Abstract
The homeobox-containing gene Hex is expressed in several cell types, including thyroid follicular cells, in which it regulates the transcription of tissue- specific genes. In this study the regulation of Hex promoter activity was investigated. Using co- transfection experiments, we demonstrated that the transcriptional activity of the Hex gene promoter in rat thyroid FRTL-5 cells is ∼10-fold greater than that observed in HeLa and NIH 3T3 cell lines (which do not normally express the Hex gene). To identify the molecular mechanisms underlying these differences, we evaluated the effect of the thyroid- specific transcription factor TTF-1 on the Hex promoter activity. TTF-1 produced 3–4-fold increases in the Hex promoter activity. Gel- retardation assays and mutagenesis experiments revealed the presence of functionally relevant TTF-1 binding sites in the Hex promoter region. These in vitro data may also have functional relevance in vivo, since a positive correlation between TTF-1 and Hex mRNAs was demonstrated in human thyroid tissues by means of RT–PCR analysis. The TTF-1 effect, however, is not sufficient to explain the difference in Hex promoter activity between FRTL-5 and cells that do not express the Hex gene. For this reason, we tested whether Hex protein is able to activate the Hex promoter. Indeed, co-transfection experiments indicate that Hex protein is able to increase the activity of its own promoter in HeLa cells ∼4-fold. TTF-1 and Hex effects are additive: when transfected together in HeLa cells, the Hex promoter activity is increased 6–7-fold. Thus, the contemporary presence of both TTF-1 and Hex could be sufficient to explain the higher transcriptional activity of the Hex promoter in thyroid cells with respect to cell lines that do not express the Hex gene. These findings demonstrate the existence of direct cross-regulation between thyroid-specific transcription factors.
INTRODUCTION
Regulation of gene expression occurs primarily at the transcriptional level. Promoter-specific transcription factors are a major class of transcriptional regulators (1,2). Some of these factors are also tissue-specific, their expression being restricted to a few cell types (3), and they play a major role in the control of cell type-specific gene expression (4,5). Therefore, these proteins are extremely important in the control of differentiation, and they have also been implicated in certain pathological states, such as cancer, which are characterized by altered differentiation (6). Maintenance of a differentiated phenotype is the result of integrated effects of multiple tissue-specific transcriptional regulators (7). There fore, to understand the molecular mechanisms underlying the differentiation of a given cell type, all or most of the tissue-specific transcription factors operating in the cell must first be identified and their functional relationships with one another must be characterized.
The thyroid follicular cell (TFC) is an excellent model for studies on the molecular mechanisms of cell type-specific transcriptional regulation and differentiation. In fact, several thyroid-specific transcription factors have been identified and characterized, including TTF-1, TTF-2, Pax8 and Hex (8,9). Although also expressed in other tissues, their combination is exclusive to TFC (8). These genes are expressed from the beginning of thyroid development to the adult state and inactivation of the genes in mice causes gross defects in thyroid gland morphogenesis and differentiation (8,10). Moreover, inactivating mutations of these genes in humans determine congenital hypothyroidism (11). Recent reports have shown that thyroid-specific transcription factors establish functional interactions with each other. For example, Hex is able to decrease the TTF-1 and Pax8 activating effects in the context of the thyroglobulin promoter (9). However, no information is available on cross-regulation of thyroid-specific transcription factors at the level of expression of their respective genes.
The homeobox-containing gene Hex is expressed in several tissues, including the thyroid gland (12,13). The promoter of the mouse Hex gene has recently been identified and characterized (14–16). Studies in transgenic mice have shown that distinct enhancer elements control Hex expression during gastrulation and early organogenesis (17). Cell culture studies have revealed a DNA element that mediates the activating effects of HNF3β and GATA-4 (15), and it appears to play a key role for Hex expression in the liver.
The objective of this study was to determine whether thyroid-specific transcription factors are capable of regulating Hex promoter activity. Our findings show that this activity is up-regulated by TTF-1 and by the Hex protein itself. We also found a positive correlation between TTF-1 and Hex mRNA levels in normal human thyroid tissues, suggesting that the functional relationship observed in vitro may also be relevant in vivo.
MATERIALS AND METHODS
Plasmids
The construct containing the mouse Hex promoter fragment –235/+22 has been previously described (15). In this construct, the fragment –235/+22 of the Hex promoter was cloned in the plasmid pGL3B, 5′ to the luciferase (LUC) gene. Plasmids containing mutations of the A and B TTF-1 binding sites of the Hex promoter were generated with the QuickChange site-directed mutagenesis kit (Stratagene, Milan, Italy), according to the manufacturer’s instructions. RSV-CAT and CMV-LUC plasmids contained the Rous sarcoma virus and the cytomegalovirus promoters linked to the chloranphenicol acetyltransferase (CAT) and LUC genes, respectively. The TTF-1 and Hex expression vectors have been previously described (9,18).
