Abstract
The methyl-CpG binding domain (MBD) family of proteins was defined based on sequence similarity in their DNA binding domains. In light of their high degree of conservation, it is of inherent interest to determine the genomic distribution of these proteins, and their associated co-repressor complexes. One potential determinant of specificity resides in differences in the intrinsic DNA binding properties of the various MBD proteins. In this report, we use a capillary electrophoretic mobility shift assay (CEMSA) with laser-induced fluorescence (LIF) and neutral capillaries to calculate MBD–DNA binding affinities. MBD proteins were assayed on pairs of methylated and unmethylated duplex oligos corresponding to the promoter regions of the BRCA1, MLH1, GSTP1 and p16INK4a genes, and binding affinities for each case were calculated by Scatchard analyses. With the exception of mammalian MBD3 and Xenopus MBD3 LF, all the MBD proteins showed higher affinity for methylated DNA (in the nanomolar range) than for unmethylated DNA (in the micromolar range). Significant differences between MBD proteins in the affinity for methylated DNA were observed, ranging within two orders of magnitude. By mutational analysis of MBD3 and using CEMSA, we demonstrate the critical role of specific residues within the MBD in conferring selectivity for methylated DNA. Interestingly, the binding affinity of specific MBD proteins for methylated DNA fragments from naturally occurring sequences are affected by local methyl-CpG spacing.
INTRODUCTION
Methylation at position 5 in cytosines is the most common modification of vertebrate genomes. In mammals, this modification occurs almost exclusively in CpG dinucleotides. In fact, 70% of all CpG dinucleotides are methylated in mammals, with the exception of CpG islands, which are CG-rich regions mostly coincident with the promoter of protein-coding genes. In some circumstances, many CpG islands become hypermethylated, resulting in gene silencing. For instance, promoter hypermethylation of tumor suppressor and DNA repair genes has been linked to cancer, and the study of methylation changes in cancer constitutes an important branch of study in cancer research (1). In this context, methyl-CpG binding proteins appear to be central players in the process of DNA methylation-dependent gene silencing (2). This family of proteins takes its definition from the methyl-CpG binding domain (MBD), the minimum portion with specific affinity for a single symmetrically methylated CpG pair. The MBD was characterized by deletion studies of MeCP2 (3). After the recognition of the MBD, four additional genes were found to contain this domain, namely MBD1, MBD2, MBD3 and MBD4 (4).
The biochemistry of MBD proteins has been studied at different levels. In general, all MBD proteins, except MBD4, have been reported to be associated with histone deacetylase subunits as parts of large multisubunit complexes. For instance, MBD3, the best characterized member of the MBD family from a biochemical point of view, has been determined as being a component of a complex called Mi-2/NURD that contains a chromatin-remodeling ATPase, a histone deacetylase and other subunits (5). In mammals, MBD2 recruits the MeCP1 histone deacetylase complex to DNA. The MeCP1 complex contains 10 major polypeptides, including MBD2 and all of the known NuRD components (6).
Some of the currently available data suggest that different MBD protein-containing complexes are targeted specifically to different methylated promoters (2), although it is possible that MBDs play no role in recognizing particular sequences. A few studies support the notion of selectivity in the association of MBD with particular promoters (7–9). However, the fact that MBD2 knockout mice are viable has been interpreted as if MBD proteins are at least partially redundant (10).
One of the still unresolved matters is the quantitative contribution of the MBD proteins to the association of their complexes with specific loci. While the MBD domain is highly conserved, seemingly minor sequence differences could produce alterations in affinity for methylated DNA. This could range from a high selectivity for methylated CpG sites to a complete lack of binding. Supporting this notion, DNase I footprinting of the MeCP2-MBD on DNA showed that the methyl-CpG site was ∼50% protected against DNase I at an MBD concentration of 1.25 × 10–9 M, indicating a dissociation constant close to this value (3). On the other hand, electrophoretic mobility shift asssy (EMSA) and southwestern analysis of mammalian MBD3 indicate that this protein cannot specifically recognize methylated DNA (4), in contrast to its amphibian homolog, which has been shown to bind methylated DNA selectively (5). Quantitative characterization of the binding to a single methyl-CpG pair of different MBD proteins remains a key issue in the evaluation of their contribution to the targeting to a promoter. Additional matters requiring further characterization are the influences of methyl-CpG density and local sequence context on binding. For instance, early results suggested that the MeCP1 complex, which was later demonstrated to contain MBD2 (11), requires at least 12 consecutive CpGs to bind methylated DNA (12). Although MBD2 has been demonstrated to recognize a single methylated CpG pair, it is possible that MBD2 prefers more densely methylated DNA. In other words, the density or distribution of methyl-CpGs may influence MBD2 binding. EMSAs and southwestern analysis are suitable and convenient techniques for qualitatively evaluating the association of MBDs with DNA. However, additional techniques need to be used to obtain reliable quantitative data on affinities between MBD proteins and methylated DNA. Capillary electrophoresis (CE) has become a useful technique for measuring binding constants. Compared with classical EMSA, in CE caging effects are avoided and the conditions are closer to the equilibrium; therefore, the fraction of protein–DNA complexes separated during the electrophoretic procedure is smaller than in EMSA (13).
To quantify protein–DNA binding affinities, we have used a modification of the high-performance capillary electrophoresis-based method called CEMSA (capillary electrophoretic mobility shift assay) (14). This modification, termed R-CEMSA (reverse capillary electrophoresis mobility shift assay), is a quantitative, simple and rapid method for calculating binding affinities that employs neutral-coating capillaries in order to avoid protein adsorption onto the capillary walls. We make use of this quantitative method to calculate the half-saturation concentration for DNA binding proteins to evaluate the intrinsic contribution of the binding properties of MBD proteins to their selective recruitment to promoters. Two issues have been addressed, namely whether different MBD proteins have significantly different binding affinities for a model methylated sequence, and whether an MBD protein is significantly affected by local sequence context and methyl-CpG density. Our results indicate that CpG distribution along the sequence may influence the interaction of each MBD protein with DNA.
