Abstract
A new explanation for the emergence of heavy (GC-rich) isochores is proposed, based on the study of thermostability, bendability, ability to B–Z transition and curvature of the DNA helix. The absolute values of thermostability, bendability and ability to B–Z transition correlated positively with GC content, whereas curvature correlated negatively. The relative values of these parameters were determined as compared to randomized sequences. In genes and intergenic spacers of warm-blooded animals, both the relative bendability and ability to B–Z transition increased with elevation of GC content, whereas the relative thermostability and curvature decreased. The usage of synonymous codons in GC-rich genes was also found to augment bendability and ability to B–Z transition and to reduce thermostability of DNA (as compared to synonymous codons with the same GC content). The analysis of transposable elements (Alu and B2 repeats in the human and mouse) showed that the level of their divergence from the consensus sequence positively correlated with relative bendability and ability to B–Z transition and negatively with relative thermostability. The bendability and ability to B–Z transition are known to relate to open chromatin and active transcription, whereas curvature facilitates chromatin condensation. Because heavy isochores are known to be gene-rich and show a high level of transcription, it is suggested here that isochores arose not as an adaptation to elevated temperature but because of a certain grade of general organization and correspondingly advanced level of genomic organization, reflected in genome structuring, with physical properties of DNA in the gene-rich regions being optimized for active transcription and in the gene-poor regions for chromatin condensation (‘transcription/grade’ concept).
INTRODUCTION
Organisms which form the crowns of the animal and plant kingdoms, warm-blooded animals and monocot plants, show strong base compositional heterogeneity of genomes. They consist of relatively homogeneous regions (>300 kb long), called isochores, which differ in the GC content and gene concentration (1–5). It is the heavy (GC-rich and gene-rich) regions which have evolved lately and thus brought about the heterogeneity of the genome as a whole (2–4,6). There are different views on the origin of heavy isochores. According to the neutralist viewpoint, they were formed by variation of mutation bias (in a broad sense, including gene conversion) among different genomic regions (7–12). The selectionist hypothesis assumes that their emergence was caused by a need for a higher thermostability of the DNA helix (3,13). However, the case of prokaryotes where there is no correlation between GC content and habitat temperature (notwithstanding a very wide range of temperatures with the upper margin far exceeding 40°C) undermines the ‘thermal’ hypothesis (14,15). This conclusion was supported by the latest studies on ectothermic (cold-blooded) vertebrates where no correlation between GC content and adaptation temperature was observed, with the temperature ranging from –2 to +45°C (16,17). Recently, it was shown that with elevation of GC content, the thermostability of genes in warm-blooded animals increased more slowly than in random sequences, whereas their bendability rose faster (18). This suggests that, if the emergence of heavy isochores was indeed due to selection, it was selection not for thermostability but for bendability of the DNA molecule, which may be related to active transcription in these gene-rich genomic regions.
Here this hypothesis is examined in several aspects. First, two other physical properties of the DNA helix (ability to B–Z transition and curvature), which are known to relate to transcription and chromatin condensation (19–22), were analyzed. Second, the intergenic spacers and the usage of synonymous codons in coding sequences were studied. Third, the evolution of transposable elements (TEs) was explored with regard to a change in the physical properties of DNA.
MATERIALS AND METHODS
The sequences of nuclear genes were extracted from GenBank. Only genes with complete coding sequences (CDSs) were taken. Genes were checked for duplicates on the basis of CDS similarity (>99%). All coding sequences were checked for the absence of internal stop codons to ensure they were not pseudogenes. The exon/intron boundaries were taken from annotations. Pseudogenes were not included in the gene level analysis but they were taken into account for determination of intergenic spacers. Only sequences where each nucleotide was known for at least 99% of positions were used. The intergenic spacers were studied only in the (better presented) human, mouse and rice genomes. The gene level analysis was also done for the chicken and maize.
The TEs in introns and intergenic spacers were determined using the RepeatMasker program (A.F.A.Smit and P.Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html, with parameter ‘-gccalc’, without masking simple repeats and low complexity regions). The level of divergence (percent of nucleotide substitutions) with regard to consensus sequence for each repeat family, the libraries of which were based on the expanded versions of Repbase (23), were also calculated with this program. The transposable elements were studied in the human, mouse and rice genomes.
