites were experimentally identified, databases storing transcription regulation information were established, such as TRANSFAC (4), and TRRD (5). Consensus sequences were used to describe the sequence patterns of cis-elements, and later positional weight matrices (PWMs) were developed to describe the sequence motifs more precisely.
Computational methods to identify transcription factor binding sites were also developed based on consensus sequences or PWMs. Quandt et al. (6) developed a program to detect consensus matches in nucleotide sequence data named MatInspector. MatInspector uses the TRANSFAC matrices (PWMs) to identify motifs presenting high similarity with the matrix in DNA sequences. Threshold of the similarity should be given when user submits sequences for analysis with the original version of MatInspector.
A high threshold would prevent some noise signals from being included with the computational result. In other words, the number of false positives (FP) will decrease when the threshold increases. However, when a high threshold is applied, some real binding sites with low consensus motifs will be ignored. Therefore, the number of false negatives (FN, real binding sites with similarity score below the threshold) increases when the threshold increases. Hence, the thresholds should be carefully selected for algorithms based on PWMs (7). A well selected threshold will restrict the FP value to a small value without losing too many real sites.
There are also computational methods aiming to identify unknown signals by a significant local multiple alignment of all sequences (8). Two important methods of this type are Gibbs sampling (9) and expectation maximization in the MEME system (10). MEME is able to analyze a group of sequences for similarities among them and produce a description (motif) for each pattern it discovers. After high frequency sequence motifs were detected with these methods, a further analysis on the motifs to find their corresponding transcription factors was very helpful in comprehending the analysis results (8,11).
This paper describes a new method to identify over-represented oligonucleotides in the promoters of a same protein family or a group of functionally related genes with the TRANSFAC matrices. These oligonucleotides are putative cis-elements that may perform important regulatory roles in the transcription of at least part of the investigated sequences. After the motifs are detected with this method, they can be directly associated with their corresponding transcription factors by the annotations of TRANSFAC. During the analysis, suitable thresholds for the putative binding sites can be automatically determined with a statistical method to ensure the significantly over-representing of the motifs in the set of closely related sequences; a similar statistical significance was used and proved to be very useful in detecting over-represented oligonucleotides (12). Three groups of sequences were used to test our method, and the positions of some putative over-represented motifs were visualized to reveal more information based on the relative positions of binding sites. A comparison was also made between the over-represented motifs detected with our approach and the motifs detected with MEME.
MATERIALS AND METHODS
Sequence sets used to detect over-represented oligonucleotides
Actin, interferon (IFN) and hemoglobin promoters were selected from the Eukaryote Promoter Database (EPD) (13) and EMBL. EPD provides the corresponding EMBL accession number for each promoter it stores, the positional information about the upstream region of the transcription start site and also hierarchical classification information about which family the corresponding gene belongs to. Actin, IFN and hemoglobin were used as keywords to query the EPD for their promoter sequences. EPD hierarchical classification information was used to manually remove those records that do not belong to the target gene families. After the EPD records were retrieved, the EMBL accession numbers and also the positions of the transcription start sites in the sequences were collected. Then the upstream region of the transcription start site was retrieved from the EMBL database with the accession number. The length of each retrieved upstream sequence ranges from ∼90 to thousands of base pairs. The upstream sequences longer than 600 bp were cut at the 3′ end, so that only the proximal 600 bases upstream of the transcription start site were used in the analysis. A redundancy analysis was applied to the three sequence groups with BLAST. Sequences with high BLAST similar scores were clustered together. Only one sequence in each cluster was kept for analysis. Thus, the upstream regulatory regions for the genes of actin, IFN and hemoglobin were collected for analysis (see Table 1). Some promoter regions were retrieved from the same EMBL record containing multiple genes. For example, the EMBL record X59989 (Gallus gallus) contains an alpha-A globin gene and an alpha-D globin gene. Upstream regions of these two genes were both collected.
Table 1. Sequence groups used to detect over-represented motifs.
| Sequence sets | Number of sequences | Average length (bp) | EMBL accession number (position of the transcription start site) |
|---|---|---|---|
| Actin | 11 | 457.36 | V01507 (92), V01218 (193), M20543 (708), X04669 (779), X05392 (417), M10607 (1091), M26773 (677), X00182 (544), V01217 (235), Y00474 (2010), X02648 (556) |
| Hemoglobin | 33 | 517.5 | X03712 (1564), X03713 (1559), X59989 (646), X59989 (3655), J00923 (331), X01831 (101), V00714 (372), M74142 (6743), M17902 (169), Z84721 (37543), X62302 (430), X04726 (216), X07053 (1419), X03234 (1451), M27933 (2028), M27934 (6775), M27932 (911), L17432 (20302), L17432 (16014), L17432 (8137), M13487 (552), X15740 (270), M63453 (10766), X14061 (38287), X14061 (53496), Y00347 (1432), U01317 (62137), U01317 (54740), U01317 (34478), U01317 (19488), M18818 (15463), X01912 (425), X01913 (423) |
| Interferon | 10 | 465.4 | X01973 (419), X01971 (616), M13710 (420), X75934 (887), X02958 (423), X02956 (369), V00533 (345), E00302 (738), V00534 (284), X14029 (1195) |
The number in parentheses after the EMBL accession number is the position of the transcription start site of the selected gene (related to the first base of the sequence).
Sets of sequences used as controls
Upstream sequences retrieved from EPD databases were used as one of the sources of control data sets. Since all the three sequence sets used in our analysis were made up of vertebrate sequences, only the vertebrate promoter sequences of EPD were collected in this EPD promoter control set. Again, the sequences longer than 600 bp were cut at their 3′ end to ensure that only the proximal 600 bases upstream of the transcription start site were used. This EPD control was used as a public control for each of the three groups of the promoters to provide a background frequency for each of the investigated motifs. 786 sequences were included in this EPD promoter control; the average length of these sequences was 515.24 bp.
In order to estimate the effect of the GC content on the background frequency of detecting a motif with a certain threshold, a set of random sequences with the same GC content and 50 times the base pairs of each of the three tested promoter sets was generated as another control. These three random control sets (one random control set for one tested set) were used to remove motifs whose over-representation was largely affected by the GC content of the tested sequences.