Cell cultures and transfections
NIH 3T3 and HeLa cells were cultured in DMEM medium with 10% calf serum (Gibco, Milan, Italy). FRTL-5 cells were maintained in F12 Coon’s modified medium with 5% calf serum and hormone mixture as previously described (19). FRT cells were grown in F12 Coon’s modified medium with 5% fetal calf serum (20). The calcium phosphate co-precipitation method was used for transfections, as described elsewhere (21). HeLa and NIH 3T3 cells were plated at 6 × 105 cells/ 100 mm culture dish, FRT cells were plated at 1.5 × 106 cells/100 mm culture dish, 20 h prior to transfection. FRTL-5 cells were plated at 1.5 × 106 cells/100 mm culture dish 48 h prior to transfection, and 3 h prior to the addition of the DNA-calcium phosphate precipitates, the medium was changed to Dulbecco’s modified Eagle’s medium containing 5% calf serum and growth factors. The plasmids were used in the following amounts: CMV-LUC, 2 µg; pGL3B, 8 µg; –235/+22 Hex promoter (and relative mutants), 8 µg; TTF-1 expression vector, 0.2, 0.5 and 2 µg; RSV-CAT, 2 µg; Hex expression vector, 2 µg. Cells were harvested 42–44 h after transfection, and cell extracts were prepared by a standard freeze and thaw procedure. CAT activity was measured by an ELISA method (Amersham, Milan, Italy). LUC activity was measured by a chemiluminescence procedure (21).
Gel-retardation assay
TTF-1 binding to the Hex promoter region –235/+22 was investigated using a series of overlapping oligonucleotides (data not shown). Double-stranded oligonucleotides, labeled at the 5′ end with 32P, were used as probes. The TTF-1 homeodomain (TTF-1HD) was used as protein, purified as described (22), and SDS–PAGE showed that it was >95% pure. In the gel-retardation assays, protein and DNA (both at a final concentration of 0.1 µM) were incubated for 30 min at room temperature in a buffer containing 20 mM Tris–HCl (pH 7.6), 75 mM KCl, 0.25 mg/ml bovine serum albumin, 5 mM dithiothreitol, 10 µg/ml calf thymus DNA and 10% glycerol. Protein-bound DNA and free DNA were separated on native 7.5% polyacrylamide gels run in 0.5× TBE for 1.5 h at 4°C. Gels were dried and exposed to a Bio-Rad GS-525 Molecular Imager. Significant TTF-1 binding was found with the oligonucleotides Wt A (corresponding to the region –162/ –146) and Wt B (corresponding to the region –102/–86), whose sense sequences are Wt A, 5′-CGGGGGTTAGTGGGGGG-3′, and Wt B, 5′-CGGCAGGAAGGGGACCG-3′.
The C site of the thyroglobulin gene promoter (sense sequence, 5′-CACTGCCCAGTCAAGTGTTCTTGA-3′) was used as a positive control (22). Nuclear extracts from HeLa cells were prepared as previously reported (18).
RT–PCR in human tissues
Twelve differentiated thyroid tumors (five follicular and seven papillary thyroid carcinomas) and nine non-nodular, normal thyroid tissues were analyzed. Informed consent was obtained from all patients. Total RNA extraction and cDNA synthesis were performed as previously described (23) and the RT–PCR amplification was performed using 3 µl of cDNA as previously described (24). As the HEX gene amplification product was produced in much greater amounts than the TTF-1 gene amplification product, to optimize the ratio of the two signals all the samples were subjected to 10 cycles of amplification with TTF-1 primers only, then the PCR was paused (<2 min at 95°C) to add HEX primers. The samples were then subjected to 25 cycles of amplification. The PCR conditions were as follows: a first step of pre-denaturation at 95°C for 10 min, denaturation at 95°C for 30 s, annealing at 58°C for 30 s and extension at 72°C for 30 s. Primer oligonucleotides for the TTF-1 gene were 5′-CTGTGCGTTTGTCGCTTACA-3′ and 5′-CGACAGGTACTTCTGTTGCT-3′. The amplification yielded a 208 bp DNA product corresponding to fragment 1654–1862 of the TTF-1 gene according to the sequence reported in GenBank (U19816.1). Primer oligonucleotides for the HEX gene were 5′-TACTCTGGAGCCCCTTCTTG-3′ and 5′-TTCAAGGTCTTCCTGGGAGG-3′. The amplification yielded a 370 bp DNA product corresponding to fragment 397–767 of the HEX gene according to the sequence reported in GenBank (XM_018176). The reaction conditions were optimized to ensure that amplification for both the TTF-1 and HEX genes remained within the exponential range. Different ratios of primers were tested to get the same efficiency of amplification and the ratio 1:1 was used (data not shown). Aliquots of 10 µl of 50 µl of the amplification products were run on 1.5 Tris–borate–EDTA agarose gels containing ethidium bromide. The bands on the positive film were scanned and the density and the width of each PCR product was measured using the NIH Image Program (Wayne Rasband, National Institutes of Health, Bethesda, MD). The correlation between Hex and TTF-1 transcript levels was studied by linear regression analysis. A level of P < 0.05 was considered statistically significant.
