Abstract
A general two step procedure for the internal labeling of l-deoxyoligonucleotides, Spiegelmers, has been developed. Through radioactive labeling oligonucleotides can easily be detected and monitored in biological samples. T4 polynucleotide kinase is shown to efficiently phosphorylate strands of l-nucleic acids which allows the labeling with phosphorous isotopes such as 32P. In order to protect the terminal phosphate label against unspecific phosphatases, one of two fragments of a Spiegelmer is enzymatically phosphorylated with [γ-32P]ATP. In a second step we used a template- directed chemical ligation reaction in order to attach the labeled oligonucleotide to the other fragment to yield the full-length Spiegelmer with an internal [32P]phosphodiester bond. It has been shown that the functionality of a chemically ligated Spiegelmer is still retained.
INTRODUCTION
In the past decade interest to develop oligonucleotides as a new substance class for therapeutic applications has increased. Several technologies have been developed to generate different classes of oligonucleotides, such as antisense oligonucleotides, ribozymes and aptamers (1,2); the latter specifically bind to and can inhibit the function of a given target molecule, for instance a protein. Spiegelmers, the latest class of potential oligonucleotide therapeutics, are enantiomeric aptamers composed of l-nucleotides (3). Due to the unnatural sugars used, Spiegelmers are resistant to host nucleases (3). No further stabilization procedures, which are mandatory to increase the biological half-life of antisense oligonucleotides, ribozymes or aptamers, are needed. This makes Spiegelmer oligonucleotides ideal for pharmaceutical applications. Spiegelmers can potentially be generated against a large variety of target structures. Recently, we reported the design of a Spiegelmer specifically binding to and inhibiting the function of the gonadotropin releasing hormone (GnRH) in vitro (4) and in vivo (5).
To perform and analyze advanced biological investigations such as pharmacokinetic and pharmacodynamic studies, sensitive assays for the quantification of mirror image l-oligonucleotides are needed. The method of choice is the incorporation of radioactive isotopes. Since the function of a Spiegelmer is mediated by its three-dimensional structure, modifications with only minimal or preferably no change to the overall structure of the Spiegelmer are a prerequisite.
In this paper, we report the synthesis of an active Spiegelmer with a 32P-labeled internal phosphodiester bond by chemical ligation of two l-deoxyoligonucleotide fragments— one of them being 5′-32P modified. An internal rather than a terminal label was chosen to avoid potential cleavage of the monophosphate esters at the termini of the oligonucleotides by unspecific enzymatic dephosphorylating activities.
It is known that T4 polynucleotide kinase (PNK) tolerates non-nucleotidic modifications at the 5′-terminus of oligonucleotides (6). In the first instance we systematically examined the efficiency of PNK to attach a radiolabeled phosphate to the four different nucleotides at the 5′-end of l-deoxyoligonucleotides. We used BrCN for the chemical ligation reaction of the l-DNA-oligonucleotide fragments on a complementary template. For this kind of reaction BrCN is described to reach high reaction kinetices without any by-products (7).
MATERIALS AND METHODS
Oligonucleotide synthesis
l-Deoxyligonucleotides were prepared on an ABI 394 DNA synthesizer (Applied Biosystems, Foster City, USA) on a 0.2 or 1.0 µmol scale using standard phosphoramidite chemistry. l-DNA-phosphoramidites were purchased from ChemGenes Corporation (Boston, MA).
Sequences of the l-DNA-oligonucleotides used: 1, 5′-GGA GTT ACA GCG CTG-3′; 2, 5′-CGA GTT ACA GCG CTG-3′; 3, 5′-AGA GTT ACA GCG CTG-3′; 4, 5′-TGA GTT ACA GCG CTG-3′; 5, 5′-CCA AGC TTG CGT AAG CAG TCT CCT CTC AGG-3′; 6, 5′-GGA GGT TGG GCG GTG CGT AAG CAC CGG TTT GCA GGG G-3′; 7, 5′-TTA CGC (rA)CC GCC CA(rA) CCT CCC CTG (rA)GA GGA GAC-3′; 8, 5′-CCA AGC TTG CGT AAG CAG TCT CCT CTC AGG GGA GGT TGG GCG GTG CGT AAG CAC CGG TTT GCA GGG G-3′.
The oligonucleotides were purified on denaturing polyacrylamide gels (PAGE).