A control set containing sequences of the second exons of vertebrate genes (based on EMBL database Release 72) was used to estimate the false positive rate for each possible threshold. Similar sequence sets were designed previously (7,14) and used as standard negative test sets. Up to now, only very few functionally relevant binding sites have been revealed experimentally in second exons; therefore the potential binding sites found in these sequences may be considered a priori as false positive (7). 8862 sequences of the second exons were collected in this Exon2 control set; the average length was 270.86 bp.
Computation of the MatInspector similarity score and similarity distribution
The algorithm of MatInspector was used to calculate the matrix similarity with TRANSFAC matrices (TRANSFAC 6.0 public). The matrix similarity thus calculated will range from 0 to 1. A higher score indicated higher similarity between the sequence scanned and the sequence pattern represented in TRANSFAC matrix (6). A segment of sequence with a similarity score larger than or equal to the defined threshold will be regarded as a binding site candidate. In order to calculate the distribution of similarity scores in the control data set and also in the tested data set, 100 threshold candidates ranging from 1 to 0 were defined, such as 1.00, 0.99, 0.98, and so on. With each threshold candidate, the total number of binding site candidates within a group of sequences was calculated. Thus, a list of similarity distributions was generated for each control data set and tested promoter groups. The similarity distributions of the EPD promoter control data set were stored for further comparison.
Some TRANSFAC matrices were derived from symmetric or partially symmetric cis-elements. The similarity score of such a symmetric matrix on both DNA strands varies simultaneously. Especially, high MatInspector similarity scores occur almost at the same position on both strands. To avoid counting two high scores for a same motif, the method to calculate the similarity distribution for symmetric matrices was specially designed.
Determination of symmetric matrices
The co-variation of the MatInspector similarity scores on both DNA strands was used as a measure to test if a matrix was symmetric or partially symmetric. For the consensus sequence derived from a symmetric or partially symmetric matrix, a center line could be defined to divide the consensus sequence into two segments. The nucleotides on one side of the line were complementary with the corresponding nucleotides on the other end. However, the center lines of such matrices were not always located at the exact middle of the matrices, which means that there were one or several extra bases on one of the two ends of the center line. The extra information for those extra bases was also recorded in the corresponding matrix.
A 20 000 bp long random sequence was analyzed with all TRANSFAC matrices, the similarity scores of both strands were recorded for each position, resulting in two serials of similarity scores. For a symmetric matrix without any extra bases, Pearson’s correlation co-efficient can be calculated to measure the co-variance of the similarity score for both strands:
where xi is the similarity score of the original DNA strand at position i, and the position i was defined as the index of the first base (5′ end) of the motif on the original strand; yi is the similarity score of the reverse complementary strand at position i. The position i on the complement strand was defined as the index of the last base (3′ end) of the motif. Thus, similarity scores of the two strands of the same DNA segments were assigned to the same position i.
For the matrices with one or several extra bases, the positions of the complementary strand should be shifted appropriately to calculate the co-variation of the two serials of similarity scores. If one extra base was located at the 3′ end of the consensus sequence, the comparison of the similarity scores should be made between xi and yi+1. In other words, the similarity score on the original strand at position i should be compared with the similarity score at position i + 1 on the complementary strand. The absolute value of this shift was determined by the number of the extra bases. A positive shift means the extra bases were located at the 3′ end of the consensus sequence derived from the matrix, and a negative shift means the extra bases were located at the 5′ end. A zero shift means there is no need to shift the positions when calculating the correlation co-efficient.
In order to determine if a matrix is symmetric or partially symmetric, all possible shifts were tested to find out the appropriate shift with the highest correlation coefficient. The range of all possible shifts was:
where LM was the length of the testing matrix. A Pearson’s correlation coefficient was calculated to measure the co-variance of the similarity score for both strands for each shift with a slightly different form of formula 1:
The shift with the highest correlation coefficient was selected as the appropriate shift for further analysis. Matrices with a highest Pearson’s coefficient >0.8 were collected in the symmetric subset of TRANSFAC matrices; the others were collected in the asymmetric subset. Figures plotting the (xi, yi+shift) pairs were generated for additional examination of the co-variation of the similarity scores for each of the matrices. The figures were manually examined, and finally 59 matrices were collected in the asymmetric subset (data not shown). The appropriate shift for each of these 59 symmetric matrices was also recorded.
Similarity distribution for symmetric matrices
The method to calculate the similarity distribution of the symmetric subset of matrices was different from that used for common matrices. Two DNA strands were scanned simultaneously; the similarity scores of the two DNA strands were compared with the positions of one strand shifted to the previously calculated shift value for that matrix. The highest one was used to count the similarity distribution for each threshold candidate (mentioned before). Hence, the total number of the positions analyzed for each sequence was the length of this sequence.
Comparison between control sets and tested groups, and the detection of the over-represented transcription factor binding sites
With the similarity distribution of the control data set available, pM,t, the probability of getting a motif with a similarity score to a matrix (M) larger than or equal to a given threshold t within the control sequences can be easily calculated:
PM,t = St/Ncontrol4
where St is the number of binding site candidates with the similarity score not less than t, and Ncontrol is the total number of sub-sequences scanned in the control data set. For common matrices, both strands of a sequence were scanned separately in our analysis. Ncontrol is actually twice the total base pairs of the control sequences, while for the symmetric matrices, Ncontrol is simply the total number of base pairs of the control sequences.
The probability of getting a motif with a similarity score to a matrix (M) larger than or equal to a given threshold t within the false positive test sequences (Exon2 set) can be similarly calculated:
FPM,t = St/NFP5
where NFP is the total number of sub-sequences scanned in the false-positive test set. Since potential cis-elements found in the sequences of second exon were regarded as false positives, FPM,t was used as the probability to get a false positive site for matrix M and at threshold t.
If k candidate binding sites with similarity score not less than t for matrix M were located in a group of sequences, the probability to observe exactly k candidate binding sites with higher similarity in a given sequence group is estimated by the binomial formula:
where N is the total number of sub-sequences scanned in the analyzed sequence group. Similarly, for the asymmetric matrices, N equals twice the total base pairs of the analyzed sequence group. For the symmetric matrices, N is simply the total base pairs of the analyzed sequences. Finally, the probability to detect k or more positions with similarity to matrix M larger than or equal to threshold t is:
For a given matrix M, the number of candidate binding sites detected within the analyzed sequence group was determined by threshold t. In other words, k is a function of t: k = K(M, t). Then the above equation can be written as:
When k is larger than the expected occurrence of candidate binding sites in the tested sequences, that is, when pM,t × N < k, a very low P(M, t) means that it is almost impossible to get k or more motifs with a similarity score for M not less than t in a randomly selected sequence group with the same number of base pairs. In other words, when pM,t × N < k, a very low P(M, t) for a given sequence group indicates the motifs similar to matrix M are over-represented in this group of sequences.