The bendability and curvature of nucleotide sequences were determined using the trinucleotide table of consensus values obtained from DNase I digestion and nucleosome positioning studies (24,25). The bendability is a local parameter representing the ability of DNA to bend (usually towards the major groove) as a result of thermal fluctuations or DNA–protein interactions (26). It was calculated in a sliding trinucleotide frame (with a 1 nt step), and averaged for each sequence. The curvature is a relatively macroscopic DNA bend, representing the intrinsic tendency of DNA to follow a non-linear pathway over an appreciable length, which is a result of variation of local bends in phase with the DNA helix (26). It is usually calculated for DNA segments of arbitrary length, which is a multiple of the number of helix turns (24,26). Different numbers of turns (one to three) were tried here; the results did not differ qualitatively. Therefore, calculations made with the shortest DNA segment (one turn, 10.5 bp) were chosen to minimize possible margin effects for short sequences. Thus, curvature was calculated in a sliding 10.5 nt frame (with a 1 nt step), and averaged for each sequence. The thermostability was determined using a unified dinucleotide table for free energy of melting (ΔG) (27), obtained from different physical studies, mainly the UV absorbance and calorimetric temperature profiles. The ability to B–Z transition was determined using the dinucleotide table for free energy of B–Z transition (the lower the free energy, the higher the ability to transition), obtained by an advanced modification of the JUMNA method (28). The thermostability and ability to B–Z transition were calculated in a sliding dinucleotide frame (with a 1 nt step), and averaged for each sequence.
For each genomic sequence (exons, introns, intergenic spacers and TEs), 10 randomizations were made and the parameters under study, calculated for these randomized sequences, were averaged. For each protein coding sequence (CDS), all synonymous codon positions, which can be permuted with conservation of the same GC content of each synonymous position (i.e. only G↔C and A↔T replacements), were randomly permuted 10 times with preservation of the mean purine content of the total set of synonymous positions, and parameters calculated for these permuted sequences were averaged. Each CDS was taken as a whole sequence, therefore curvature was not determined because of its relatively long calculation frame, which could produce considerable errors on exon junctions. The relative value of each physical parameter under study was determined for each genomic sequence as the difference between the value for the genomic sequence itself and the average for its 10 randomized or permuted sequences. (The relative parameters determined as ratios between the values for genomic and randomized/permuted sequences were also analyzed and showed qualitatively the same results.)
The conventional statistical analyses were done with the Statgraphics (Statistical Graphics Co.) and Statistica (StatSoft Inc.) software.
RESULTS
Absolute values
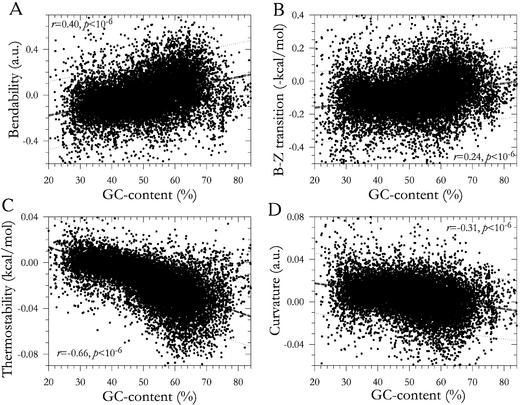
For any type of DNA sequences (genomic or randomized) studied here, the bendability, ability to B–Z transition and thermostability increased with elevation of GC content, whereas curvature decreased. Human intergenic spacers are presented as an example (Table 1). The bendability and ability to B–Z transition always correlated positively and strongly between themselves, even independently of their link to GC content (Table 1). The thermostability and curvature usually correlated negatively (and more weakly) with bendability and ability to B–Z transition if GC content was fixed (Table 1).
Table 1. Coefficients of correlation between the absolute values of the parameters under study and GC percent and coefficients of their partial correlation between themselves (at fixed GC percent) for human intergenic spacers (n = 2606).
| Parameter | GC percent | Partial correlation at fixed GC percent | ||
|---|---|---|---|---|
| Bendability | B–Z transition | Thermostability | ||
| Bendability | 0.95 | |||
| B–Z transition | 0.97 | 0.79 | ||
| Thermostability | 0.99 | –0.49 | –0.24 | |
| Curvature | –0.94 | –0.59 | –0.49 | 0.62 |
All correlations have significance P < 10–6 at least.
