Abstract
The German Neurospora Genome Project has assembled sequences from ordered cosmid and BAC clones of linkage groups II and V of the genome of Neurospora crassa in 13 and 12 contigs, respectively. Including additional sequences located on other linkage groups a total of 12 Mb were subjected to a manual gene extraction and annotation process. The genome comprises a small number of repetitive elements, a low degree of segmental duplications and very few paralogous genes. The analysis of the 3218 identified open reading frames provides a first overview of the protein equipment of a filamentous fungus. Significantly, N.crassa possesses a large variety of metabolic enzymes including a substantial number of enzymes involved in the degradation of complex substrates as well as secondary metabolism. While several of these enzymes are specific for filamentous fungi many are shared exclusively with prokaryotes.
INTRODUCTION
Fungi form a large eukaryotic kingdom comprising probably more than 100 000 species, including yeasts and molds as well as mushrooms (1). They have spread through diverse natural habitats living saprophytically or parasitically on the degradation of a large variety of organic material. Fungal activities affect us in many ways and thus fungi have been studied in great detail (2).
Due to their small genome size fungi have been especially suitable for genome analysis. The genomes of Saccharomyces cerevisiae (3) and Schizosaccharomyces pombe (4) have been sequenced completely and analyzed in detail. Efforts to expand genome sequencing to filamentous fungi date back to the late 1990s. It was reasoned that yeasts like S.cerevisiae and S.pombe would not sufficiently reflect the genetic and biochemical diversity of the fungal kingdom due to the limited metabolic and developmental capabilities needed for their specific ecological niches (5). As a step ahead to a better understanding of fungal biology, sequencing of the genome of the ascomycete Neurospora crassa was initiated (6).
Neurospora crassa has found widespread use in research laboratories as a eukaryotic model organism, as a well-understood filamentous fungus, and in addition as a valuable organism for biotechnological applications (7,8). It was chosen by Beadle and Tatum for their experiments leading to the ‘one gene one enzyme’ hypothesis (9), and later on was used to study chromosome cytology (10,11). More recent topics include circadian rhythm (12), vesicle trafficking (13), mitochondrial biogenesis (14,15) and epigenetic phenomena resulting in repeat-induced point mutation (16), gene silencing (17) and meiotic silencing (18). Neurospora crassa provides favorable growth properties on a large variety of carbon sources. Its ability to form heterokaryons comprising different haploid nuclei has made it amenable to specific genetic manipulations (19). A well-organized research community supported by the Fungal Genetics Stock Center has established a large collection of wild-type and mutant strains and a variety of tools to analyze and manipulate this organism (20).
Neurospora crassa has a genome of some 40 Mb in seven chromosomes (linkage groups LG I–LG VII). Calculated from the mobility of chromosomes in pulsed-field gel electrophoresis (PFGE) the sizes of the chromosomes range from 4 to 10 Mb (21,22). Genome sequencing started on cosmid and BAC clones ordered along individual chromosomes (23). At a later stage, a whole genome shotgun approach was initiated by the Whitehead Genome Center, Cambridge, MA (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/). In the course of the German Neurospora Genome Project cosmid and BAC clones attributed to LG II and LG V (6) were analyzed. Here we describe the database resulting from these sequences and provide a first insight into the contents and peculiarities of the genome of a filamentous fungus. We choose this point in the course of the genome project because the sequencing of cosmid and BAC clones of LG II and LG V has been completed and the database of the German Neurospora Genome Project is currently expanded to cover the entire genome, including sequence data provided by the Whitehead Genome Center.
MATERIALS AND METHODS
Library construction and mapping was done as described previously (23). Selected cosmid and BAC clones were fragmented and subcloned into plasmids. Random sequences obtained from plasmids were assembled into contigs and gaps were closed by primer walking. Depth of coverage was on average 10-fold. The sequencing error rate was <1 in 30 000 bp calculated from the number of single base differences observed in overlapping cosmid and/or BAC clones. All sequence uncertainties were resolved except for poly(G) tracts with more than 12 G residues.
Gene modeling was based on predictions obtained with FGENESH (24), trained on experimentally confirmed N.crassa genes. Predictions by GENEMARK (25), GENSCAN (26) and GENEFINDER (P.Green and L.Hillier, unpublished results) as well as significant matches to ESTs and known genes were used for corrections where appropriate. Putative functions were assigned on the basis of similarities to known genes. Deduced genes were subjected to PEDANT (Protein Extraction, Description and ANalysis Tool) (27). Secondary and tertiary structures were predicted by using PREDATOR (28) and IMPALA (29).
Sequences, predicted genes and analysis results are accessible online at http://mips.gsf.de/proj/neurospora/.
RESULTS
Sequencing
Large insert clones from several cosmid libraries and a BAC library were mapped by hybridization. Starting with probes representing single chromosomes isolated from CHEF gels (30), chromosome-specific sub-libraries were selected and ordered by mutual hybridization of the clones to each other. This eventually resulted in 34 clone contigs for the two linkage groups (23). Selected clones from these contigs were completely sequenced at MWG Biotech AG. Sequence data were assembled into larger contigs and made publicly accessible through the MIPS Neurospora crassa database (MNCDB) (6,31).
Sequences attributed to LG II and LG V based on identified genetic markers add up to 10 Mb. In addition, there are 5 Mb of sequence included in the database that are located on other linkage groups. Sequences located on LG II and LG V were assembled into 13 and 12 contigs, respectively. Cosmid and BAC clones cover the genome insufficiently leaving a considerable number of gaps. An exhaustive search for clones extending the current contigs by hybridization of cosmid and BAC libraries did not succeed.
The combined lengths of the contigs located on LG II add up to 4.7 Mb and thus already exceed the size of 4.6 Mb expected from the electrophoretic mobility of the chromosomes. In contrast, the combined 5.3 Mb of the contigs on LG V are far below its estimated 9.2 Mb. In part this is due to the large rDNA cluster located at the far left arm of this chromosome, which accounts for up to 1.8 Mb (32) and is not covered by contigs. In addition, the electrophoretic mobility of this chromosome may be altered by the rDNA cluster resulting in an erroneous size estimate. This assumption is supported by the fact that the overall ratio of physical size to genetic dimension for LG V is 53 kb per map unit, which is much higher than ratios obtained for the other LGs ranging between 30 and 45 kb per map unit (33).
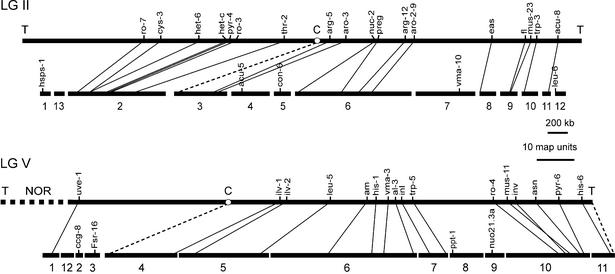
Figure 1 shows a tentative alignment of sequence contigs and genetic maps of LG II and LG V. All genetic loci linked to known sequence information were identified in the present set of sequence contigs. Two small contigs, attributed to LG II and LG V by hybridization data, lack mapped loci and were positioned arbitrarily. Sequence gaps not covered by cosmid or BAC clones include centromeres and telomeres as well as the nucleolar organizer with the rDNA cluster on LG V. Contig 3 on LG II and contig 4 on LG V are supposedly close to the centromeres. Sequences at the centromere-proximal ends of these contigs are devoid of genes and reveal an AT-rich, repetitive sequence similar to the centromeric sequence reported for LG VII (34). However, because neither includes genes mapping at both sides of the centromeric region(s) it is unlikely that either or both include a complete centromere. Similarly, only part of the telomere at the right of LG V (35) is found in contig 11 of LG V.
Figure 1.
Alignment of genetic map of LG II and LG V and sequence contigs. Genetic loci are positioned as published by Perkins (87). The scale of the genetic maps is based on 150 map units for LG II and 140 map units for LG V (33). Gap sizes are not drawn to scale. Positions of contig 13 LG II and contig 12 LG V are arbitrary. The positions of centromere and telomere like sequences in the contigs are indicated by dashed lines. hsps-1, close to left telomere; acu-5, linked to arg-5 and aro-3; con-6, left of arg-12; vma10, linked near arg-12; leu-6, between trp-3 and right telomere; ccg-8 and Fsr-16, linked to centromere; nuo21.3a, linked to inl and inv; ppt-1, linked to inl; NOR, nucleolar organizer.