MATERIALS AND METHODS
Reagents
DNAs, purchased as single-stranded oligonucleotides (Operon-Qiagen), were as follows: forward GAC, GAT CCG ACG ACG ACG ACG ACG ACG ACG ACG ACG ACG ACG ATC; reverse GAC, GAT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGT CGG ATC; forward GAM1 (monomethylated GAC), GAT CCG ACG ACG ACG ACG AXG ACG ACG ACG ACG ACG ACG ATC; reverse GAM1 (monomethylated GAC), GAT CGT CGT CGT CGT CGT CGT XGT CGT CGT CGT CGT CGG ATC, where X stands for 5-methyl-C; forward GSTP1, CCC TCC AGA AGA GCG GCC GGC GCC GTG ACT CAG CAC TGG GGC GGA GCG GG; reverse GSTP1, CCC GCT CCG CCC CAG TGC TGA GTC ACG GCG CCG GCC GCT CTT CTG GAG GG; forward MLH1, GAA CGT GAG CAC GAG GCA CTG AGG TGA TTG GCT GAA GGC ACT TCC GTT GA; reverse MLH1, TCA ACG GAA GTG CCT TCA GCC AAT CAC CTC AGT GCC TCG TGC TCA CGT TG; forward p16INK4a, GCG CTC GGC GGC TGC GGA GAG GGG GAG AGC AGG CAG CGG GCG GCG GGG AG; reverse p16INK4a, CTC CCC GCC GCC CGC TGC CTG CTC TCC CCC TCT CCG CAG CCG CCG AGC GC; forward BRCA1, AAA ACT GCG ACT GCG CGG CGT GAG CTC GCT GAG ACT TCC TGG ACG GGG GA; and reverse BRCA1, TCC CCC GTC CAG GAA GTC TCA GCG AGC TCA CGC CGC GCA GTC GCA GTT TT. Oligonucleotides corresponding to natural promoter CpG islands were obtained as fully methylated and unmethylated versions. In order to perform a systematic analysis of the influence of the methyl-CpG density and spacing on binding of MBD proteins, six different versions of the BRCA1 oligos were obtained: a non-methylated version, a monomethylated oligo with the methyl-CpG at the third CpG (BRCA1-M), and two different dimethylated oligos with methyl-CpG at the third and fourth CpGs (BRCA1-D1) or the third and sixth positions (BRCA1-D2). Finally, two different trimethylated oligos were obtained with the methyl-CpGs at positions 2, 3 and 4 (BRCA1-T1) or 1, 3 and 6 (BRCA1-T2) (Fig. 4E).
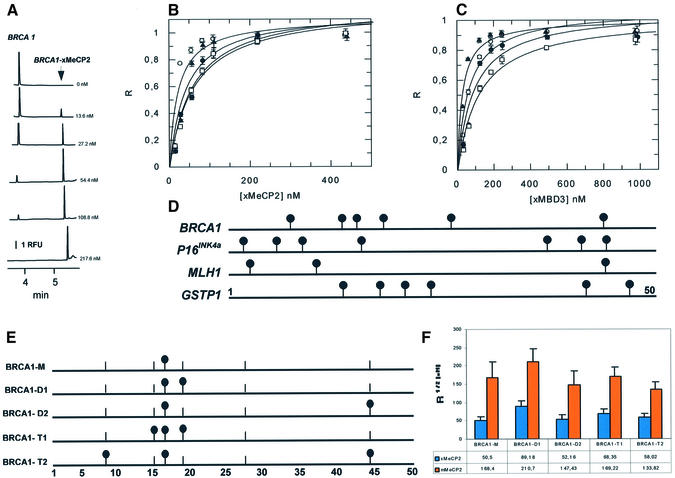
Figure 4.
Binding of xMeCP2 and MBD3 with synthetic methylated human CpG islands. (A) Electropherograms for mixtures of BRCA1 DNA oligo (24 nM) and increasing concentrations of xMeCP2 in 10 mM Tris–HCl pH 8.0, 3 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, 2 mM DTT, 5% glycerol and 0.4 mg/ml BSA. Analytical conditions as in Figure 2A. RFU, relative fluorescence units. (B) Single-site ligand binding fit for xMeCP2 with synthetic oligos corresponding to the promoter region of BRCA1 (white circles), p16INK4a(black circles), MLH1 (white squares) and GSTP1 (black triangles) using GraFit 3.1 software. R, saturation ([complex]/[complex] + [DNA]). Results are expressed as mean ± SD. Analytical conditions as described in Figure 2A. (C) Single-site ligand binding fit for xMBD3 with synthetic oligos corresponding to the promoter region of BRCA1, p16INK4a, MLH1 and GSTP1. Symbols as in (B). (D) Schematic representation of the location of methylcytosines (black circles) in the synthetic oligos BRCA1, p16INK4a, MLH1 and GSTP1. (E) Schematic representation of the location of methylcytosines (black circles) in the synthetic oligos BRCA1-M, BRCA1-D1, BRCA1-D2, BRCA1-T1 and BRCA1-T2. A base pair scale is shown at the bottom. A vertical bar lacking a black circle shows the location of an unmethylated CpG. (F) R1/2 values for Xenopus (x) (blue bar) and mammalian (m) (orange bar) MeCP2 and synthetic oligos BRCA1-M, BRCA1-D1, BRCA1-D2, BRCA1-T1 and BRCA1-T2 calculated by fitting the data obtained from R-CEMSA to non-cooperative binding behavior using GraFit software. Results are expressed as mean ± SD.
Forward oligonucleotides were labeled at their 5′ ends with 6-FAM. Complementary oligonucleotides were mixed at equimolar concentrations, and annealed by bringing the solution to 95°C and allowing it cool down slowly to room temperature.
Mouse MeCP2 was subcloned in pET23b from the IMAGE clone 4948925 using NdeI and XhoI.
