Abstract
Ribosome biogenesis requires a vast number of trans-acting factors many of which are required for the chemical modification and processing of the pre-rRNA component. The U3 snoRNP complex is required for the early cleavage steps in pre-rRNA processing. We have cloned cDNAs encoding the human and mouse homologs of the yeast U3 snoRNP-associated proteins Imp3 and Imp4. Both human proteins localize to nucleoli and interact with the U3 snoRNA. The results of complementation experiments show that, in contrast to mouse Imp4, mouse Imp3 can partially alleviate the growth defect of the corresponding yeast null strain, indicating that the role of Imp3 in pre-rRNA processing is evolutionarily conserved. The results of density gradient centrifugation experiments show that, in contrast to hU3-55K, the human Imp3 and Imp4 proteins predominantly interact with the U3 snoRNA in 60–80S ribonucleoprotein complexes. In addition, we have found that hImp3, hImp4 and hMpp10 can form a stable hetero-trimeric complex in vitro, which is generated by direct interactions of both hImp3 and hImp4 with hMpp10. The analysis of hImp3 and hImp4 mutants indicated that their binding to hMpp10 correlates with their nucleolar accumulation, strongly suggesting that the formation of the ternary complex of hImp3, hImp4 and hMpp10 is required for their association with nucleolar components.
INTRODUCTION
The U3 small nucleolar RNA (snoRNA), a box C/D type snoRNA, is required for the early cleavage steps in pre-rRNA processing, which are essential for the formation of the small ribosomal subunit RNA (18S rRNA) in both fungi and vertebrates (1–3). In yeast, the U3 snoRNA is essential for processing at sites A0, A1 and A2 (reviewed in 4). Base-pairing interactions of the U3 snoRNA with the 5′ external transcribed spacer (5′ETS) of the precursor ribosomal RNA (pre-rRNA) and the 5′ terminal region of the 18S rRNA have been demonstrated to play an essential role in this process.
The U3 snoRNA contains several evolutionarily conserved sequence elements, designated box GAC, A, A′, B, C, C′ and D. The box GAC, box A and box A′ display sequence complementarity to regions of the 5′ETS and 18S rRNA sequences within the pre-rRNA (5–14). The boxes B/C and C′/D are located in the 3′ part of the U3 snoRNA and function as protein binding sites (10,15,16).
The U3 snoRNA interacts with proteins that are common to all box C/D snoRNPs: 15.5K (Snu13p in yeast), Nop56, Nop58 and fibrillarin (Nop1p) (16–24). In addition, a number of proteins have been identified in a yeast system that specifically associate with the U3 snoRNA: Rrp9p (hU3-55K in human), Dhr1p, Lcp5p, Rcl1p, Sof1p, Imp3p, Imp4p and Mpp10p (hMpp10 in human) (reviewed in 25).
Purification of the yeast ∼80S SSU processome has led to the identification of a vast number of novel proteins that interact with the U3 snoRNA (26). With the exception of Rcl1p and Lcp5p, these complexes contained all U3 snoRNA-associated proteins mentioned above and a large number of other non-ribosomal proteins that are associated with the U3 snoRNA. It has been proposed that the U3 snoRNA, together with the proteins that are associated with U3 in the SSU processome, is required to assist in the proper folding of the pre-rRNA.
The human U3 snoRNA is associated with both 12S and 60–80S complexes (27,28). Nop56, Nop58, 15.5K, fibrillarin and hU3-55K are most likely to be the first proteins to interact with the U3 snoRNA during its assembly and together may constitute the 12S U3 complex (16–18). Base-pairing interactions of the U3 snoRNA with pre-rRNA, which may require a number of auxiliary factors, may lead to the formation of 60–80S complexes, equivalent to the SSU processome described in yeast (26).
Imp3p and Imp4p were identified in a yeast two-hybrid screen using yeast Mpp10p as a bait (29). Both the human and yeast Mpp10 proteins contain several putative coiled-coil regions that have been suggested to mediate intramolecular and/or intermolecular interactions (30–32). Imp3p contains a putative S4 RNA binding domain (29). Imp4p is a member of the Imp4 superfamily of proteins, which are characterized by the σ70-like motif, a sequence motif first identified in the σ70 family of prokaryotic transcription factors (33). Interestingly, the σ70-like motif in the eukaryotic Imp4 superfamily confers binding to RNA and all the proteins in this family interact with pre-rRNA processing intermediates (33). In contrast to other members of this family, Imp4p associates with the U3 snoRNA and is required for the early cleavage steps in pre-rRNA processing (29,33). In this study we have addressed the question whether the Mpp10-Imp3-Imp4 complex also exists in human cells and we have investigated its association with the human U3 snoRNP. For these studies cDNAs encoding the human homologs of the yeast Imp3 and Imp4 proteins were cloned and characterized. Besides data on their association with U3 snoRNA-containing complexes, details on their interactions with the human Mpp10 protein will be presented.
MATERIALS AND METHODS
Oligonucleotides used in this study
mImp3A: 5′-GCGAGATCTCTCGAGATGGTGCGGAAGCTTAAGTTC-3′; hImp3: 5′-GCGAGATCTAGGCCTCTAGCTAGCGGCTTCTAGATCAAAGTCATC-3′; hImp4a: 5′-GCGAGATCTCTCGAGATGCTGCGCCGCGAGGCCCGC-3′; hImp4b: 5′-GCGAGATCTAGGCCTTCAGCTAGCCTCGGTGCTCAGGAAGCATCT-3′; hImp3DS4: 5′-GCGAGATCTAGGCTTCTAGCTAGCGGCGGGGTCGGTGACCAC-3′; hImp3DCCRev: 5′-CGAAGCGCGCACGCGGAACTGCAGCCGGTAACGCCCGCAGCAC-3′; hImp4 DCCFor: 5′-GCGCTCGAGCGCCTGATTCCCACTGAGTTA-3′; hImp4Dσ70For: 5′-AAGAAGACAGACCACCGCAACCTGGGCACGCTGGAGCAGGAG-3′; hImp4σ70ERAA: 5′-GTGGAGCTCACTGCGGTCGGGCCCGCGTTTG AGCTGAAGCTGTACATG-3′; hImp4TFAAFor: 5′-AGCCACCTGCCCTTTGGTCCTGCGGCCTACGCGACGCTGTGCAATGTGGTCATG-3′; hImp4TFAARev: 5′-CATGA CCACATTGCACAGCGTCGCGTAGGCCGCAGGACCAAAGGGCAGGTGGCT-3′; mImp3Bam: 5′-CGCGGATCCATGGTGCGGAAGCTTAAGTTC-3′.
DNA manipulations
The human Imp3 and Imp4 cDNAs were cloned by PCR using a teratocarcinoma cDNA library and oligonucleotides mImp3a and hImp3, hImp4a and hImp4b, respectively. The PCR products were cloned into the pCR4-TOPO vector according to manufacturer’s procedure (Invitrogen). The resulting constructs contain in-frame BglII restriction sites at both the 5′ and 3′ end, a XhoI restriction site upstream of the translational start codon and an NheI and StuI site at the 3′ end in-frame with the coding sequence. The integrity of each construct was verified by DNA sequencing.
