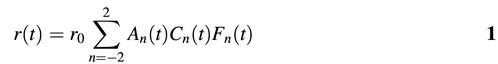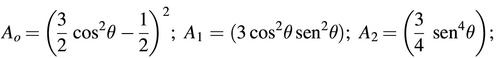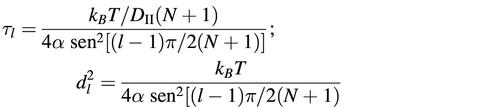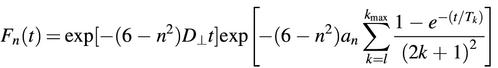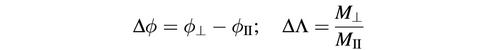Abstract
The human protein OGG1 (hOGG1) targets the highly mutagenic base 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxodG) and shows a high specificity for the opposite DNA base. Abasic sites can arise in DNA in close opposition to 8-oxodG either during repair of mismatched bases (i.e. 8-oxodG/A mismatches) or, more frequently, as a consequence of ionizing radiation exposure. Bistranded DNA lesions may remain unrepaired and lead to cell death via double-strand break formation. In order to explore the role of damaged-DNA dynamics in recognition/excision by the hOGG1 repair protein, specific oligonucleotides containing an 8-oxodG opposite an abasic site, at different relative distances on the complementary strand, were synthesized. Rotational dynamics were studied by means of fluorescence polarization anisotropy decay experiments and the torsional elastic constant as well as the hydrodynamic radius of the DNA fragments were evaluated. Efficiency of excision of 8-oxodG was tested using purified human glycosylase. A close relation between the twisting flexibility of the DNA fragment and the excision efficiency of the oxidative damage by hOGG1 protein within a cluster was found.
INTRODUCTION
7,8-Dihydro-8-oxo-2′-deoxyguanosine (8-oxodG) is a powerful mutagenic lesion formed spontaneously in the genome of aerobic organisms and under the action of ionizing radiation.
The repair mechanism requires a multi-step process in which, first, a DNA glycosylase, hOGG1 in human cells, recognizes the lesion and catalyzes its removal by cleavage of the N-glycosylic bond between the base and the sugar phosphate backbone, producing an abasic (AP) site. Secondly, strand breakage at the AP site is catalyzed by the AP lyase activity of the same enzyme, leaving a 3′-α,β-unsaturated aldehyde and a 5′ phosphate. The intrinsic 3′ phosphodiesterase activity of human AP endonuclease (APE1) is required for removing the 3′ blocking group (reviewed in 1). Alternatively, APE1 can substitute for the AP lyase activity of hOGG1 by directly cleaving the generated AP site (2). In either case the resulting 1 nt gap is repaired mainly by short-patch base excision repair (BER) (reviewed in 3).
Other lesions might occur in close vicinity to 8-oxodG, either in the course of repair events or due to ionizing radiation exposure (clustered damage). Replication of unrepaired 8-oxodG generates 8-oxodG/A mismatches that are also a substrate for BER by a process initiated, in human cells, by the monofunctional DNA glycosylase hMYH. The removal of the mismatched A leads to the formation of the repair intermediate 8-oxodG/AP site (4). Clustered damage is formed when different lesions [oxidative damage, single-strand break (SSB), abasic site] are present on both DNA strands at a distance of a few nucleotides. This kind of damage is preferentially due to ionizing radiation exposure and in particular to high linear energy transfer (high LET) radiation (5,6). It has been recently reported that 8-oxodG repair is strongly inhibited when clustered damage occurs in the genome. In particular, the presence of an AP site near to 8-oxodG on the complementary strand greatly reduces 8-oxodG removal by the Escherichia coli formamidopyrimidine DNA glycosylase (Fpg) (7). Similarly, the efficiency of cleavage by yeast OGG1 (yOGG1) or hOGG1 is inhibited by the presence of another 8-oxodG or of an abasic site on the complementary strand, especially when the second lesion is localized one base 3′ opposite 8-oxodG (8,9). This inhibitory effect might be biologically relevant by providing the cells of a strategy to minimize the likelihood of formation of double-strand breaks (DSBs).
In a previous study (10), we reported that a single 8-oxodG modifies the twisting motion of a 30mer duplex with respect to the unmodified control and we suggested that this modification might signal the recognition and repair of the oxidized base. In this study we have investigated whether the presence of additional lesions in close opposition to an 8-oxodG influences the elasticity properties of DNA. This might account for reduced repair of the oxidized base. Duplexes containing an 8-oxodG and a synthetic AP site on the complementary strand at different relative positions were synthesized and their internal motion was analyzed by fluorescence polarization anisotropy (FPA). In addition, the cleavage and binding efficiency of hOGG1 on duplexes containing the repair intermediate 8-oxodG/AP site were determined.