The order of markers on the genetic map is mostly conserved in the contigs, whereas the ratio of genetic distance to physical distance varies considerably. It ranges from 6 kb per map unit for the pair arg-5/aro-3 on LG II to 115 kb per map unit for the pair am/his-1 on LG V. A centromere effect resulting in a significant increase in the physical/genetic ratio close to the centromere as reported for LG III (36) is not apparent for LG II and LG V. According to the genetic distance of markers located on different sequence contigs, the gaps between these contigs are expected to be small. However, the variability of the physical/genetic ratio does not allow a prediction of single gap sizes. The location of sequence contigs as well as gap sizes is ambigous on the far left part of LG II and the entire left arm of LG V. This is due to the scarcity of mapped genetic loci linked to known sequence in these regions.
Repeated DNA elements and duplications in the N.crassa genome
Repeated DNA elements identified readily on LG II and LG V, apart from micro- or minisatellites and homopolymeric stretches, are copies of the 5S rRNA genes present on both chromosomes. The 5S rRNA genes show some sequence variations and several variants were mapped genetically (e.g. Fsr-16 in Fig. 1) (37). On LG II 21 copies have been identified, while LG V comprises 12 copies.
In order to search for large-scale or segmental duplications we applied an exhaustive TBLASTX comparison between and within the chromosomes of LG II and LG V and identified clusters of collinear hits. Only small DNA sequence stretches in the range 4–10 kb were found, which are duplicated either on the same chromosome or between the two chromosomes. No indications of chromosome or genome duplication were apparent. Sequence identities of duplicated regions range between 57 and 74% (Table 1). Three of the four duplicated sequences are found at three idependent locations. The duplicated sequence stretches are all found in non-coding DNA and are very AT rich. None of the repeat regions encode transposon-like elements.
Table 1. Duplicated sequence stretches on linkage groups II and V.
| Position 1 | Position 2 | Sequence identity |
|---|---|---|
| 7k21 (Contig 5 LG V) 23117–32319 | 9a66 (Contig 10 LG V) 51272–60682 | 57% |
| 23889–30528 | b14h13 (Contig 6 LG II) 4594–11233 | 57% |
| 9a45 (Contig 10 LG V) 91978–96382 | b14h13 (Contig 6 LG II) 13626–17996 | 58% |
| 9a82 (Contig 7 LG II) 47486–53701 | 9a48 (Contig 4 LG II) 123116–129306 | 69% |
| 48969–56878 | 9a63 (Contig 4 LG II)164064–156136 | 74% |
| b23e9 (Contig 2 LG II) 22126–26569 | 9a28 (Contig 2 LG V) 52341–47862 | 72% |
| 22614–26536 | 9a52 (Contig 4 LG V) 88337–92261 | 68% |
Mobile elements are rare in N.crassa, nevertheless close scrutiny revealed transposon-related sequences on LG II and LG V (Table 2). A reading frame, encoding a 270 amino acid polypeptide, shares sequence similarity to the first 130 amino acids of the putative transposase Fot1, originally detected in Fusarium oxysporum (38). The flanking genomic region is very similar to the previously described Punt transposable element of N.crassa (39). The 270 amino acid sequence may thus represent the remains of a Punt element, which itself is closely related to the Fot1 transposon. Fot1 is a putative eukaryotic class II transposon (40). It has a conserved open reading frame (ORF) of more than 500 amino acids, as well as terminal inverted repeats, and generates an ‘AT’ target site duplication. The related putative transposase in N.crassa is not only much shorter but, more importantly, only fragments of the Fusarium homolog can be identified and the terminal inverted repeats are missing. Therefore, it appears to be an inactive element which has likely fallen victim to the N.crassa RIP mechanism. Repeat-induced point mutations were found to inactivate repeated DNA sequences during or prior to meiosis in N.crassa (16,41). Often these mutations are G:C→A:T transitions. Other remnants of transposases appear to have been inactivated by RIP, as well. Confirming findings published some years ago by Kinsey et al. (42) a total of five copies of the LINE-like element Tad are present on LG II and LG V. Three copies are located within 220 kb of one sequence contig (Table 2). In addition, a retrotransposon-like sequence similar to DAB1 (43) is present on LG V. Likely to escape the RIP mechanism due to their shortness are Guest elements, which are very short inverted repeat transposon-like elements that do not encode a transposase (44). However, copies of Guest appear not to be present on either chromosome II or V, unless sequences differ significantly from the originally published sequence (44).
Table 2. Transposon-related sequences on linkage groups II and V.
Gene prediction and annotation
MNCDB not only provides sequence data to be searched and/or downloaded, but also an annotation of the sequence, i.e. a curated prediction of the genes encoded in the genome. The analysis presented here is based on the results of the manual annotation of 12 Mb of sequence data mostly of LG II and LG V. The database contains 3218 proteins at the time of writing. Extrapolating the number of postulated genes a total of 11 000 genes for the entire genome is estimated. In collaboration with the Whitehead Genome Center the database will be expanded to the entire genome, merging the cosmid and BAC sequences with the assembled shotgun sequences. It will be continuously updated according to the progress of manual gene prediction and annotation.
Gene models predicted by software tools were manually corrected after homology searches using a non-redundant protein database and a Neurospora EST database to include extrinsic information. Only EST matches with >98% identity as well as very similar matches to experimentally known proteins were used to correct gene models. We did not correct gene models according to predicted or hypothetical proteins derived from other genomic sequencing projects. In uncertain cases the gene model predicted by Neurospora trained FGENESH was used. Gene prediction was restricted to ORFs larger than 100 codons. Smaller ORFs were included only if similarities to other proteins or EST matches confirm their existence or if a coding region is postulated by all prediction programs used. Therefore, only 102 proteins smaller than 100 amino acids are currently present in the database.
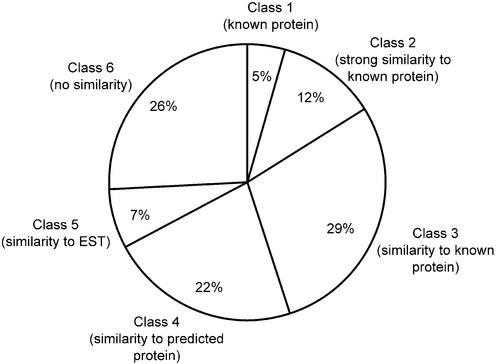
Protein sequences were derived from the final gene models and annotated. They are classified according to the MIPS classification catalog (Fig. 2). Class 1 proteins are previously known Neurospora proteins which have been experimentally validated and retain the original title in the database. Proteins in class 2 yield at least 1/3 of FASTA self-score when compared to experimentally characterized proteins. Class 2 proteins are named according to a well-characterized homolog with the prefix ‘probable’. If the FASTA score is lower but still above 200, a protein is ranked into class 3 carrying the prefix ‘related’ in addition to the title of the similar protein. Similarities or strong similarities to proteins lacking experimental evidence lead to class 4 proteins, which are termed conserved hypothetical proteins. All proteins in class 5 considered to be putative proteins exhibit no similarity to a protein but have an EST match. The remaining proteins (class 6) have no similarity to any protein and no EST match and are hypothetical proteins.
Figure 2.
Distribution of classified ORFs in MNCDB. Classes 1, 2 and 3 represent ORFs with known relatives. Class 4 represents ORFs with relatives deduced from nucleic acid sequence. Classes 5 and 6 represent ORFs without relatives in other organisms.
All proteins with a known, characterized homolog, e.g. proteins in classes 1–3, were manually assigned to functional categories using the MIPS functional catalog (FunCat) (31). The most closely related protein with a known function served as the basis for the functional assignment. Proteins in classes 4–6 cannot be functionally classified due to the lack of any experimentally characterized homolog. After manual gene modeling and annotation steps, the extracted ORFs were subjected to an extensive automatic analysis and annotation routine, which is performed by PEDANT, a collection of software tools for protein sequence analysis (27).