Relative values in introns, exons and intergenic spacers
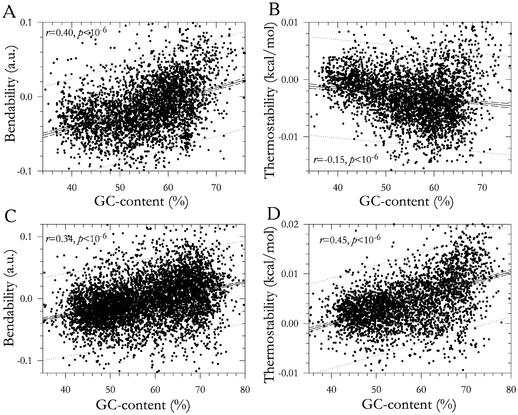
In the human, mouse and chicken, the relative bendability and ability to B–Z transition increased with elevation of GC percent in all types of genomic sequences (Table 2 and Fig. 1). The relative thermostability and curvature showed quite the opposite trend. All correlations were stronger in the introns as compared to exons and intergenic spacers (Table 2). In exons of rice and maize, the three parameters (relative bendability, ability to B–Z transition and thermostability) increased with elevation of GC content, whereas relative curvature decreased (Table 2). For introns and intergenic spacers of monocots, the data were inconsistent.
Table 2. Coefficients of correlation between the relative values of the parameters under study and GC percent for different genomic sequences.
| Sequence | n | Bendability | B–Z transition | Thermostability | Curvature | |
|---|---|---|---|---|---|---|
| Human | Exons | 29 423 | 0.25 | 0.22 | –0.19 | –0.13 |
| Introns | 23 212 | 0.40 | 0.24 | –0.66 | –0.31 | |
| Intergenic spacers | 2606 | 0.29 | 0.09 | –0.48 | –0.25 | |
| Mouse | Exons | 9601 | 0.16 | 0.15 | –0.15 | –0.07 |
| Introns | 7074 | 0.20 | 0.10 | –0.54 | –0.17 | |
| Intergenic spacers | 520 | 0.11 | NS | –0.35 | NS | |
| Chicken | Exons | 1483 | 0.13 | 0.19 | –0.08 | –0.12 |
| Introns | 1230 | 0.15 | 0.32 | –0.35 | –0.28 | |
| Rice | Exons | 29 009 | 0.06 | 0.11 | 0.17 | –0.13 |
| Introns | 16 756 | –0.04 | –0.06 | NS | –0.11 | |
| Intergenic spacers | 6089 | –0.11 | –0.18 | NS | 0.04 | |
| Maize | Exons | 1266 | 0.08 | 0.08 | 0.11 | –0.16 |
| Introns | 1017 | 0.12 | 0.13 | 0.13 | –0.15 |
All correlations have significance P < 0.01 at least, mostly much higher. NS, not significant.
Figure 1.
Regression of relative values of parameters under study on GC percent for human introns.
Synonym-relative values in coding sequences
In the human, mouse and chicken, the synonym-relative values, obtained by permutation of synonymous codon positions with preservation of GC content of each position, behave in a similar way to the relative values obtained by complete randomization of their genomic sequences. The synonym-relative bendability and ability to B–Z transition increased with elevation of GC percent, whereas the synonym-relative thermostability decreased (Table 3 and Fig. 2). In the monocots, the synonym-relative values of coding sequences behave similarly with the relative values for exons, i.e. bendability, ability to B–Z transition and thermostability increased with elevation of GC content (Table 3 and Fig. 2).
Table 3. Coefficients of correlation between the synonym-relative values of the parameters under study and GC percent for protein coding sequences.
| Organism | n | Bendability | B–Z transition | Thermostability |
|---|---|---|---|---|
| Human | 3733 | 0.40 | 0.37 | –0.15 |
| Mouse | 1317 | 0.39 | 0.27 | NS |
| Chicken | 281 | 0.29 | 0.31 | NS |
| Rice | 5968 | 0.34 | 0.20 | 0.45 |
| Maize | 235 | 0.52 | 0.44 | 0.61 |
All correlations have significance P < 10–5 at least. NS, not significant.
Figure 2.
Regression of synonym-relative values of parameters under study on GC percent for protein coding sequences. (A) and (B) Human; (C) and (D) maize.
Relative values in the transposable elements
The most numerous transposable elements in the human and mice genomes are Alu and B2 repeats. The level of their divergence from the consensus sequence positively correlated with the relative bendability and ability to B–Z transition, and negatively with relative thermostability (Table 4). The most numerous transposable elements in rice (among those presented in Repbase) were the TNR1 and Wanderer repeats. With the growth of divergence from the consensus sequence, the TNR1 repeats showed an increase in three parameters, bendability, ability to B–Z transition and thermostability (Table 4). The Wanderer repeats showed a picture similar to transposons of warm-blooded vertebrates, i.e. an increase in relative bendability and ability to B–Z transition and a decrease in relative thermostability (Table 4).