MATERIALS AND METHODS
The synthesis of 5′-O-dimethoxytrityl-1′,2′-dideoxyribose- 3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite and 8-oxodG-phosphoramidite were performed using standard methods (11,12). Automatic synthesis of the oligomers was performed on a Biosearch Expedite 8909 by the cyanoethyl-phosphoramidite method, at the 1 µmol scale. Reagents for automatic synthesis were from Proligo Biochemie GmbH (Hamburg, Germany) and Beckman Coulter S.p.A. (Milan, Italy). Synthetic oligonucleotides were cleaved from the solid support with ammonium hydroxide-methylamine (Beckman Coulter) by 10 min incubation at room temperature (r.t.) followed by 15 min at 55°C. In the case of oligonucleotides containing 8-oxodG, cleavage from support (1 h at r.t.) and deprotection (17 h at 55°C) were performed with ammonium hydroxide–0.25 M 2-mercaptoethanol. The oligonucleotides were purified first by HPLC on a Dynamax Microsorb C4 column, according to standard procedures, and then by denaturing preparative polyacrylamide gel electrophoresis (PAGE) (13). Oligonucleotides to be used for optical measurements were hybridized with their complementary strands (1:1 molar ratio) and the annealing efficiency was verified by non-denaturing 15% PAGE. The sequences of the various oligonucleotides are presented in Figure 1. Oligonucleotides to be used for enzymatic assays were 3′-end-labeled. In particular, the top strands of the G/C, oxodG/C and oxodG/AP duplexes were 3′-end-labeled by incubation with 25 U of terminal transferase (Roche Diagnostics GmbH, Mannheim, Germany) in the presence of [α-32P]ddATP (Amersham Pharmacia Biotech Italia) (110 TBq/mmol) for 1 h at 37°C. The annealing reaction was performed with an excess of the complementary strands and verified by 15% PAGE.
Figure 1.
Sequences of synthetic duplexes. G and X (in bold) indicate the position of the 8-oxodG and the abasic site, respectively.
Purification of hOGG1
Escherichia coli strain BK3004 (FPG::kan) was transformed by pUC18-HOGH1-30 (13) and in 5 l of LB medium at 37°C to a density of 8 × 108 cells/ml; isopropylthio-β-d-galactoside (0.5 mM) was then added and the incubation continued for 2 h. The cell pellet was resuspended in 100 ml of buffer A (50 mM Tris, pH 7.0, 100 mM NaCl and 10 mM 2-mercaptoethanol) and cell-free extract was prepared by sonication. An assay for enzymatic activity was used to monitor hOGG1 purification. The cell extract was applied to an SP Sepharose column (15 ml; Amersham) equilibrated with buffer A and eluted with a linear gradient of 0–1 M NaCl in buffer A. Active fractions (eluting at 400 mM NaCl) were pooled and applied to a ACA44 gel filtration column (20 ml; Ultrogel, LKB) equilibrated with 1 M NaCl in buffer A. Active fractions were pooled, dialyzed against buffer B (50 mM MES, pH 6.0, 10 mM 2-mercaptoethanol) and applied to a MonoS column (HR 5/5; Pharmacia). The MonoS column was eluted with a linear gradient of 0–1 M NaCl in buffer B and active fractions (eluting at 0.4 M NaCl) were collected.
Cleavage and band shift assays
3′-32P-end-labeled DNA duplexes were incubated with increasing amounts of hOGG1 protein and the formation of SSBs was monitored by 15% denaturing PAGE. In particular, 0.3 pmol of the labeled duplexes were incubated for 30 min in 5 µl of buffer R (20 mM HEPES–KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA) with various amounts of hOGG1 at 37°C, as indicated in the legends to the figures. The addition of ∼10 ng of protein was necessary to reach the maximum cleavage of ∼50% of the substrate molecules. Six microliters of denaturing stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) were then added to the samples which were loaded on 15% denaturing polyacrylamide gels containing 7 M urea in TBE buffer. The gels were run at 28 V/cm for 1 h at r.t. and then exposed to Kodak X-OMATUV films. The densitometric analysis of the autoradiographs was performed by an ULTROSCAN XL laser densitometer (Pharmacia LKB) and the subsequent band area evaluation was performed using the public domain NIH Image program (developed at the US National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
The band shift assays were performed by incubating 0.3 pmol of the labeled duplexes with 0.5–5 ng of hOGG1 in buffer R at 37°C for 30 min. Aliquots of the reaction were loaded on non-denaturing 10% polyacrylamide gels in TBE buffer and gels were run at 10 V/cm for 3 h 30 min at 4°C. The gels were then exposed to films and quantified as described above.
Lifetimes and FPA measurements
Dynamic fluorescence measurements were performed by means of a frequency domain fluorometer K2-ISS (Urbana, IL, USA), at modulation frequencies between 1 and 40 MHz. The excitation light was the 514 nm 0.5 W output of a Coherent Innova 90C Argon laser. The modulation ratio was always in the 60–70% range and the detection cross-correlation frequency was 80 Hz.