Construction of MBD3 mutations
The coding sequence of mouse MBD3 was amplified from pET MBD3 (4) using the following primers: sense, CAT ATG GAG CGG AAG AGG TGG GAG; antisense, CTC GAG CAC TCG CTC TGG CTC CGG CTC. The amplification product was T/A cloned (Topo T/A, Invitrogen) and sequenced on both strands. The coding sequence was then subcloned into pET21a using NdeI and XhoI. Mutations were introduced into the resulting MBD3 pET21a clone by oligonucleotide-directed mutagenesis (QuickChange, Stratagene). The following mutagenic oligonucleotides were used: F34Y sense, GCC GGC CAC AGG GAT GTC TAT TAC TAT AGC CCC AGC; F34Y antisense, GCT GGG GCT ATA GTA ATA GAC ATC CCT GTG GCC GGC; H30K sense, GGG CTG TCG GCC GGC AAA AGG GAT GTC TTT TAC; H30K antisense, GTA AAA GAC ATC CCT TTT GCC GGC CGA CAG CCC; H30K, F34Y sense; GGG CTG TCG GCC GGC AAA AGG GAT GTC TAT TAC TAT AGC CCC AGC GGG; and H30K, F34Y antisense, CCC GCT GGG GCT ATA GTA ATA GAC ATC CCT TTT GCC GGC CGA CAG CCC. All mutagenized clones were verified by DNA sequencing.
Purification of recombinant protein
Recombinant mouse MeCP2, MBD2b and MBD3, and Xenopus MeCP2, MBD3 and MBD3 LF were expressed in Escherichia coli BL21 (DE3). A 500 ml aliquot of LB was inoculated with 5 ml of an overnight culture and incubated at 37°C to an A600 of 0.7. Induction was performed by the addition of isopropyl-β-d-thiogalactosidase to 0.5 mM and incubation at 37°C for a further 4 h. Cells were harvested and resuspended in 10 ml of extraction/wash buffer (50 mM sodium phosphate pH 7.0, 300 mM NaCl, 0.1% NP-40, 1 mM 2-mercaptoethanol). Purification of the soluble His-tagged protein was performed with TALON resin (Clontech), according to the manufacturer’s protocol. Protein was dialyzed against 20 mM Tris–HCl pH 8.0, 300 mM NaCl. Quantification was performed using the BioRad protein assay and routinely checked by 12% SDS–PAGE.
Gel mobility shift and southwestern assays
Binding reactions for mobility shift assays were performed in binding buffer as described by Wade et al. (5). 6-FAM-labeled DNAs were first radiolabeled. Increasing amounts of MBD proteins were added to 6-FAM-labeled DNAs in binding buffer [10 mM Tris-HCl pH 8.0, 3 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, 2 mM dithiothreitol (DTT), 5% glycerol and 0.4 mg/ml bovine serum albumin (BSA)] and incubated overnight at 4°C. Gel mobility shifts were performed in 10% polyacrylamide gels run in 0.5× TBE buffer (45 mM Tris pH 8.0, 45 mM boric acid, 1 mM EDTA) using GAC or GAM1 double-stranded oligonucleotide probes. One picomole of 6-FAM-labeled probe was mixed with purified recombinant protein, as indicated in the figure legends, in 10 mM Tris–HCl pH 8.0, 3 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, 2 mM DTT, 5% glycerol and 0.4 mg/ml BSA. The samples were incubated for 30 min at 37°C. A total of 30 pmol of competitor DNA (GAC or GAM1) was used per binding reaction. Gels were scanned on a PhosphorImager (Molecular Dynamics). The procedure used for southwestern assay was as described (15), except for the use of 6-FAM-labeled oligos.
Capillary electrophoresis mobility shift assay
A neutral-coating capillary (Beckman Coulter S.A.) (32.5 cm × 50 µm, effective length 20 cm) was used in a P/ACE MDQ capillary electrophoresis system (Beckman Coulter S.A.) connected to a Karat Software® data-processing station. The running buffer (40 mM Tris-borate, 0.95 mM EDTA, pH 8.0) was chosen to provide a low current when working at high voltage (30 kV, 923 V/cm) in order to maintain the stability of protein–DNA complexes during separation. Laser-induced fluorescence (LIF) was detected by excitation at 488 nm (3 mW argon ion laser provided by Beckman Coulter S.A.), and emissions were collected through a 520 nm emission filter (Beckman Coulter S.A.). Samples were injected under pressure (0.2 p.s.i.) for 2 s and the run temperature was maintained at 20°C. Before each run, the capillary was conditioned by washing with running buffer for 2 min. Buffers and running solutions were filtered through 0.2 µm pore-size filters. Three replicates of each concentration were prepared, and each was run twice.
Binding reactions were performed in binding buffer as described by Wade et al. (5). Increasing amounts of all MBDs were added to 6-FAM-labeled DNAs in binding buffer (10 mM Tris–HCl pH 8.0, 3 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, 2 mM DTT, 5% glycerol and 0.4 mg/ml BSA) and incubated overnight at 4°C.
Binding affinities were quantified by Scatchard analyses using GraFit 3.1 software (Fig. 2). In brief, the saturation of the oligo (R = [complex]/([complex] + [MBD])) was plotted against increasing quantities of each MBD protein. The concentration required for 50% saturation of binding (R½) was then calculated, seeking the best fit of the data to different binding models/curves.
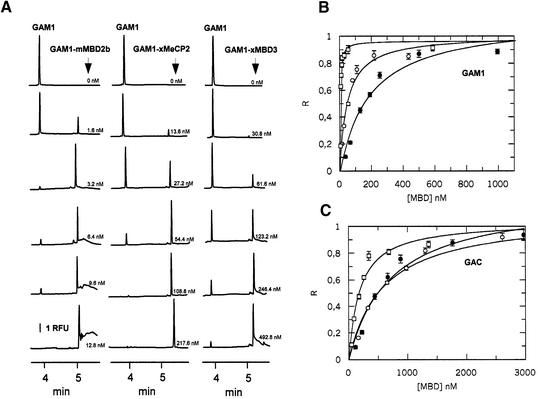
Figure 2.