Table 4. Coefficients of correlation between the relative values of the parameters under study and the level of divergence from the consensus sequence for different transposable elements.
| Transposable element | n | Bendability | B–Z transition | Thermostability | Curvature |
|---|---|---|---|---|---|
| Human Alu | 54 573 | 0.37 | 0.20 | –0.26 | –0.15 |
| Mouse Alu | 5492 | 0.38 | 0.37 | –0.22 | –0.52 |
| Mouse B2 | 2837 | 0.25 | 0.27 | NS | NS |
| Rice TNR1 | 840 | 0.29 | 0.21 | 0.11 | NS |
| Rice Wanderer | 1018 | 0.44 | 0.37 | –0.51 | –0.25 |
All correlations have significance p < 0.01 at least, mostly much higher. NS, not significant.
DISCUSSION
The obtained results indicate that, if heavy isochores of warm-blooded vertebrates were indeed formed by selection for some physical properties of DNA, it was selection for traits associated with transcription (higher bendability and ability to B–Z transition and lower curvature) rather than for thermostability. In the case of selection not for GC content itself but for a certain physical property related to it (which is quite reasonable because non-coding sequences of heavy isochores are also involved in the elevation of GC content), the enhancement of this physical property should overtake the increase in GC content. Therefore, the statistically significant deviation from random can serve as an indicator of this selective force. The conclusion about the increase in relative bendability and ability to B–Z transition with elevation of GC content pertains not only to introns and intergenic spacers (and to the evolution of TEs located in them) but also to synonymous positions in protein-coding DNA as well. An unequal usage of synonymous codons is a long-standing problem (see for example 29–31). In some organisms, it is associated with the level of gene expression and can be explained by relative abundance of corresponding isoacceptor tRNAs (29,32,33). However, in the warm-blooded vertebrates the causes of unequal codon usage remain unclear (29,30,34). In these organisms, codon usage is generally related to isochore structure (2,3,35). The data obtained here show that the usage of codons in the GC-rich genes of warm-blooded vertebrates augments bendability and ability to B–Z transition of DNA (even as compared to synonymous codons with the same GC content).
With its non-neutralized electric charges, the DNA helix behaves as a rigid, steel-like rod, and even after its neutralization with histones, bending of such a wire into the orderly, compact chromatin structure remains a challenging task (36,37). In the multicellular eukaryotes, the main problem of the genetic machinery is to control repression and activation of genes, which is connected with bending and unbending of the DNA molecule. Although the GC-rich regions make up only a minor part of the mammalian genome (10–15%), gene concentration in the heaviest isochore family (H3) is 20-fold higher than in the GC-poor regions (2,6). The heavy isochores constitute early replicated, actively transcribed chromatin (38,39). The expression level of genes located in the heavy isochores is on average higher (40). It is known that an increase in DNA bendability promotes the transition from nucleosome-wrapped to extended DNA (21,22), thereby it should facilitate transcription. In the warm-blooded vertebrates, all correlations found here are stronger in introns as compared to exons and intergenic spacers (Table 2). This is in agreement with the proposed concept, because exons are under strong selection for informational content, while intergenic spacers should be involved in transcription-related decondensation to a lesser extent as compared to genes.
The ability to B–Z transition is also linked very tightly to transcription. It is known that antibodies to Z-DNA bind preferentially to actively transcribed genes (20). The association between the amount of Z-DNA and transcription activity has even been shown for individual genes (c-myc, corticotropin releasing-hormone gene). When the gene is up-regulated, the level of Z-form increases, when it is down-regulated, it decreases (20). At the same time, the level of Z-DNA is not affected by replication. A wave of Z-DNA is formed after the moving RNA polymerase, which may prevent the following RNA polymerase from transcribing that region of a gene (19,20). It should also facilitate untwisting of the DNA helix, absorbing supercoils and thereby buffering torsional stress generated by RNA polymerase until this stress is relieved by topoisomerases. In any case, the strong association between Z-DNA and transcription is well established.