Ethidium bromide (EB) was used as fluorophore at a ratio of 1 EB molecule per 300 bp. The DNA samples were diluted at the concentration of 5.5 × 10–4 M in 10 mM Na-phosphate buffer pH 7.2, 100 mM NaCl, 0.1 mM EDTA and then 2 ml were placed in quartz cells (1 cm path length). A long pass filter (550 nm) was used to minimize the scattering radiation that reaches the photomultiplier. The samples were thermostated at temperatures ranging between 4 and 40°C. Lifetime measurements were performed with polarizers positioned at ‘magic angle’ conditions, using a latex scattering sample as a reference. Ten frequencies logarithmically spaced in the 2–40 MHz range were acquired. For each sample, at least four different lifetimes were measured in the 10–40°C range. Subsequent analysis was performed by means of the K2-ISS software. The temperature dependence of lifetime values, obtained through linear fit of the data, was then used for the anisotropy data analysis.
FPA measurements were performed on the same samples, acquiring data at 20 frequencies in the 2–40 MHz range. The experimental set-up as well as the theoretical background are detailed elsewhere (14,15).
Briefly, the measured time-dependent fluorescence anisotropy is:
where III and I⊥ are the emission fluorescence intensities, respectively, detected with the emission polarizer at 0° and 90° with respect to the vertical exciting beam.
As intercalated ethidium may be considered firmly bound to the DNA, the decay of the fluorescence anisotropy is strictly correlated to DNA internal motion. The anisotropy decay may be described by the product of correlation functions related to internal, twisting and bending motion (16) as reported:
In equation 1, r0 is the EB limiting anisotropy, An(t) are the internal correlation functions related to the angle (θ) between the EB transition dipole and the helical axis (e.g. θ = 70.5°). An(t) are fully described by the following trigonometric function:
Cn(t) are the correlation functions related to torsional motion. Through the relaxation time τl of the lth normal mode (14) and its corresponding mean square amplitude d2l, they are related to the torsional rigidity constant α and to the rotational diffusion coefficient DII:
Fn(t), the bending angle correlation functions, are described by the equation:
where D⊥ is the rotational diffusion coefficient in the perpendicular axis direction.
The fluorescence emission of the different samples was measured by FPA. Due to the different motion of the samples, the emission phases were shifted with respect to the excitation phase and a demodulation was observed. The phase difference and the demodulation ratio are expressed by the equations:
related to the Laplace transforms of the fluorescence intensity time decay (14).
FPA data were analyzed using a Fortran routine that minimizes phases and modulations according to the Marquardt algorithm (14). In the fitting procedure, the elastic torsional constant (α) and the hydrodynamic radius (Rh) and the rise (b) were free parameters.
UV thermal denaturation measurements
The thermal denaturation of the DNA duplexes was monitored by analysis of the absorbance at 260 nm (A260), by means of a Cary 3 UV/VIS spectrophotometer. The samples, as a rule at a fixed concentration of 50 µg/ml in 10 mM Na-phosphate buffer, pH 7.0, 100 mM NaCl, 0.1 mM EDTA, were thermostated using a Peltier device. The temperature was raised at a rate of 0.5°C/min from 5 to 95°C and monitored in the sample compartment. Absorbance values were corrected for water thermal expansion and normalized at A260 = 1 at the lowest temperature. The thermodynamic analysis was performed according to Breslauer (17).
RESULTS
Enzymatic assays
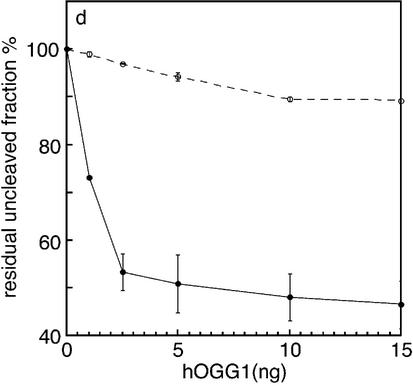
An abasic site opposite to 8-oxodG is introduced during repair of the highly mutagenic 8-oxodG/A mismatch by the DNA glycosylase hMYH. The effect of a synthetic abasic site opposite to 8-oxodG on hOGG1 cleavage was tested by comparing the cleavage efficiency of the oxodG/AP duplex with that of the oxodG/C duplex (Fig. 2). The AP tetrahydrofuran analog was employed, as the natural abasic site is intrinsically unstable. Since the oligonucleotides containing the 8-oxodG residue are 3′-end-labeled, the expected product of a complete cleavage reaction by hOGG1 is an 18 nt fragment. As shown in Figure 2, efficient cleavage is detected when oxodG/C duplexes are used as substrate (Fig. 2a) whereas the presence of an abasic site opposite to 8-oxodG strongly inhibits the cleavage reaction (Fig. 2b). As expected, hOGG1 is completely ineffective towards the G/C duplex (Fig. 2c). The quantitative analysis of the uncleaved fraction of DNA as a function of hOGG1 concentration is displayed in Figure 2d. Under conditions where the maximum cleavage of the 8-oxodG substrate was achieved (53%), only 11% of the oxodG/AP substrate was cleaved.
Figure 2.