A mouse EST bearing a portion of the putative mouse Imp3 homolog (mImp3) was identified by searching with the yeast Imp3 protein sequence. The clone was obtained from Research Genetics (cDNA ID: 634066), and the insert was used to screen a mouse myeloid leukemia cDNA library cloned into the λ-Zap vector (LTR6; provided by Hui Zhang, Yale University) to obtain a full-length mImp3 clone. For subcloning, primers for PCR amplification were made (mImp3Bam and a T7 primer). The BamHI site in the 5′ primer and an existing XhoI site in the 3′ UTR were used to clone mImp3 into yeast expression vectors. The mouse Imp4 cDNA was cloned as described previously (33). The yeast U3 snoRNA expression vector used in this study has been described previously (34). The pEGFPhImp3 and pEGFPhImp4 constructs were generated by inserting XhoI–BglII fragments of the cDNA constructs into the pEGFP-C3 vector (Clontech) digested with XhoI and BamHI. The pCI-neoVSVhImp constructs were generated by insertion of the XhoI–StuI fragments in plasmid pCI-neo5′VSV (35) digested with XhoI and SmaI. The resulting constructs contained a VSV-G sequence in-frame with the translational start codon. The pGEX-2T GST-fusion constructs were generated using the BglII and StuI digested cDNA plasmids. hImp3ΔS4 mutant was generated by PCR using oligonucleotides hImp3DS4 and mImp3a. The PCR product was gel purified and cloned into pCR4-TOPO (Invitrogen). The hImp3ΔCC mutant was generated by two sequential PCRs. In the first round of PCR, oligonucleotide hImp3DCCRev was used in combination with oligonucleotide mImp3a. The PCR product was gel purified and used as a primer in a second reaction in combination with oligonucleotide hImp3. The resulting PCR product was cloned into pCR4-TOPO (Invitrogen). hImp4ΔCC was generated using oligonucleotides hImp4DCCFor and hImp4b and cloned into pCI-neo5′VSV and pEGFP-C3 using the XhoI and SmaI restriction sites. The hImp4Δσ70 mutant was generated using oligonucleotides hImp4Dσ70For, hImp4b and hImp4a via the megaprimer procedure as described above.
To generate the hImp4σ70ERAA mutant, a PCR was performed with oligonucleotides hImp4σ70ERAA and hImp4b. The PCR product was subsequently cloned into pCR4-TOPO, isolated by SstI and StuI digestion and ligated into pCR4-TOPOhImp4 digested with the same enzymes. The hImp4TFAA mutant was generated using the oligonucleotides hImp4TFAAFor and hImp4TFAARev and the QuikChange mutagenesis kit according to the manufacturer’s procedure (Stratagene)
All mutant hImp3 and hImp4 constructs were subsequently digested with XhoI and StuI and cloned into pCI-neo5′VSV and pEGFP-C3 using the XhoI and SmaI restriction sites. The hImp3ΔS4 VSV and GFP-fusion constructs were generated by digesting pCR4TOPOhImp3ΔS4 with XhoI and EcoRI followed by ligation of the gel purified cDNA in the pCI-neo5′VSV and pEGFPC3 vectors digested with the same enzymes
To generate pCI-neoVSVhMpp10, a XhoI site was introduced by PCR [pBS SK+ Mpp10 as template (32)] upstream of the translational start codon and XbaI and SmaI sites at the 3′ end. A XhoI–SmaI digested cDNA was inserted into pCI-neo5′VSV digested with the same enzymes. The resulting construct contained a VSV-G tag in frame with the Mpp10 cDNA. The GFPhMpp10 deletion mutant constructs were generated by digestion with appropriate restriction sites. The following internal restriction sites of the hMpp10 cDNA were used: BamHI (ΔN58); PvuII (ΔC567); NdeI (ΔC456); HindIII (ΔC325); EcoRV and HindIII (Δ103–326). The integrity of each construct was verified by DNA sequencing.
Yeast manipulations
The mImp3 gene was placed under the control of a constitutive yeast promoter on both the p415GPD (LEU2 gene as marker) and p416GPD (URA3 gene as marker) vectors. The p415GPD-mImp3 was shuffled into yeast strain (pGAL1::Imp3) as previously described (29) and grown at 22°C. This strain was designated mouse Imp3 (mImp3).
Mpp10p homologs were expressed from p415GPD, a single copy plasmid, or p425GPD, a multicopy plasmid (30,36).
In order to concurrently overexpress various homologs of IMP3 and MPP10 in the same strain the following strains were created by switching the p415GPD mImp3 plasmid for the p416GPD mImp3 or p416GPD yeast Imp3p plasmid by a modified plasmid shuffle. The mImp3 strain was transformed with either p416GPD Imp3 plasmids and grown on SD–Ura media. Individual colonies were then struck out on SD. Cells that had lost p415GPD mImp3 and now only carried p416GPD mImp3 or p416GPD yImp3 were then transformed with the plasmids containing the MPP10 genes and selected on SD–Leu, Ura media. For serial dilutions yeast were grown to saturation in SD–Leu or SD–Leu, Ura, to select for strains with plasmids containing homologs of MPP10 or U3 gene. Cells (2 × 107) were diluted 10-fold and spotted onto plates containing the appropriate selective media. The plates were then grown at 22 or 30°C for 2 days.
Fluorescence microscopy
HEp-2 monolayer cells were grown to 70% confluency in DMEM supplemented with 10% fetal calf serum (FCS). Cells (2 × 106) were transfected with 20 µg of DNA in a total volume of 800 µl DMEM/10% FCS. Electroporation was performed at 260 V at a capacity of 950 µF with a Gene Pulser II (BioRad). After electroporation, cells were resuspended in 5 ml DMEM/10% FCS, grown for 16 h on coverslips before further processing. Coverslips were washed twice with phosphate buffered saline (PBS), placed in methanol for 5 min at –20°C and rinsed with acetone at room temperature. GFP-fusion proteins were visualized by fluorescence microscopy directly after cells were fixed and permeabilized. Images were obtained using an Olympus BH2 microscope in combination with an Olympus DP10 digital camera and analySIS software (Soft Imaging System GmbH).
Glycerol gradient sedimentation and immunoprecipitation experiments
A T150 flask with HEp-2 cells was grown to 70% confluency using the conditions described above. Cells were harvested and disrupted in 1 ml gradient buffer [20 mM HEPES–KOH (pH 7.9), 150 mM NaCl, 0.5 mM DTT] by sonication (Branson microtip setting 2–3) for 3 × 20 s. Triton X-100 was added to 0.2% (v/v) and insoluble material was removed by centrifugation at 10 000 g in a microcentrifuge. The supernatant (1 ml) was loaded on a 10–30% glycerol gradient (v/v) prepared in gradient buffer containing 0.2% Triton X-100. Gradients were centrifuged in a Th674 rotor (Sorvall) for 15 h at 25 000 r.p.m. Gradients were manually fractionated in 22 fractions each. Fractions were subjected to phenol/chloroform/isoamylalcohol (25:24:1) extraction and the RNA was isolated by ethanol precipitation. Proteins were precipitated by the addition of 5 vol of acetone to the organic phase. RNAs were resolved on 10% denaturing polyacrylamide gels and transferred to Hybond N+ (Amersham). Hybridization of blots with antisense snoRNA probes was performed as described previously (35). The positions of the 18S and 28S rRNAs were determined by agarose gel electrophoresis and ethidium bromide staining.