When the threshold t decreases, more motifs in control data sets and also in other data sets with a similarity score ≥t will be detected, so when t decreases, pM,t will increase. In order to restrict the number of false positives, the thresholds for each matrix M were selected from the set:
{t | FPM,t × 1000 ≤ 1.5&pM,t > 0&pM,t × N < k} ∩ {t | P(M,t) ≤ 10–4}.9
Thus, in the false positive test set, the average number of similar motifs detected with threshold t per 1000 bp will be no more than 1.5 (the probability of FP is no more than 0.0015). Notably, when no similar motifs are detected for matrix M at threshold t in the control data set, that is, when pM,t = 0, k = K(M, t) will usually range at 1 or 2 for the compared data set. Thus, threshold t with pM,t = 0 will be excluded from the above threshold set, since this study just focused on the over-represented motifs; motifs that just occur only once or twice in the tested sequence group will not be of interest in this analysis. However, in that situation, with pM,t = 0, P(M, t) will always be 0 for any k > 0. This candidate threshold set can also be empty, which means there is no suitable threshold for matrix M when dealing with the tested sequences.
When several threshold values were available in the above set for a matrix M, the t with the smallest P(M, t) will be used as the final threshold to detect putative binding sites for the transcription factors associated with matrix M. When multiple t values share the same P(M, t), the t value with the lowest pM,t, that is, the biggest t, will be used (to reduce false positives).
The comparison between control sequences and tested sequences was automatically carried out. If the candidate threshold set defined in equation 9 for a matrix was not empty, an appropriate threshold t for the matrix would be selected according to the rules defined above. Then the over-represented cis-elements located with this matrix were recorded for each tested sequence. The transcription factors associated with the matrix may have an important role in the transcriptional regulation for the tested sequences by binding to the putative cis-elements. Finally, with the threshold thus determined, a detailed report was generated which includes the TRANSFAC matrix accession (M) associated with each motif, the positions of the putative cis-elements, the similarity scores, on which strand the element was located, and K(M, t) and P(M, t) for each of the selected thresholds.
The frequency of some motifs in the sequences was largely affected by the GC-content of the sequences. If a motif was only selected with the EPD control and failed to be selected with the random control, the over-representation of such a motif in the tested sequences would probably be an effect of the special GC-content of the tested sequence. Finally, with the EPD control used to get the appropriate threshold value for the matrices, the random control was used as an additional control to remove those GC-content related motifs.
Visualization of the distribution of the over-represented motifs in promoters
After the report file was generated for a group of sequences, a JAVA program was used to visualize the distribution of the over-represented motifs in the tested sequence group. Positional information may be useful for further analysis.
Comparison between the motifs detected with the new approach and MEME
The three sequence sets were analyzed with MEME, which is available as a web service on http://meme.sdsc.edu/meme/website/meme.html. Any number of repetitions of each motif was allowed within each sequence. The minimum width of the motifs was 6 and the maximum width of the motifs was set to 30, because the maximum length of the matrices in the TRANSFAC was 30. The maximum number of different motifs found within each group of sequences was tested from 6 to 16. The total number of different motifs stopped increasing, after the value assigned for the maximum number became large enough. Finally, the maximum number of different motifs was set to 12 for each sequence set, because no more than 12 different motifs were detected with MEME in any of the sequence sets.
The motifs detected with MEME (MEME motif) were then compared with the putative cis-elements detected with our approach. Two motifs that share more than half of the bases of the smaller one were regarded as overlap motifs. Each motif detected with MEME was analyzed to locate all the overlapped cis-elements (detected with our approach) in each sequence set. MEME motifs without any overlapped putative cis-elements and putative cis-elements without any overlapping MEME motifs were also counted. Then the proportion of the copies of MEME Motif-i with at least one overlapped cis-element was calculated:
OPi = Oi/Ni10
where the Oi is the number of copies of MEME Motif-i with at least one overlapped cis-element in a sequence set, and Ni is the total number of the copies of MEME Motif-i. Similarly, the proportion of the copies of MEME Motif-i with at least one overlapped cis-element of TRANSFAC matrix M was calculated:
OPi,M = Oi,M/Ni11
where the Oi,M is the number of copies of MEME Motif-i with at least one overlapped cis-element detected with TRANSFAC matrix M. For each of the cis-elements detected with TRANSFAC matrices, the proportion of overlapped copies was also calculated similarly:
OPM = OM/KM12
OPM,i = OM,i/KM13
where OPM represents the proportion of the cis-elements for TRANSFAC matrix M with at least one overlapped MEME motif, and OPM,i is the proportion of the cis-elements for matrix M with at least one overlapped MEME Motif-i. OM is the number of overlapped cis-elements for matrix M, and OM,i represents the number of cis-elements with at least one overlapped MEME Motif-i. KM is the number of all cis-elements located with TRANSFAC matrix M within the sequence group. OPi, OPi,M, OPM and OPM,i were used to compare the result of MEME and the putative cis-elements detected with TRANSFAC matrices.
RESULTS AND DISCUSSION
Over-represented motifs and their corresponding transcription factors
For each of the three test sequence sets, two groups of over-represented cis-elements were detected with the two control sets mentioned above (the EPD control and the random control). A list of the corresponding TRANSFAC matrices and transcription factors is shown in Table 2. Only the motifs detected by both EPD control and random control are shown. Thresholds and also the corresponding P(M, t) value for each transcription factor are also given, which can be a useful reference for further research based on the same TRANSFAC matrices. A zero probability means that the probability was too small to fit the program’s computational ability. Motifs with a high MatInspector similarity score for the matrices associated with the listed transcription factors occur more frequently than expected. Thus, the listed transcription factors may be of great importance for the regulation of at least part of the genes in the group.