Protein domain statistics
Known homologs were identified for 1446 proteins (classes 1–3 in Fig. 2). A total of 730 different Pfam domains (45) were described for 1206 proteins and 1028 ORFs were found to contain 385 different Prosite patterns (46). Combining these data, 1682 proteins (52% of annotated ORFs) were assigned to a functional category (Table 3). Several proteins are listed in more than one functional category, since more than one protein domain or pattern with different functional implications are present or the function of a close homolog is listed with more than one category. Compared to S.cerevisiae a high proportion of deduced proteins involved in metabolism as well as cellular communication and signal transduction is apparent for N.crassa (Table 3). Among the sub-categories of metabolism by far the most significant difference between N.crassa and S.cerevisiae is found for the sub-category ‘secondary metabolism’. While five entries are found for the entire yeast genome, 23 entries are already listed in MNCDB with one-third of the N.crassa genome analyzed. Accordingly, few N.crassa genes attributed to secondary metabolism have a S.cerevisiae relative (7 out of 23), as well as other yeasts. None of the genes in this sub-category is specifically found in fungi, while a significant number have relatives exclusively among eubacterial genes (Table 3).
Table 3. Distribution of deduced ORFs among functional categories.
| FunCat category | S.cerevisiae | N.crassa | N.crassa ORFs specific for | ||
|---|---|---|---|---|---|
| 6 | Total | Total | Filamentous fungi | Fungi | Bacteria |
| 1 Metabolism (all sub-categories) | 1066 | 390 | 12 | 12 | 33 |
| 1.05 Carbohydrate metabolism | 415 | 149 | 8 | 6 | 19 |
| 1.20 Secondary metabolism | 5 | 23 | 6 | ||
| 2 Energy | 252 | 79 | 6 | 1 | |
| 3 Cell cycle and DNA processing | 628 | 142 | 13 | 6 | |
| 4 Transcription | 771 | 207 | 22 | ||
| 5 Protein synthesis | 359 | 97 | 4 | ||
| 6 Protein fate | 595 | 195 | 1 | 12 | 2 |
| 8 Cellular transport and transport mechanism | 495 | 125 | 1 | 8 | |
| 10 Cellular communication/signal transduction | 59 | 62 | 1 | ||
| 11 Cell rescue, defense and virulence | 278 | 90 | 2 | 5 | 6 |
| 13 Interaction with cellular environment | 199 | 22 | 3 | ||
| 14 Cell fate (all sub-categories) | 427 | 116 | 2 | 15 | |
| 14.01 Cell growth and morphogenesis | 96 | 26 | 2 | 4 | |
| 29 Transposable elements, viral and plasmid proteins | 116 | 10 | 2 | ||
| 30 Control of cellular organization | 209 | 47 | 4 | ||
| 67 Transport (all sub-categories) | 313 | 96 | 9 | 1 | |
| 67.07 Carbohydrate transporter | 46 | 19 | 5 | 1 |
Listed are the number of ORFs attributed to a functional category from S.cerevisiae and N.crassa (current data set). Filamentous fungi, ORFs of N.crassa with homologs exclusively among filamentous fungi; Fungi, ORFs of N.crassa with homologs exclusively among fungi; Bacteria, ORFs of N.crassa with homologs exclusively among bacteria and filamentous fungi.
Table 4 lists the 20 most common protein domains identified in MNCDB compared to the abundance of protein domains in a yeast, a plant and an animal. For a better comparison the numbers for N.crassa have been extrapolated to the expected total set of 11 000 genes in Table 4. To avoid extrapolations of very small numbers the analysis is restricted to the most common domains. Not surprisingly many abundant domains are involved in signal transduction, protein– protein interaction and transcriptional regulation. Domains of signal switching proteins include the eukaryotic protein kinase domain at the top of the list, as in other eukaryotes, and the ras family domain present in small GTPases. Compared to the other organisms listed in Table 4, the number of kinase domains identified for N.crassa is rather low. The proteins comprising these domains are common to fungi, found in filamentous fungi as well as yeasts. Tyrosine- or serine/threonine-specific receptor kinases abundant in plants and animals are rare in N.crassa as well as in yeasts. Proteins with WD/G-β repeat domains, ankyrin repeats and SH3 domains are the most common protein–protein interaction domains in the N.crassa genome. Transcription factors are mainly from the zinc finger superfamily, where the fungal-specific family of Zn2-Cys6 binuclear clusters are the most abundant, followed by zinc fingers of the C2H2 type and bZIP transcription factors.
Table 4. Abundance of most common protein domains.
| Pfam domain | InterPro no. | No. of proteins with Pfam domain | |||
|---|---|---|---|---|---|
| N.crassaa | S.cerevisiae | A.thaliana | D.melanogaster | ||
| Protein kinase domain | 0719 | 81 | 115 | 1038 | 248 |
| Short chain dehydrogenase | 2198 | 69 | 13 | 86 | 52 |
| WD domain, G-β repeat | 1680 | 57 | 97 | 240 | 175 |
| Helicase C-terminal domain | 1650 | 51 | 72 | 142 | 75 |
| Mitochondrial carrier protein | 1993 | 51 | 34 | 55 | 49 |
| RNA recognition motif | 0504 | 48 | 55 | 249 | 140 |
| DEAD/DEAH box helicase | 1410 | 45 | 73 | 138 | 75 |
| Fungal Zn2-Cys6 binuclear cluster | 1138 | 45 | 57 | 1 | 0 |
| Ras family | 1806 | 45 | 24 | 101 | 75 |
| Sugar (and other) transporter | 3662 | 45 | 52 | 91 | 97 |
| Monooxygenase | 0733 | 42 | 3 | 16 | 2 |
| AMP-binding enzyme | 0873 | 36 | 11 | 45 | 30 |
| Zinc finger, C2H2 type | 0822 | 36 | 53 | 169 | 347 |
| Ankyrin repeat | 2110 | 33 | 18 | 113 | 86 |
| Actin | 4000 | 30 | 10 | 19 | 15 |
| bZIP transcription factor | 4827 | 30 | 16 | 74 | 27 |
| DnaJ domain | 1623 | 30 | 21 | 100 | 34 |
| Cytochrome P450 | 1128 | 27 | 3 | 246 | 86 |
| SH3 domain | 1452 | 27 | 24 | 4 | 76 |
| Phox-like domain | 1683 | 24 | 15 | 9 | 16 |
The occurrence of the 20 most common protein domains in MNCDB is compared to that of fully sequenced eukaryotes. The data for S.cerevisiae, A.thaliana and Drosophila melanogaster are from the EBI Proteome website (http://www.ebi.ac.uk/proteome) (88).
aThe numbers for N.crassa have been extrapolated to a total of 11 000 ORFs for better comparison.
A significantly high abundance among N.crassa ORFs is apparent for domains specific for short chain dehydrogenases, FAD monooxygenases and AMP-binding enzymes. The short chain dehydrogenase domains are the second most abundant domains among ORFs of N.crassa in contrast to other organisms, in which these domains are far less prominent. Short chain dehydrogenases participate in a variety of catabolic as well as anabolic pathways involving redox reactions of hydroxy or keto functions (47). Substrates include fatty acids, amino acids, polyketides, sugars and steroids. Sequence analysis does not reveal clear cut allocations of deduced ORFs to a specific function. Identifiable are five acyl carrier protein-directed ketoacyl reductases which are involved in the synthesis of specific acyl moeties. At least one of these is located in the mitochondrial matrix (ORF b2a19_180). Another four short chain dehydrogenases of N.crassa are likely to be involved in secondary metabolism participating in the synthesis of alkaloids, anthraquinone derivatives and sugar alcohols. The only known short chain dehydrogenase is bli-4, a mitochondrial, light-induced dehydrogenase (48). Exceptionally high is the number of identified flavoprotein monooxygenase domains (49). Neurospora crassa comprises more of these domains than any other organism listed in Table 4. Most of the N.crassa enzymes forming such a monooxygenase domain are supposed to be involved in the catabolism of aromatic substrates. On the one hand these enzymes are needed for the utilization of complex substrates, on the other hand they are involved in the modification of toxic compounds. Last but not least, monooxygenases participate in the biosynthesis of steroids, ubiquinone and pigments (see Table 6). Another type of monooxygenases comprises cytochrome P450 domains (50). Though rather frequent in N.crassa, animals and especially plants have even more P450 monooxygenases. While no N.crassa P450 monooxygenase has so far been described, a number of these enzymes in other fungi are known. Most are involved in the synthesis of polyketides, like aflatoxin (51), or isoprenoid compounds, for example gibberellin (52) (see Table 6), or in the degradation of aromatic compounds (53). Most short chain dehydrogenases and monooxygenases lack a close yeast homolog. While most of the P450 monooxygenases have their closest non-fungal relative among animal proteins, short chain dehydrogenases as well as FAD monooxygenases are frequently most closely related to bacterial enzymes. The AMP-binding enzymes are mostly acyl-CoA ligases and synthetases participating in the degradation and synthesis of amino acids and lipids. Homologs of the AMP-binding enzymes are found in eukaryotes, including yeasts, as well as prokaryotes.