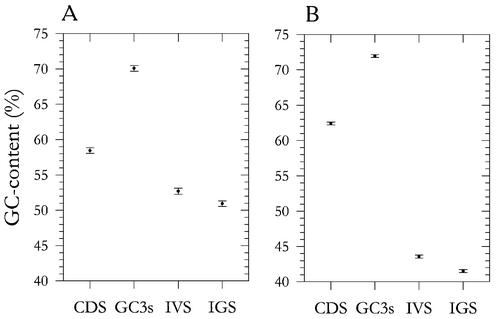
In monocot plants, the picture is more complicated and selection for thermostability cannot be excluded. The coding DNA might have been selected both for bendability/ability to B–Z transition and thermostability. Different TEs probably located predominantly in different genomic segments evolved either similarly to monocot CDSs (i.e. in the direction of both bendability/ability to B–Z transition and thermostability) or similarly to the genomes of warm-blooded vertebrates (in the direction of bendability/ability to B–Z transition only). It is noteworthy that although exons of GC-rich genes usually show a higher GC content as compared to corresponding introns in both groups of organisms, the exon–intron contrast in GC content is much higher in monocots than in warm-blooded vertebrates (41,42). The same picture was observed for the coding DNA–intergenic spacer contrast and for the silent codon position–non-coding DNA contrast (Fig. 3). This can explain why non-coding DNA behaves more similarly to exons in warm-blooded vertebrates as compared to monocots (Table 2). The heavy isochores of monocots are not the same as in warm-blooded vertebrates: in the former they are more heterogeneous in GC content [although they have a similar base compositional ‘smoothness’ on the intron–exon margins (42)].
Figure 3.
Mean GC content of coding sequences (CDS), intervening sequences (introns, IVS), third silent codon positions (GC3s) and adjacent intergenic spacers (IGS) in the heavy isochores of human (A) and rice (B) genomes. A cut-off GC3s >50% was used for attribution of a gene to the heavy isochores.
In general, the GC-rich DNA seems to be a ‘good’ DNA. In the highest multicellular organisms, it constitutes gene-rich, actively transcribed genomic regions. It is thermostable, bendable and able to B–Z transition. Then why are there the GC-poor parts of the genome and why are introns and intergenic spacers in the heavy isochores still GC-poorer than exons? The substitution pressure in the human genome is generally AT-biased (4,5,12,43). This helps explain why the most part of the genome is GC-poor. The recombination rate is much higher in the heavy isochores (44–46), which should facilitate selection for a ‘good’ DNA in these regions. One possible cause for a lower GC content in non-coding DNA is bombardment by transposable elements, since purifying selection against the integration of TEs should be much stronger in the coding DNA (47). The functional significance of a lower GC content can be related to a dual role of curvature and bendability in chromatin condensation. The curved DNA segments serving as primers for DNA–histone interaction are necessary for nucleosome formation (21,22). Since curvature is a long-range parameter (as compared to bendability), this property cannot be furnished in exons which are under strong pressure for informational content (and where silent positions do not constitute a continuous sequence). Therefore, it is introns which should provide the necessary segments of curvature. Several cases have been described where the experimental removal of an intron resulted in a failure of nucleosome formation (48,49). Bendability and curvature, which are shown here to correlate disparately with the GC content, affect chromatin condensation in the opposite manner. Bendability is more prone to destabilize nucleosomes, whereas curvature acts in the opposite direction (21,22). In the GC-poor isochores, which constitute the late replicated (i.e. rarely decondensed) chromatin, there should be a greater need in the curved DNA segments for a tight chromatin condensation, as compared to GC-rich regions.
The concept fostered here suggests that GC-rich isochores (and hence the compositional heterogeneity of the genome as a whole) arose in warm-blooded animals not as an adaptation to elevated temperature but because of the high level of their general organization and the corresponding level of genomic organization, with the physical properties of DNA in the gene-rich regions being optimized for active transcription and in the gene-poor regions for chromatin condensation. Thus, the structuring of the genome reflects its advanced organization.
The less pronounced isochores in rodents as compared to other mammals (50–52) can be related to a lower organization of these r-selected, short-living animals [which is expressed, for instance, in a relatively smaller brain (53)]. It is known that rodents have a less efficient DNA repair (54,55), which suggests that precise genomic organization is not as important for them as for other mammals. Similarly, some lesser pronounced isochoric structure probably exists in the cold-blooded vertebrates (16,56), which may reflect the respective levels of their general organization.
For the isochoric structure to emerge, selection need not necessarily be on each nucleotide (especially in non-coding DNA) but could be selection of DNA-handling proteins, which in their turn provide an appropriate mutation bias. Recently, support was found for a biased gene conversion (BGC) explanation for elevation of GC content in highly recombining regions of the mammalian genome (11,57–61). [At least some of these facts can be equally interpreted as a result of nucleotide selection (11,57–60), albeit this would suggest a high genetic load (11).] More exactly, the elevation of GC content is assumed to be due to biased DNA repair after gene conversion (11,59), i.e. gene conversion simply plays a role of mutation. Recombination, which is the main cause of gene conversion, was shown to be generally mutagenic (62). One of the problems for a purely neutralist explanation of the origin of GC-rich isochores (e.g. due to BGC) is the increasing deviation of DNA physical properties from randomized sequences with the elevation of GC content, revealed here and in a previous work (18). This directional deviation is due to the sequence specificity of GC-rich regions, which suggests that even if it indeed arose due to some mutation/repair bias, a whole complex of mutation/repair pathways should be involved in this process. The proteins belonging to this complex could be under selection to provide an appropriate bias in different genomic regions. If so, such indirect selection should probably have an inertia, which could drive the process beyond the optimum. This may explain the recent proposal that in mammals (especially rodents) the isochores are lately deteriorating (35,59,63).