Kinase reaction
400 pmol of the 15mer DNA-oligonucleotides 1–4 or 37mer DNA-oligonucleotide 6 were dissolved in 20 µl of 1× kinase buffer (MBI-Fermentas, St. Leon-Rot, Germany), 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM dithiothreitol, 0.1 mM spermidine, 0.1 mM EDTA and incubated at 37°C in the presence of 5–80 µCi [γ-32P]ATP (3000 Ci/mmol; Hartmann-Analytik, Braunschweig, Germany) and 40 U T4 PNK (MBI Fermentas). After 3 h of incubation the reactions were analyzed on a 20 or 10% denaturing gel, respectively. Products were visualized by auto-radiography. The yield of the phosphorylation reactions was calculated by dividing the radioactivity of the respective oligonucleotide (excised band from gel) by the input radioactivity.
Chemical ligation
Strand 5 (0.8 nmol), the product of the kinase reaction of strand 6 (0.5 nmol), and template 7 (0.8 nmol) were dissolved in morpholinoethylsulfonate (MES) triethylamine buffer (18 µl, 0.55 M, pH 7.75) containing 55 mM MgCl2 and denatured at 98°C for 4 min. The mixture was cooled slowly (10 min) to 0°C and the ligation reaction was started by an addition of 2 µl of 5 M BrCN in DMF. After 15 min at 0°C, 1 M NaOH was added to a final concentration 0.5 M and incubated at 98°C for 10 min. The mixture was neutralized with an equal volume of 0.5 M HCl, precipitated with ethanol and purified by 10% denaturing PAGE.
Affinity gel capillary electrophoresis
Separations were carried out on a BioFocus 3000 Capillary Electrophoresis instrument (Bio-Rad, Munich, Germany). An AAEE-coated fused-silica capillary (BioCap Oligonucleotide Capillary 75 µm I.D. and 375 µm O.D.; Bio-Rad) was used with an effective length of 24 cm and a total length of 27 cm. The electrophoresis buffer CE Oligonucleotide Run Buffer (Bio-Rad) was used both with and without 5 µM l-GnRH. Before each analysis the capillary was first rinsed with water at 30°C for 3 min and then with the electrophoresis buffer for 80 s. The applied voltage was 10 kV and the capillary temperature was maintained at 30°C. The oligonucleotides were detected at 260 nm.
The concentration of the samples was 5.5 µM for the ligate 8 and 6 µM for the completely chemically synthesized 67mer Spiegelmer 8. A 20mer d-oligothymidine (pdT20); not interacting with l-GnRH was added and used as internal standard. Injection of the samples occurred electro-kinetically with 6 kV for 5 s. The relative migration times (rMTs) of the Spiegelmers were calculated according to the following equation:
rMTSpiegelmer = MTSpiegelmer / MTpdT20
RESULTS AND DISCUSSION
In previous publications it was reported that T4 PNK would not be able to phosphorylate l-DNA (8,9); data were only described with a 6mer l-oligocytidine (8), but no experimental details were shown. To circumvent the problem of directly labeling l-oligonucleotides, mixed oligonucleotides that contained a small tail of two d-thymidine nucleotides at the 5′-end were synthesized (9). These mixed oligonucleotides allowed enzymatic phosphorylation, but no further studies were undertaken to examine the ability of the kinase to recognize other enantiomeric DNA. To address the question if non-natural l-oligonucleotides are substrates for an enzymatic phosphorylation reaction more systematically, a set of four different 15mer l-DNA oligonucleotides that differed only in the terminal 5′-nucleotide were synthesized. These l-DNA oligonucleotides 1–4 were then subjected to a kinase reaction using [γ-32P]ATP.
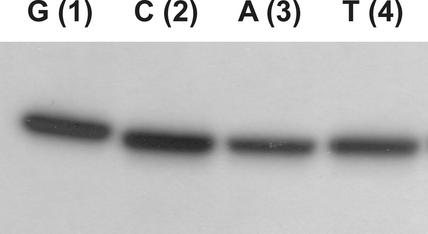
In contrast to previous publications (8,9), we found that l-DNA is a substrate for T4 PNK and can be enzymatically labeled (Fig. 1). We observed only a slight influence of the 5′-terminal nucleotide (C ≈ G > T ≈ A). The yield of the phosphorylation reactions ranged from a 27 to 48% incorporation of the radiolabeled phosphate (Table 1). Compared to the kinase reaction of d-DNA-oligonucleotides, no significant dependence of the phosphorylation yield on the 5′-nucleotide was observed (10). For d-DNA-oligonucleotides a terminal cytidine showed a 6-fold lower phosphorylation yield compared to guanosine, which turned out to be the best terminal nucleotide for labeling.