Table 6. Neurospora crassa ORFs related to biosynthesis of pigments and mycotoxins.
| Closest homolog(s) | Sequence identity | Function | Metabolite | Reference | |
|---|---|---|---|---|---|
| 80a10_310 | abr-1 A.fumigatus | 42% | Dioxigenase | DHN melanin | (64) |
| 1nc310_160 | ayg-1 A.fumigatus | 48% | unknown | DHN melanin | (64) |
| 90c4_150 | tyrosinase Podospora anserina | 58% | Monooxygenase | DOPA melanin | (65) |
| 1nc800_090 | tyrosinase P.anserina | 33% | Monooxygenase | DOPA melanin | |
| b23b10_250 | lac3 Gaeumannomyces graminis | 50% | Dioxygenase | melanin? | (89) |
| 2nc610_110 | PKS1 C.heterostrophus | 37% | PKS | T-toxin | (70) |
| lovF A.terreus | 35% | Lovastatin | Genes | (71) | |
| b13b3_040 | amt A.alternata | 34% | NRPS | AM-toxin | (72) |
| b10d6_150 | ordA Aspergillus parasiticus | 30% | P450 MO | Aflatoxin | (51) |
| b24m22_100 | tri4 Fusarium sporotrichioides | 33% | P450 MO | Trichothecene | (90) |
| 1nc200_350 | tri4 F.sporotrichioides | 31% | P450 MO | Trichothecene | (90) |
| 1nc570_360 | tri11 Gibberella zeae | 35% | P450 MO | Trichothecene | |
| b1d4_280 | P450I G.fujikuori | 48% | P450 MO | Gibberellin | (52) |
| Genes | lovA A. terreus | 39% | Lovastatin | Genes | (71) |
| 2nc610_160 | fum6 Gibberella moniliformis | 33% | P450 MO | Fumonisin | (91) |
| 2nc610_090 | paxP P.paxilli | 30% | P450 MO | Paxillin | (75) |
| b24n4_150 | moxY A.parasiticus | 37% | FAD MO | Aflatoxin | (51) |
| b20j13_070 | moxY Aspergillus flavus | 34% | FAD MO | Aflatoxin | (51) |
| b5k2_230 | Bcmfs1 Botryotinia fuckeliana | 39% | Transporter | (92) | |
| aflT A.parasiticus | 37% | Aflatoxin | |||
| b19c19_140 | cefT Acremonium chrysogenum | 28% | Transporter | Cephalosporin | (93) |
| b14a21_060 | stcC A.nidulans | 31% | Peroxidase | Sterigmatocystin | (94) |
| b9j10_120 | lovD A.terreus | 30% | Transesterase | Lovastatin | (71) |
| 64c2_020 | paxU P.paxilli | 40% | Unknown | Paxillin | (75) |
| 1nc580_650 | pclA Penicillium chrysogenum | 55% | PCL | Penicillin |
Comparative genomics
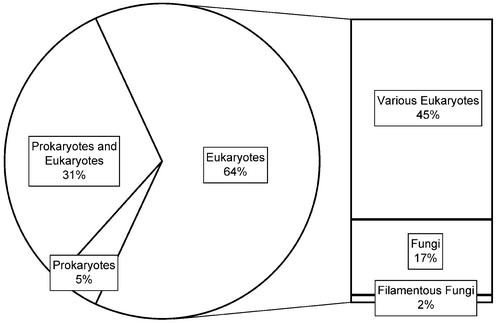
A classification of genes according to their distribution among phylogenetic divisions is provided by Figure 3. The analysis is based on BLASTP results against known and predicted ORFs in the EMBL database. Included are hits with an expected value below 10–8. Of ORFs with a sufficient BLAST hit, about one-third have hits in prokaryotes as well as eukaryotes, while two-thirds yield relatives in eukaryotes only (Fig. 3). Among ORFs with prokaryotic relatives two-thirds have hits to bacterial genes, while half pick archaeal genes. A small number of ORFs have hits in bacteria only. No genes were identified matching an archaeal gene without having a eukaryotic relative. While the total number of homologs identified is directly affected by the threshold used, relative distributions of ORFs in the categories presented in Figure 3 are affected only slightly by the threshold, e.g. a change in the maximal expect value from 10–2 to 10–8 results in a change in the percentage of ORFs with hits exclusively in prokaryotes from 4.3 to 4.8% and a change in the percentage of ORFs with hits among eukaryotes exclusively from 60.5 to 63.9%.
Figure 3.
Distribution of ORFs according to the phylogenetic association of relatives. The percentage of N.crassa ORFs having homologs among prokaryotes exclusively, prokaryotes and eukaryotes and eukaryotes exclusively are given. The right column shows the percentage of N.crassa ORFs with homologs exclusively among filamentous fungi, fungi or various eukaryotes, respectively.
The group of genes specific for filamentous fungi is rather small in number. Table 3 gives the distribution of the genes termed specific for filamentous fungi according to the attributed functional classification. The largest fraction of genes with predicted functions is supposed to be involved in metabolism. Most of the genes code for proteins hydrolyzing complex carbohydrates. This reflects the well-known ability of filamentous fungi to use a variety of carbon sources and to degrade plant material by secreting hydrolases. Other categories have two entries at most and the number of functionally characterized genes of filamentous fungi is still too small for a comprehensive analysis. Thus the data in Table 3 concerning ORFs specific for filamentous fungi are representative but only preliminary.
A much broader basis for the analysis is obtained if yeasts are included to reveal genes specific for fungi in general. In the current data set, 252 genes of N.crassa produce hits with fungal genes including yeasts solely. Of these, 123 are attributed to at least one functional category. As listed in Table 3, the most populated sub-category is mRNA transcription. Most of the 20 genes listed in this sub-category are involved in transcriptional control. A high number of entries is also found in the sub-categories cell cycle and cell differentiation, indicating a specific conservation of mechanisms directing processes in cellular development among fungi. While no proteins involved in transport were found to be specific for filamentous fungi, several are apparently fungal specific. The number of entries in the metabolism categories is rather small in light of the abundance of this category.
Several genes are related to bacterial genes but lack known relatives in yeasts, plants or animals. A total of 71 genes was found to belong to this group. This is more than the number of genes N.crassa shares exclusively with plants or animals. Fifty-six ORFs are new to the eukaryotic kingdom, while filamentous fungal homologs to 15 ORFs were known already. Of those ORFs with a known or proposed function three-quarters are involved in metabolism (Table 3). As mentioned, many short chain dehydrogenases and monooxygenases of N.crassa are most closely related to bacterial proteins. In addition, many hydrolases acting on complex carbohydrates are found among the bacterial relatives and lack close relatives in yeasts, plants or animals. The only sub-categories outside of metabolism with more than one entry are found in the category ‘cellular rescue and defense’. Most genes in this category code for enzymes degrading or modifying toxins.
DISCUSSION
The annotation of chromosomes II and V of N.crassa is a preview of what is unique to a filamentous fungal genome. It reveals the differences between N.crassa and all other genomes sequenced to date, both in the landscape and the coding potential. It highlights the small degree of redundancy, the high gene diversity and the wide metabolic capabilities.