Table 2. Over-represented motifs and their corresponding TRANSFAC matrices/transcription factors.
| Set | Matrix name | Matrix access | Threshold (t) | Motifs located in the group (k) | P(M, t) | Overlapped motifs detected with other TRANSFAC matrices (matrix name/access) |
|---|---|---|---|---|---|---|
| Actin | Adf-1 | M00171 | 0.86 | 25 | 1.17E–09 | |
| Hb | M00022 | 0.91 | 67 | 5.03E–14 | STE11/M00274 | |
| RAP1 | M00213 | 0.81 | 26 | 9.82E–09 | ||
| Sp1 | M00008 | 0.87 | 59 | 3.56E–07 | GC/M00255 | |
| M00196 | 0.83 | 73 | 6.83E–10 | |||
| SRF | M00152 | 0.72 | 17 | 3.09E–11 | AG/M00151, MCM1/M00125 AGL3/M00392, AGL3/M00393 | |
| M00186 | 0.91 | 16 | 0 | |||
| M00215 | 0.89 | 15 | 0 | |||
| Hemoglobin | GATA-2 | M00348 | 0.96 | 17 | 8.69E–06 | |
| GATA-X | M00203 | 0.95 | 24 | 7.16E–07 | ||
| Oct-1 | M00135 | 0.72 | 110 | 9.53E–07 | CF2-II/M00012, CF2-II/M00013 Croc/M00266 XFD-1/M00267, XFD-2/M00268 | |
| M00138 | 0.8 | 156 | 0 | |||
| TATA | M00252 | 0.85 | 153 | 0 | ||
| Interferon | BR-C | M00094 | 0.86 | 68 | 0 | |
| dl | M00120 | 0.83 | 45 | 1.42E–07 | ||
| Dof2 | M00353 | 0.96 | 41 | 3.30E–08 | ||
| FOXJ2 | M00422 | 0.82 | 85 | 4.44E–16 | ||
| IRF-1 | M00062 | 0.77 | 53 | 0 | ||
| IRF-2 | M00063 | 0.71 | 72 | 2.22E–16 | ||
| ISRE | M00258 | 0.72 | 47 | 2.11E–09 | ||
| Oct-1 | M00138 | 0.84 | 39 | 0 |
The ‘overlapped motifs’ column lists the overlapped motifs detected with other TRANSFAC matrices.
Overlapped motifs detected with other TRANSFAC matrices were listed in the last column of Table 2. These overlapped motifs detected with different matrices were caused by the similarity between these matrices. For example, most of the motifs detected with the matrices for MCM1, AGL3 and AG within actin promoters overlapped with motifs detected with the matrix for SRF (data not shown). MCM1 is an SRF-like transcription factor of yeast. AGL3 and AG are transcription factors presenting a strong sequence similarity with SRF and MCM1 (15) which were found in Arabidopsis.
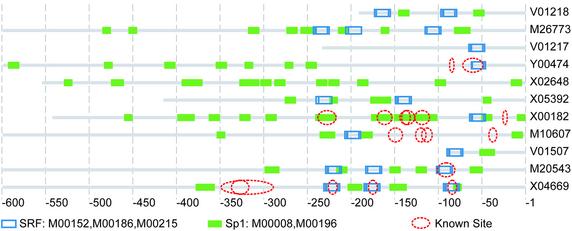
For the putative transcription factor binding sites located in actin promoters, SRF and Sp1 were previously reported in many articles to perform functional regulation on muscle gene regulation (16,17). Almost every sequence in the actin set contains multiply putative binding sites of Sp1. Sp1 was also reported to be able to mediate the fibroblast growth factor receptor 1 (FGFR1) gene in chicken skeletal muscle cells (18). Multiple binding sites of Sp1 were also detected in the promoter of FGFR1, indicating Sp1 might have an important function in regulating the muscle-specific gene and the multiple copies of binding sites may be important for proper regulation mediated by Sp1. Positions of the putative binding sites for SRF and Sp1 are shown in Figure 1. Known sites derived from EPD annotation are also shown.
Figure 1.
Distribution of two over-represented motifs and known binding sites in actin promoters. The corresponding TRANSFAC names and matrix accesses of the two over-represented motifs are: SRF (M00152, M00186, M00215) and Sp1 (M00252). The EMBL accession of each sequence was also included with the image behind each line representing a sequence. Known sites derived from the annotation of EPD and TRANSFAC records were also shown in the figure. The promoters of beta-actin (V1217, Y00474 and X00182) present a different distribution of the two kinds of putative cis-elements compared with other promoters of the actin set. The distribution of the two putative motifs in the sequence of V01218, M26773, M20543 and X04669 were conserved, especially the distribution of the putative binding sites of SRF.
Most of the known binding sites were covered by the putative cis-elements detected with our method (Fig. 1). Four actin promoters (Y00474, X00182, M10607 and X04669) contain known motifs that were not detected with the new approach. For all the known SRF binding sites in the actin promoter set, only one known SRF site was not detected (Y00474). Known binding sites for MyoD in the promoter region of M10607 and X04669 were not covered by any putative binding sites detected with our method. Putative binding sites for Sp1 in the promoter region of X00182 overlap with four known binding sites for ETF (TRANSFAC annotation) and one known binding site for GCF (TRANSFAC annotation). Only one known ETF binding site was not overlapped by putative binding sites of Sp1. Three known binding sites (TRANSFAC accession numbers R01750 and R01751) were not discovered with our method.
The promoters of beta-actins (V1217, Y00474 and X00182) represent a different distribution of the two kind of putative cis-elements compared with the other promoters of actin (Fig. 1). The locations of the putative binding sites of SRF in these three sequences were almost the same. Different motif distributions of different types of actin genes may indicate that due to the different functions of different actin genes, different regulatory mechanisms were developed for each kind of actin during the evolution. The distribution of the two putative motifs in the sequence of V01218, M20543, M26773 and X04669 were conserved, especially the distribution of the putative binding sites of SRF (Fig. 1). V01218 and M20543 are both alpha-actin sequences of skeletal muscle, while M26773 and X04669 are both cardiac alpha-actin sequences. V01507 is another skeletal alpha-actin sequence. The putative binding sites of SRF and Sp1 on V01507 are also similar to that on the proximal regions of the promoters in V01218, M26773 and M20543. Multiple copies of putative Sp1 binding sites were located in almost every sequence of the actin promoter set.