Figure 1.
Comparative analysis of the T4 PNK with l-DNA 1–4 (the terminal nucleotides are indicated). 400 pmol of the respective l-DNA was incubated in kinase buffer with 40 U T4 PNK in the presence of [γ-32P]ATP in a 20 µl reaction. Incubation was carried out at 37°C for 3 h. Products were visualized by autoradiography (20% denaturing polyacrylamide gel).
Table 1. Yield of kinase reaction for l-DNA 15mers 1–4 (incorporation of the 32P label, two independent labeling experiments).
| Terminal nucleotide (l-DNA 1–4) | G (1) | C (2) | A (3) | T (4) |
|---|---|---|---|---|
| Yield, labeling 1 (%) | 43.9 | 46.8 | 27.6 | 31.0 |
| Yield, labeling 2 (%) | 43.5 | 49.3 | 25.9 | 30.1 |
| Average yield (%) | 44 | 48 | 27 | 31 |
The yield of each phosphorylation reaction was calculated by dividing the radioactivity of the respective labeled oligonucleotide (excised band from gel) by the input radioactivity.
Unexpectedly, T4 PNK is able to phosphorylate l-oligonucleotides as demonstrated for different l-DNAs. l-RNA is also substrate for this kinase (data not shown). T4 PNK is not the only enzyme that shows a lack of stereodifferentiation between d- and l-nucleotides and thus a limited specificity. It has been shown previously that DNA polymerase α, Escherichia coli DNA polymerase I, terminal transferase and HIV-reverse transcriptase can utilize l-dNTP as a substrate (11). Interestingly, human deoxycytidine kinase and HSV-1-thymidine kinase also lack enantiospecificity (12). Moreover, l-ATP seems to interact with the ATP-binding site of T4 DNA ligase and it has been suggested that l-ATP can perform the adenylation step of the enzyme reaction, although it could not be used as a cofactor in the ligase-catalyzed joining reaction and is therefore a strong inhibitor of that enzyme (13).
For incorporation of the 32P-label into the GnRH-binding l-DNA-oligonucleotide, the 67mer [8, 5′-CCA AGC TTG CGT AAG CAG TCT CCT CTC AGG GGA GGT TGG GCG GTG CGT AAG CAC CGG TTT GCA GGG G-3′ (5)] was dissected into two fragments 5 and 6. In order to identify a suitable ligation site, a set of oligonucleotides was synthesized and subjected to phosphorylation and subsequent ligation reactions. Convincing results were obtained with a G-G ligation site (data not shown). A tendency of reduced phosphorylation and chemical ligation yield for longer and more structured oligonucleotides was observed.
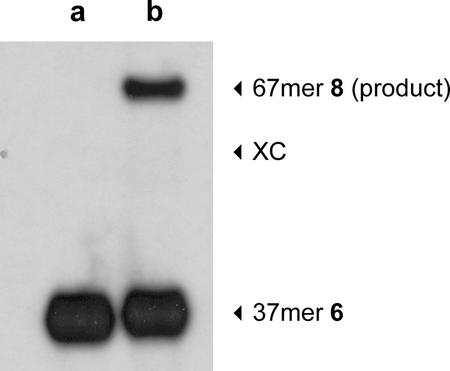
The l-DNA 37mer fragment 6 was radioactively labeled in a PNK phosphorylation reaction (Fig. 2, lane a). The efficiency of the label incorporation tended to be between 10 and 15% of the total [32P]ATP. In comparison to oligonucleotides 1–4 the yield was lower, thus indicating that the efficiency of the phosphorylation reaction might be dependent on the structure of the l-oligonucleotide. The yield for the corresponding 37mer in the d-configuration was in the range of 40–50%.
Figure 2.
Autoradiogram of a 10% denaturing polyacrylamide gel of the phosphorylation reaction of 6 (lane a) and the internal radioactive labeling using chemical ligation of 30mer l-DNA 5 and phosphorylated 37mer l-DNA 6 on a 33mer template 7 (lane b) with BrCN. XC, xylene cyanol.