Only a few small segmental duplications are apparent in the two chromosomes analyzed. In contrast to budding yeast and Arabidopsis thaliana, which still reveal ancient duplications of the entire genomes (54,55), large-scale duplication events are not evident in the N.crassa genome. In this respect it rather resembles fission yeast (4). As expected, only a very limited number of transposon-related sequences were found. All appear to have been subjected to RIP. However, in N.crassa and other organsims mobile elements are particularly frequent in regions close to telomeres and centromeres (34,55,56). Since these sequences are not present in the current data set the existence of active transposons on LG II and LG V cannot be excluded.
The predicted number of 11 000 ORFs is significantly higher than the count in the yeasts S.cerevisiae (6500 ORFs) and S.pombe (5000 ORFs). In addition, the level of redundancy among ORFs of N.crassa is very low. In our data set, consisting of about one-third of the expected N.crassa ORFs, few closely related paralogs are apparent. If a pair of paralogs is defined as two ORFs having at least 40% sequence identity and differing in their length by not more than 20%, then just 36 of 3218 analyzed ORFs find a paralog in MNCDB. Extrapolation to a total of 11 000 ORFs yields up to 400 ORFs (3.1%) with at least one paralog (Table 5). In comparison, the same criteria applied to a search in the genome of S.cerevisiae yields 1139 ORFs (including more than 80 copies of Ty). In the genomes of S.pombe and Candida albicans a similar percentage of 9.5% genes with a paralog is found. Significantly higher frequencies of paralogs are found in the genomes of animals and A.thaliana. Comparing the numbers in Table 5 it has to be considered that the high frequencies of paralogs in S.cerevisiae and A.thaliana certainly reflect the ancient duplications of the respective genomes (54,55). Nevertheless it is apparent that the genome of N.crassa comprises especially few paralogs. Whether this is caused solely by the RIP mechanism or is a peculiarity of filamentous fungi in general will be revealed as soon as additional genomes of filamentous fungi are analyzed.
Table 5. Frequency of paralogs in eukaryotic genomes.
| Genes | Genes | Paralogs No. | Percent |
|---|---|---|---|
| S.pombe | 5010 | 477 | 9.5 |
| C.albicans | 6165 | 586 | 9.5 |
| S.cerevisiae | 6449 | 1139 | 17.7 |
| N.crassaa | 11000 | 396 | 3.6 |
| D.melanogaster | 14148 | 2617 | 18.5 |
| C.elegans | 20391 | 3418 | 16.8 |
| A.thaliana | 25000 | 11283 | 45.1 |
Paralogs are defined as a pair of genes sharing at least 80% identity in their deduced amino acid sequence and differing by no more than 20% in their length.
aThe numbers for N.crassa are extrapolated to the entire genome.
The current database of annotated ORFs still includes a large number of orphans, hypothetical or putative ORFs lacking any ortholog (Fig. 2). At least in part this is due to the fact that very few genes of filamentous fungi have been deposited in the databases so far. This causes the small number of ORFs specific for filamentous fungi, as revealed in Figure 2 and Table 3. Filamentous fungi are the subject of several large-scale sequencing projects in progress. However, genes have not yet been extracted from the genomic sequences and are not available in the databases. A TBLASTX search of hypothetical and putative N.crassa genes against a six frame translation of the current Aspergillus fumigatus genome sequence (www.tigr.org/tdb/e2k1/afu1) revealed that almost half of the putative genes and about one-fourth of the hypothetical genes cause hits with expect values below 10–8. Thus, with the expected ORFs of the A.fumigatus genome alone the number of N.crassa genes specific for filamentous fungi will increase to 16%. In turn, the number of genes with no detectable sequence relative (classes 5 and 6) will drop below 20%.
As expected from the high number of ORFs and the low redundancy almost half of the annotated genes of N.crassa lack a readily detectable yeast homolog. Most of these have no supposed function so far and are classified as hypothetical, putative or conserved hypothetical. Of ORFs with a homolog (E < 10–8), 186 (13%) lack a comparable hit with yeasts proteins. In general, proteins missing in yeasts tell more about the yeasts than about specific features of N.crassa. In contrast to the filamentous fungi, yeasts have streamlined their metabolism resulting in the loss of numerous genes (57). Examples are the more than 30 subunits of respiratory complex I (58) or enzymes involved in the synthesis of the cofactor molybdopterin and molybdoenzymes.
In contrast, filamentous fungi are adapted to a changing environment. They are capable of growing on many different substrates and dealing with a variety of detrimental and toxic chemicals. Though the large number of different filamentous fungi have developed a variety of specific capabilities to thrive in their ecological niche, these properties are reflected in the genome of N.crassa as well. Highlighted especially are the multi-faceted metabolic potentialities evident from the total number of relevant enzymes and the high number of pertinent protein domains (Tables 3 and 4).
In which metabolic pathways are the many short chain dehydrogenases, monooxygenases and AMP-binding proteins involved? Very few are known in Neurospora, a few more are known in other fungi and a substantial number are known so far only in bacteria. Well-known secondary fungal metabolites are pigments and mycotoxins. These substances are important elements of fungal pathogenicity and thus have been of substantial interest (59,60). Often the enzymes involved in these pathways are coded by gene clusters (61,62). Related gene clusters are unknown in N.crassa and secondary metabolism has been studied in pathogenic fungi rather than N.crassa. However, several non-clustered homologs of enzymes involved in the formation of toxins and pigments are present in MNCDB (Table 6).
The black pigment of pathogenic fungi is generally dihydroxynaphthalene (DHN) melanin formed by the polyketide pathway (63). Enzymes catalyzing the formation of DHN melanin include a polyketide synthase, a scytalone dehydratase and a hydroxynaphthalene reductase. While homologs of these enzymes are so far not present in MNCDB, two ORFs indicate a DHN-related pigment formation in N.crassa (Table 6). The closest homologs to these ORFs are abr1 (aspergillus brown 1) and ayg1 (aspergillus yellowish green 1), respectively, which have been identified in a pigment biosynthesis gene cluster in A.fumigatus (64). The gene cluster comprises four other genes coding for a putative polyketide synthase, a scytalone dehydratase, a hydroxynaphthalene reductase and a laccase. The formation of dihydroxyphenylalanine (DOPA) melanin, which is synthezised in the skins of animals, is questionable in fungi (63). Its synthesis starts with tyrosine, which is converted to DOPA by tyrosinase (phenol monooxygenase). DOPA is subsequently cyclized and polymerized in several steps to DOPA melanin. The polymerization is again catalyzed by tyrosinase but occurs spontaneously as well (63). MNCDB reveals two tyrosinases sharing 32% sequence identity (Table 6). One tyrosinase has been extensively studied, serving as a model system (65). The other is less closely related to known tyrosinases. It may catalyze a specific step in the formation of DOPA melanin, as has been described for tyrosine-related proteins of mammals (66). In some instances laccase (polyphenol oxidase) has been reported to be involved in the formation of melanin (67). Like other filamentous fungi N.crassa possesses more than one laccase. In addition to a known N.crassa laccase (68), MNCDB contains a cluster of four laccase-related ORFs including the abr1 homolog.