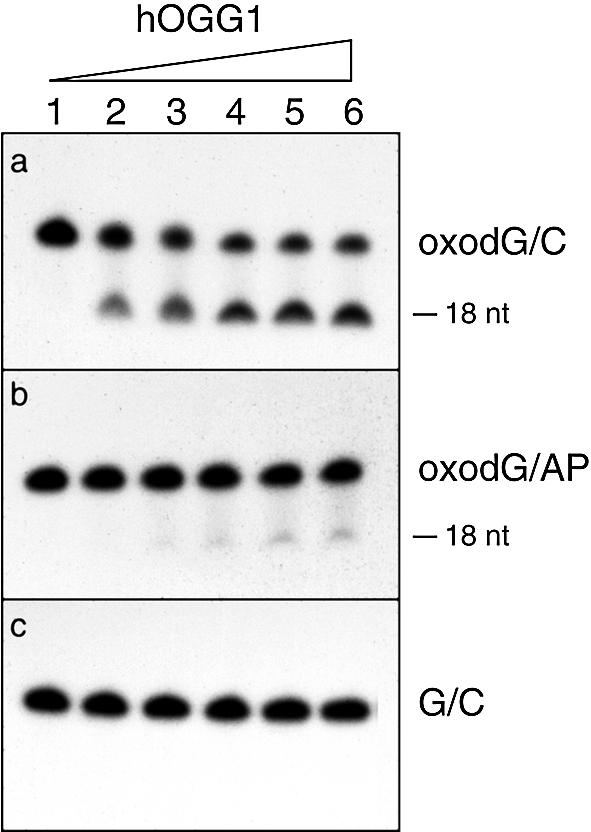
Effect of the presence of an opposing AP site on 8-oxodG cleavage by hOGG1. The duplex oligonucleotides oxodG/C, oxodG/AP and G/C (0.3 pmol) containing the 8-oxodG or G on the 3′-end-labeled strand were incubated with increasing amount of hOGG1 for 30 min at 37°C. Aliquots were electrophoresed on 15% denaturing polyacrylamide gels. (a–c) Electrophoretic profile of the reaction products. Lanes 1, control with no enzyme; lanes 2–6, after incubation with 1.0, 2.5, 5.0, 10 and 15 ng of hOGG1, respectively. (d) The residual uncleaved fraction of the oxodG/C (filled circles) and oxodG/AP (open circles) duplexes was measured by densitometric analysis of the autoradiographs. The fraction of the intact strand was calculated as percentage of the total (cleaved + uncleaved fractions). The values are the means of three independent experiments.
To gain insights into this mechanism the effect of an abasic site opposite to 8-oxodG on hOGG1 binding efficiency was investigated. As shown in Figure 3, incubation of oligonucleotides containing the oxodG/AP site with increasing concentrations of hOGG1 results in a band shift that is less intense when compared with that obtained with the oxodG/C duplexes (compare lanes 1–5 with lanes 6–10). The extent of inhibition on hOGG1 binding, obtained from two independent experiments, was ∼3.5-fold.
Figure 3.
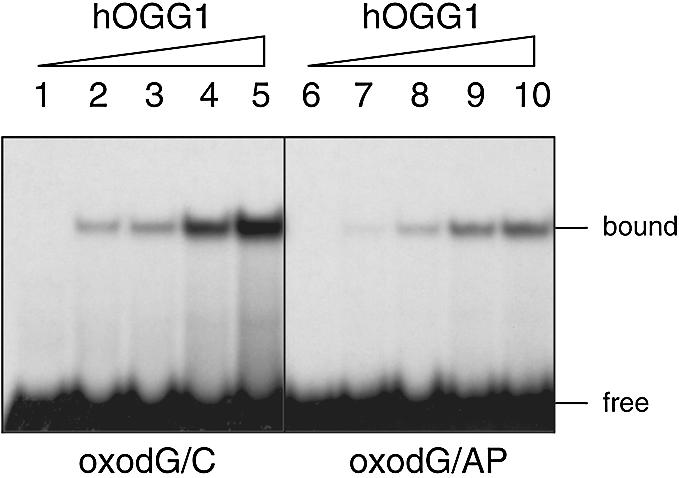
Effect of the presence of an opposing AP site on 8-oxodG binding by hOGG1. The duplex oligonucleotides oxodG/C and oxodG/AP (0.3 pmol) containing the 8-oxodG on the 3′-end-labeled strand were incubated with increasing amounts of hOGG1 for 30 min at 37°C. Aliquots were electrophoresed on 10% non-denaturing polyacrylamide gels. Lanes 1–5, oxodG/C substrate; lanes 6–10, oxodG/AP substrate. Lanes 1 and 6, control samples with no enzyme; lanes 2–5 and 7–10, after incubation with 0.5, 1.0, 2.5 and 5.0 ng of hOGG1, respectively.
These results indicate that the introduction of an abasic site opposite to the oxidized base determines both a poor binding of the enzyme and an inhibition of its catalytic activity.