Binding of MBD2b, xMeCP2 and MBD3 with methylated (GAM1) and unmethylated (GAC) synthetic oligos. (A) Electropherograms for mixtures of GAM1 (24 nM) and increasing concentrations of MBD2b, xMeCP2 and MBD3 in 10 mM Tris–HCl pH 8.0, 3 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, 2 mM DTT, 5% glycerol and 0.4 mg/ml BSA. Analytical conditions: 32.5 cm × 50 µm capillary (effective length 20 cm); low pressure injection at 0.2 p.s.i. for 2 s; 20°C; 30 kV voltage; reverse polarity (anode at the detector end); buffer, 40 mM Tris-borate, 0.95 mM EDTA pH 8.0. LIF detection: excitation at 488 nm, emission at 532 nm. RFU, relative fluorescence units. (B) Single-site ligand binding fit for mMBD2b (white squares), xMeCP2 (white circles) and xMBD3 (black circles) with GAM1 using GraFit 3.1 software. R, saturation ([complex]/[complex] + [DNA]). [MBD], methylation binding protein concentration. (C) Single-site ligand binding fit for mMBD2b (white squares), xMeCP2 (white circles) and xMBD3 (black circles) with GAC using GraFit 3.1 software. R, saturation ([complex]/[complex] + [DNA]). Results in (B) and (C) are expressed as the mean ± SD.
Molecular modeling
On the basis of the close homology between different MBD domains, models of murine MBD2- and MBD3- and Xenopus MBD3-binding domains were developed using the NMR structure of human MBD1 as reference, with the program Whatif (16), following the program manual.
MBD domain boundaries were identified after sequence alignment using ClustalX (17), and the BLDPIR routine of Whatif was used to build the models.
RESULTS AND DISCUSSION
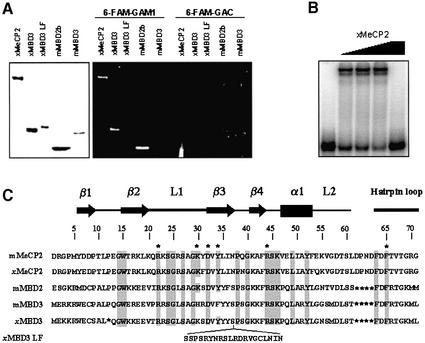
Prior to initiating quantitative DNA binding experiments using CEMSA, we performed classical DNA binding experiments (mobility shift assays and southwestern blot analysis) to determine whether introduction of a fluorescent probe altered MBD–DNA interactions (Fig. 1). Southwestern blots were performed with both unmethylated and methylated probes, (GAC and GAM1; described in Materials and Methods). In agreement with previous results (4,5), Xenopus MeCP2, Xenopus MBD3 and murine MBD2b selectively bind 6-FAM-GAM1 (Fig. 1A, central panel). In contrast, murine MBD3 and Xenopus MBD3 LF, a splice variant of MBD3, with its MBD disrupted by an insertion (Fig. 1C), showed no affinity for this probe (Fig. 1A, central panel). No binding to the unmethylated probe, 6-FAM-GAC, was observed in any case (Fig. 1A, right panel). Mobility shift assays produced results similar to the analysis by southwestern assays. As an example, MeCP2 binding to 6-FAM-GAM1 is shown (Fig. 1B).
Figure 1.
Checking MBD–DNA binding analysis with 6-FAM-labeled probes by standard methods. (A) The left panel shows a Coomassie Blue-stained gel with recombinant Xenopus MeCP2, Xenopus MBD3, Xenopus MBD3 LF, murine MBD2b and murine MBD3. The central and right panel depict a southwestern assay, with the same proteins, probed with the 6-FAM-labeled methylated oligo, GAM1 (central panel), and unmethylated oligo, GAC (right panel). (B) Mobility shift assay with xMeCP2 and 6-FAM-labeled GAM1 DNA probe. Binding reactions were performed as described in Materials and Methods. All lanes contain 6-FAM-labeled methylated GAM1. The first lane contains only the probe, without any added protein. The subsequent lanes contain 50, 100, 200 and 200 nM xMeCP2. GAM1 (30 pmol) was added in the last lane as competitor for binding. (C) Sequence alignment of the MBD of murine MeCP2, Xenopus MeCP2, murine MBD2 and MBD3, and Xenopus MBD3 and MBD3 LF. The positions of β-strands (arrows), loops (thick lines) and the α-helix (rectangle), defined by the solution structures of MeCP2 (19) and MBD1 (22), are indicated above the alignment. General numbering for MBDs is located above the sequence. Conserved residues are shaded, and those that proved to be essential for binding to methylated DNA are indicated with an asterisk.
MBD proteins bind to methylated DNA with significantly different affinities
To quantitate binding of each MBD to a single methylated CpG pair, we used a synthetic duplex oligo with 12 GAC repeats, where the central CpG is methylated. The methylated CpG pair is located at the central repeat in order to ensure that the MBD protein establishes all the contacts required for its correct positioning along the DNA. An unmethylated version of this oligonucleotide was also used to evaluate non-specific contributions of binding to DNA. Several proteins were analyzed, namely Xenopus MeCP2, MBD3 and MBD3 LF, and murine MeCP2, MBD2b and MBD3. Addition of MBD proteins to the 6-FAM-labeled duplex oligo GAM1 produced a discrete, sharp peak on capillary electrophoresis. Excellent resolution of free DNA and DNA–MBD complexes was obtained (Fig. 2A), with no gel matrix required, thus avoiding putative caging effects on the calculated binding affinities (18). Under these conditions, the migration time of the free oligo was ∼4 min while for the complex it was ∼5 min, regardless of the MBD protein type. The appearance and subsequent increase of the protein–DNA complex was concomitant with the decrease in the area of the peak corresponding to the free probe. BSA, used as a negative control, did not produce any retardation peak (not shown). A discrete complex also appeared when the unmethylated oligo GAC1 was used. However, the amount of MBD protein required to obtain a retardation peak was significantly greater.