RESULTS
Activity of the Hex promoter in thyroid cells
The mouse Hex promoter has recently been identified (15). In our experiments we used the construct Hex –235/+22, which contains 235 bp of the mouse Hex 5′-flanking sequence. In a first set of experiments, we tested whether the transcriptional activity of this construct was different between differentiated thyroid cell lines (FRTL-5 and FRT cells) and other cell lines, which do not express the Hex gene (HeLa and NIH 3T3). As shown in Table 1, when the activity of the RSV promoter is normalized to that of the CMV promoter, it is similar in the four cell lines. In contrast, transcription of the Hex –235/+22 construct in FRTL-5 cells was 10 times greater than that observed in HeLa and NIH 3T3 cells. However, in FRT cells, which express a different pattern of thyroid-specific transcription factors (25), the activity of the Hex –235/+22 promoter was 2.5 times lower than that observed in FRTL-5, but still higher than HeLa and NIH 3T3 cells. These results indicate that mechanisms capable of enhancing the transcriptional activity of the Hex promoter are operative in thyroid cells but not in cell lines that do not express the Hex gene.
Table 1. Comparison of Hex promoter activity in different cell lines.
| Cell line | Promoter activitya | ||
|---|---|---|---|
| CMV | RSV | Hex | |
| HeLa | 100 | 53.5 | 33.8 |
| NIH 3T3 | 100 | 34.6 | 39.6 |
| FRTL-5 | 100 | 37.0 | 373.8 |
| FRT | 100 | 36.5 | 144.0 |
aPromoter activity is expressed as a percentage of CMV promoter activity.
Effects of TTF-1 on the Hex promoter
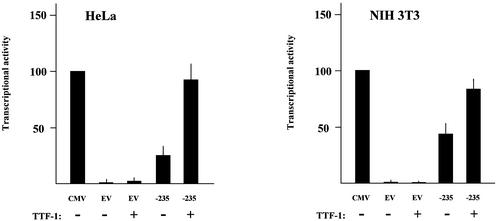
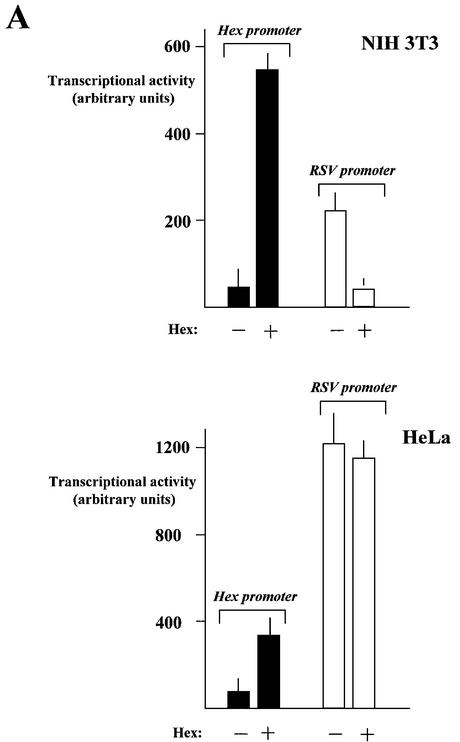
One of the simplest explanations for the difference described above is that Hex promoter activity in thyroid cells is affected by tissue-specific transcriptional activators that are not found in the other cells lines we examined. TTF-1 seemed to be a likely candidate for this role, since it has been shown to activate most of the thyroid-specific promoters assayed thus far (8), and it is expressed in FRTL-5 cells but not in FRT cells (25). To test this hypothesis, we evaluated the effects of TTF-1 on Hex promoter activity in non-thyroid cells. HeLa cells and NIH 3T3 cells were transfected with the Hex –235/+22 construct, with and without a TTF-1 expression vector. The CMV promoter was used to normalize the promoter activities. The results are shown in Figure 1. In HeLa cells TTF-1 increased the transcriptional activity of the Hex promoter 3- or 4-fold. The TTF-1 effect was much more modest in NIH 3T3 cells; in fact, an increase in transcriptional activity was present, but <1-fold. These data indicate that TTF-1 increases activity of the Hex promoter when expressed in non-thyroid cells. However, the extent of the up-regulation induced by TTF-1 is cell type-dependent. Nevertheless, these data suggest that the increased transcriptional activity of the Hex promoter in thyroid cells with respect to non-thyroid cells was due, at least partially, to the action of TTF-1.
Figure 1.
TTF-1 effect on the –235/+22 Hex promoter. HeLa and NIH 3T3 cell lines were transfected as described in Materials and Methods. The activity of each promoter was normalized for the efficiency of transfection using the RSV-CAT construct. Results are expressed as percentages of the activity of the CMV promoter. EV, empty pGL3B vector; –235, –235/+22 mouse Hex promoter. Each bar represents the mean value (±SD) of three independent transfections.