Lifetimes and fluorescence polarization anisotropy measurements
Lifetime and FPA measurements were performed by monitoring the fluorescence emission of the EB probe intercalated into the duplexes. The duplex oligonucleotides which were analyzed contained either the oxodG/AP structure or the AP site positioned one base 5′ or 3′ to 8-oxodG on the complementary strand. These last constructs were included since the introduction of an abasic site at position +1 opposite 8-oxodG [oxodG/AP(+1)] is known to strongly reduce the cleavage ability of hOGG1, whereas the presence of an abasic site one base 5′ opposite to 8-oxodG [oxodG/AP(–1)] does not significantly affect cleavage (8; our unpublished data). For comparison, all the measurements were also undertaken on the oligonucleotides containing only the abasic site.
Phase differences and demodulations were recorded at different temperatures and the data were fitted according to Allison and Schurr’s model (16) as modified by Collini et al. (14). In this model, DNA fragments are assumed to be a string of disks, formed by joining two successive base pairs, connected by springs that allow both bending and twisting motion. Due to the short length of our samples the changes that we observed in the phase and in the modulation are essentially due to the torsional motion. In the fitting procedure, the dependence of the lifetime on temperature was taken into account (15,18); the limiting anisotropy ro was kept constant at 0.36, while the torsional constant (α), the hydrodynamic radius (Rh) and the rise (b) were free parameters. The lifetimes of the bound EB for the various oligonucleotides were quite similar and ranged between ∼21.3 and ∼22.6 ns at 293 K, showing that the intercalation site is probably the same in all the samples.
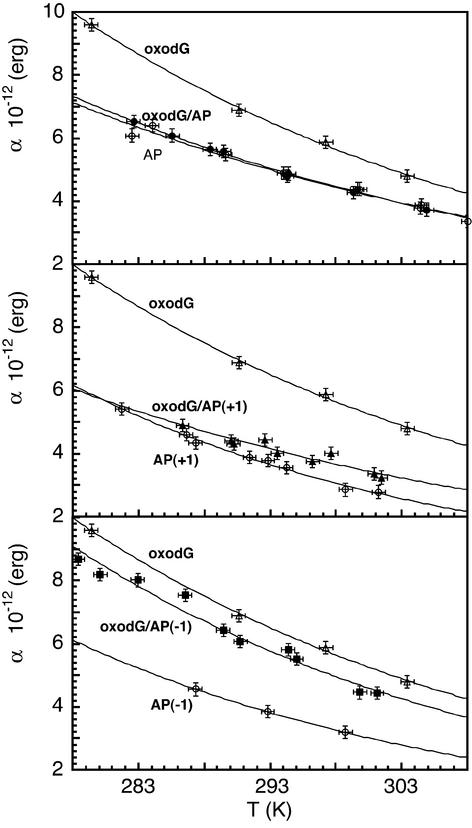
In Figure 4, the torsional rigidity constants (α) of the damaged oligonucleotides are reported as a function of temperature. The experimental data were empirically fitted with an exponential function. A strong reduction of the α values was observed at all temperatures when an abasic site was positioned either opposite (oxodG/AP) or 1 nt 3′ to the oxidized base [oxodG/AP(+1)] on the complementary strand. On the contrary, the AP site located 1 nt 5′ to the site opposite to 8-oxodG [oxodG/AP(–1)] slightly affected the α values.
Figure 4.
Effect of the presence of 8-oxodG with opposing AP sites at various relative positions on DNA torsional rigidity constant as a function of temperature. oxodG/AP (filled circles), oxodG/AP(+1) (filled triangles), oxodG /AP(–1) (filled squares). In all three panels the rigidity constant values of the duplexes with a single abasic site (open circles) and with a single 8-oxodG lesion (open triangles) are reported as a reference.
To identify the contribution of the abasic site alone to the fragment internal motion the functions describing the temperature dependence of the rigidity of the DNA duplexes containing only the abasic site are also reported.
From the equation:
where kB is the Boltzmann constant and T the absolute temperature, the average fluctuation of the twist angle (Δξ) between two successive base pairs steps was estimated. These values, together with the rigidity constant at 293 K, the hydrodynamic radius and the rise are reported in Table 1. The comparison among the multiple damaged fragments, the corresponding duplexes containing a single lesion (an 8-oxodG or an AP site) and the fragment without any damage shows that the torsional rigidity constant is strongly reduced in both the oxodG/AP and oxodG/AP(+1) samples, while in the oxodG/AP(–1) duplex there is only a slight effect. All the corresponding duplexes containing a single AP site manifest a reduction of the torsional rigidity with respect to the unmodified fragment.