The template-directed chemical ligation of the phosphorylated 37mer l-DNA fragment 6 and the 30mer l-DNA acceptor strand 5 with BrCN proceeded rapidly. The efficiency was determined to be 29 ± 4% based on the radioactivity of the full-length ligation product (67mer) compared to the input radioactivity of the phosphorylated 37mer l-DNA fragment 6 (Fig. 2, lane b).
To facilitate the dissociation of the template–product duplex 8–7 after the ligation reaction, a 33mer chimeric l-DNA/RNA oligonucleotide 7 containing three riboadenosines was used as a template. Under alkaline pH conditions and high temperature the chimeric template was hydrolyzed at the ribonucleotide positions (14) and the smaller template fragments were easily separated from the labeled product strand by PAGE. The resulting labeled 67mer Spiegelmer was extracted from the gel for further analyses.
To test the functionality of such a chemically ligated GnRH binding Spiegelmer, the BrCN ligation was performed with oligonucleotide 6 that was chemically 5′-phosphorylated during solid-phase synthesis. The resulting 67mer 8 was analyzed for its functionality to bind to l-GnRH using affinity gel capillary electrophoresis as a method to study oligonucleotide–ligand interactions (15–17).
In the capillary electrophoresis experiment without peptide the rMT of the two samples were in accordance with 1.562 for the chemically ligated 8 and 1.572 for the completely chemically synthesized oligonucleotide of the same sequence (Table 2). Under affinity gel capillary electrophoresis conditions the Spiegelmer peak split into two peaks, one nearly unaffected by the peptide and one with shorter rMT for the oligonucleotide–peptide complex that shifted to 1.507 for the ligate 8 and to 1.495 for the chemically synthesized 67mer (control). Although the oligonucleotide–peptide complex should have a higher size/charge ratio in affinity gel shift assays, earlier experiments revealed that in this case the oligonucleotide–peptide complex eluted unexpectedly at a shorter migration time than the free oligonucleotide (unpublished results). This behavior is probably due to a conformational binding fit of the large 67mer oligonucleotide (∼21 kDa) to the small GnRH. The predicted increase in size/charge ratio of the complex (∼+1 kDa with one positive charge for GnRH) was more than compensated by the decrease in size of the folded oligonucleotide.
Table 2. rMTs of the ligated 8 and a control Spiegelmer of the same sequence (completely chemically synthesized) in capillary gel electrophoresis shift experiments both with and without GnRH.
| rMT of ligated 8 | rMT of control | |
|---|---|---|
| – GnRH | 1.562 | 1.572 |
| + GnRH | 1.507 (60%) | 1.495 (64%) |
The experiments were performed on a BioCap Oligonucleotide Capillary (75 µM I.D. and 375 µM O.D.; Bio-Rad) at 30°C. The rMT values were calculated using the mobility time of an internal standard pdT20. The rMT of the Spiegelmer–peptide complex is indicated together with the percentage of the active conformation in percent.
The activity determined for the chemically ligated Spiegelmer (60% binding conformation, Table 2) is in accordance with the activity of the chemically synthesized control (64%). We therefore conclude that the internally labeled Spiegelmer retains its binding properties to GnRH and is consequently fully functional.
Our results demonstrate that l-oligodeoxynucleotides are substrates for T4 PNK. The phosphorylation yield is relatively independent on the nature of the 5′-terminal nucleotide. However, compared to shorter l-oligonucleotides the phosphorylation yield of longer and probably more structured l-oligonucleotides was reduced. Subsequently, the 5′- phosphorylated l-DNA-fragment was attached to another l-DNA oligonucleotide in a template-directed chemical ligation reaction, resulting in a ligation product with an internal 32P-label. It has been shown that the resulting full-length Spiegelmer retains its full functionality compared to an l-oligonucleotide control prepared by solid-phase synthesis. The methods presented for the preparation of 32P-internally labeled l-oligonucleotides provide a useful and convenient tool to detect and to analyze Spiegelmers in biological samples.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Marcus Janssen for performing the capillary electrophoresis assays, Thomas Schoetzau for helpful discussions and Michael Courtney and Lara Zilberkweit for critically reading the manuscript.