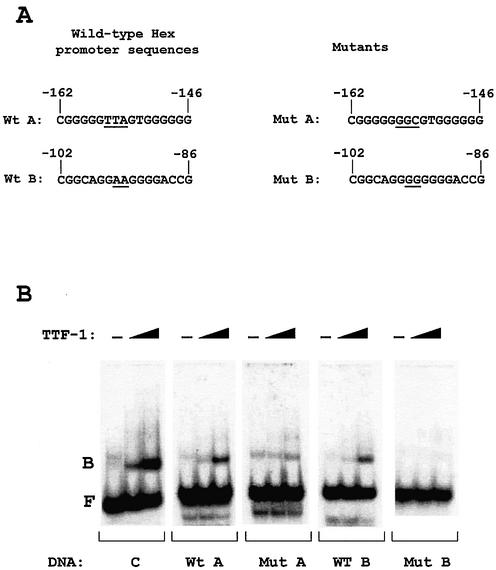
TTF-1 binding sites on the Hex promoter
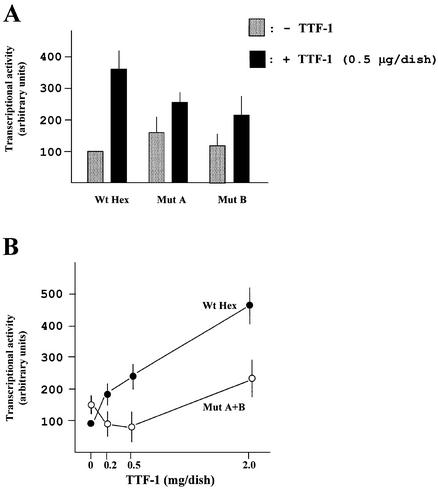
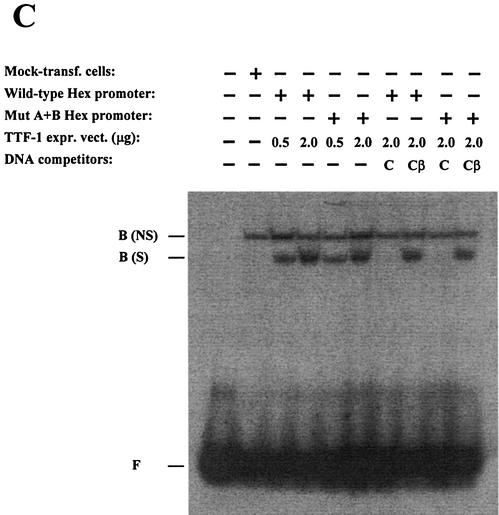
In order to identify TTF-1 binding sites on the Hex promoter, overlapping oligonucleotides covering the entire –235/+22 DNA sequence were tested in gel-retardation assays, using TTF-1HD as the bound protein (data not shown). The C site of the thyroglobulin promoter (known to contain a high-affinity TTF-1 binding site) (22) was used as a positive control. Two TTF-1 binding sites were found within the –235/+22 sequence, the first located between nucleotides –162 and –146 (site A) and the second between nucleotides –102 and –86 (site B) (Fig. 2A). To assess the transcriptional relevance of these TTF-1 binding sequences, mutants were designed to abolish TTF-1 binding at each of these sites (Fig. 2A). Gel-retardation assays demonstrated that the introduced nucleotide changes abolished TTF-1HD binding to both sites (Fig. 2B). Each of the mutant sequences was then incorporated into the Hex –235/+22 promoter construct, and the activity of the mutant promoters was tested by co-transfecting HeLa cells with the TTF-1 expression vector. Mutations at either site diminished the activating effect of TTF-1 on the Hex –235/+22 promoter (Fig. 3A). Mutant A has a basal activity significantly higher than the Hex wild-type promoter. Both the A and B mutant promoters are transactivated by TTF-1 <2-fold (using 0.5 µg of TTF-1 expression vector). A Hex –235/+22 construct containing both the A and B mutations was then generated (Mut A+B) and its activity assayed in HeLa cells, using different amounts of TTF-1 expression vector. Figure 3B shows that 0.2 and 0.5 µg of TTF-1 expression vector activated the wild-type Hex promoter but not the Mut A+B Hex promoter. At a higher dose of TTF-1 expression vector (2 µg) the Hex A+B mutant was also transactivated, but much less than the wild-type promoter. A possible explanation of this latter finding could be the presence of additional, lower affinity TTF-1 binding sites (data not shown). Figure 3C shows gel-retardation assays of HeLa cells transfected with either the wild-type or the A+B mutant Hex promoter and different amounts of TTF-1 expression vector. As shown, the A+B mutant promoter did not modify the amount of TTF-1 protein, excluding inhibitory effects of the Hex A+B mutant promoter on the expression of TTF-1. These data indicate that the activating effect exerted by TTF-1 on the Hex promoter is due, at least in part, to a direct protein–DNA interaction.
Figure 2.