Whatever the actual mechanisms, it is the final result that matters. The independent origin of strongly pronounced isochores in mammals and birds (2), even not considering monocots, and their functional significance upheld here suggest that the isochoric structure of the genome is a grade trait, and even if actual mechanisms do involve mutation/repair bias, the emergence of isochores was not accidental.
Acknowledgments
ACKNOWLEDGEMENTS
I am grateful to Giorgio Bernardi and two anonymous reviewers for helpful comments. This work was supported by the Russian Foundation for Basic Research (RFBR).
REFERENCES
- 1.Bernardi G., Olofsson,B., Filipski,J., Zerial,M., Salinas,J., Cuny,G., Meunier-Rotival,M. and Rodier,F. (1985) The mosaic genome of warm-blooded vertebrates. Science, 228, 953–958. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi G. (2000) Isochores and the evolutionary genomics of vertebrates. Gene, 241, 3–17. [DOI] [PubMed] [Google Scholar]
- 3.Bernardi G. (2000) The compositional evolution of vertebrate genomes. Gene, 259, 31–43. [DOI] [PubMed] [Google Scholar]
- 4.Bernardi G. (2001) Misunderstandings about isochores. Part 1. Gene, 276, 3–13. [DOI] [PubMed] [Google Scholar]
- 5.Eyre-Walker A. and Hurst,L.D. (2001) The evolution of isochores. Nature Rev. Genet., 2, 549–555. [DOI] [PubMed] [Google Scholar]
- 6.D'Onofrio G., Jabbari,K., Musto,H., Alvarez-Valin,F., Cruveiller,S. and Bernardi,G. (1999) Evolutionary genomics of vertebrates and its implications. Ann. N. Y. Acad. Sci., 870, 81–94. [DOI] [PubMed] [Google Scholar]
- 7.Wolfe K., Sharp,P.M. and Li,W.H. (1989) Mutation rates differ among regions of the mammalian genome. Nature, 337, 441–456. [DOI] [PubMed] [Google Scholar]
- 8.Francino M.P. and Ochman,H. (1999) Isochores result from mutation not selection. Nature, 400, 30–31. [DOI] [PubMed] [Google Scholar]
- 9.Fryxell K.J. and Zuckerkandl,E. (2000) Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol. Biol. Evol., 17, 1371–1383. [DOI] [PubMed] [Google Scholar]
- 10.Sueoka N. and Kawanishi,Y. (2000) DNA G+C content of the third codon position and codon usage biases of human genes. Gene, 261, 53–62. [DOI] [PubMed] [Google Scholar]
- 11.Galtier N., Piganeau,G., Mouchiroud,D. and Duret,L. (2001) GC content evolution in mammalian genomes, the biased gene conversion hypothesis. Genetics, 159, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 13.Bernardi G. and Bernardi,G. (1986) Compositional constraints and genome evolution. J. Mol. Evol., 24, 1–11. [DOI] [PubMed] [Google Scholar]
- 14.Galtier N. and Lobry,J.R. (1997) Relationships between genomic G+C content, RNA secondary structures and optimal growth temperature in prokaryotes. J. Mol. Evol., 44, 632–636. [DOI] [PubMed] [Google Scholar]
- 15.Hurst L.D. and Merchant,A.R. (2001) High guanine–cytosine content is not an adaptation to high temperature, a comparative analysis amongst prokaryotes. Proc. R. Soc. Lond. B Biol. Sci., 268, 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belle E.M., Smith,N. and Eyre-Walker,A. (2002) Analysis of the phylogenetic distribution of isochores in vertebrates and a test of the thermal stability hypothesis. J. Mol. Evol., 55, 356–363. [DOI] [PubMed] [Google Scholar]
- 17.Ream R.A., Johns,G.C. and Somero,G.N. (2003) Base compositions of genes encoding alpha-actin and lactate dehydrogenase-A from differently adapted vertebrates show no temperature-adaptive variation in G + C content. Mol. Biol. Evol., 20, 105–110. [DOI] [PubMed] [Google Scholar]
- 18.Vinogradov A.E. (2001) Bendable genes of warm-blooded vertebrates. Mol. Biol. Evol., 18, 2195–2200. [DOI] [PubMed] [Google Scholar]
- 19.Herbert A. and Rich,A. (1996) The biology of left-handed Z-DNA. J. Biol. Chem., 271, 11595–11598. [DOI] [PubMed] [Google Scholar]
- 20.Herbert A. and Rich,A. (1999) Left-handed Z-DNA, structure and function. Genetica, 106, 37–47. [DOI] [PubMed] [Google Scholar]