Titrations were repeated three times for each of the MBD proteins. The results were plotted as saturation fraction against MBD protein concentration (Fig. 2B and C). The data follow classical saturation plots, and the protein concentrations required for 50% saturation of binding (R1/2) were calculated using GraFit software. We observed striking differences among MBD proteins, particularly in the case of mammalian MBD2b, which showed the highest affinity for methylated DNA (Table 1), 2.7 ± 0.8 nM. In contrast, Xenopus MBD3 showed a significantly lower affinity for GAM1, 186.5 ± 42.5 nM. Non-specific binding to the unmethylated versions of these oligonucleotides was also observed. In fact, addition of MBDs to the oligonucleotide resulted in the appearance of a discrete complex, similar to that observed with methylated DNA. However, the R1/2 values of the unmethylated oligos were all within the same micromolar range (Fig. 2C), indicating that the relative contribution of non-specific binding was significantly lower than that of specific interaction. Xenopus MBD3 LF failed to produce a retardation peak for either methylated or unmethylated DNA.
Table 1. Fractional saturation values.
| GAC | BRCA1 | p16INK4a | MLH1 | GSTP1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GAM1 (M) | GAC (U) | U/M | M | U | U/M | M | U | U/M | M | U | U/M | M | U | U/M | |
| xMeCP2 | 48.2 ± 8.5 | 555.9 ± 77.3 | 11.5 | 21.7 ± 7.7 | 394.6 ± 99.7 | 18.2 | 57.6 ± 15.4 | – | – | 61.3 ± 11.9 | 473.2 ± 117.6 | 7.7 | 43.8 ± 16.0 | 527.7 ± 106.1 | 12.0 |
| xMBD3 | 186.5 ± 42.5 | 778.6 ± 256.8 | 4.2 | 62.9 ± 15.9 | 555.0 ± 127.7 | 8.8 | 90.5 ± 15.4 | – | – | 121.6 ± 23.5 | 676.6 ± 250.3 | 5.6 | 28.8 ± 7.2 | 536.9 ± 162.7 | 18.6 |
| mMeCP2 | 171.8 ± 23.2 | 458.4 ± 88.2 | 2.7 | 161.3 ± 39.9 | 441.0 ± 138.5 | 2.7 | 150.5 ± 23.2 | – | – | 192.0 ± 49.1 | 640.0 ± 49.3 | 3.3 | 118.5 ± 25.3 | 349.2 ± 87.8 | 2.9 |
| mMBD3 | 579.5 ± 105.3 | 683.81 ± 124.3 | 1.1 | – | – | – | – | – | – | - | – | – | – | – | – |
| mMBD2b | 2.7 ± 0.8 | 188.7 ± 46.8 | 69.8 | 3.5 ± 1.5 | 200.4 ± 53.6 | 57.3 | 1.6 ± 0.2 | – | – | 7.9 ± 1.9 | 257.4 ± 125.4 | 32.5 | 1.09 ± 0.1 | 136.5 ± 37.3 | 125.2 |
| xMBDR LF | n.b. | n.b. | – | n.b. | n.b. | - | n.b. | – | – | n.b. | n.b. | – | n.b. | n.b. | – |
R1/2 values for Xenopus (x) and mammalian (m) MBD proteins and synthetic oligos (GAC, BRCA1, p16INK4a, MLH1 and GSTP1) in their methylated (M) and unmethylated (U) forms calculated by fitting the data obtained from R-CEMSA to non-cooperative binding behavior using GraFit software. Ratios of R1/2 unmethylated values and R1/2 methylated values are shown (U/M). Results are expressed as mean ± SD. n.b., no-retardation peak detected. Values are expressed in molar units and are multiplied by 109.
Interestingly, the comparison of binding properties of Xenopus MeCP2 versus murine MeCP2 indicates that the former has a higher affinity for the methylated probe than the latter, indicating that minor sequence differences can result in substantial changes in DNA binding properties. It is unlikely that differences in affinity could be attributed to the four changes within the MBD sequence, namely amino acid residues at positions 7, 31, 39 and 54 (Fig. 1C), since structural data (19) have not assigned an essential role for any of these residues. Instead, it is possible that differences in the binding properties of these two proteins could be due to their differences in the sequence outside the MBD.
We also analyzed the binding properties of mammalian MBD3. In agreement with previous observations (4), mMBD3 showed no ability to recognize methylated DNA selectively (see Fig. 3B and C). For both the unmethylated and methylated oligo, the retardation peak appeared only at micromolar concentrations and the R1/2 values were very similar, suggesting that MBD3 only interacts non-specifically with DNA.
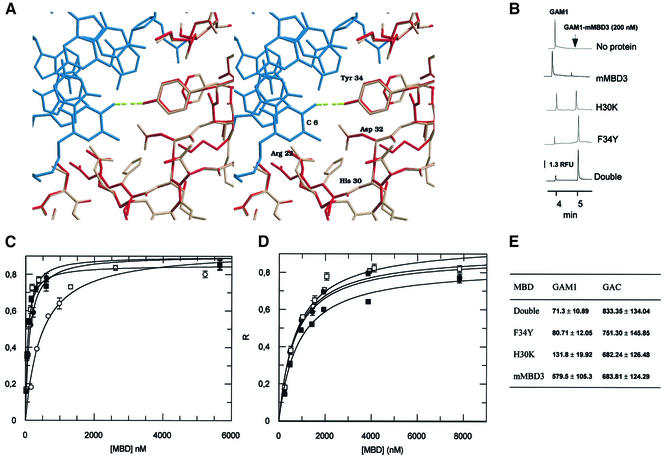
Figure 3.