TTF-1 binding sites in the –235/+22 Hex promoter. (A) Wild-type and mutant sequences of the mouse Hex promoter. (B) Gel-retardation assays of oligonucleotides containing sequences indicated in (A). TTF-1HD was used as the protein. B, protein-bound DNA; F, free DNA.
Figure 3.
Effect of mutation of TTF-1 binding sites on the activity of Hex promoter. (A) Effect of single mutations indicated in Figure 2 on the –235/+22 Hex promoter activity. The activity of each promoter was normalized for the efficiency of transfection using the RSV-CAT construct. Results are expressed as percentage of the –235/+22 Hex promoter in the absence of TTF-1. Each bar represents the mean value (±SD) of four independent transfections. (B) Effect of the double mutation (Mut A+B). Each dot represents the mean value (±SD) of three independent transfections. (C) Effects of the wild-type and Mut A+B promoters on Hex expression in transfected HeLa cells. The gel-retardation assay was performed by using the C site of the thyroglobulin promoter as labeled probe and 6 µg of HeLa nuclear extracts. Competitor DNAs were used at 50-fold molar excess with respect to the labeled C probe. Cβ is a mutant of the C site, in which the TTF-1 binding site has been abolished (9). F, free DNA; B(NS), non-specific binding activity; B(S), specific TTF-1 binding. Note that B(NS) is present also in untransfected HeLa cells.
Correlation between TTF-1 and HEX gene expression in human thyroid
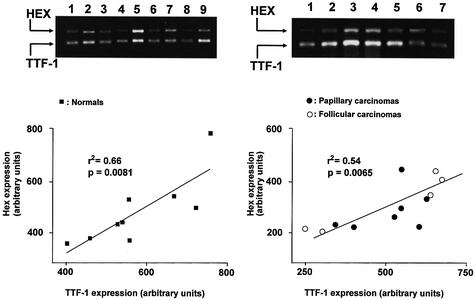
To verify the existence of a functional interaction between TTF-1 and the Hex promoter in vivo, we measured TTF-1 and the Hex mRNA levels in both normal and neoplastic human thyroid tissues by means of multiplex PCR, as described in Materials and Methods. As shown in Figure 4, TTF-1 mRNA levels displayed a significant correlation with those of HEX both in normal thyroid tissues (r2 = 0.66, P = 0.0081) and in differentiated thyroid neoplasms (papillary and follicular carcinomas) (r2 = 0.54, P = 0.0065), which is compatible with our hypothesis that TTF-1 regulates HEX promoter activity in vivo. The absence of such a correlation would have indicated that the functional interaction observed in vitro had no in vivo relevance.
Figure 4.
HEX and TTF-1 mRNA detection by semi-quantitative RT–PCR in normal human thyroid tissues. (Left) Normal tissues; (right) papillary and follicular carcinomas. (Top) Example of agarose gel electrophoresis of RT–PCR amplified human HEX and TTF-1 mRNA genes. Expected bands are indicated by black arrows. To determine the relative expression levels of these genes the positive bands were scanned and the density and the width of each PCR product was measured as described in Materials and Methods. Values of Hex and TTF-1 expression were correlated in the graphs at the bottom of the figure.
Effect of Hex protein on the Hex promoter
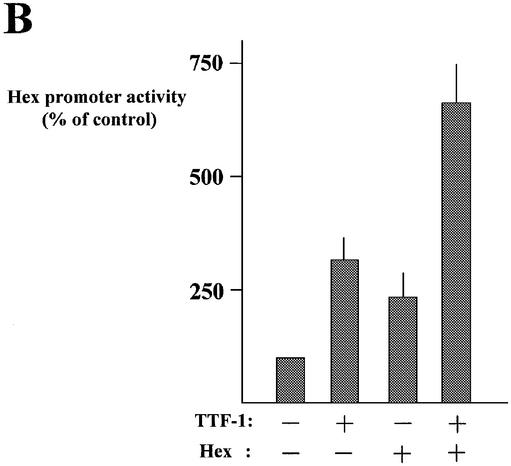
Data obtained in the co-transfection experiments (Fig. 1) indicated that TTF-1 increases Hex promoter activity by 3–4-fold. However, as shown in Table 1, the activity of the Hex promoter in thyroid cells was roughly 10 times higher than that of other cell lines, suggesting that TTF-1 is not the only tissue-specific factor that influences Hex promoter activity in thyroid cells. Since the Hex protein itself is a transcription factor (26), we suspected that it might also exert modulatory effects on its own gene promoter. We tested this hypothesis by transfecting NIH 3T3 and HeLa cells with the Hex –235/+22 promoter, with and without a Hex expression vector. Figure 5 shows the results. In agreement with previous observations indicating Hex as a transcriptional repressor (9 and references therein), the Hex protein was found to suppress the activity of the RSV promoter in NIH 3T3 cells. However, in our experimental conditions, Hex protein had no inhibitory effects on the RSV promoter in HeLa cells (Fig. 5A). Both in NIH 3T3 and in HeLa cells, Hex protein increased the transcriptional activity of the Hex –235/+22 promoter (10- and 4-fold, respectively). Hence, a positive regulatory loop between the Hex gene promoter and the protein product may be hypothesized.