For immunoprecipitations, antibodies were coupled to protein A agarose beads either directly [anti-hU3-55K, anti-hMpp10, anti-(tri)methylguanosine cap (H20)] or via rabbit anti-chicken IgY antibodies (Jackson Immunoresearch) (anti-hImp3, anti-hImp4). Immunoprecipitations shown in Figure 2 were performed using fresh HEp-2 cell lysates prepared in gradient buffer as described above. After incubation with extracts (2 h, 4°C), beads were washed four times with gradient buffer and co-precipitated RNAs were isolated by phenol:chloroform:isoamylalcohol extraction and ethanol precipitation. RNA was resolved on 6% PAA/7 M urea gels. For the immunoprecipitation experiment shown in Figure 4, fractions 4–7 and 16–20 of the glycerol gradient were pooled and 450 µl was incubated with the antibodies coupled to protein A beads or beads alone for 2 h at 4°C. Wash steps and extraction of co-precipitated RNAs was performed as described above and RNA was resolved on 10% PAA/7 M urea gel.
Figure 2.
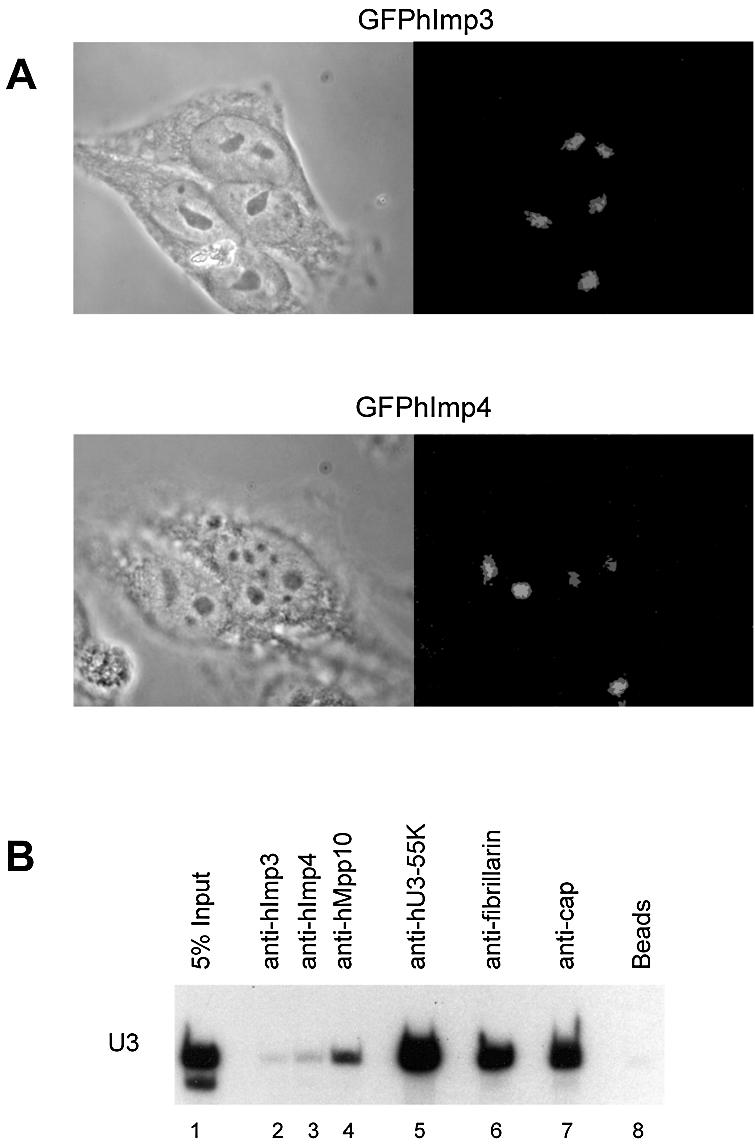
hImp3 and hImp4 localize to nucleoli in HEp-2 cells and interact with the U3-snoRNA in vivo. (A) HEp-2 cells were transfected with constructs encoding GFPhImp3 and GFPhImp4 fusion proteins and after 16 h cells were fixed and the subcellular localization of the fusion proteins was determined by fluorescence microscopy (right panels). The corresponding phase-contrast images are shown in the left panels. (B) HEp-2 total cell extracts were subjected to immunoprecipitation using antibodies against hImp3, hImp4, hMpp10, hU3-55K, fibrillarin (27B9) and anti-cap antibodies (H20) (lanes 2–7). Co-precipitated RNAs were separated on denaturing polyacrylamide gels and analyzed by northern blot hybridization using a U3 snoRNA specific probe. In lane 1, RNA isolated from the total cell extract was loaded (5% of the amount used for immunoprecipitation) and as a negative control an immunoprecipitation was performed in the absence of antibodies (beads, lane 8).
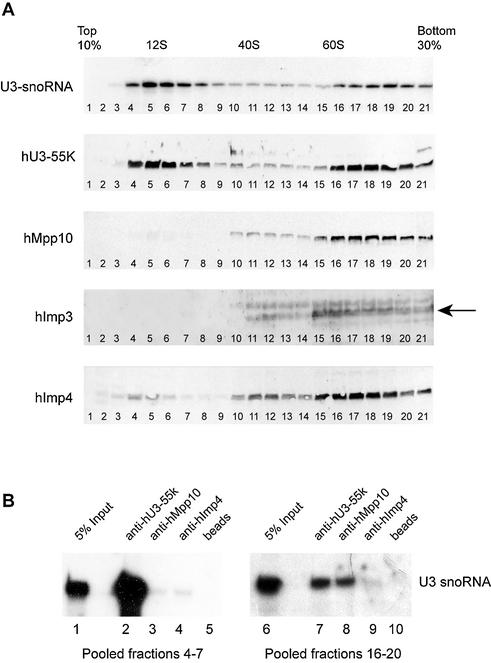
Figure 4.
hImp3, hImp4 and hMpp10 predominantly sediment at 60–80S in glycerol gradients. (A) HEp-2 cell lysates were loaded on a 10–30% (v/v) glycerol gradient, and the sedimentation of U3 snoRNA, hU3-55K, hMpp10, hImp3 and hImp4 was analyzed by northern blot hybridization and immunoblotting, respectively. The sedimentation of the large ribosomal RNAs was determined by agarose gel electrophoresis and ethidium bromide staining and these were used as markers for 40S and 60S particles in the gradient. The U1 snRNA was used as a marker for 12S complexes (data not shown). The arrow marks the hImp3 band. (B) The fractions containing either the 12S or 60–80S U3 snoRNP complexes (4–7 and 16–20) were pooled and used for immunoprecipitation experiments with anti-hU3-55K (lanes 2 and 7), anti-hMpp10 (lanes 3 and 8) and anti-hImp4 (lanes 4 and 9) antibodies. RNAs isolated from the pooled fractions (5% input, lanes 1 and 6) and co-precipitated RNAs were separated on denaturing polyacrylamide gels and analyzed by northern blot hybridization using a U3 snoRNA specific probe. As a negative control an immunoprecipitation was performed in the absence of antibodies (beads, lanes 5 and 10).
Expression and purification of GST-fusion proteins
GST-fusion proteins were expressed and purified essentially as described previously (37). The proteins were eluted in the presence of 0.2% Triton X-100 and stored at –70°C after the addition of glycerol (10%).
In vitro transcription and translation
All pCI-neoVSV constructs were transcribed and translated in reticulocyte lysate using either the TnT coupled transcription/translation kit (Promega), or by run-of transcription and translation using manufacturers procedures (Promega). To generate hMpp10 deletion mutant translates, pCI-neoVSVhMpp10 was digested with the corresponding enzymes as described above.