Different copies for the same cis-element with overlapped bases were all counted. Therefore, some long tandem repeat sequences may significantly increase the number of putative cis-elements located with some matrices. For example, 18 of the motifs detected with Rap1/M00213 (yeast) were located in a GT-tandem repeat segment in the sequence M26773. Five of the other motifs located with Rap1 overlap with the motifs detected with other matrices. Therefore, the motifs located with M00213 within actin promoters may not be the actual binding sites of Rap1. The possible function of the GT-tandem repeat needs further investigation.
Most of the motifs detected with the matrices of Hb/M00022 (fruit fly) and STE11/M00274 (fission yeast) within the actin promoters are poly(A) (M00022) or poly(T) (M00274) segments. The sequence of V01217 contains a long segment of poly(T) segment made up of 47 T bases. This segment contributes more than half of the motifs detected with STE11/M00274. These poly(A)/poly(T) segments in the actin promoters may not be the actual binding sites of Hb and STE11. The possible function of the poly(A)/poly(T) segments need to be further investigated using an experimental approach.
In hemoglobin promoters, only three kinds of over- represented cis-elements were located with this method within the 33 promoter sequences. The corresponding transcription factors are: GATA, Oct-1 and TATA-binding protein. The GATA family comprises key transcription factors in stimulating the syntheses of hemoglobins (19). GATA-1 is an important regulator of erythrocyte differentiation. GATA-1 can stimulate the syntheses of alpha- and beta-globins, and the enzymes of heme biosynthesis. Recently, Horak et al. (20) reported that by using mammalian chIp–chip analysis, GATA-1 binding sites were mapped in the beta-globin locus. The binding sites of Oct-1 were also reported previously (21,22). Mutation at the Oct-1 binding site within the promoter region of gamma-globin can lead to activation of gamma-globin gene (22), suggesting Oct-1 may play an important role in regulating the switch between gamma-globin and beta-globin during the development of hematopoietic system.
The hemoglobin group was made up of the promoters of alpha-like hemoglobins and beta-like hemoglobins; the relatively fewer number of over-represented cis-elements thus detected within the hemoglobin group may result from different regulation patterns of different kinds of hemoglobins.
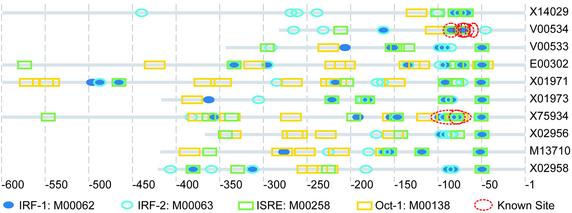
In the IFN group, putative binding sites of interferon regulatory factor (IRF-1 and IRF-2) were successfully recognized with our method. Putative IFN-stimulated response elements (ISREs) were also detected in the IFN group, part of the ISRE motifs overlapped with the motifs detected with matrices for IRF-1 and IRF-2. These over-represented motifs are all potential binding sites of the IRFs. The distribution of putative cis-elements detected in IFN promoters with the TRANSFAC matrices of IRF-1, IRF-2, ISRE and Oct-1 is shown in Figure 2. Known binding sites derived from the EPD and TRANSFAC annotation are marked as red circles.
Figure 2.
The distribution of putative cis-elements detected in IFN promoters and known binding sites (the red circles). Putative binding sites detected with the TRANSFAC matrix IRF-1 (M00062), IRF-2 (M00063), ISRE (M00258) and Oct-1 (M00138) are shown. The putative binding sites detected with IRF-1, IRF-2 and ISRE are partially overlapped.
IRF-1 is a transcription factor that regulates IFN-induced genes and type I IFNs (23). IRF-2 is a transcription repressor of IFN signaling and thereby acts as an IRF-1 antagonist (24). IFNs induce the expression of a number of different proteins that mediate the antiviral, antiproliferative and immunomodulatory effects of IFNs (25). IRF-1 and IRF-2 are both important transcription factors in the IFN signaling system, acting as intermediate signals in the IFN signal pathway. The binding sites of these two transcription factors in the IFN promoters indicate a potential auto regulatory pathway of IFN. In hepatoma cells, a positive feedback mechanism in the IFN signaling system was discovered (25). The feedback regulation of IFN may be of great importance in precisely controlling the expression of such an important protein.
Multiple copies of putative binding sites of Oct-1 were also detected in almost every sequence in the IFN set (Fig. 2). Oct-1 was previously reported as an important transcription factor in regulating the transcription of IFN-γ, and the binding sites of Oct-1 were also discovered in the promoter of IFN-γ (26).
No literature information could be found that presents some relationship between IFN and the other four over-represented motifs detected in IFN promoters. The corresponding TRANSFAC matrices were BR-C (M00094), dl (M00120), Dof2 (M00353) and FOXJ2 (M00422). The potential function of these motifs needs further investigation. For the known binding sites, only the binding site for NF-kappaB in the sequence V00534 located at ∼65–55 bp upstream of the transcription binding site was not covered by the putative cis-elements. All known binding sites of IRF-1 and IRF-2 were covered by the putative cis-elements detected with TRANSFAC matrices (Fig. 2).
Comparison between the MEME motifs and putative cis-elements
With the parameters of MEME set as mentioned above, four motifs were detected in the actin set by MEME, 12 in the hemoglobin set, and eight in the IFN set. The motifs detected with MEME and the result of comparison between the MEME motif and the putative cis-elements are shown in Tables 3–5. The result of MEME contains no overlapping copies of the same motif, while different copies for the same cis-element detected with the same TRANSFAC matrix with overlapped bases were counted independently. Therefore the Oi,M value does not always equal the value of OM,i for different MEME Motif-i and TRANSFAC matrix M.