REFERENCES
- 1.Famulok M. and Mayer,G. (1999) Aptamers as tools in molecular biology and immunology. Curr. Top. Microbiol. Immunol., 243, 123–136. [DOI] [PubMed] [Google Scholar]
- 2.Usman N. and Blatt,L.M. (2000) Nuclease-resistant synthetic ribozymes: developing a new class of therapeutics. J. Clin. Invest., 106, 1197–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klussmann S., Nolte,A., Bald,R., Erdmann,V.A. and Fürste,J.-P. (1996) Mirror-image RNA that binds D-adenosine. Nat. Biotechnol., 14, 1112–1115. [DOI] [PubMed] [Google Scholar]
- 4.Leva S., Lichte,A., Burmeister,J., Muhn,P., Jahnke,B., Fesser,D., Erfurth,J., Burgstaller,P. and Klussmann,S. (2002) GnRH-binding RNA and DNA Spiegelmers—a novel approach towards GnRH antagonism. Chem. Biol., 9, 351–359. [DOI] [PubMed] [Google Scholar]
- 5.Wlotzka B., Leva,S., Eschgfäller,B., Burmeister,J., Kleinjung,F., Kaduk,C., Muhn,P., Hess-Stumpp,H. and Klussmann,S. (2002) In vivo properties of an anti-GnRH Spiegelmer: an example of an oligonucleotide-based therapeutic substance class. Proc. Natl Acad. Sci. USA, 99, 8898–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontanel M.-L., Bazin,H. and Teoule,R. (1993) 32P labeling of nonnucleosidic moieties 5′-attached to oligonucleotides. Anal. Biochem., 214, 338–340. [DOI] [PubMed] [Google Scholar]
- 7.Dolinnaya N.G., Sokolova,N.I., Ashirbekova,D.T. and Shabarova,Z.A. (1991) The use of BrCN for assembling modified DNA duplexes and DNA–RNA hybrids; comparison with water-soluble carbodiimide. Nucleic Acids Res., 19, 3067–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damha M.J., Giannaris,P.A., Marfey,P. and Reid,L-S. (1991) Oligodeoxynucleotides containing unnatural L-2′-deoxyribose. Tetrahedron Lett., 32, 2573–2576. [Google Scholar]
- 9.Williams K.P., Liu,X.H., Schumacher,T.N., Lin,H.Y., Ausiello,D.A., Kim,P.S. and Bartel,D.P. (1997) Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc. Natl Acad. Sci. USA, 94, 11285–11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Houten V., Denkers,F., van Dijk,M., van den Brekel,M. and Brakenhoff,R. (1998) Labeling efficiency of oligonucleotides by T4 polynucleotide kinase depends on 5′-nucleotide. Anal. Biochem., 265, 386–389. [DOI] [PubMed] [Google Scholar]
- 11.Focher F., Maga,G., Bendiscioli,A., Capobianco,M., Colonna,F., Garbesi,A. and Spadari,S. (1995) Stereospecificity of human DNA polymerases alpha, beta, gamma, delta and epsilon, HIV-reverse transcriptase, HSV-1 DNA polymerase, calf thymus terminal transferase and Escherichia coli DNA polymerase I in recognizing D- and L-thymidine 5′-triphosphate as substrate. Nucleic Acids Res., 23, 2840–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verri A., Focher,F., Priori,G., Gosselin,G., Imbach,J.L., Capobianco,M., Garbesi,A. and Spadari,S. (1997) Lack of enantiospecificity of human 2′-deoxycytidine kinase: relevance for the activation of beta-l-deoxycytidine analogs as antineoplastic and antiviral agents. Mol. Pharm., 51, 132–138. [DOI] [PubMed] [Google Scholar]
- 13.Verri A., Montecucco,A., Gosselin,G., Boudou,V., Imbach,J.L., Spadari,S. and Focher,F. (1999) L-ATP is recognized by some cellular and viral enzymes: does chance drive enzymic enantioselectivity? Biochem. J., 337, 585–590. [PMC free article] [PubMed] [Google Scholar]
- 14.Hausch F. and Jäschke,A. (2000) Multifunctional DNA conjugates for the in vitro selection of new catalysts. Nucleic Acids Res., 28, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamdan I., Skellern,G.G. and Waigh,R.D. (1998) Use of capillary electrophoresis in the study of ligand–DNA interactions. Nucleic Acids Res., 12, 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman A. and Cooke,N. (1991) Capillary gel affinity electrophoresis of DNA fragments. Anal. Chem., 63, 2038–2042. [DOI] [PubMed] [Google Scholar]
- 17.German I., Buchanan,D.D. and Kennedy,R.T. (1998) Aptamers as ligands in affinity probe capillary electrophoresis. Anal. Chem., 70, 4540–4545. [DOI] [PubMed] [Google Scholar]