GST pull-down experiments
GST-fusion protein (500 ng) was incubated with radiolabeled in vitro translated protein in buffer A [20 mM HEPES–KOH (pH 7.9), 150 mM KCl, 0.2% Triton X-100, 1.5 mM MgCl2, 0.2 mM EDTA] in a final volume of 20 µl. Reactions were incubated for 1 h on ice. Subsequently, the reaction mixtures were diluted to 250 µl with buffer A, 10 µl of glutathione– Sepharose beads were added and the mixture was incubated by end-over-end rotation for 2 h at 4°C. Beads were washed four times with buffer A. Proteins were eluted from the beads by adding SDS sample buffer and heating for 5 min at 100°C. Co-precipitated proteins were separated by SDS–PAGE and visualized by autoradiography.
Generation of antisera
Female white chickens were immunized with 50 µg of purified GST-fusion protein and boosted every two weeks with the same amount of protein. Two chickens were used for immunization with GSThImp3 (006 and 007) and two for immunization with GSThImp4 (008 and 009). The 007 and 008 bleeds showed significant reactivity on western blots with the corresponding HeLa cell and recombinant proteins as well as in immunoprecipitation of the recombinant in vitro translated proteins and thus were used for further experiments.
Affinity-purification of antibodies
Anti-hU3-55K antibodies (38) were affinity-purified using the peptide that was used for the immunizations coupled to Sulfolink resin as described by the manufacturer’s procedure (Pierce).
Western blot analysis
Western blots loaded with gradient fractions were blocked in MP buffer [PBS, 5% non-fat milk powder, 0.1% NP-40 (v/v)] for 1 h at room temperature. Primary antibody was diluted in MP buffer and incubated for 1 h at room temperature. Blots were washed three times for 10 minutes with MP buffer, followed by a 1 h incubation with the secondary antibody. Antibodies used: chicken anti-hImp4, diluted 1:10 000; chicken anti-hImp3, diluted 1:5000; affinity purified anti-hU3-55K antibodies 1:100; guinea pig anti-hMpp10 antibodies, 1:2000 (a kind gift from Dr Larry Gerace) (32). As secondary antibodies, horseradish peroxidase conjugated rabbit anti-mouse (1:2500), rabbit anti-guinea pig (1:2500), swine anti-rabbit (1:2500) or rabbit anti-chicken IgY (1:20 000) were used. Except for the rabbit anti-chicken antibodies (Jackson Immunoresearch), all secondary antibodies were purchased from Dako (Glostrup, Denmark).
RESULTS
Cloning of cDNAs encoding the mammalian Imp3p and Imp4p homologs
Database searches performed with the yeast Imp3 and Imp4 sequences led to the identification of putative Imp3p and Imp4p homologs in a number of species, several of which have been reported before (29,33,39,40). Human and mouse Imp3 and Imp4 cDNAs were isolated and sequenced. The amino acid sequence derived from the cloned human Imp4 cDNA (hImp4) was identical to the putative human Imp4p homolog recently described (40) (accession number: AF054996). While this work was in progress, a protein designated MRPS4, the sequence of which is identical to the sequence derived from the cloned human Imp3 cDNA (hImp3), was described as a mitochondrial ribosomal subunit component (accession numbers: BAB54955 and NP_060755), although no experimental evidence for its association with the mitochondrial ribosome was presented (41). Figure 1 shows an alignment of Imp3 and Imp4 amino acid sequences. Both the human and mouse Imp3 and Imp4 proteins have predicted molecular masses of 21.8 and 33.7 kDa, respectively. Both mammalian Imp proteins display ∼50% identity and 65% similarity to their yeast counterparts.
Figure 1.
Amino acid sequence alignment of Imp3 and Imp4. Amino acid sequences derived from cDNAs encoding Imp3 (A) and Imp4 (B) of Homo sapiens (hs), Mus musculus (mm), D.melanogaster (dm), Caenorhabditis elegans (ce), Arabidopsis thaliana (at) and S.cerevisiae (sc) were aligned by ClustalW and shaded using Boxshade. Identical amino acids (in at least three of the sequences) are marked by black boxes and similar amino acids are marked by gray boxes. The boxed regions mark the predicted coiled-coil domains. The black bar in (A) underlines the putative S4 RNA binding domain of Imp3. The black bar in (B) underlines the putative σ70-like RNA binding domain of Imp4. The broken line in (B) indicates the position of the Imp4 domain. Accession numbers: dmImp3, AAL49156; ceImp3, NM_059571, atImp3, NM_121580; mmImp3, BC009145; hsImp3, BAB54955 and NP_060755; scImp3p, AA184637; dmImp4, AE003750; ceImp4, AF334609; atImp4, AC011622; hsImp4, AF054996; mmImp4, XP_129730; scImp4p, AA404192.
Besides a putative S4 domain in hImp3 and a σ70-like motif in hImp4, coiled-coil regions are predicted to reside in both human proteins (Fig. 1). The Interproscan program (http://www.ebi.ac.uk/interpro/scan.html) revealed the so-called Imp4 domain (InterProScan ID: Q96G21) that was recently identified in Imp4 homologs from several species (40).
The hImp3 and hImp4 proteins localize to nucleoli and interact with the human U3 snoRNP complex
To investigate whether the hImp3 and hImp4 cDNAs encoded the functional counterparts of the yeast Imp3p and Imp4p proteins, their subcellular localization and association with the U3 snoRNP were determined. HEp-2 cells were transfected with constructs encoding GFP-fusion proteins of hImp3 and hImp4 and after 18 h the subcellular localization of the GFP-tagged proteins was examined by fluorescence microscopy. The results show that both fusion proteins accumulate in the nucleoli (Fig. 2A). Thus, the subcellular localization of hImp3 and hImp4 is consistent with U3 snoRNP association and a role in ribosomal RNA maturation. To investigate whether these proteins indeed associate with U3, immunoprecipitations were performed with anti-hImp3 and anti-hImp4 antibodies on HEp-2 total cell extracts (Fig. 2B, lanes 3 and 4). As controls anti-hMpp10, anti-hU3-55K, anti-fibrillarin and anti-(tri)methylguanosine cap antibodies were used (Fig. 2B, lanes 4–7). After immunoprecipitation, co-precipitated RNAs were analyzed by northern blot hybridization using various riboprobes. We observed U3 snoRNA co-precipitation with both the anti-hImp3 and anti-hImp4 antibodies, although the amounts were reproducibly low (Fig. 2B, lanes 2 and 3). Co-precipitation of U3 with the anti-hImp4 antibodies was always more efficient than with anti-hImp3 antibodies. As expected, the U3 snoRNA efficiently co-precipitated with the anti-fibrillarin and anti-cap antibodies (Fig. 2B, lanes 6 and 7).
The mouse Imp3 protein partially complements a yeast imp3 null strain
The association of hImp3 and hImp4 with the U3 snoRNP complex and their subcellular localization suggests that these proteins are involved in pre-rRNA processing and thus indeed represent functional counterparts of the respective yeast proteins. To substantiate this, we investigated whether the expression of the mammalian Imp3 and Imp4 proteins in yeast could complement the respective yeast null alleles. As evident from Figure 3A, the expression of mouse Imp3 (mImp3) partially complemented an imp3 null allele in yeast at 22°C, whereas mouse Imp4 (mImp4) did not complement the imp4 null allele.