Influence of F34Y and H30K mutations on the DNA binding affinity of murine MBD3. (A) Stereoview of the interaction between Tyr34 of human MBD1 (red) and the C6 amino group of the methylated DNA (blue); PDB entry 1IG4. The model of murine MBD3 (pink) has been superimposed on the MBD1 structure (red). The change of tyrosine/phenylalanine promotes the disruption of the hydrogen bond between the amino acid at position 34 and the methylated C6 in the major groove. The figure was produced using Raster3D (29). (B) Electropherograms for mixtures of GAC DNA oligo (24 nM) and 200 nM of wild-type mMBD3, H30K, F34Y and double mutant MBD3 in 10 mM Tris–HCl pH 8.0, 3 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, 2 mM DTT, 5% glycerol and 0.4 mg/ml BSA. Analytical conditions as in Figure 2A. RFU, relative fluorescence units. (C) Single-site ligand binding fit for mMBD3 (white circles), F34Y (black circles), H30K (white squares) and double mutant (black squares) with the synthetic oligo GAM1 using GraFit 3.1 software. R, saturation ([complex]/[complex] + [DNA]). Results are expressed as mean ± SD. Analytical conditions are as described in Figure 2A. (D) Single-site ligand binding fit for wild-type MBD3 (white circles), F34Y (black circles), H30K (white squares) and double mutant (black squares) with the synthetic oligo GAC using GraFit 3.1 software. R, saturation ([complex]/[complex] + [DNA]). Results are expressed as mean ± SD. Analytical conditions as described in Figure 2A. (E) R1/2 values for wild-type mMBD3, H30K, F34Y and double mutant MBD3 and synthetic oligos GAM1 and GAC forms calculated by fitting the data obtained from R-CEMSA to non-cooperative binding. Results are expressed as mean ± SD. Values are expressed in molar units and are multiplied by 109.
Amino acid substitutions alter the affinity of mammalian MBD3 for methylated DNA
Our results concerning the lack of selectivity to bind methylated DNA by murine MBD3 can be interpreted in terms of current structural data. On the basis of the close homology between different MBD domains, models of murine MBD2- and MBD3- and Xenopus MBD3 binding domains were produced with the program Whatif (16), taking the NMR structure of human MBD1 as reference. These structural models shed light on the binding mechanism of different proteins that contain MBDs. Furthermore, the models based on the NMR structure of MBD1 support the proposed molecular mechanism. In the case of mammalian MBD3, the absence of binding has been attributed to the presence of phenylalanine instead of a highly conserved tyrosine in position 34 (20). The disruption of the hydrogen bond between the hydroxyl group of Tyr34 and the amino group of C6 can be observed in the model of murine MBD3 (Fig. 3A). This change should not disturb important interactions such as that of the guanidinium group of Arg44 with the π-electron cloud of the aromatic ring of the amino acid in position 34, which ensures the correct positioning of the Arg44 side chain (Fig. 3A). However, some additional changes, due to the presence of histidine and arginine in positions 30 and 31 in the murine MBD3 sequence instead of the invariant lysine/arginine and serine conserved in other MBDs, might contribute to the loss of selectivity for methylated DNA.
To test our hypothesis, we prepared point mutations of murine MBD3 (described in Materials and Methods). In particular, we focused on Phe34 and His30, since these residues are the only two conserved in all MBD proteins except for mMBD3, and prepared single mutants for each of these residues and a double mutant form of MBD3. Recombinant proteins were produced and purified as described above. Binding properties of the mutants were tested by CEMSA with GAM1 and GAC oligos and compared with wild-type MBD3. In all three mutant forms, an increase in affinity for methylated DNA was observed. Figure 3B shows the appearance of the retardation peak at the same concentration of each MBD3 form. Titrations for each MBD3 form were performed as described above. The affinity for unmethylated probe was similar in all three mutants and comparable with wild-type mammalian MBD3 (Fig. 3D), indicating that the point mutations did not alter non-specific binding. In contrast, the R1/2 values for methylated DNA were decreased substantially (Fig. 3C). Both the F34Y and H30K mutants exhibited increased selectivity for the methylated substrate, with the F34Y mutant demonstrating marginally improved specificity. The double mutation (F34Y, H30K) resulted in a small improvement in differential interaction compared with the F34Y mutation alone (R1/2 value of 71.3 versus 80.71 nM; see Fig. 3C). These results suggest that the tyrosine hydroxyl interaction with cytosine is crucial for selective interaction with methylated DNA and can only be partially compensated for by the histidine to lysine change at position 30. Our results are in agreement with data recently published by Saito and Ishikawa (21), which also found that both residues H30 and F34 are responsible for the inability of mammalian MBD3 to bind mCpG. However, in contrast to their results, which indicate that H30K itself has no stimulatory effect on binding of MBD3 to methylated DNA, we observed that this single mutation is able to enhance the specific binding affinity for methylated DNA ∼4.5-fold. It is conceivable that this difference can be due to the different separation technique employed in each case.
Surprisingly, the point mutant forms of mMBD3 had better affinity for methylated DNA than Xenopus MBD3. A possible explanation for this is that xMBD3 has a deletion of the proline residue at position 12 (see Fig. 1C). The NMR structures of the MBDs of MeCP2 and MBD1 (19,22) show that this residue provides a sharp turn between β-strands 1 and 2, and thus contributes to the organization of the MBD fold. Mutations at the equivalent position (P101) in human MeCP2 have been correlated with Rett syndrome (23).
MBD proteins are influenced by both sequence features and methyl-CpG density
An additional experimental question of relevance for MBD–DNA interactions is whether the density of methyl-CpGs or local sequence context affect binding affinity. We designed oligos corresponding to four different CpG islands that become methylated in cancer (Fig. 4D). The selected sequences correspond to 50 bp portions of CpG islands of the BRCA1, GSTP1, p16INK4a and MLH1 genes. These oligos differ both in methyl-CpG density and in DNA sequence.
In CEMSA, we observed the appearance of retardation peaks concomitant with the decrease of the free DNA peak (Fig. 4A). Considering the resolution of this technique, the appearance of a single peak can be interpreted as resulting from the formation of a complex comprising one molecule each of DNA and of protein. The incorporation of additional molecules of MBD proteins should result in the appearance of additional peaks, as is apparent on the addition of excess amounts of xMeCP2 to the methylated BRCA1 DNA (Fig. 4A). We therefore interpreted our results as representing the formation of a single complex, even when more than one CpG pair was present for each oligo.