Figure 5.
Hex transactivates its own promoter and has an additive effect with TTF-1. (A) The –235/+22 Hex promoter construct (containing the LUC gene as reporter) and the RSV promoter construct (containing the CAT gene as reporter) were co-transfected in NIH 3T3 and HeLa cells with and without 2 µg of the Hex expression vector. LUC and CAT were measured as described in Materials and Methods. (B) The –235/+22 Hex construct was used as test promoter. TTF-1 and Hex expression vectors were used at concentrations of 0.5 and 2 µg/dish, respectively. Transfections and measurement of the reporter gene were performed as described in Materials and Methods. Each bar represents the mean value (±SD) of four independent transfections.
Once it had been demonstrated that Hex protein was able to activate its own promoter, the effect of the concurrent presence of TTF-1 and Hex proteins on Hex promoter activity was tested. Co-transfection experiments were performed in the HeLa cell line. In this cell line, in fact, the RSV promoter (used to normalize for transfection efficiency) was not inhibited by Hex protein (Fig. 5B). The results are shown in Figure 5B. Both TTF-1 (using 0.5 µg of expression vector) and Hex protein (using 2.0 µg of expression vector) increased Hex promoter activity by ∼3- and 2-fold, respectively. When TTF-1 and Hex expression vectors were transfected together, the Hex promoter activity increased 6- or 7-fold. Therefore, the TTF-1 and Hex proteins exert additive effects on the Hex promoter. Interestingly, in HeLa cells expressing both TTF-1 and Hex proteins the Hex promoter reached a transcriptional activity similar to that observed in FRTL-5 cells (compare results shown in Table 1 and Fig. 5). Thus, the contemporary presence of both TTF-1 and Hex could be sufficient to explain the higher transcriptional activity of the Hex promoter in FRTL-5 cells with respect to HeLa cells.
DISCUSSION
Several examples in a wide range of organisms indicate that tissue-specific gene expression is the result of functional interactions between distinct tissue-specific transcriptional regulators (7,27,28). Cross-talk of this type could theoretically occur at three levels: (i) protein–protein interactions between the transcription factors themselves; (ii) protein– DNA interactions involving the binding of different combinations of transcription factors to promoter/enhancer regions of target genes; (iii) protein–DNA interactions between one or more transcription factors and the regulatory regions of genes encoding other transcription factors. In a previous study (9), we demonstrated functional interaction among tissue-specific transcription factors in thyroid cells at the level of common target genes, i.e. Hex-induced repression of the thyroglobulin promoter activating effects of TTF-1 or Pax8. Our present findings indicate that cross-regulation of thyroid-specific transcription factors also involves interaction between the factors themselves and the promoter regions of genes for other transcription factors.
It has been previously shown that in Hex-null mice development of the thyroid is arrested at the budding stage at 9.5 days post coitum (10) and, at this stage, the thyroid primordium is characterized by the absence of TTF-1. These findings indicate that Hex controls TTF-1 gene expression. The results of our study indicate that the reverse also occurs, i.e. TTF-1 controls Hex expression. Collectively, these observations suggest the existence of a positive feedback loop between TTF-1 and Hex.
We have recently shown that Hex expression is retained in most human thyroid undifferentiated carcinomas (29), although TTF-1 expression is abolished (30,31). In these neoplasms, therefore, Hex promoter activity seems to be maintained by factors other than TTF-1, which is consistent with our observation in the present study of significant basal Hex promoter activity in the absence of TTF-1, both in HeLa and NIH 3T3 cells (see Fig. 1). Therefore, under certain conditions, the presence of TTF-1 may not be essential for the activation of the Hex promoter. Within this context, it is interesting to recall that Kikkawa et al. have recently demonstrated that ubiquitous transcription factors are also involved in the activation of the Hex promoter (16).
Nevertheless, the correlation between HEX and TTF-1 mRNA levels, which we found in both normal and neoplastic human thyroid tissue, strengthens the hypothesis that TTF-1 does indeed regulate Hex expression. The absence of this correlation would have indicated that the TTF-1 effect on the Hex promoter is not relevant in normal human thyroid tissue. It must be pointed out, however, that the correlation between TTF-1 and Hex might just as well be a reflection of other types of control mechanisms. For example, TTF-1 gene expression might be regulated by Hex, rather than vice versa, and this possibility is also suggested by the disappearance of TTF-1 mRNA in Hex-null mice (10). Alternatively, there might be an upstream control mechanism that modulates the expression of both the TTF-1 and Hex genes in the same direction, without any form of functional interaction between the two factors themselves. In our opinion, however, this latter mechanism is unlikely: the fact that TTF-1 expression, but not that of Hex, is suppressed in most undifferentiated thyroid carcinomas (29,30) suggests separate mechanisms of control for the expression of these two transcription factors.