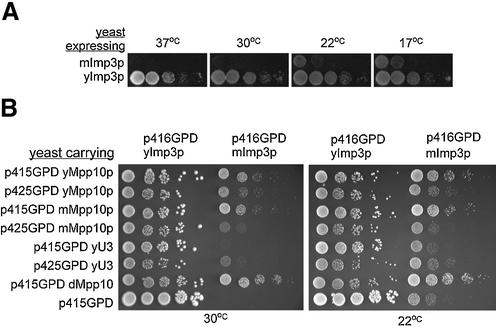
Figure 3.
mImp3 partially complements the growth defect of a yeast imp3 null strain. (A) mImp3 complements the null allele at cold temperatures. Serial dilutions of yeast expressing only mImp3 or yImp3 were grown at 37, 30, 22 and 17°C. (B) The growth defect conferred by mImp3 can be suppressed by over-expression of related components. Growth of mImp3-dependent strains was assessed when either Mpp10 from different species or the yeast U3 snoRNA was co-expressed from either a low copy vector (p415GPD) or high copy vector (p425GPD) at 30 and 22°C.
Interestingly, the growth defect conferred by mImp3 could be partially alleviated by over-expressing other members of the yeast SSU processome, including Mpp10p and the yeast U3 snoRNA (Fig. 3B). Homologs of Mpp10p from different species and the U3 snoRNA were over-expressed at different levels in a yeast strain expressing either mImp3 or yeast Imp3. Most notably, Drosophila melanogaster Mpp10 (dMpp10p) suppressed the growth defect the best at both 22 and 30°C (Fig. 3B). This is most likely due to the fact that dMpp10p and yeast Mpp10p share a higher degree of conservation than yeast and mouse Mpp10 (33). Over-expression of the U3 snoRNA also alleviated the growth defect of the yeast mImp3 strain, although to a lesser extent than dMpp10p. Very high levels of Mpp10p (expressed from p425GPD) had a negative effect on growth suggesting that the Mpp10p must be tightly regulated. Likewise depletion of Mpp10p in yeast leads to a decrease of Imp3p and Imp4p protein levels (33) and over-expression of Mpp10p also increases the levels of Imp3p and Imp4p (S.Wormsley and S.J.Baserga, unpublished). Co-over expression of all three is lethal (data not shown) suggesting that this dominant negative affect could be due to dilution of other factors associated with these proteins.
hImp3, hImp4 and hMpp10 sediment at 60–80S in glycerol gradients
The human U3 snoRNA is associated with 12S and 60–80S complexes of which the latter probably represent the U3 snoRNA associated with pre-rRNA processing complexes (26–28,42). Since most of the yeast U3 snoRNA-associated proteins have been reported to interact with the RNA in higher order complexes (26,42,43), the association of hImp3 and hImp4 with the U3 snoRNP was further studied by density gradient sedimentation. Since yeast Imp3p and Imp4p interact with Mpp10p (29), we included the human Mpp10 protein in these analyses. In addition, the sedimentation of hU3-55K, a U3 snoRNP specific protein, was investigated (35,44). HEp-2 cell extracts were fractionated in 10–30% glycerol gradients. Northern blotting revealed that the bulk of the U3 snoRNA sedimented at 12S and 60–80S, in agreement with previous observations (27,28). The U3-specific protein hU3-55K co-sedimented with the U3 snoRNA, and peaked with the U3 snoRNA in fractions 5–6 (∼12S) and fractions 17–19 (∼70S). Interestingly, the bulk of hImp3, hImp4 and hMpp10, co-sedimented with the U3 snoRNA at 60–80S (Fig. 4A, fractions 17–19), whereas only very small amounts sedimented near 12S (Fig. 4A, fractions 4–7). These results suggested that, in contrast to hU3-55K, the hImp3, hImp4 and Mpp10 proteins are not associated with the 12S U3 complex, but that they only interact with the larger, 60–80S U3 snoRNA-containing complexes. Immunoprecipitation experiments performed with anti-hU3-55K, anti-hMpp10 and anti-hImp4 antibodies (Fig. 4B, lanes 7–9) confirmed the interaction between these proteins and the U3 snoRNA in the 60–80S fractions. Furthermore, only very low amounts of U3 snoRNA were co-precipitated by anti-hImp4 and anti-hMpp10 antibodies from 12S fractions (Fig. 4B, lanes 3–5).
hMpp10, hImp3 and hImp4 form a hetero-trimeric complex in vitro
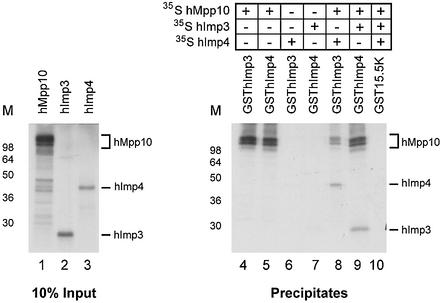
In yeast, the Imp3p and Imp4p proteins were identified by virtue of their interaction with Mpp10p (29). To study whether the human proteins also directly bind to each other, GST pull-down experiments were performed using GSThImp3, GSThImp4 and 35S-labeled, in vitro translated hImp3, hImp4 and hMpp10 proteins. The GST-fusion proteins and radiolabeled proteins were incubated and the complexes that were formed were precipitated using glutathione–Sepharose beads. The results in Figure 5 show that GSThImp3 and GSThImp4 efficiently interacted with hMpp10 (lanes 4 and 5). In contrast, no detectable interaction was observed between GSThImp3 and hImp4 and between GSThImp4 and hImp3 (Fig. 5, lanes 6 and 7). These results suggest that, like their yeast counterparts, hImp3 and hImp4 interact with Mpp10 but not with each other. In agreement with these interactions, hetero-trimeric complexes could be reconstituted in vitro when either GSThImp3 was incubated with radiolabeled hMpp10 and hImp4 (Fig. 5, lane 8) or when GSThImp4 was incubated with radiolabeled hMpp10 and hImp3 (Fig. 5, lane 9). When GST15.5K was used in these experiments, no detectable co-precipitation of radiolabeled hImp3, hImp4 or hMpp10 (Fig. 5, lane 10) was observed.
Figure 5.
hImp3 and hImp4 interact with hMpp10 but not with each other in vitro. Radiolabeled hMpp10, hImp3 and hImp4 were expressed in an in vitro translation system (lanes 1–3). These polypeptides were incubated with GST-tagged hImp3 or hImp4 as indicated, followed by precipitation with immobilized glutathione (lanes 4–9). A GST15.5K fusion protein was used as a control (lane 10). Note that hImp4 is only co-precipitated with GSThImp3 in the presence of hMpp10 and that, vice versa, hImp3 is only co-precipitated with GSThImp4 in the presence of hMpp10. The positions of radiolabeled hMpp10, hImp3 and hImp4 in the gel are indicated on the right. On the left of each panel the positions of molecular mass markers are shown.