Mycotoxins include derivatives of polyketides (e.g. aflatoxin), peptides (e.g. penicillin) and isoprenoids (e.g. paxillin). Accordingly, key enzymes in the synthesis of mycotoxins are polyketide synthases (PKS), non-ribosomal peptide synthetases (NRPS) and geranylgeranyl pyrophosphate synthetases (GGPS). Pathogenic fungi appear to produce a substantial variety of these enzymes, while obligate saprophytic fungi like Neurospora encode few or none of these (69). The only PKS present in MNCDB is most closely related to PKS1 of Cochliobolus heterostrophus, forming T-toxin (70), and lovF of Aspergillus terreus, involved in the synthesis of lovastatin (71) (Table 6). A probable NRPS is related to enzymes synthesizing toxic peptides in Metarhizium anisopliae (72) and Alternaria alternata (73). So far only one GGPS is known in N.crassa which participates in the primary synthesis of carotenoids (74). Penicillium paxilli as well as Gibberella fujikuori have a second, specific GGPS encoded by gene clusters attributed to the synthesis of paxillin and gibberellin, respectively (52,75). Besides the key synthetases, mycotoxin gene clusters frequently include several P450 monooxygenases as well as FAD monooxygenases and drug transporters. A number of ORFs in MNCDB are most closely related to these enzymes participating in the synthesis of mycotoxins (Table 6). Homologs to additional enzymes found in clusters of secondary metabolism in other fungi include a peroxidase and a transesterase. Short chain dehydrogenases are also known to be required for the synthesis of fungal toxins and pigments (64,76). Homologs of these enzymes are, however, not yet found in MNCDB. Sequence identities of ORFs listed in Table 6 to their fungal homologs are mostly not sufficient to conclude a participation in a specific pathway and it is not yet possible to define the metabolites produced by these enzymes. Thus, the genomic database of N.crassa provides access to metabolic pathways so far unknown in this fungus.
Gene clusters frequent in other fungi, particularly for biosynthetic enzymes, appear to be generally rare in N.crassa. A well-known example of a catabolic gene cluster in N.crassa is the qa cluster (77). Just one additional example of a gene cluster is present in MNCDB. Two genes, acl1 and acl2, coding for two subunits of ATP citrate lyase (78), are clustered (ORFs b14d6_310 and b14d6_320). In contrast, genes clustered in other fungi are separated in N.crassa. The genes sAT and sCT needed for sulfur assimilation are clustered in A.terreus and Aspergillus nidulans (79), but are separated by 100 kb in the genome of N.crassa. Similarly, genes of carotenoid synthesis carB and carRA are clustered in G.fujikuori (80) and are separated by 80 kb in N.crassa. The small distance between N.crassa sAT and sCT as well as carB and carRA indicates a rather recent separation of these genes.
The genome of N.crassa comprises a significant number of metabolic enzymes so far unknown in fungi as well as other eukaryotes (Table 3). For example, besides the multi-domain cytosolic fatty acid synthetase, N.crassa possesses a substantial number of putative single domain ketoacyl-acyl carrier protein reductases typical for prokaryotic fatty acid synthases. These enzymes are likely to catalyze the formation of specific derivatives of fatty acids or polyketides. The closest homolog to ORF b10k17_110 is phaB, a ketoacyl reductase needed for the synthesis of polyhydroxyalkonates, which are bacterial storage compounds, in Bacillus megaterium (81). ORF b9b15_060 is closely related to rhlG, encoding a ketoacyl reductase involved in the formation of glycolipids in Pseudomonas aeruginosa (82). Several examples for enzymes lacking close eukaryotic relatives are found among oxygenases. MNCDB comprises seven putative phenol hydroxylases which are FAD monooxygenases involved in the degradation of aromatic compounds. While several bacterial enzymes have been studied (83), very few examples of fungal phenol hydroxylases are known (84,85). No phenol hydroxylase is found in S.cerevisiae or S.pombe. For two phenol hydroxylases identified in MNCDB no significant eukaryotic relative is known. These are related to pentachlorphenol oxygenases (80a10_290) and salicylate monooxygenases (1nc100_020). Close bacterial homologs to a probable dioxygenase (b13d15_170) are involved in the degradation of chlorinated phenols (86).
Analysis of the N.crassa genome sequence provides a first insight into the genetic endowment of a filamentous fungus. On the one hand this will allow a more comprehensive characterization of fungal biology. In addition, the genomic sequence provides access to many novel genes. Neurospora crassa has a long history as a model organism for biochemical genetics. In particular, the low degree of redundancy in its gene complement makes it especially amenable to study the effect of mutations on biochemical pathways. Already Beadle and Tatum have taken advantage of this when establishing the ‘one gene one enzyme’ hypothesis (9). The availability of the entire genome sequence will make N.crassa an even more attractive system.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to the members of the German Neurospora Genome Project Wolf-H. Kunau, Walter Neupert, Helga Ninnemann, Ludger Rensing, Manfred Schliwa, Maximilian Tropschug and Hanns Weiss for their support. Very helpful suggestions to improve the manuscript by Martha Merrow and the referees of NAR are gratefully acknowledged. This work was supported by the Deutsche Forschungsgemeinschaft (Schu 698/3).
REFERENCES
- 1.Hawksworth D.L. (1991) The fungal dimension of biodiversity: magnitude, significance and conservation. Mycol. Res., 95, 641–655. [Google Scholar]
- 2.Alexopoulos C.J., Mims,C.W. and Blackwell,M. (1996) Introductory Mycology, 4th Edn. John Wiley and Sons, New York, NY.
- 3.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M., Louis,E.J., Mewes,H.W., Murakami,Y., Philippsen,P., Tettelin,H. and Oliver,S.G. (1996) Life with 6000 genes. Science, 274, 546–567. [DOI] [PubMed] [Google Scholar]
- 4.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 414, 871–880. [DOI] [PubMed] [Google Scholar]
- 5.Bennett J.W. (1997) White paper: genomics for filamentous fungi. Fungal Genet. Biol., 21, 3–7. [DOI] [PubMed] [Google Scholar]
- 6.Schulte U., Becker,I., Mewes,H.W. and Mannhaupt,G. (2002) Large scale analysis of sequences from Neurospora crassa. J. Biotechnol., 94, 3–13. [DOI] [PubMed] [Google Scholar]
- 7.Davis R.H. and Perkins,D.D. (2002) Neurospora: a model of model microbes. Nature Rev. Genet., 3, 397–403. [DOI] [PubMed] [Google Scholar]
- 8.Perkins D.D. and Davis,R.H. (2000) Neurospora at the millennium. Fungal Genet. Biol., 31, 153–167. [DOI] [PubMed] [Google Scholar]
- 9.Beadle G.W. and Tatum,E.L. (1941) Genetic control of biochemical reaction in Neurospora. Proc. Natl Acad. Sci. USA, 27, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClintock B. (1945) Neurospora. I. Preliminary observations of the chromosomes of Neurospora crassa. Am. J. Bot., 32, 671–678. [Google Scholar]
- 11.Perkins D.D. and Barry,E.G. (1977) The cytogenetics of Neurospora. Adv. Genet., 19, 133–285. [DOI] [PubMed] [Google Scholar]
- 12.Lee K., Loros,J.J. and Dunlap,J.C. (2000) Interconnected feedback loops in the Neurospora circadian system. Science, 289, 107–110. [DOI] [PubMed] [Google Scholar]
- 13.Henningsen U. and Schliwa,M. (1997) Reversal in the direction of movement of a molecular motor. Nature, 389, 93–96. [DOI] [PubMed] [Google Scholar]
- 14.Kunkele K.P., Heins,S., Dembowski,M., Nargang,F.E., Benz,R., Thieffry,M., Walz,J., Lill,R., Nussberger,S. and Neupert,W. (1998) The preprotein translocation channel of the outer membrane of mitochondria. Cell, 93, 1009–1019. [DOI] [PubMed] [Google Scholar]
- 15.Mohr S., Stryker,J.M. and Lambowitz,A.M. (2002) A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell, 109, 769–779. [DOI] [PubMed] [Google Scholar]