A subset of the primary data for these experiments is shown in Figure 4; Table 1 shows the R1/2 values for each MBD on each fragment. In general, the R1/2 values for methylated DNA followed the pattern observed on the model GAM1 substrate. MBD2b had the strongest affinity for the substrate, reaching half-saturation at single digit nanomolar concentrations (Table 1). xMeCP2, mMeCP2 and xMBD3 reach half-saturation on methylated substrates at somewhat higher concentrations, ranging from 20 to 120 nM (Fig. 4 and Table 1). The ability of a given MBD to discriminate between unmethylated and methylated versions of the same DNA fragment can be determined by calculating the ratio of the R1/2 values for unmethylated DNA divided by the same value for methylated DNA. MBD2b displays the greatest capacity to differentiate, with 30- to 130-fold differences between half-saturation values for unmethylated versus methylated DNA. In contrast, xMeCP2 and xMBD3 displayed only 5- to 20-fold differences. Curiously enough, mMeCP2 only showed around 3-fold differences between half-saturation values for unmethylated versus methylated oligos. The molecular basis for this increased ability to differentiate between MBD2b and the other three proteins currently is unclear. It has to be mentioned that MBD2b is a minor fraction of the total MBD2 protein. The major form of MBD2, MBD2a, contains an N-terminal extension of 152 amino acids proximal to the MBD which may significantly affect the binding affinity of the protein.
Two variables determine methylation density, the absolute number of methyl-CpG pairs and how tightly they are clustered on a given substrate. The DNA fragments used in this study have three (MLH1), six (BRCA1 and GSTP1) or seven (p16INK4a) methyl-CpG dinucleotides. MBD2b has roughly equivalent affinity for the p16INK4a and GSTP1 fragments, with the lowest affinity for MLH1, the fragment with the fewest methyl-CpG pairs (Table 1). xMeCP2, on the other hand, had the highest affinity for the BRCA1 fragment, followed by GSTP1, p16INK4a and MLH1. Finally, xMBD3 preferred GSTP1, followed by BRCA1, p16INK4a and MLH1. It is thus apparent that the absolute number of CpGs affects binding affinity, as expected. MLH1 has roughly half the number of methyl-CpGs and is the poorest substrate for all three proteins.
It is informative to compare the R1/2 values of these MBD proteins for the two fragments with the same number of methyl-CpGs, GSTP1 and BRCA1. MBD2b, mMeCP2 and xMBD3 bind more avidly to GSTP1, while xMeCP2 binds better to BRCA1 (Table 1). Differences in affinity for these two fragments with the same number of CpGs could be due to two different effects, local sequence or CpG distribution.
In order to assess the relative effects of methyl-CpG spacing and density in the context of a natural sequence in a more systematic manner, we prepared five different oligos with the BRCA1 sequence in which one, two or three of its CpG dinucleotides were methylated with different spacing (Fig. 4E). Figure 4F summarizes the results obtained with the five differentially methylated BRCA1 oligos and the two versions of MeCP2. We observed that xMeCP2 and mMeCP2 have a lower affinity for any of the partially methylated BRCA1 oligos than the affinity observed for the fully methylated BRCA1 oligo (Table 1). This result supports the notion that the absolute number of CpGs affects binding. Furthermore, although only small differences were observed among the different partially methylated BRCA1 oligos, ranging within 2-fold, the behavior of both Xenopus and mouse MeCP2 was similar, suggesting that these differences were significant. The poorest affinity observed for both xMeCP2 and mMeCP2 was obtained with the dimethylated oligo with the shortest spacing (BRCA1-D1). It is possible that the existence of an additional methyl-CpG within the 12 bp protected by the MBD (24) acts as a discouraging feature for binding.
Finally, our results do not allow local sequence effects on affinity to be ruled out. However, the binding affinities of MeCP2 for monomethylated oligos (GAM1 and BRCA1-M) and trimethylated oligos (MLH1 and BRCA1-T2) were virtually identical regardless of the sequence, suggesting that at least in the above examples no sequence effects occur.
The major conclusions of this work are consistent with the likelihood that, at least in some instances, the binding properties of a given MBD protein can influence its targeting to a specific locus. We have observed that small sequence differences in the MBD domain are manifested in rather striking changes in DNA recognition properties. The finding that the F34Y mutant version of mMBD3 selectively recognizes methylated DNA significantly better than xMBD3 strongly supports this notion. We also observed that, on naturally occurring sequences, increases in the total number of methyl-CpG pairs resulted in increased binding affinity. Of course, increasing the total number of methyl-CpGs per fragment serves to increase the number of available binding sites. Most interesting of all, there appears to be an additional determinant of binding affinity related to either the spacing of methyl CpGs or to local sequence context. Whatever the molecular determinants of the differences observed in this work, they suggest that MBD proteins are likely to be differentially localized within the genome, at least in part, as a direct reflection of their inherent binding preferences.
Obviously, additional factors associated with MBD complexes should be involved in their specific recruitment to particular promoters. Therefore, based on our data, it seems that all potential substrates will bind MBD2 in preference to other MBD proteins. However, this is not the situation in vivo. Additional factors (the N-terminal extension in MBD2a, other proteins complexed to the MBD proteins; chromatin context) may well have effects in vivo. The current results concerning the targeting of MBD to hypermethylated promoters support this model. In fact, the specific association of the MBD complex with diverse loci has been demonstrated in a few cases (7–9). Although determinants of specific targeting remain unknown, sequence-specific transcription factors are likely to be involved in the selective recruitment of MBD-containing complexes by interaction with any of their subunits. For instance, Drosophila Mi-2, a component of a fly MBD-containing complex, interacts with the Hunchback transcriptional repressor, which binds directly to regulatory sequences of HOX genes (25). Also, the zinc finger protein Ikaros interacts with Mi-2 in erythroid cells (26). The observation that dMBD-like, the Drosophila MBD2/3 homolog, which is unable to bind methylated DNA, is associated with discrete loci suggests that this protein is targeted within the genome by interactions with sequence-specific DNA binding proteins (27).