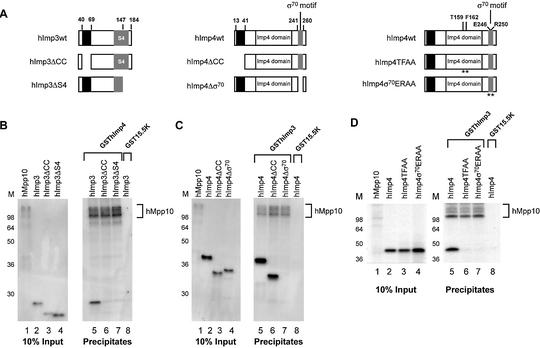
Domains of hImp3, hImp4 and hMpp10 involved in mutual interactions
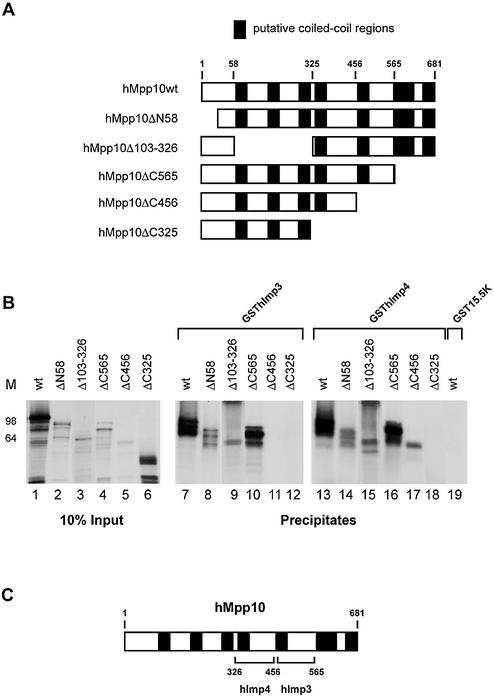
The efficient interactions between hImp3, hImp4 and hMpp10 prompted us to examine the regions involved in their mutual interactions. Five hMpp10 deletion mutants were generated (Fig. 6A) and analyzed by GST pull-down experiments. The results show that GSThImp3 efficiently bound to hMpp10ΔN58, hMpp10Δ103–326 and hMpp10ΔC565, but not to hMpp10ΔC456 and hMpp10ΔC325 (Fig. 6B, lanes 8–12). GSThImp4 bound to all mutants (Fig. 6B, lanes 14–18) with the exception of hMpp10ΔC325. These results suggest that the region of hMpp10 bordered by amino acids 457 and 565 is required for its interaction with hImp3, whereas the region bordered by amino acids 327–456 is involved in the binding to hImp4 (Fig. 6C).
Figure 6.
Regions of hMpp10 involved in its interaction with hImp3 and hImp4. (A) Schematic representation of the hMpp10 deletion mutants. The black boxes represent putative coiled-coil regions in hMpp10 (32). (B) Binding of GSThImp3 and GSThImp4 to hMpp10 deletion mutants. GST pull-down experiments were performed with GSThImp3, GSThImp4 and GST15.5K. Lanes 1–6 show 10% of the input material. Precipitation of hMpp10 mutants by GSThImp3 is shown in lanes 7–12, by GSThImp4 in lanes 13–18 and precipitation of wild type hMpp10 by GST15.5K in lane 19. On the left of the panel the positions of molecular mass markers are shown. (C) Schematic representation of the results shown in (B).
To investigate the regions of hImp3 and hImp4 involved in the interaction with hMpp10, the capacity of hImp3 and hImp4 deletion mutants (Fig. 7A) to interact with in vitro translated hMpp10 bound to GSThImp4 and GSThImp3, respectively, was determined. Deletion of either the putative coiled-coil domain of hImp3 or the C-terminal region including part of the S4 domain abrogated the interaction with hMpp10 (Fig. 7B, lanes 6 and 7). Deletion of the putative coiled-coil region of hImp4 did not affect its interaction with hMpp10 (Fig. 7C, lane 6). However, deletion of the σ70-like motif abrogated its interaction with hMpp10 (Fig. 7C, lane 7). These data indicate that the deletion of spatially separated regions of hImp3 interfere with its binding to hMpp10 and that the σ70-like motif of hImp4 is required for its association with hMpp10. To substantiate the importance of the σ70-like motif for this interaction and to investigate the possibility that (also) the Imp4 domain plays a role in this interaction, we generated amino acid substitution mutants. Two highly conserved amino acids in the σ70-like motif (E246 and R250) or the Imp4 domain (T159 and F162) were substituted by alanines (Fig. 7A, hImp4σ70ERAA and hImp4TFAA) (33,40). The results show that both mutants did not efficiently interact with hMpp10 (Fig. 7D, lanes 6–8), suggesting that the σ70-like motif and conserved amino acids in the hImp4 domain are important for binding to hMpp10 in vitro.
Figure 7.
Regions of hImp3 and hImp4 required for their interaction with hMpp10 in vitro. (A) Schematic representation of the hImp3 and hImp4 deletion and substitution mutants. The black boxes indicate the putative coiled-coil regions in hImp3 and hImp4. The putative RNA binding domains in hImp3 (S4) and hImp4 (σ70-like) are depicted as gray boxes. The Imp4 domain is marked with a white box. The asterisks indicate the positions in hImp4 that were changed in the substitution mutants. (B–D) Binding of hImp3 (B) and hImp4 (C) deletion mutants and hImp4 substitution mutants (D) to hMpp10. Reconstituted complexes were precipitated using glutathione–Sepharose beads as described in Materials and Methods and analyzed by SDS–PAGE. The hImp3 and hImp4 (mutant) translates are indicated above the respective lanes and radiolabeled hMpp10 was added to all reconstitution reactions. The GST- fusion proteins used in this experiment are indicated on top of the panels. Lanes 1–4 show 10% of the input material used in the pull-down experiments. Lanes 5–8 show the precipitates (in lane 8 GST15.5K was used as a negative control). On the left of each panel the positions of molecular mass markers are shown. On the right the position of hMpp10 in the gel is indicated.
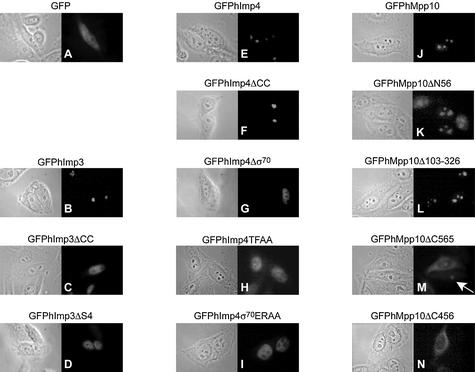
Complex formation deficient hImp3, hImp4 and hMpp10 mutants fail to localize to nucleoli in HEp-2 cells
To determine the effects of the deletions and substitutions in the hImp3, hImp4 and hMpp10 proteins on their nucleolar accumulation, HEp-2 cells were transiently transfected with constructs encoding GFP-fusion proteins of these mutants. Interestingly, all hImp3 and hImp4 mutants that failed to interact with hMpp10 in vitro predominantly accumulated in the nucleoplasm (Fig. 8C, D, G, H and I), whereas the hImp4 mutant that retained the ability to interact with hMpp10 accumulated in the nucleoli like the wild-type proteins (Fig. 8B, E and F). These results suggest that the interaction of hImp3 and hImp4 with hMpp10 is required for their entry into the nucleolus.
Figure 8.
Effect of mutations in hImp3, hImp4 and hMpp10 on their subcellular localization. HEp-2 cells were transiently transfected with constructs encoding GFP fusion proteins of hImp3, hImp4, hMpp10 and mutants thereof and the localization of the fusion proteins was determined by fluorescence microscopy. The mutants used in these experiments are outlined in Figures 6A and 7A. The corresponding phase contrast image is shown on the left of each fluorescence image. (A) Results for GFP alone; (B–D) GFP-tagged hImp3 and hImp3 mutants; (E–I) GFP-tagged hImp4 and hImp4 mutants; (J–N) GFP-tagged hMpp10 and hMpp10 mutants. The arrow indicates the nucleolar accumulation of GFPhMpp10Δ565 in a cell expressing relatively low levels of this mutant.