- 16.Singer M. and Selker,E.U. (1995) Genetic and epigenetic inactivation of repetitive sequences in Neurospora crassa: RIP, DNA methylation and quelling. Curr. Top. Microbiol. Immunol., 197, 165–177. [DOI] [PubMed] [Google Scholar]
- 17.Cogoni C. (2001) Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol., 55, 381–406. [DOI] [PubMed] [Google Scholar]
- 18.Shiu P.K., Raju,N.B., Zickler,D. and Metzenberg,R.L. (2001) Meiotic silencing by unpaired DNA. Cell, 107, 905–916. [DOI] [PubMed] [Google Scholar]
- 19.Perkins D.D. (1975) The use of duplication-generating rearrangements for studying heterokaryon incompatibility genes in Neurospora. Genetics, 80, 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fungal Genetics Stock Center (2002) Catalogue of strains 9th Edition. Fungal Genet. Newsl., 49, Supplement. [Google Scholar]
- 21.Orbach M.J. (1992) Fungal Genet. Newsl., 39, 92. [Google Scholar]
- 22.Orbach M.J., Vollrath,D., Davis,R.W. and Yanofsky,C. (1988) An electrophoretic karyotype of Neurospora crassa. Mol. Cell. Biol., 8, 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aign V., Schulte,U. and Hoheisel,J.D. (2001) Hybridization-based mapping of Neurospora crassa linkage groups II and V. Genetics, 157, 1015–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salamov A.A. and Solovyev,V.V. (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res., 10, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borodowsky M. and Peresetzky,A. (1994) Deriving non-homogenous DNA Markov chain models by cluster analysis algorithm minimizing multiple alignment entropy. Comput. Chem., 18, 259–267. [DOI] [PubMed] [Google Scholar]
- 26.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 27.Frishman D., Albermann,K., Hani,J., Heumann,K., Metanomski,A., Zollner,A. and Mewes,H.W. (2001) Functional and structural genomics using PEDANT. Bioinformatics, 17, 44–57. [DOI] [PubMed] [Google Scholar]
- 28.Frishman D. and Argos,P. (1997) 75% accuracy in protein secondary structure prediction. Proteins, 27, 329–335. [DOI] [PubMed] [Google Scholar]
- 29.Schaffer A.A., Wolf,Y.I., Ponting,C.P., Koonin,E.V., Aravind,L. and Altschul,S.F. (1999) IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics, 15, 1000–1011. [DOI] [PubMed] [Google Scholar]
- 30.Kelkar H.S., Griffith,J., Case,M.E., Covert,S.F., Hall,R.D., Keith,C.H., Oliver,J.S., Orbach,M.J., Sachs,M.S., Wagner,J.R., Weise,M.J., Wunderlich,J.K. and Arnold,J. (2001) The Neurospora crassa genome: cosmid libraries sorted by chromosome. Genetics, 157, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mewes H.W., Frishman,D., Guldener,U., Mannhaupt,G., Mayer,K., Mokrejs,M., Morgenstern,B., Munsterkotter,M., Rudd,S. and Weil,B. (2002) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 30, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler S.K. and Metzenberg,R.L. (1989) Premeiotic change of the nucleolus organiser size in Neurospora. Genetics, 122, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radford A. and Parish,J.H. (1997) The genome and genes of Neurospora crassa. Fungal Genet. Biol., 21, 285–266. [DOI] [PubMed] [Google Scholar]
- 34.Cambareri E.B., Aisner,R. and Carbon,J. (1998) Structure of the chromosome VII centromere region in Neurospora crassa: degenerate transposons and simple repeats. Mol. Cell. Biol., 18, 5465–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schechtman M.G. (1990) Characterization of telomere DNA from Neurospora crassa. Gene, 88, 159–165. [DOI] [PubMed] [Google Scholar]
- 36.Davis C.R., Kempainen,R.R., Srodes,M.S. and McClung,C.R. (1994) Correlation of the physical and genetic maps of the centromeric region of the right arm of linkage group III of Neurospora crassa. Genetics, 136, 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Free S., Rice,P.W. and Metzenberg,R.L. (1979) Arrangement of the genes coding for ribosomal RNA in Neurospora crassa. J. Bacteriol., 137, 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daboussi M.J., Langin,T. and Brygoo,Y. (1992) Fot1, a new family of fungal transposable elements. Mol. Gen. Genet., 232, 12–16. [DOI] [PubMed] [Google Scholar]
- 39.Margolin B.S., Garrett-Engele,P.W., Stevens,J.N., Fritz,D.Y., Garrett-Engele,C., Metzenberg,R.L. and Selker,E.U. (1998) A methylated Neurospora 5S rRNA pseudogene contains a transposable element inactivated by repeat-induced point mutation. Genetics, 149, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daboussi M.J. and Langin,T. (1994) Transposable elements in the fungal plant pathogen Fusarium oxysporum. Genetica, 93, 49–59. [Google Scholar]
- 41.Cambareri E.B., Jensen,B.C., Schabacht,E. and Selker,E.U. (1989) Repeat-induced G-C to A-T mutations in Neurospora. Science, 244, 1571–1575. [DOI] [PubMed] [Google Scholar]
- 42.Kinsey J.A., Garrett-Engele,P.W., Cambareri,E.B. and Selker,E.U. (1994) The Neurospora transposon Tad is sensitive to repeat-induced point mutation (RIP). Genetics, 138, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bibbins M., Cummings,N.J. and Connerton,I.F. (1998) DAB1: a degenerate retrotransposon-like element from Neurospora crassa. Mol. Gen. Genet., 258, 431–436. [DOI] [PubMed] [Google Scholar]
- 44.Yeadon P.J. and Catcheside,D.E.A. (1995) Guest: a 98 bp inverted repeat transposable element in Neurospora crassa. Mol. Gen. Genet., 247, 105–109. [DOI] [PubMed] [Google Scholar]
- 45.Bateman A. Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam Protein Families Database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falquet L. Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joernvall H., Persson,B., Krook,M., Atrian,S., Gonzalez-Duarte,R., Jeffery,J. and Ghosh,D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry, 34, 6003–6013. [DOI] [PubMed] [Google Scholar]
- 48.Bruchez J.J., Eberle,J., Kohler,W., Kruft,V., Radford,A. and Russo,V.E. (1996) bli-4, a gene that is rapidly induced by blue light, encodes a novel mitochondrial, short-chain alcohol dehydrogenase-like protein in Neurospora crassa. Mol. Gen. Genet., 252, 223–229. [DOI] [PubMed] [Google Scholar]
- 49.Harayama S., Kok,M. and Neidle,E.L. (1992) Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol., 46, 565–601. [DOI] [PubMed] [Google Scholar]
- 50.Nelson D.R., Kamataki,T., Waxman,D.J., Guengerich,F.P., Estabrook,R.W., Feyereisen,R., Gonzalez,F.J., Coon,M.J., Gunsalus,I.C., Gotoh,O., Okudam,K. and Nebert,D.W. (1993) The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes and nomenclature. DNA Cell Biol., 12, 1–51. [DOI] [PubMed] [Google Scholar]
- 51.Yu J., Chang,P.K., Cary,J.W., Wright,M., Bhatnagar,D., Cleveland,T.E., Payne,G.A. and Linz,J.E. (1995) Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol., 61, 2365–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tudzynski B. and Hoelter,K. (1998) Gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet. Biol., 25, 157–170. [DOI] [PubMed] [Google Scholar]
- 53.Mingot J.M., Penalva,M.A. and Fernandez-Canon,J.M. (1999) Disruption of phacA, an Aspergillus nidulans gene encoding a novel cytochrome P450 monooxygenase catalyzing phenylacetate 2-hydroxylation, results in penicillin overproduction. J. Biol. Chem., 274, 14545–14550. [DOI] [PubMed] [Google Scholar]
- 54.Wolfe K.H. and Shields,D.C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 55.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 56.Levis R.W., Ganesan,R., Houtchens,K., Tolar,L.A. and Sheen,F.M. (1993) Transposons in place of telomeric repeats at a Drosophila telomere. Cell, 75, 1083–1093. [DOI] [PubMed] [Google Scholar]
- 57.Braun E.L., Halpern,A.L., Nelson,M.A. and Natvig,D.O. (2000) Large scale comparison of fungal sequence information: mechanism of innovation in Neurospora crassa and gene loss in Saccharomyces cerevisiae. Genome Res., 10, 416–430. [DOI] [PubMed] [Google Scholar]