- 21.Anselmi C., Bocchinfuso,G., De Santis,P., Savino,M. and Scipioni,A. (1999) Dual role of DNA intrinsic curvature and flexibility in determining nucleosome stability. J. Mol. Biol., 286, 1293–1301. [DOI] [PubMed] [Google Scholar]
- 22.Anselmi C., Bocchinfuso,G., De Santis,P., Savino,M. and Scipioni,A. (2000) A theoretical model for the prediction of sequence-dependent nucleosome thermodynamic stability. Biophys. J., 79, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurka J. (2000) Repbase update, a database and an electronic journal of repetitive elements. Trends Genet., 16, 418–420. [DOI] [PubMed] [Google Scholar]
- 24.Gabrielian A. and Pongor,S. (1996) Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Lett., 393, 65–68. [DOI] [PubMed] [Google Scholar]
- 25.Munteanu M.G., Vlahovicek,K., Parthasarathy,S., Simon,I. and Pongor,S. (1998) Rod models of DNA, sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem. Sci., 23, 341–347. [DOI] [PubMed] [Google Scholar]
- 26.Goodsell D.S. and Dickerson,R.E. (1994) Bending and curvature calculations in B-DNA. Nucleic Acids Res., 22, 5497–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SantaLucia J. Jr (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafontaine I. and Lavery,R. (2000) Optimization of nucleic acid sequences. Biophys. J., 79, 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp P.M., Averof,M., Lloyd,A.T., Matassi,G. and Peden,J.F. (1995) DNA sequence evolution, the sounds of silence. Phil. Trans. R. Soc. Lond. B Biol. Sci., 349, 241–247. [DOI] [PubMed] [Google Scholar]
- 30.Urrutia A.O. and Hurst,L.D. (2001) Codon usage bias covaries with expression breadth and the rate of synonymous evolution in humans, but this is not evidence for selection. Genetics, 159, 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinogradov A.E. (2001) Intron length and codon usage. J. Mol. Evol., 52, 2–5. [DOI] [PubMed] [Google Scholar]
- 32.Xia X. (1996) Maximizing transcription efficiency causes codon usage bias. Genetics, 144, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiapello H., Lisacek,F., Caboche,M. and Henaut,A. (1998) Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene, 209, GC1–GC38. [DOI] [PubMed] [Google Scholar]
- 34.Hurst L.D. and Williams,E.J. (2000) Covariation of GC content and the silent site substitution rate in rodents, implications for methodology and for the evolution of isochores. Gene, 261, 107–114. [DOI] [PubMed] [Google Scholar]
- 35.Galtier N. and Mouchiroud,D. (1998) Isochore evolution in mammals, a human-like ancestral structure. Genetics, 150, 1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maher L.J. III (1998) Mechanisms of DNA bending. Curr. Opin. Chem. Biol., 2, 688–694. [DOI] [PubMed] [Google Scholar]
- 37.Williams L.D. and Maher,L.J.,III (2000) Electrostatic mechanisms of DNA deformation. Annu. Rev. Biophys. Biomol. Struct., 29, 497–521. [DOI] [PubMed] [Google Scholar]
- 38.Federico C., Saccone,S. and Bernardi,G. (1998) The gene-richest bands of human chromosomes replicate at the onset of the S-phase. Cytogenet. Cell Genet., 80, 83–88. [DOI] [PubMed] [Google Scholar]
- 39.Saccone S., Federico,C., Solovei,I., Croquette,M.F., Della Valle,G. and Bernardi,G. (1999) Identification of the gene-richest bands in human prometaphase chromosomes. Chromosome Res., 7, 379–386. [DOI] [PubMed] [Google Scholar]
- 40.Konu O.O. and Li,M.D. (2002) Correlations between mRNA expression levels and GC contents of coding and untranslated regions of genes in rodents. J. Mol. Evol., 54, 35–41. [DOI] [PubMed] [Google Scholar]
- 41.Carels N., Hatey,P., Jabbari,K. and Bernardi,G. (1998) Compositional properties of homologous coding sequences from plants. J. Mol. Evol., 46, 45–53. [DOI] [PubMed] [Google Scholar]
- 42.Vinogradov A.E. (2001) Within-intron correlation with base composition of adjacent exons in different genomes. Gene, 276, 143–151. [DOI] [PubMed] [Google Scholar]