Table 1. Hydrodynamic parameters derived by the FPA measurements.
| Sample | α293K 10–12 (erg) | Δξ (°) | Rh 293K (Å) | b (Å) |
|---|---|---|---|---|
| oxodG/C | 6.4 ± 0.2 | 4.6 ± 0.1 | 11.0 ± 0.3 | 3.4 |
| oxodG/AP | 5.0 ± 0.2 | 5.2 ± 0.1 | 11.2 ± 0.3 | 3.2 |
| oxodG/AP(+1) | 4.1 ± 0.2 | 5.7 ± 0.1 | 11.2 ± 0.3 | 3.4 |
| oxodG/AP(–1) | 5.7 ± 0.2 | 4.8 ± 0.1 | 10.6 ± 0.3 | 3.2 |
| G/C | 4.9 ± 0.2 | 5.2 ± 0.1 | 11.0 ± 0.3 | 3.4 |
| AP | 4.9 ± 0.2 | 5.2 ± 0.1 | 11.7 ± 0.3 | 3.4 |
| AP(+1) | 3.7 ± 0.2 | 6.0 ± 0.1 | 9.6 ± 0.3 | 3.6 |
| AP(–1) | 3.8 ± 0.2 | 5.9 ± 0.1 | 10.4 ± 0.3 | 3.4 |
The reduction in the rigidity constant involves the increase of the fluctuation of the torsion angle by 0.6° and 1.2° in the oxodG/AP and in the oxodG/AP(–1) sample, while no significant changes are present in the oxodG/AP(+1) duplex. It is important to take into account that α as well as Δξ are average values so that, depending on the extension of influence of the damage on the duplex, the effective modification at the damage site is possibly underestimated. However, it can be inferred also from the data in Table 1 that the multiple damaged DNAs retain their B-form, with the rise between 3.2 and 3.4 Å and the mean value of the radius equal to 11.0 Å.
Thermodynamic characterization
Experiments of UV denaturation were performed on the duplexes containing the 8-oxodG base damage and a single AP site at various relative positions on the complementary strand. The enthalpic changes of the transitions were calculated from the UV melting derivative curves. These values, together with the melting temperatures (Tm), are reported in Table 2. Tm values are the mean of two independent experiments. All three samples with multiple lesions exhibit substantial destabilization with respect to the control, which contains only the 8-oxodG lesion. A Tm reduction of 6–7°C, with respect to the oxodG/C sample, is observed when an abasic site is also present on the complementary strand. This effect can be ascribed to the AP site since the 8-oxodG alone does not destabilize the duplex, as inferred by the comparison with the unmodified duplex, whereas the AP site is well known to destabilize the duplex oligonucleotides as a function of both the missing and the neighbouring bases (19,20). The oxodG/AP(+1) sample, where the abasic site is flanked by two pyrimidines and is located opposite to an adenine, exhibited the greatest enthalpy difference with respect to the oxodG/C sample. This result agrees with the finding that oligonucleotides in which two cytosines flank an abasic site (CXC) present a very large enthalpic destabilization (21). The destabilizing effects observed following the introduction of the additional AP site seem to be independent of the relative position of the lesions.
Table 2. Melting temperatures and enthalpy changes in samples containing a single 8-oxodG with or without an additional abasic site.
| Sample | Tm (°C) | ΔH (kcal/mol) | ΔΔH (kcal/mol) |
|---|---|---|---|
| oxodG/C | 70.5 ± 0.5 | –146.5 ± 8 | – |
| oxodG/AP | 63.5 ± 0.5 | –89.2 ± 4 | –56.8 |
| oxodG/AP(+1) | 64.5 ± 0.5 | –77.5 ± 4 | –69.0 |
| oxodG/AP(–1) | 64.3 ± 0.5 | –105.9 ± 5 | –40.6 |
| G/C | 71.5 ± 0.5 | –151.0 ± 8 | +4.5 |
DISCUSSION
In this study the relationship between the DNA torsional rigidity and the excision efficiency by specific repair enzymes of multiple damaged substrates was examined.
We present evidence that hOGG1 cleavage and binding are strongly inhibited by the 8-oxodG/AP substrate which is produced as repair intermediate during the processing of 8-oxodG/A mismatches. In contrast, the bacterial protein that excises 8-oxodG, Fpg, cleaves very efficiently these mismatches, thus creating a DSB (22). It seems that eukaryotic cells have developed a further level of surveillance from DSB formation by using an enzyme, hOGG1, that is unable to cleave the oxidized base when the repair intermediate, the AP site, is present on the opposite strand. The abasic site is another example of distinct preference for the paired base displayed by Fpg and hOGG1 (23). Differences in the architecture of the DNA binding domain of these two DNA glycosylases (24,25) might explain the distinctive effect of the paired base on 8-oxodG cleavage. In particular, hOGG1 establishes strong and specific interactions with the opposite cytosine (25) and this might account for the strong inhibition of cleavage that we have observed when an abasic site is located opposite to 8-oxodG.