The overall picture can accommodate not only specific targeting but also partial redundancy. Fine-scale regulation of the expression levels of MBD proteins may contribute to the selection of the MBD complex involved in the repression at specific sites (28). Differences in the affinity of different MBD proteins for methylated DNA, as demonstrated in the current study, must surely play a role in the selective recruitment of MBDs to promoters.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by I+D+I project SAF 2001-0059 and the International Rett Syndrome Association. M.F. and E.B. are funded by a University educational staff Postdoctoral Fellowship and the Ramón y Cajal Program, respectively. P.T. and P.W. were supported by research grants from the National Institute of Child Health and Human Development and from the Rett Syndrome Research Foundation. P.W. gratefully acknowledges patronage of the Massachusetts Rett Syndrome Association.
REFERENCES
- 1.Esteller M., Corn,P.G., Baylin,S.B. and Herman,J.G. (2001) A gene hypermethylation profile of human cancer. Cancer Res., 61, 3225–3229. [PubMed] [Google Scholar]
- 2.Ballestar E. and Wolffe,A.P. (2001) Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem., 268, 1–6. [DOI] [PubMed] [Google Scholar]
- 3.Nan X., Meehan,R.R. and Bird,A. (1993) Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res., 21, 4886–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrich B. and Bird,A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- 6.Feng Q. and Zhang,Y. (2001) The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev., 15, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magdinier F. and Wolffe,A.P. (2001) Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc. Natl Acad. Sci. USA, 98, 4990–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Osta A., Kantharidis,P., Zalcberg,J.R. and Wolffe,A.P. (2002) Precipitous release of methyl-CpG binding protein 2 and histone deacetylase 1 from the methylated human multidrug resistance gene (MDR1) on activation. Mol. Cell. Biol., 22, 1844–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakker J., Lin,X. and Nelson,W.G. (2002) Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J. Biol. Chem., 277, 22573–22580. [DOI] [PubMed] [Google Scholar]
- 10.Hendrich B., Guy,J., Ramsahoye,B., Wilson,V.A. and Bird,A. (2001) Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev., 15, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng H.H., Zhang,Y., Hendrich,B., Johnson,C.A., Turner,B.M., Erdjument-Bromage,H., Tempst,P., Reinberg,D. and Bird,A. (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature Genet., 23, 58–61. [DOI] [PubMed] [Google Scholar]
- 12.Lewis J.D., Meehan,R.R., Henzel,W.J., Maurer-Fogy,I., Jeppesen,P., Klein,F. and Bird,A. (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell, 69, 905–914. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka Y. and Terabe,S. (2002) Estimation of binding constants by capillary electrophoresis. J. Chromatogr. B, 768, 81–92. [DOI] [PubMed] [Google Scholar]
- 14.Foulds G.J. and Etzkorn,F.A. (1998) A capillary electrophoresis mobility shift assay for protein–DNA binding affinities free in solution. Nucleic Acids Res., 26, 4304–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballestar E., Yusufzai,T.M. and Wolffe,A.P. (2000) Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry, 39, 7100–7106. [DOI] [PubMed] [Google Scholar]
- 16.Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- 17.Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- 18.Fried M. and Crothers,D.M. (1981) Equilibria and kinetics of lac repressor–operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res., 9, 6505–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakefield R.I., Smith,B.O., Nan,X., Free,A., Soteriou,A., Uhrin,D., Bird,A.P. and Barlow,P.N. (1999) The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol., 291, 1055–1065. [DOI] [PubMed] [Google Scholar]
- 20.Ohki I., Shimotake,N., Fujita,N., Jee,J., Ikegami,T., Nakao,M. and Shirakawa,M. (2001) Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell, 105, 487–497. [DOI] [PubMed] [Google Scholar]
- 21.Saito M. and Ishikawa,F. (2002) The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J. Biol. Chem., 277, 35434–35439. [DOI] [PubMed] [Google Scholar]
- 22.Ohki I., Shimotake,N., Fujita,N., Nakao,M. and Shirakawa,M. (1999) Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J., 18, 6653–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheadle J.P., Gill,H., Fleming,N., Maynard,J., Kerr,A., Leonard,H., Krawczak,M., Cooper,D.N., Lynch,S., Thomas,N., Hughes,H., Hulten,M., Ravine,D., Sampson,J.R. and Clarke,A. (2000) Long-read sequence analysis of the MECP2 gene in Rett syndrome patients: correlation of disease severity with mutation type and location. Hum. Mol. Genet., 9, 1119–1129. [DOI] [PubMed] [Google Scholar]
- 24.Nan X., Meehan,R.R. and Bird,A. (1993) Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res., 21, 4886–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehle J., Beuchle,D., Treuheit,S., Christen,B., Kennison,J.A., Bienz,M. and Muller,J. (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science, 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill D.W., Schoetz,S.S., Lopez,R.A., Castle,M., Rabinowitz,L., Shor,E., Krawchuk,D., Goll,M.G., Renz,M., Seelig,H.P., Han,S., Seong,R.H., Park,S.D., Agalioti,T., Munshi,N., Thanos,D., Erdjument-Bromage,H., Tempst,P. and Bank,A. (2000) An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol., 20, 7572–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballestar E., Pile,L.A., Wassarman,D.A., Wolffe,A.P. and Wade,P.A. (2001) A Drosophila MBD family member is a transcriptional corepressor associated with specific genes. Eur. J. Biochem., 268, 5397–5406. [DOI] [PubMed] [Google Scholar]
- 28.Billard L.M., Magdinier,F., Lenoir,G.M., Frappart,L. and Dante,R. (2002) MeCP2 and MBD2 expression during normal and pathological growth of the human mammary gland. Oncogene, 21, 2704–2712. [DOI] [PubMed] [Google Scholar]
- 29.Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]