The analysis of the GFPhMpp10 fusion proteins suggested that also hMpp10 requires the interaction with hImp3, and possibly hImp4, for nucleolar localization. The wild-type protein (GFPhMpp10), the N-terminal deletion mutant (GFPhMpp10ΔN58) and the internal deletion mutant (GFPhMpp10Δ103–326) accumulated in nucleoli (Fig. 8J–L). Thus a large portion of the N-terminal part of hMpp10 can be deleted without affecting its subcellular distribution. Interestingly, a C-terminal deletion of 116 amino acids (GFPhMp10ΔC565) resulted in both nucleolar and cytoplasmic accumulation, while only weak staining was observed in the nucleoplasm (Fig. 8M). The most pronounced nucleolar accumulation with this mutant was observed in cells expressing relatively low levels of the fusion protein (Fig. 8M; indicated by an arrow). The cytoplasmic accumulation might be related to the amino acid composition of the region that was deleted, which contains clusters of basic amino acids that might be involved in nuclear import (32). The larger C-terminal deletion mutant GFPhMpp10ΔC456 predominantly accumulated in the cytoplasm (Fig. 8N), whereas only very small amounts appeared to reside in the nucleoplasm. Similar observations were made with the GFPhMpp10ΔC325 mutant (data not shown). With these mutants no significant staining was observed in the nucleoli. Taken together, these results suggest that the region in hMpp10 that is important for binding hImp3 (amino acids 456–565) is also required for nucleolar localization, while the C-terminal region comprising amino acids 565–681 is required for nuclear import.
DISCUSSION
In this paper we describe the cloning and characterization of mammalian Imp3p and Imp4p homologs. Our data show that like in the yeast Saccharomyces cerevisiae the hImp3 and hImp4 proteins both interact with hMpp10, but not with each other. Furthermore, they indicate that hImp3, hImp4 and hMpp10 predominantly associate with the U3 snoRNA in 60–80S complexes, which are likely to represent pre-ribosomal complexes. The results obtained with hImp3, hImp4 and hMpp10 mutants suggest that the nucleolar accumulation of these proteins is dependent on formation of the ternary complex.
hImp3 and hImp4 interact with the U3 snoRNA in 60–80S U3 snoRNA-containing particles
Previously it has been shown that the yeast Imp3p and Imp4p proteins specifically interact with the U3 snoRNP complex (29). Our immunoprecipitation data indicate that this also occurs in the human system. Despite the fact that a considerable amount of U3 snoRNA co-sedimented with hImp3 and hImp4 in glycerol gradients, we reproducibly observed a relatively inefficient U3 snoRNA co-precipitation, indicating that only a small amount of U3 snoRNA is stably associated with these proteins in HEp-2 cells. We obtained similar results with anti-GFP immunoprecipitations using extracts prepared from transiently transfected HEp-2 cells expressing GFP-tagged hImp3 or hImp4 (data not shown). The possibility that the inefficient co-precipitation was due to a poor accessibility of the hImp3 and hImp4 proteins for the antibodies in the 60–80S U3 complexes cannot be excluded.
The results of gradient sedimentation analyses suggested that hImp3, hImp4 and hMpp10 are efficiently associated with 60–80S complexes, at least part of which also contain the U3 snoRNA. U3-containing complexes with a similar sedimentation behavior have been reported by Tyc and Steitz using HeLa cell extracts (28). Components of the yeast ∼80S SSU processome are associated with pre-rRNA (J.E.G.Gallagher and S.J.Baserga, unpublished results) (42) and these complexes have been suggested to represent the terminal balls seen on nascent pre-rRNA transcripts (26,42,46,47). Attempts to demonstrate the association of pre-rRNA by immunoprecipitation with anti-hImp and anti-hMpp10 antibodies from 60–80S gradient fractions were not successful. This is most likely due to the low abundance of these precursors in the cell and low stability of these RNAs. However, based on recent studies performed in yeast (26,42), it is likely that the human U3 complexes sedimenting at 60–80S are associated with pre-rRNA precursors.
Our results are consistent with the formation of a relatively stable trimeric complex of Imp3, Imp4 and Mpp10, which interacts with the U3 snoRNA only when it is associated with higher order complexes. The formation of such a trimeric complex is supported by affinity-purifications using tagged Mpp10p expressed in yeast cells, which yielded Mpp10p, Imp3p and Imp4p and no other SSU processome components (F.Dragon, S.Wormsley and S.J.Baserga, unpublished observations). The efficient association of hImp3, hImp4 and hMpp10 with large complexes suggests a model in which U3 is recruited to the pre-rRNA by virtue of its interaction with this trimeric complex. Alternatively, the association of U3 with pre-rRNA may create or expose the binding site for the hImp3-hImp4-hMpp10 complex.
Mpp10 association with U3 snoRNA requires sequence elements that are important for base pairing with the 5′ ETS of the pre-rRNA (S.Granneman, N.J.Watkins, J.Vogelzangs, R.Lührmann, W.J.van Venrooij and G.J.M.Pruijn, manuscript in preparation) (34). In addition, a C-terminal truncation of the yeast Mpp10p led to cold sensitivity and pre-rRNA processing defects at A1 and A2 (36,45). A similar processing defect and growth phenotype has been observed in a yeast strain bearing mutations in box A that block base-pairing with 18S rRNA sequences (13). Thus hMpp10, together with hImp3 and hImp4, might function by facilitating or stabilizing base-pairing interactions between U3 snoRNA and the pre-rRNA.
Imp3 is believed to mediate the association of the heterotrimeric complex with the U3 snoRNA (36). A possible explanation for the partial complementation of the yeast imp3 null allele by mImp3 would be that this mouse protein is incapable of interacting with the yeast Mpp10p. However, mImp3 interacts in a two-hybrid system with yeast Mpp10p to the same degree as yeast Imp3p (data not shown). The observation that over-expression of the yeast U3 snoRNA also partially alleviated the growth defect indicates that the interactions between mImp3 and the yeast U3 snoRNA may be inefficient, resulting in a growth defect at permissive temperatures.
Formation of the hImp3-hMpp10-hImp4 complex correlates with the nucleolar localization of these proteins
Surprisingly, the analysis of deletion mutants showed that the putative RNA binding domains of hImp3 (S4 domain) and hImp4 (σ70-like motif) are required for their binding to hMpp10 in vitro. Although it is possible that the observed effects are (in part) due to conformational changes in the mutant proteins caused by the deletions or substitutions, the correlation between their interactions in vitro and their nucleolar accumulation is intriguing and strongly suggests that the entry of hImp3, hImp4 and hMpp10 into the nucleolus is dependent on formation of the ternary complex.
The human SSU processome
The presence of the U3 snoRNA in both 12S and 60–80S complexes raises the question of whether the label U3 snoRNP is entirely accurate. Like in yeast (16,48), the hU3-55K and the common box C/D snoRNP proteins are associated with the human 12S U3 complex (this work; S.Granneman, N.J.Watkins, J.Vogelzangs, R.Lührmann, W.J.van Venrooij and G.J.M.Pruijn, manuscript in preparation). However, the majority of the U3-specific proteins in yeast, including Imp3p, Imp4p and Mpp10p, seem to be preferentially or exclusively associated with the U3 snoRNA in higher order complexes (26,42,43,49). Based on the relatively well defined composition of the 12S complex, we propose to name this complex the U3 snoRNP and the 60–80S U3-containing complexes the human SSU processome.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Larry Gerace for providing the anti-hMpp10 antibodies and Michael Pollard for providing the anti-fibrillarin antibodies. We would like to thank Nick Watkins for providing the GST15.5K construct, Reinout Raijmakers for critically reading the manuscript and Antoon Huiskens for cloning the hImp3 and hImp4 cDNAs. This work was supported by grants from the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW), and from the National Institutes of Health (R01GM52581 to S.J.B. and GM20905 to J.E.G.).