Table 3. Motifs detected with MEME with each sequence set.
| Sequence set | MEME motif | Consensus sequence | Nia | Oib | OPic (%) |
|---|---|---|---|---|---|
| Actin | Motif-1 | AAAAAAAAAAA | 16 | 12 | 75 |
| Motif-2 | CCCTCTCCCCACCCC | 29 | 26 | 89.6 | |
| Motif-3 | TCCATATTTGG | 22 | 20 | 90.9 | |
| Motif-4 | TATAAAAA | 6 | 2 | 33.3 | |
| Hemoglobin | Motif-1 | GGGGGGGCAGGGCGGGGGCCAGGGCTGGGG | 67 | 3 | 4.4 |
| Motif-2 | ATATTTATTTGTATTTATTTTTTTTATTTT | 50 | 47 | 94 | |
| Motif-3 | ACCCTAACCCCAACCCCAGCTCATGCCGGG | 13 | 0 | 0 | |
| Motif-4 | ACACCCTGGCCTTGGCCAATCTGCTCACAG | 14 | 8 | 57.1 | |
| Motif-5 | GGGGAGCCAGGGGGCTGAGC | 21 | 2 | 9.5 | |
| Motif-6 | GGCTGTCATCACTGAAGCCTCACCCTGTAG | 8 | 1 | 12.5 | |
| Motif-7 | GAATAAAAGGCCGCGCCGTGCAGCAGCTGC | 12 | 9 | 75 | |
| Motif-8 | ATAAAAGGCAGGGCAGAGTCAGCTGCTGC | 8 | 7 | 87.5 | |
| Motif-9 | GTGGAGGATAAAGAAGAGGGTAGAGATGG | 12 | 6 | 50 | |
| Motif-10 | CTCTTAAGCCAGTGCCAGGGCGGCCAAGGA | 9 | 0 | 0 | |
| Motif-11 | CAAGGAGGATGTTTTTAGTAGCAATTTGT | 9 | 4 | 44.4 | |
| Motif-12 | TGAGCGGCGCCCCGCCGGGC | 15 | 0 | 0 | |
| Interferon | Motif-1 | AAAGAAAGCCCAAACAGAAGTGGAAAGTG | 17 | 16 | 94.1 |
| Motif-2 | CTATTTAAGACCCATGCACAGAGCAAGGTC | 9 | 2 | 22.2 | |
| Motif-3 | CATTCAGAAAGTGGAAACTAGTATGTGCC | 9 | 9 | 100 | |
| Motif-4 | GGGCAGGGAAAGGGGAGGCAATAATGAAAA | 7 | 7 | 100 | |
| Motif-5 | ATGGTATATCTGTGTTATTAAAATTTCATG | 8 | 8 | 100 | |
| Motif-6 | TTCCAATTAGGAAGAAATTCCCTAAAAGCC | 10 | 10 | 100 | |
| Motif-7 | ACACGGCCCTACCCCCATGGGGAGAGGGC | 5 | 2 | 40 | |
| Motif-8 | AGGGTTTCTCTGTGAAGTCC | 9 | 6 | 66.6 |
The number of the MEME motifs overlapped with the putative cis-elements detected with our approach with the same sequence set was given.
aNi, the total number of the copies of MEME Motif-i in the sequence set.
bOi, the number of copies of MEME Motif-i with at least one overlapped cis-element in a sequence set.
cOPi, the proportion of the copies of MEME Motif-i with at least one overlapped cis-element detected with our approach.
Table 5. The proportion of putative cis-elements without any overlapped MEME motifs (1 – OPM).
| Sequence set | Corresponding TRANSFAC matrix name (accession) | KM – OM / KMa | 1 – OPM (%) |
|---|---|---|---|
| Actin | Sp1 (M00008) | 31/59 | 52 |
| Adf-1 (M00171) | 25/25 | 100 | |
| RAP1 (M00213) | 21/25 | 84 | |
| Hemoglobin | Oct-1 (M00135) | 63/108 | 58 |
| GATA-X (M00203) | 16/24 | 66 | |
| GATA-2 (M00348) | 10/17 | 58 | |
| Interferon | Oct-1 (M00138) | 21/39 | 53 |
| Dof-2 (M00353) | 21/41 | 51 | |
| FOXJ2 (M00422) | 43/85 | 50 |
Putative cis-elements detected with TRANSFAC matrix with an overlapping rate (OPM) >0.5 are not shown.
aNumber of putative cis-elements without overlapped MEME motifs / total number of the putative cis-elements detected with TRANSFAC matrix M.
In actin promoters, most of the copies of MEME Motif-2 and Motif-3 were covered by the putative cis-elements detected with the matrices for Sp1 and SRF, respectively. Seventy-five percent of the copies of Motif-1 were covered by the Poly(A)/Poly(T) related motif detected by the matrices of M00022 and M00274. Motif-4 is a conserved sequence of TATA-box. Ten putative TATA-boxes were also detected with the matrix of M00216 with the random control set (data not shown). TATA-box is a commonly used motif that can bind with TATA-binding proteins. The result indicates that the number of putative TATA-boxes was not significantly higher than the average level in all the vertebrate promoters, so TATA-boxes were not included as over-represented motifs when using the EPD vertebrate promoters as control set.
Similarly, the OPi values of five motifs detected with MEME in IFN promoters (Motif-1, -3, -4, -5 and -6) were >90.0%, indicating a high overlapping rate between these five MEME motifs and the putative cis-elements located with our approach. The corresponding putative cis-elements overlapped with these five motifs were detected with matrices for IRF-1 (M00062), IRF-2 (M00063), Oct1 (M00138) and BR-C (M00094). Four MEME motifs (Motif-1, -3, -4, -6 and -8) contain copies that were overlapped with the putative binding sites of IRF-1 and IRF-2.
MEME motifs with high OPi values were relatively fewer in the hemoglobin promoters. Only three MEME motifs (Motif-2, -7 and -8) have OPi values >0.75. Most of the copies of Motif-7 and Motif-8 overlap with the putative binding sites detected with the TRANSFAC matrices TATA (M00252). Part of the copies of Motif-2 and Motif-9 overlap with the over-represented cis-elements detected with M00266, M00267 and M00268. Only two putative cis-elements with highest OPi,M value are shown for each MEME Motif-i in Table 4. Actually, part of the copies of Motif-2 and Motif-9 were overlapped with putative cis-elements detected by the matrices of Oct-1 (most of the putative cis-elements detected with M00266, M00267 and M00268 were found to be overlapped with the putative Oct-1 binding sites, see Table 2).