- 43.Petrov D.A. and Hartl,D.L. (1999) Patterns of nucleotide substitution in Drosophila and mammalian genomes. Proc. Natl Acad. Sci. USA, 96, 1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisenbarth I., Striebel,A.M., Moschgath,E., Vogel,W. and Assum,G. (2001) Long-range sequence composition mirrors linkage disequilibrium pattern in a 1.13 Mb region of human chromosome 22. Hum. Mol. Genet., 10, 2833–2839. [DOI] [PubMed] [Google Scholar]
- 45.Hurst L.D. (2000) Crossing over a boundary. Trends Genet., 16, 542. [DOI] [PubMed] [Google Scholar]
- 46.Eisenbarth I., Vogel,G., Krone,W., Vogel,W. and Assum,G. (2000) An isochore transition in the NF1 gene region coincides with a switch in the extent of linkage disequilibrium. Am. J. Hum. Genet., 67, 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duret L. and Hurst,L.D. (2001) The elevated G and C content at exonic third sites is not evidence against neutralist models of isochore evolution. Mol. Biol. Evol., 18, 757–762. [DOI] [PubMed] [Google Scholar]
- 48.Lauderdale J.D. and Stein,A. (1992) Introns of the chicken ovalbumin gene promote nucleosome alignment in vitro. Nucleic Acids Res., 20, 6589–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu K., Sandgren,E.P., Palmiter,R.D. and Stein,A. (1995) Rat growth hormone gene introns stimulate nucleosome alignment in vitro and in transgenic mice. Proc. Natl Acad. Sci. USA, 92, 7724–7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabeur G., Macaya,G., Kadi,F. and Bernardi,G. (1993) The isochore patterns of mammalian genomes and their phylogenetic implications. J. Mol. Evol., 37, 93–108. [DOI] [PubMed] [Google Scholar]
- 51.Robinson M., Gautier,C. and Mouchiroud,D. (1997) Evolution of isochores in rodents. Mol. Biol. Evol., 14, 823–828. [DOI] [PubMed] [Google Scholar]
- 52.Douady C., Carels,N., Clay,O., Catzeflis,F. and Bernardi,G. (2000) Diversity and phylogenetic implications of CsCl profiles from rodent DNAs. Mol. Phylogenet. Evol., 17, 219–230. [DOI] [PubMed] [Google Scholar]
- 53.Lewin R. (1988) Living in the fast track makes for small brains. Science, 242, 513–514. [DOI] [PubMed] [Google Scholar]
- 54.Adelman R., Saul,R.L. and Ames,B.N. (1988) Oxidative damage to DNA: relation to species metabolic rate and life span. Proc. Natl Acad. Sci. USA, 85, 2706–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ames B.N. (1989) Endogenous DNA damage as related to cancer and aging. Mutat. Res., 214, 41–46. [DOI] [PubMed] [Google Scholar]
- 56.Hughes S., Zelus,D. and Mouchiroud,D. (1999) Warm-blooded isochore structure in Nile crocodile and turtle. Mol. Biol. Evol., 16, 1521–1527. [DOI] [PubMed] [Google Scholar]
- 57.Eyre-Walker A. (1999) Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics, 152, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith N.G. and Eyre-Walker,A. (2001) Synonymous codon bias is not caused by mutation bias in human. Mol. Biol. Evol., 18, 982–986. [DOI] [PubMed] [Google Scholar]
- 59.Duret L., Semon,M., Piganeau,G., Mouchiroud,D. and Galtier,N. (2002) Vanishing GC-rich isochores in mammalian genomes. Genetics, 162, 1837–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lercher M.J., Smith,N.G., Eyre-Walker,A. and Hurst,L.D. (2002) The evolution of isochores. Evidence from snp frequency distributions. Genetics, 162, 1805–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galtier N. (2003) Gene conversion drives GC content evolution in mammalian histones. Trends Genet., 19, 65–68. [DOI] [PubMed] [Google Scholar]
- 62.Lercher M.J. and Hurst,LD. (2002) Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet., 18, 337–340. [DOI] [PubMed] [Google Scholar]
- 63.Smith N.G. and Eyre-Walker,A. (2002) The compositional evolution of the murid genome. J. Mol. Evol., 55, 197–201. [DOI] [PubMed] [Google Scholar]