We show that DNA duplexes containing the 8-oxodG/AP structure or the AP site located one base 3′ opposite to 8-oxodG, which both display a strong inhibitory effect on hOGG1 activity (8 and this study), present a significant reduction in the torsional rigidity constants with respect to the oxodG/C sample. A high twisting rigidity characterizes the sample containing the 8-oxodG lesion alone (10 and this study), whereas the presence of a single abasic site slightly reduces the DNA torsional rigidity as compared to the unmodified duplex. This last finding is in agreement with NMR and theoretical studies where a reduction of the twisting rigidity has been evaluated, independently of the missing base, if the unpaired base remained stacked in the double helix (26–28). In the case of the AP and AP(+1) samples, the unpaired purine is likely to assume an intra-helical conformation, independently of the flanking bases (26–28). In the case of the AP(–1) sample, an equilibrium between the intra- and the extra-helical form is still possible for the unpaired pyrimidine, depending on the flanking bases (26–28). Some differences observed in the absolute values of the rigidity constant might be ascribed to the different sequence context, both the opposite and the flanking bases, of the abasic site. The combination of the abasic site and the oxidized base on complementary strands leads to an effect which is strongly affected by the relative position of these two lesions. The α values equal to ∼6.0 × 10–12 erg, as determined at r.t., characterize the duplexes oxodG/C and oxodG/AP(+1) which are a good substrate for hOGG1, whereas lower α values are associated with poor or no repair [duplexes oxodG/AP, oxodG/AP(–1) and G/C]. Then, FPA measurements together with the enzymatic assay indicate that an optimum value of the twisting rigidity is necessary for binding and cleavage of 8-oxodG by hOGG1.
A difference in the reduction of the α values was observed when comparing oxodG/AP and oxodG/AP(+1) samples. This effect might be again ascribed to the sequence context. In fact in the oxodG/AP(+1) sample, the abasic site interrupts a homopurinic/homopyrimidinic sequence as AGAG/CXCT. dApdGp dinucleotide repeats have been found to influence the dynamic characteristics of short duplexes, increasing their average torsional rigidity values (18). It is possible that the effect of a missing base is hampered when it lies in a region where strong stacking interactions occur. From the thermodynamic analysis no relation between the relative distance of the lesions and enzyme efficiency has been revealed.
DNA glycosylases play the important role of excising abnormal bases thus preventing lethal events or misreplication. An open question remains how they locate damaged bases within the cell genome. Structural studies indicate that the mechanism of excision of 8-oxodG by hOGG1 involves the extrusion of the oxidized base from the helix and the insertion of the opposite cytosine intra-helically (25). In addition, in the crystal structure of the DNA-hOGG1 complex the DNA has a pronounced curvature due to a kink at the damaged site. However, it has been recently shown that the 8-oxodG flipping out follows the DNA-hOGG1 complex formation (29). Therefore, the recognition of the lesion by hOGG1 cannot be ascribed to these conformational changes. Moreover, studies on protein-free DNA containing a single oxidative lesion showed that, apart from some local changes in the backbone geometry near the lesion, neither DNA kink nor global conformational changes are induced by the presence of DNA damage (10). On the contrary, differences between damaged and undamaged duplexes have been shown in the elastic properties of the protein-free DNA (10). It has been proposed that the increase in flexibility associated with DNA damage might favour the recognition of damaged DNAs (10) and a similar mechanism has been proposed for hOGG1 when searching for 8-oxodG (29).
In this study, we show a direct correlation between the changes of the torsional elastic constant and the cleavage efficiency by hOGG1 of DNA duplexes containing 8-oxodG and abasic sites in close vicinity on complementary strands. Our findings add another important factor to the DNA damage recognition process: an optimal DNA twisting is required for lesion recognition. This property might be one of the ‘sensors’ that repair enzymes use in searching for damage.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank F. Pedone (Dipartimento di Genetica e Biologia Molecolare, Università ‘La Sapienza’, Roma) and E. Seeberg (Department of Molecular Biology, University of Oslo, Norway) for helpful discussions and critical reading of the manuscript. This work was partially supported by a Collaborative Linkage NATO grant (F.M.).
REFERENCES
- 1.Boiteux S. and Radicella,J.P. (2000) The human OGG1 gene: structure, functions and its implication in the process of carcinogenesis. Arch. Biochem. Biophys., 377, 1–8. [DOI] [PubMed] [Google Scholar]
- 2.Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dogliotti E., Fortini,P., Pascucci,B. and Parlanti,E. (2001) Multiple pathways for DNA base excision repair. The mechanism of switching among multiple BER pathways. Prog. Nucleic Acid Res. Mol. Biol., 68, 1–28. [DOI] [PubMed] [Google Scholar]