Early investigations indicated that the Hex protein is a transcriptional repressor (32). Indeed, the activity of the thyroglobulin gene promoter is inhibited by Hex expression (9). However, more recent data indicate that Hex protein may also act as a transcriptional activator (26). Altogether, our data indicate that the Hex protein transcriptional effects are dependent on cell types and target promoters. The interaction between Hex and the promoter region of its gene reflects a positive feedback loop, which is probably important for maintenance of Hex expression. Interestingly, a similar phenomenon has been described for TTF-1 (33), suggesting that positive feedback of this type is common in thyroid cells. A mechanism of this type might contribute to the maintenance of the differentiated phenotype once other events have triggered initial expression of tissue-specific transcription factors.
Acknowledgments
ACKNOWLEDGEMENTS
This work was funded by a grant to G.D. from Consiglio Nazionale delle Ricerche (CNR, Target Project on Biotechnology) and grants from MURST-COFIN to G.D. and S.F.
REFERENCES
- 1.Kingston R.E. and Green,M.R. (1994) Modelling eukaryotic transcriptional activation. Curr. Biol., 4, 325–332. [DOI] [PubMed] [Google Scholar]
- 2.Struhl K. (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell, 98, 1–4. [DOI] [PubMed] [Google Scholar]
- 3.Lai E. and Darnell,J.E.,Jr (1991) Transcriptional control in hepatocytes: a window on development. Trends Biochem. Sci., 16, 427–430. [DOI] [PubMed] [Google Scholar]
- 4.Dasen J.S. and Rosenfeld,M.G. (2001) Signalling and transcriptional mechanisms in pituitary development. Annu. Rev. Neurosci., 24, 327–355. [DOI] [PubMed] [Google Scholar]
- 5.Andrews N.C. and Orkin,S.H. (1994) Transcriptional control of erythropoiesis. Curr. Opin. Hematol., 1, 119–124. [PubMed] [Google Scholar]
- 6.Chi N. and Epstein,J.A. (2002) Getting your Pax straight: Pax proteins in development and disease. Trends Genet., 18, 41–47. [DOI] [PubMed] [Google Scholar]
- 7.Herskowitz I. (1989) A regulatory hierarchy for cell specialization in yeast. Nature, 342, 749–757. [DOI] [PubMed] [Google Scholar]
- 8.Damante G., Tell,G. and Di Lauro,R. (2001) A unique combination of transcription factors controls differentiation of thyroid cells. Prog. Nucleic Acid Res. Mol. Biol., 66, 307–356. [DOI] [PubMed] [Google Scholar]
- 9.Pellizzari L., D’Elia,A., Rustighi,A., Manfioletti,G., Tell,G. and Damante,G. (2000) Expression and function of the homeodomain-containing protein Hex in thyroid cells. Nucleic Acids Res., 28, 2503–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez Barbera J.P., Clements,M., Thomas,P., Rodriguez,T., Meloy,D., Kioussis,D. and Beddington,R.S. (2000) The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development, 127, 2433–2445. [DOI] [PubMed] [Google Scholar]
- 11.Macchia P.E. (2000) Recent advances in understanding the molecular basis of primary congenital hypothyroidism. Mol. Med. Today, 6, 36–42. [DOI] [PubMed] [Google Scholar]
- 12.Thomas P.Q., Brown,A. and Beddington,R.S.P. (1998) Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development, 125, 85–94. [DOI] [PubMed] [Google Scholar]
- 13.Bogue C.W., Ganea,G.R., Sturm,E., Ianucci,R. and Jacobs,H.C. (2000) Hex expression suggests a role in the development and function of organs derived from foregut endoderm. Dev. Dyn., 219, 84–89. [DOI] [PubMed] [Google Scholar]
- 14.Myint Z., Inazu,T., Tanaka,T., Yamada,K., Keng,V.W., Inoue,Y., Kuriyama,M. and Noguchi,T. (1999) Genomic organization and promoter analysis of a mouse homeobox gene, Hex. J. Biochem., 125, 795–802. [DOI] [PubMed] [Google Scholar]
- 15.Denson L.A., McClure,M.H., Bogue,C.W., Karpen,S.J. and Jacobs,H.C. (2000) HNF3beta and GATA-4 transactivate the liver-enriched homeobox gene, Hex. Gene, 246, 311–320. [DOI] [PubMed] [Google Scholar]
- 16.Kikkawa E., Hinata,M., Keng,V.W., Myint,Z., Sato,A., Yamada,K., Tanaka,T. and Noguchi,T. (2001) Sp family members stimulate transcription of the hex gene via interactions with GC boxes. J. Biochem., 130, 885–891. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez T.A., Casey,E.S., Harland,R.M., Smith,J.C. and Beddington,R.S. (2001) Distinct enhancer elements control Hex expression during gastrulation and early organogenesis. Dev. Biol., 234, 304–316. [DOI] [PubMed] [Google Scholar]