- 58.Schulte U. and Weiss,H. (1995) Generation and characterization of NADH:ubiquinone oxidoreductase mutants in Neurospora crassa. Methods Enzymol., 260, 3–14. [DOI] [PubMed] [Google Scholar]
- 59.Henson J.M., Butler,M.J., Day,A.W. and Henson,J.M. (1999) The dark side of the mycelium: melanins of phytopathogenic fungi. Annu. Rev. Phytopathol., 37, 447–471. [DOI] [PubMed] [Google Scholar]
- 60.Oliver R. and Osbourn,A. (1995) Molecular dissection of fungal phytopathogenicity. Microbiology, 141, 1–9. [DOI] [PubMed] [Google Scholar]
- 61.Smith D.J., Burnham,M.K., Bull,J.H., Hodgson,J.E., Ward,J.M., Browne,P., Brown,J., Barton,B., Earl,A.J. and Turner,G. (1990) Beta-lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J., 9, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura N. and Tsuge,T. (1993) Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol., 175, 4427–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bell A.A. and Wheeler,M.H. (1986) Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol., 24, 411–451. [Google Scholar]
- 64.Tsai H.F., Wheeler,M.H., Chang,Y.C. and Kwon-Chung,K.J. (1999) A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol., 181, 6469–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kupper U., Niedermann,D.M., Travaglini,G. and Lerch,K. (1989) Isolation and characterization of the tyrosinase gene from Neurospora crassa. J. Biol. Chem., 264, 17250–17258. [PubMed] [Google Scholar]
- 66.del Marmol V. and Beermann,F. (1996) Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett., 381, 165–168. [DOI] [PubMed] [Google Scholar]
- 67.Williamson P.R., Wakamatsu,K. and Ito,S. (1998) Melanin biosynthesis in Cryptococcus neoformans. J. Bacteriol., 180, 1570–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Germann U.A., Muller,G., Hunziker,P.E. and Lerch,K. (1988) Characterization of two allelic forms of Neurospora crassa laccase. Amino- and carboxyl-terminal processing of a precursor. J. Biol. Chem., 263, 885–896. [PubMed] [Google Scholar]
- 69.Yoder O.C. and Turgeon,B.G. (2001) Fungal genomics and pathogenicity. Curr. Opin. Plant Biol., 4, 315–321. [DOI] [PubMed] [Google Scholar]
- 70.Yang G., Rose,M.S., Turgeon,B.G. and Yoder,O.C. (1996) A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell, 8, 2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kennedy J., Auclair,K., Kendrew,S.G., Park,C., Vederas,J.C. and Hutchinson,C.R. (1999) Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science, 284, 1368–1372. [DOI] [PubMed] [Google Scholar]
- 72.Bailey A.M., Kershaw,M.J., Hunt,B.A., Paterson,I.C., Charnley,A.K., Reynolds,S.E. and Clarkson,J.M. (1996) Cloning and sequence analysis of an intron-containing domain from a peptide synthetase-encoding gene of the entomopathogenic fungus Metarhizium anisopliae. Gene, 173, 195–197. [DOI] [PubMed] [Google Scholar]
- 73.Johnson R.D., Johnson,L., Itoh,Y., Kodama,M., Otani,H. and Kohmoto,K. (2000) Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM-toxin synthesis and pathogenicity. Mol. Plant Microbe Interact., 13, 742–753. [DOI] [PubMed] [Google Scholar]
- 74.Carattoli A., Romano,N., Ballario,P., Morelli,G. and Macino,G, (1991) The Neurospora crassa carotenoid biosynthetic gene (albino 3) reveals highly conserved regions among prenyltransferases. J. Biol. Chem., 266, 5854–5859. [PubMed] [Google Scholar]
- 75.Young C., McMillan,L., Telfer,E. and Scott,B. (2001) Molecular cloning and genetic analysis of an indole-diterpene gene cluster from Penicillium paxilli. Mol. Microbiol., 39, 754–764. [DOI] [PubMed] [Google Scholar]
- 76.Keller N.P., Kantz,N.J. and Adams,T.H. (1994) Aspergillus nidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol., 60, 1444–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geever R.F., Huiet,L., Baum,J.A., Tyler,B.M., Patel,V.B., Rutledge,B.J., Case,M.E. and Giles,N.H. (1989) DNA sequence, organization and regulation of the qa gene cluster of Neurospora crassa. J. Mol. Biol., 207, 15–34. [DOI] [PubMed] [Google Scholar]
- 78.Nowrousian M., Kück,U., Loser,K. and Weltring,K.M. (2000) The fungal acl1 and acl2 genes encode two polypeptides with homology to the N- and C-terminal parts of the animal ATP citrate lyase polypeptide. Curr. Genet., 37, 189–193. [DOI] [PubMed] [Google Scholar]
- 79.Borges-Walmsley M.I., Turner,G., Bailey,A.M., Brown,J., Lehmbeck,J. and Clausen,I.G. (1995) Isolation and characterisation of genes for sulphate activation and reduction in Aspergillus nidulans: implications for evolution of an allosteric control region by gene duplication. Mol. Gen. Genet., 247, 423–429. [DOI] [PubMed] [Google Scholar]
- 80.Linnemannstons P., Prado,M.M., Fernandez-MartinR., Tudzynski,B. and Avalos,J. (2002) A carotenoid biosynthesis gene cluster in Fusarium fujikuroi: the genes carB and carRA. Mol. Genet. Genomics, 267, 593–602. [DOI] [PubMed] [Google Scholar]
- 81.McCool G.J. and Cannon,M.C. (2001) PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J. Bacteriol., 183, 4235–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campos-Garcia J., Caro,A.D., Najera,R., Miller-Maier,R.M., Al-Tahhan,R.A. and Soberon-Chavez,G. (1998) The Pseudomonas aeruginosa rhlG gene encodes an NADPH-dependent beta-ketoacyl reductase which is specifically involved in rhamnolipid synthesis. J. Bacteriol., 180, 4442–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Powlowski J. and Shingler,V. (1994) Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation, 5, 219–236. [DOI] [PubMed] [Google Scholar]
- 84.Kalin M., Neujahr,H.Y., Weissmahr,R.N., Sejlitz,T., Johl,R., Fiechter,A. and Reiser,J. (1992) Phenol hydroxylase from Trichosporon cutaneum: gene cloning, sequence analysis and functional expression in Escherichia coli. J. Bacteriol., 174, 7112–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eppink M.H., Cammaart,E., Van Wassenaar,D., Middelhoven,W.J. and van Berkel,W.J. (2000) Purification and properties of hydroquinone hydroxylase, a FAD-dependent monooxygenase involved in the catabolism of 4-hydroxybenzoate in Candida parapsilosis CBS604. Eur. J. Biochem., 267, 6832–6840. [DOI] [PubMed] [Google Scholar]
- 86.Takizawa N., Yokoyama,H., Yanagihara,K., Hatta,T. and Kiyohara,H. (1995) A locus of Pseudomonas pickettii DTP0602, had, that encodes 2,4,6-trichlorophenol-4-dechlorinase with hydroxylase activity and hydroxylation of various chlorophenols by the enzyme. J. Ferment. Bioeng., 80, 318–326. [Google Scholar]
- 87.Perkins D.D. (2000) Neurospora crassa genetic maps and mapped loci. Fungal Genet. Newsl., 47, 40–58. [Google Scholar]
- 88.Apweiler R. Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Litvintseva A.P. and Henson,J.M. (2002) Cloning, characterization and transcription of three laccase genes from Gaeumannomyces graminis var. tritici, the take-all fungus. Appl. Environ. Microbiol., 68, 1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hohn T.M., Desjardins,A.E. and McCormick,S.P. (1995) The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet., 248, 95–102. [DOI] [PubMed] [Google Scholar]
- 91.Seo J.A., Proctor,R.H. and Plattner,R.D. (2001) Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol., 34, 155–165. [DOI] [PubMed] [Google Scholar]
- 92.Hayashi K., Schoonbeek,H.J. and De Waard,M.A. (2002) Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol., 68, 4996–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ullan R.V., Liu,G., Casqueiro,J., Gutierrez,S., Banuelos,O. and Martin,J.F. (2002) The cefT gene of Acremonium chrysogenum C10 encodes a putative multidrug efflux pump protein that significantly increases cephalosporin C production. Mol. Genet. Genomics, 267, 673–683. [DOI] [PubMed] [Google Scholar]
- 94.Brown D.W., Yu,J.H., Kelkar,H.S., Fernandes,M., Nesbitt,T.C., Keller,N.P., Adams,T.H. and Leonard,T.J. (1996) Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl Acad. Sci. USA, 93, 1418–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.