Table 4. Putative cis-elements overlapped with motifs detected with MEME in each sequence set.
| Sequence set | MEME motif | Corresponding matrix of the overlapped cis-elements | Oi,Ma | OPi,Mb (%) | OM,i / KM = OPM,ic |
|---|---|---|---|---|---|
| Actin | Motif-1 | M00022 | 10 | 62.5 | 53/67 = 79.1% |
| M00274 | 9 | 56.2 | 25/31 = 80.6% | ||
| Motif-2 | M00196 | 24 | 82.7 | 42/73 = 57.5% | |
| M00255 | 24 | 82.7 | 42/77 = 54.5% | ||
| Motif-3 | M00151 | 18 | 81.8 | 28/38 = 73.6% | |
| M00186 | 16 | 72.7 | 16/16 = 100% | ||
| Hemoglobin | Motif-2 | M00267 | 39 | 78 | 93/179 = 51.9% |
| M00266 | 37 | 74 | 110/184 = 59.7% | ||
| Motif-4 | M00203 | 3 | 21.4 | 5/24 = 20.8% | |
| Motif-7 | M00252 | 9 | 75 | 9/153 = 5.8% | |
| Motif-8 | M00252 | 7 | 87.5 | 8/153 = 5.2% | |
| Motif-9 | M00267 | 5 | 41.6 | 5/179 = 2.7% | |
| Motif-11 | M00268 | 3 | 33.3 | 4/172 = 2.3% | |
| M00267 | 3 | 33.3 | 3/179 = 1.6% | ||
| Interferon | Motif-1 | M00094 | 11 | 64.7 | 12/68 = 17.6% |
| M00063 | 11 | 64.7 | 25/72 = 34.7% | ||
| Motif-2 | M00353 | 2 | 22.2 | 2/41 = 4.8% | |
| Motif-3 | M00063 | 9 | 100 | 11/72 = 15.2% | |
| M00062 | 9 | 100 | 9/53 = 16.9% | ||
| Motif-4 | M00094 | 4 | 57.1 | 5/68 = 7.3% | |
| M00063 | 4 | 57.1 | 7/72 = 9.7% | ||
| Motif-5 | M00094 | 4 | 50 | 4/68 = 5.8% | |
| M00138 | 4 | 50 | 6/39 = 15.3% | ||
| Motif-6 | M00094 | 8 | 80 | 13/68 = 19.1% | |
| M00062 | 5 | 50 | 7/53 = 13.2% | ||
| Motif-7 | M00138 | 1 | 20 | 1/39 = 2.5% | |
| M00422 | 1 | 20 | 1/85 = 1.1% | ||
| Motif-8 | M00258 | 5 | 55.5 | 5/47 = 10.6% | |
| M00063 | 4 | 44.4 | 4/72 = 5.5% |
Motifs with OPi,M values <20% were not shown, and only two putative cis-elements with the highest OPi,M values were shown for each MEME Motif-i.
aOi,M, the number of copies of MEME Motif-i with at least one overlapped cis-element detected with TRANSFAC matrix M.
bOPi,M, the proportion of the copies of MEME Motif-i with at least one overlapped cis-element of TRANSFAC matrix M.
cOPM,i, the proportion of the cis-elements for matrix M with at least one overlapped MEME Motif-i.
Three motifs without any overlapped putative cis-elements were also detected in hemoglobin promoters by MEME (Motif-3, -10 and -12). These motifs detected by MEME may be potential binding sites of unknown transcription factors. The copies of putative binding sites detected with TRANSFAC matrices without any overlapped MEME motifs were also counted for each matrix in the three promoter sets. The binding sites with an overlapping rate (OPM) <50% are listed in Table 5. All of the three sets of promoters contain putative cis-elements detected with TRANSFAC matrices with an overlapping rate <50%. Some of these were putative binding sites of some transcription factors known to be important in transcription regulation of the genes, e.g. Sp1 in the actin set, Oct-1 and GATA in the hemoglobin set, Oct-1 in the IFN set. This result indicates that the analysis with our approach may also detect some signals that were not detected with MEME.
CONCLUSIONS
Based on the PWM data provided by TRANSFAC, a method to recognize putative over-represented cis-elements in a group of related sequences was developed. With the promoters of a group of possibly co-regulated genes available, a number of over-represented motifs with a high similarity score to some PWMs were located in the submitted promoters. Thus, a list of corresponding transcription factors for these putative cis-elements can be recognized with our method and the proper thresholds for some PWMs of TRANSFAC can be also determined automatically with the comparison with the pre-defined control data set. These thresholds can be used for further investigation based on TRANSFAC PWMs.
The observation of high frequency motifs in a sequence group indicates that these over-represented motifs probably have an important function, and their corresponding transcription factors may play important roles in the regulation of genes in this sequence group. Because of this, the pattern of these motifs was highly conserved after years of evolution.
The analysis to detect putative over-represented cis- elements is largely affected by the size of the sequence group and the length of the promoter sequences one can get from the database. If the full length of each promoter is available, more information should be retrieved with this method. Only the 600 bases upstream of the transcription start site were used in the analysis, avoiding collecting too much sequences without transcription factor binding sites. Although there were known binding sites of some transcription factors located, the distribution of the binding sites in the remote upstream region were relatively small.
PEG, a tool to extract promoters from GenBank, was developed by Theresa Zhang (27). With the ability to get promoters belonging to the same family, our method can be further tested with other data sets.
The over-represented motifs detected with our approach were compared with the motifs detected with MEME. Some motifs were detected by both methods. Several motifs detected by MEME were not detected by our approach; these motifs were probably binding sites of some unknown transcription factors. Our method was based on PWM data derived from known binding sites. Only the over-represented motifs that were similar to some PWMs in the database could be detected. An overlapping rate analysis for the motifs detected with TRANSFAC matrices indicated that our approach may be able to detect some signals on the sequences that were not presented in the output of MEME. Moreover, over-represented cis-elements detected with TRANSFAC matrices can be directly connected with their corresponding binding factors with TRANSFAC data.
Recently, the number of matrices collected by TRANSFAC version 6.0 had increased to more than 500. When more matrices become available, this method should be more useful to detect the over-representing binding sites of transcription factors within a group of related promoters. However, the PWMs used in this method are not necessarily restricted to TRANSFAC matrices. This method can also be used to define proper thresholds for other customized PWMs and locate putative binding motifs in promoters with similarity scores.