- 18.De Felice M., Damante,G., Zannini,M., Francis-Lang,H. and Di Lauro,R. (1995) Redundant domains contribute to the transcriptional activity of the thyroid transcription factor 1. J. Biol. Chem., 270, 26649–26656. [DOI] [PubMed] [Google Scholar]
- 19.Ambesi-Impiombato F.S., Parks,L.A. and Coon,H.G. (1980) Culture of hormone-dependent functional epithelial cells from rat thyroids. Proc. Natl Acad. Sci. USA, 77, 3455–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitsch L., Tramontano,D., Ambesi-Impiombato,F.S., Quarto,N. and Bonatti,S. (1985) Morphological and functional polarity of an epithelial thyroid cell line. Eur. J. Cell Biol., 38, 57–66. [PubMed] [Google Scholar]
- 21.Francis-Lang H., Zannini,M., De Felice,M., Berlingieri,M.T., Fusco,A. and Di Lauro,R. (1992) Multiple mechanisms of interference between transformation and differentiation in thyroid cells. Mol. Cell. Biol., 12, 5793–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damante G. and Di Lauro,R. (1991) Several regions of Antennapedia and thyroid transcription factor 1 homeodomains contribute to DNA binding specificity. Proc. Natl Acad. Sci. USA, 88, 5388–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arturi F., Russo,D., Bidart,J.M., Scarpelli,D., Schlumberger,M. and Filetti,S. (2001) Expression pattern of the pendrin and sodium/iodide symporter genes in human thyroid carcinoma cell lines and human thyroid tumors. Eur. J. Endocrinol., 145, 129–135. [DOI] [PubMed] [Google Scholar]
- 24.Arturi F., Russo,D., Schlumberger,M., du Villard,JA., Caillou,B., Vigneri,P., Wicker,R., Chiefari,E., Suarez,HG. and Filetti,S. (1998) Iodide symporter gene expression in human thyroid tumors. J. Clin. Endocrinol. Metab., 83, 2493–2496. [DOI] [PubMed] [Google Scholar]
- 25.Mascia A., De Felice,M., Lipardi,C., Gentile,R., Cali,G., Zannini,M., Di Lauro,R. and Nitsch,L. (1997) Transfection of TTF-1 gene induces thyroglobulin gene expression in undifferentiated FRT cells. Biochim. Biophys. Acta, 1354, 171–181. [DOI] [PubMed] [Google Scholar]
- 26.Sekiguchi K., Kurabayashi,M., Oyama,Y., Aihara,Y., Tanaka,T., Sakamoto,H., Hoshino,Y., Kanda,T., Yokoyama,T., Shimomura,Y., Iijima,H., Ohyama,Y. and Nagai,R. (2001) Homeobox protein Hex induces SMemb/nonmuscle myosin heavy chain-B gene expression through the cAMP-responsive element. Circ. Res., 88, 52–58. [DOI] [PubMed] [Google Scholar]
- 27.Rosenfeld M.G., Briata,P., Dasen,J., Gleiberman,A.S., Kioussi,C., Lin,C., O’Connell,S.M., Ryan,A., Szeto,D.P. and Treier,M. (2000) Multistep signaling and transcriptional requirements for pituitary organogenesis in vivo. Recent Prog. Horm. Res., 55, 1–13. [PubMed] [Google Scholar]
- 28.Duncan S.A. (2000) Transcriptional regulation of liver development. Dev. Dyn., 219, 131–142. [DOI] [PubMed] [Google Scholar]
- 29.D’Elia A.V., Tell,G., Russo,D., Arturi,F., Puglisi,F., Manfioletti,G., Gattei,V., Mack,D.L., Cataldi,P., Filetti,S., Di Loreto,C. and Damante,G. (2002) Expression and localization of the homeodomain-containing protein HEX in human thyroid tumors. J. Clin. Endocrinol. Metab., 87, 1376–1383. [DOI] [PubMed] [Google Scholar]
- 30.Fabbro D., Di Loreto,C., Beltrami,C.A., Belfiore,A., Di Lauro,R. and Damante,G. (1994) Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Res., 54, 4744–4749. [PubMed] [Google Scholar]
- 31.Miettinen M. and Franssila,K.O. (2000) Variable expression of keratins and nearly uniform lack of thyroid transcription factor 1 in thyroid anaplastic carcinoma. Hum. Pathol., 31, 1139–1145. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka T., Inazu,T., Yamada,K., Myint,Z., Keng,V.W., Inoue,Y., Taniguchi,N. and Noguchi,T. (1999) cDNA cloning and expression of rat homeobox gene, Hex and functional characterization of the protein. Biochem. J., 339, 111–117. [PMC free article] [PubMed] [Google Scholar]
- 33.Oguchi H. and Kimura,S. (1998) Multiple transcripts encoded by the thyroid-specific enhancer-binding protein (T/EBP)/thyroid-specific transcription factor-1 (TTF-1) gene: evidence of autoregulation. Endocrinology, 139, 1999–2006. [DOI] [PubMed] [Google Scholar]