- 4.Slupska M.M., Luther,W.M., Chiang,J.H., Yang,H. and Miller,J.H. (1999) Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J. Bacteriol., 181, 6210–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutherland B.M., Bennet,P.V., Sidorkina,O. and Laval,J. (2000) Clustered DNA damages induced in isolated DNA and human cells by low doses of ionizing radiation. Proc. Natl Acad. Sci. USA, 97, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutherland B.M., Bennet,P.V., Sidorkina,O. and Laval,J. (2000) Clustered damages and total lesions induced in DNA damages by ionizing radiation: oxidized bases and strand breaks. Biochemistry, 39, 8026–8031. [DOI] [PubMed] [Google Scholar]
- 7.David-Cordonnier M.H., Laval,J. and O’Neill,P. (2001) Recognition and kinetics for excision of a base lesion within clustered DNA damage by the Escherichia coli proteins Fpg and Nth. Biochemistry, 40, 5738–5746. [DOI] [PubMed] [Google Scholar]
- 8.David-Cordonnier M.H., Boiteux,S. and O’Neill,P. (2001) Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein. Nucleic Acids Res., 29, 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David-Cordonnier M.H., Boiteux,S. and O’Neill,P. (2001) Efficiency of excision of 8-oxo-guanine with DNA clustered damage by XSR5 nuclear extracts and purified human OGG1 protein. Biochemistry, 40, 11811–11818. [DOI] [PubMed] [Google Scholar]
- 10.Barone F., Cellai,L., Giordano,C., Matzeu,M., Mazzei,F. and Pedone,F. (2000) γ-Ray footprinting and fluorescence polarization anisotropy of a 30-mer synthetic DNA fragment with one 2′-deoxy-7-hydro-8-oxoguanosine lesion. Eur. Biophys. J., 28, 621–628. [DOI] [PubMed] [Google Scholar]
- 11.Takeshita M., Chang,C.N., Johnson,F., Will,S. and Grollman,A.P. (1987) Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 12.Bodepudi V., Shibutani,S. and Johnson,F. (1992) Synthesis of 2′-deoxy-7,8-dihydro-8-oxoguanosine and 2′-deoxy-7,8-dihydro-8-oxoadenosine and their incorporation into oligomeric DNA. Chem. Res. Toxicol., 5, 608–617. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Collini M., Chirico,G. and Baldini,G. (1995) Influence of ligands on the fluorescence polarization anisotropy of ethidium bound to DNA. Biophys. Chem., 53, 227–239. [DOI] [PubMed] [Google Scholar]
- 15.Barone F., Chirico,G., Matzeu,M., Mazzei,F. and Pedone,F. (1998) Triple helix DNA oligomer melting measured by fluorescence polarization anisotropy. Eur. Biophys. J., 27, 137–146. [DOI] [PubMed] [Google Scholar]
- 16.Allison S.A. and Schurr,J.M. (1979) Torsion dynamics and depolarization of fluorescence of linear macromolecules. I. Theory and application to DNA. Chem. Phys., 41, 35–39. [DOI] [PubMed] [Google Scholar]
- 17.Breslauer K.J. (1994) Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transition. In Agraval,S. (ed.), Methods in Molecular Biology. Humana, Totowa, pp. 347–372. [DOI] [PubMed]
- 18.Pedone F., Mazzei,F., Matzeu,M. and Barone,F. (2001) Torsional constant of 27-mer DNA oligomers of different sequences. Biophys. Chem., 94, 175–184. [DOI] [PubMed] [Google Scholar]
- 19.Gelfaland C.A., Plum,G.E., Grollman,A.P., Johnson,F. and Breslauer,K.J. (1998) Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry, 37, 7321–7327. [DOI] [PubMed] [Google Scholar]
- 20.Sàgi J., Guliaev,A.B and Singer,B. (2001) 15-mer DNA duplexes containing an abasic site are thermodynamically more stable with adjacent purine than with pyrimidines. Biochemistry, 40, 3859–3868. [DOI] [PubMed] [Google Scholar]
- 21.Coppel Y., Berthet,N., Coulombeau,C., Coulombeau,C., Garcia,J. and Lhomme,J. (1997) Solution conformation of an abasic DNA undecamer duplex d(CGCACXCACGC).d(GCGTGTGTGCG): the unpaired thymine stacks inside the helix. Biochemistry, 36, 4817–4830. [DOI] [PubMed] [Google Scholar]
- 22.Tchou J. Bodepudi,V., Shibutani,S., Anthoshechkin,I., Miller,J., Grollman,A.P. and Johnson,F. (1994) Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J. Biol. Chem., 269, 15318–15324. [PubMed] [Google Scholar]
- 23.Asagoshi K., Yamada,T., Terato,H., Ohyama,Y., Monden,Y., Arai,T., Nishimura,S., Aburatani,H., Lindahl,T. and Ide,H. (2000) Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J. Biol. Chem., 275, 4956–4964. [DOI] [PubMed] [Google Scholar]
- 24.Serre L., Pereira de Jesus,K., Boiteux,S., Zelwer,C. and Castaing,B. (2002) Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J., 21, 2854–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruner S.D., Norman,D.P.G. and Verdine,G.L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 26.Lhomme J., Constant,J.F. and Demeunynck,M. (1999) Abasic DNA structure, reactivity and recognition. Biopolymers, 52, 65–83. [DOI] [PubMed] [Google Scholar]
- 27.Cuniasse P., Fazakerley,G.V., Guschlbauer,W., Kaplan,B.E. and Sowers,L.C. (1990) The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C and T opposite a model abasic site. J. Mol. Biol., 213, 303–314. [DOI] [PubMed] [Google Scholar]
- 28.Ayadi L., Coulombeau,C. and Lavery,R. (2000) The impact of abasic sites on DNA flexibility. J. Mol. Struct. Dyn., 17, 645–653. [DOI] [PubMed] [Google Scholar]
- 29.Chen L., Haushalter,K.A., Lieber,C.M. and Verdine,G.L. (2002) Direct visualization of a DNA glycosylase searching for damage. Chem. Biol., 9, 345–350. [DOI] [PubMed] [Google Scholar]