The result of the analysis was also affected by the quality of the collection of promoters of possibly co-regulated genes. Currently, co-regulated genes were selected in the same protein family from EPD. However, different proteins belonging to the same family may not have the same function. Therefore, transcription regulation of these proteins may not be actually conserved, even though the proteins still belong to the same family. An analysis with the promoters of carefully selected co-regulated genes should reveal more useful information. Gene expression data can be used to cluster genes into different co-regulated groups. Co-regulated genes thus clustered can be used to detect potential transcription factor binding sites located within most of the promoters. Gene expression data of yeast has already been used in detecting putative transcription factor binding sites (28). Twenty-six TRANSFAC matrices (TRANSFAC 6.0 public) were derived from cis-elements identified in yeast. With these 26 yeast matrices, our approach can be easily applied to the yeast promoters. The analysis of the promoters of co-regulatory genes determined by yeast expression data may reveal interesting results.
Acknowledgments
ACKNOWLEDGEMENTS
This work was funded by the National Natural Science Grant in China (no. 19947006) and the National Key Foundation Research Grant in China (863).
REFERENCES
- 1.Fickett J.W. and Wasserman,W.W. (2000) Discovery and modeling of transcriptional regulatory regions. Curr. Opin. Biotechnol., 11, 19–24. [DOI] [PubMed] [Google Scholar]
- 2.Halfon M.S., Grad,Y., Church,G.M. and Michelson,A.M. (2002) Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res., 12, 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stormo G.D. and Fields,D.S. (1998) Specificity, free energy and information content in protein-DNA interactions. Trends Biochem Sci., 23, 109–113. [DOI] [PubMed] [Google Scholar]
- 4.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Pruss,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolchanov N.A., Podkolodnaya,O.A., Ananko,E.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N., Busygina,T.V., Kolpakov,F.A., Podkolodny,N.L., Naumochkin,A.N., Korostishevskaya,I.M., Romashchenko,A.G. and Overton,G.C. (2000) Transcription regulatory regions database (TRRD): its status in 2000. Nucleic Acids Res., 28, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector—new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kel A.E., Kel-Margoulis,O.V., Farnham,P.J., Bartley,S.M., Wingender,E., and Zhang,M.Q. (2001) Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J. Mol. Biol., 309, 99–120. [DOI] [PubMed] [Google Scholar]
- 8.Ohler U. and Niemann,H. (2001) Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet., 17, 56–60. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 10.Bailey T.L. and Elkan,C.P. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Altman,R., Brutlag,D., Karp,P., Lathrop,R. and Searls,D. (eds), Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, USA, pp. 28–36. [PubMed]
- 11.GuhaThakurta D. and Stormo,G.D. (2001) Identifying target sites for cooperatively binding factors. Bioinformatics, 17, 608–621. [DOI] [PubMed] [Google Scholar]
- 12.van Helden J., Andre,B. and Collado-Vides,J. (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- 13.Praz V., Perier,R., Bonnard,C. and Bucher,P. (2002) The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression data. Nucleic Acids Res., 30, 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickert L., Reuter,I., Klawonn,F. and Wingender,E. (1998) Transcription regulatory region analysis using signal detection and fuzzy clustering. Bioinformatics, 14, 244–251. [DOI] [PubMed] [Google Scholar]
- 15.Ma H, Yanofsky,M.F. and Meyerowitz,E.M. (1991) AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev., 5, 484–495. [DOI] [PubMed] [Google Scholar]
- 16.Frech K., Quandt,K. and Werner,T. (1998) Muscle actin genes: a first step towards computational classification of tissue specific promoters. In Silico Biol., 1, 29–38. [PubMed] [Google Scholar]
- 17.Biesiada E., Hamamori,Y., Kedes,L. and Sartorelli,V. (1999) Myogenic basic helix–loop–helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac alpha-actin promoter. Mol. Cell. Biol., 19, 2577–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parakati R. and DiMario,J.X. (2002) Sp1- and Sp3-mediated transcriptional regulation of the fibroblast growth factor receptor 1 gene in chicken skeletal muscle cells. J. Biol. Chem., 277, 9278–9285. [DOI] [PubMed] [Google Scholar]
- 19.Sieweke M.H. and Graf,T. (1998) A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev., 8, 545–551. [DOI] [PubMed] [Google Scholar]
- 20.Horak C.E., Mahajan,M.C., Luscombe,N.M., Gerstein,M., Weissman,S.M. and Snyder,M. (2002) GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp–chip analysis. Proc. Natl Acad. Sci. USA, 99, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan T.M., Sun,C.W., Ren,J. and Townes,T.M. (2000) Human gamma-globin gene promoter element regulates human beta-globin gene developmental specificity. Nucleic Acids Res., 28, 2736–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X.S., Glazer,P.M. and Wang,G. (2000) Activation of human gamma-globin gene expression via triplex-forming oligonucleotide (TFO)-directed mutations in the gamma-globin gene 5′ flanking region. Gene, 242, 219–228. [DOI] [PubMed] [Google Scholar]
- 23.Tada Y., Ho,A., Matsuyama,T. and Mak,T.W. (1997) Reduced incidence and severity of antigen-induced autoimmune diseases in mice lacking interferon regulatory factor-1. J. Exp. Med., 185, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Der Fits L., Van Der Wel,L.I., Laman,J.D., Prens,E.P. and Verschuren,M.C. (2003) Psoriatic lesional skin exhibits an aberrant expression pattern of interferon regulatory factor-2 (IRF-2). J. Pathol., 199, 107–114. [DOI] [PubMed] [Google Scholar]
- 25.Melen K., Keskinen,P., Lehtonen,A. and Julkunen,I. (2000) Interferon-induced gene expression and signaling in human hepatoma cell lines. J. Hepatol., 33, 764–772. [DOI] [PubMed] [Google Scholar]
- 26.Penix L.A., Sweetser,M.T., Weaver,W.M., Hoeffler,J.P., Kerppola,T.K. and Wilson,C.B. (1996) The proximal regulatory element of the interferon-gamma promoter mediates selective expression in T cells. J. Biol. Chem., 271, 31964–31972. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T. and Zhang,M. (2001) Promoter extraction from GenBank (PEG): automatic extraction of eukaryotic promoter sequences in large sets of genes. Bioinformatics, 17, 1232–1233. [DOI] [PubMed] [Google Scholar]
- 28.Jakt L.M., Cao,L., Cheah,K.S. and Smith,D.K. (2001) Assessing clusters and motifs from gene expression data. Genome Res., 11, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]