REFERENCES
- 1.Hughes J.M. and Ares,M.,Jr (1991) Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J., 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kass S., Tyc,K., Steitz,J.A. and Sollner-Webb,B. (1990) The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell, 60, 897–908. [DOI] [PubMed] [Google Scholar]
- 3.Savino R. and Gerbi,S.A. (1990) In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J., 9, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 5.Beltrame M. and Tollervey,D. (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame M. and Tollervey,D. (1995) Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borovjagin A.V. and Gerbi,S.A. (1999) U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol., 286, 1347–1363. [DOI] [PubMed] [Google Scholar]
- 8.Borovjagin A.V. and Gerbi,S.A. (2000) The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol., 300, 57–74. [DOI] [PubMed] [Google Scholar]
- 9.Borovjagin A.V. and Gerbi,S.A. (2001) Xenopus U3 snoRNA GAC-box A′ and box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol., 21, 6210–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Méreau A., Fournier,R., Gregoire,A., Mougin,A., Fabrizio,P., Lührmann,R. and Branlant,C. (1997) An in vivo and in vitro structure–function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein–RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol., 273, 552–571. [DOI] [PubMed] [Google Scholar]
- 11.Samarsky D.A. and Fournier,M.J. (1998) Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyc K. and Steitz,J.A. (1992) A new interaction between the mouse 5′ external transcribed spacer of pre-rRNA and U3 snRNA detected by psoralen crosslinking. Nucleic Acids Res., 20, 5375–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes J.M. (1996) Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]
- 14.Sharma K. and Tollervey,D. (1999) Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol., 19, 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker K.A. and Steitz,J.A. (1987) Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol. Cell. Biol., 7, 2899–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watkins N.J., Segault,V., Charpentier,B., Nottrott,S., Fabrizio,P., Bachi,A., Wilm,M., Rosbash,M., Branlant,C. and Lührmann,R. (2000) A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- 17.Lafontaine D.L. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafontaine D.L. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyman S.K., Gerace,L. and Baserga,S.J. (1999) Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA, 5, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman D.R., Kuhn,J.F., Shanab,G.M. and Maxwell,E.S. (2000) Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E.C. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu P., Brockenbrough,J.S., Metcalfe,A.C., Chen,S. and Aris,J.P. (1998) Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szewczak L.B., DeGregorio,S.J., Strobel,S.A. and Steitz,J.A. (2002) Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol., 9, 1095–1107. [DOI] [PubMed] [Google Scholar]
- 24.Watkins N.J., Dickmanns,A. and Lührmann,R. (2002) Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol., 22, 8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terns M.P. and Terns,R.M. (2002) Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr., 10, 17–39. [PMC free article] [PubMed] [Google Scholar]
- 26.Dragon F., Gallagher,J.E., Compagnone-Post,P.A., Mitchell,B.M., Porwancher,K.A., Wehner,K.A., Wormsley,S., Settlage,R.E., Shabanowitz,J., Osheim,Y. et al. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature, 417, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein P., Reddy,R. and Busch,H. (1984) Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry, 23, 5421–5425. [DOI] [PubMed] [Google Scholar]
- 28.Tyc K. and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S.J. and Baserga,S.J. (1999) Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol., 19, 5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunbar D.A., Wormsley,S., Agentis,T.M. and Baserga,S.J. (1997) Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol., 17, 5803–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupas A. (1996) Coiled coils: new structures and new functions. Trends Biochem. Sci., 21, 375–382. [PubMed] [Google Scholar]
- 32.Westendorf J.M., Konstantinov,K.N., Wormsley,S., Shu,M.D., Matsumoto-Taniura,N., Pirollet,F., Klier,F.G., Gerace,L. and Baserga,S.J. (1998) M phase phosphoprotein 10 is a human U3 small nucleolar ribonucleoprotein component. Mol. Biol. Cell., 9, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wehner K.A. and Baserga,S.J. (2002) The sigma(70)-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell, 9, 329–339. [DOI] [PubMed] [Google Scholar]
- 34.Wormsley S., Samarsky,D.A., Fournier,M.J. and Baserga,S.J. (2001) An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA, 7, 904–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pluk H., Soffner,J., Lührmann,R. and van Venrooij,W.J. (1998) cDNA cloning and characterization of the human U3 small nucleolar ribonucleoprotein complex-associated 55-kilodalton protein. Mol. Cell. Biol., 18, 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wehner K.A., Gallagher,J.E. and Baserga,S.J. (2002) Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol., 22, 7258–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raijmakers R., Noordman,Y.E., van Venrooij,W.J. and Pruijn,G.J. (2002) Protein–protein interactions of hCsl4p with other human exosome subunits. J. Mol. Biol., 315, 809–818. [DOI] [PubMed] [Google Scholar]
- 38.Granneman S., Pruijn,G.J., Horstman,W., Van Venrooij,W.J., Lührmann,R. and Watkins,N.J. (2002) The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in vitro. J. Biol. Chem., 227, 48490–48500. [DOI] [PubMed] [Google Scholar]
- 39.Fatica A., Cronshaw,A.D., Dlakic,M. and Tollervey,D. (2002) Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell, 9, 341–351. [DOI] [PubMed] [Google Scholar]
- 40.Mayer C., Suck,D. and Poch,O. (2001) The archaeal homolog of the Imp4 protein, a eukaryotic U3 snoRNP component. Trends Biochem. Sci., 26, 143–144. [DOI] [PubMed] [Google Scholar]
- 41.Kenmochi N., Suzuki,T., Uechi,T., Magoori,M., Kuniba,M., Higa,S., Watanabe,K. and Tanaka,T. (2001) The human mitochondrial ribosomal protein genes: mapping of 54 genes to the chromosomes and implications for human disorders. Genomics, 77, 65–70. [DOI] [PubMed] [Google Scholar]
- 42.Grandi P., Rybin,V., Bassler,J., Petfalski,E., Strauss,D., Marzioch,M., Schafer,T., Kuster,B., Tschochner,H., Tollervey,D. et al. (2002) 90S Pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell, 10, 105–115. [DOI] [PubMed] [Google Scholar]
- 43.Billy E., Wegierski,T., Nasr,F. and Filipowicz,W. (2000) Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J., 19, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukowiak A.A., Granneman,S., Mattox,S.A., Speckmann,W.A., Jones,K., Pluk,H., van Venrooij,W.J., Terns,R.M. and Terns,M.P. (2000) Interaction of the U3-55k protein with U3 snoRNA is mediated by the box B/C motif of U3 and the WD repeats of U3-55k. Nucleic Acids Res., 28, 3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S.J. and Baserga,S.J. (1997) Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl Acad. Sci USA, 94, 13536–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller O.L. Jr and Beatty,B.R. (1969) Visualization of nucleolar genes. Science, 164, 955–957. [DOI] [PubMed] [Google Scholar]
- 47.Mougey E.B., O’Reilly,M., Osheim,Y., Miller,O.L.,Jr, Beyer,A. and Sollner-Webb,B. (1993) The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes.l. Genes Dev., 7, 1609–1619. [DOI] [PubMed] [Google Scholar]
- 48.Venema J., Vos,H.R., Faber,A.W., van Venrooij,W.J. and Raué,H.A. (2000) Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA, 6, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wegierski T., Billy,E., Nasr,F. and Filipowicz,W. (2001) Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA, 7, 1254–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]