Abstract
Mitochondrial transcription factor A (TFAM), a member of the high mobility group proteins, is essential for maintenance of mitochondrial DNA (mtDNA). Most TFAM and mtDNA (both of which are normally soluble) was recovered from the particulate fraction of human placental mitochondria when extracted with the non-ionic detergent Nonidet P-40. mtDNA and TFAM were co-immunoprecipitated by anti-TFAM antibodies. TFAM was released into the supernatant by DNase I digestion of mtDNA in the particulate fraction. Thus, TFAM and mtDNA are tightly associated with each other, and it is likely that few TFAM or mtDNA molecules exist in an unbound form in mitochondria. Based on the fact that TFAM is abundant enough to wrap mtDNA entirely, these results suggest that human mtDNA is packaged with TFAM.
INTRODUCTION
Mitochondrial transcription factor A (TFAM) was first purified and cloned as a transcription factor for mitochondrial DNA (mtDNA) (1,2). TFAM shows a high affinity to the light and heavy strand promoters, LSP and HSP, respectively (1,2). TFAM indeed enhances mtDNA transcription by mitochondrial RNA polymerase in a promoter-specific fashion in the presence of a mitochondrial transcription factor B (3,4). Because replication of mammalian mtDNA is proposed to be coupled with transcription (5), TFAM is thought to be essential for replication of mtDNA. Consistent with this notion, targeted disruption of the mouse TFAM gene is an embryonic lethal mutation causing depletion of mtDNA (6). TFAM is a member of the high mobility group (HMG) proteins and contains two HMG-box domains. Many HMG-family proteins are capable of binding, wrapping, bending and unwinding DNA regardless of sequence specificity (7–9). Abf2p, a TFAM homolog of Saccharomyces cerevisiae, is also an HMG-family protein. Abf2p abundantly exists in mitochondria, every 15 bp of mtDNA (10). Unlike mammalian TFAM, Abf2p is not required for transcription initiation of mtDNA (10). Saccharomyces cerevisiae devoid of Abf2p loses mtDNA when cultured in the presence of fermentable carbon sources (11), but this mtDNA depletion is rescued by a bacterial histone-like protein HU (12), suggesting that Abf2p maintains mtDNA as an architectural factor. Because TFAM can substitute for Abf2p as well (2), human TFAM may share common properties with Abf2p and HU. Despite of its ability to package mtDNA, TFAM has not been considered to package mtDNA in human mitochondria because its amount was estimated to be only 15 molecules per mtDNA (1).
mtDNA is postulated to have a nucleoid structure in lower eukaryotes, S.cerevisiae (13–15) and Physarum polycephalum (16). However, its structure is not well elucidated at a molecular level. Recently, human mtDNA was also proposed to exist in a nucleoid structure, but the proposal is simply based on a dotted pattern of mtDNA staining (17,18). The human mtDNA nucleoid has never been isolated and thus its structure is much less characterized than those of lower eukaryotes. Recently, we have reported that human TFAM is abundant enough to wrap entire mtDNA (19). In this study, we show that most TFAM is indeed associated with mtDNA and thus organizes a protein–DNA complex. The misunderstanding that mtDNA is rather naked has not been experimentally denied in mammals to date. The results in this study are the first to experimentally show that human mtDNA is far from naked. Also, we propose that TFAM functions as a main constitutive factor of nucleoid structure in mammals. In addition, we discuss the association of the mtDNA/TFAM complex with mitochondrial membranes.
MATERIALS AND METHODS
Materials and miscellaneous methods
Anti-human TFAM antibodies were raised by immunizing rabbits with recombinant GST-TFAM fusion protein (20). The antibodies were purified using an affinity column in which histidine-tagged TFAM protein (20) was immobilized on the gel using an UltraLink™ kit (Pierce). Anti-human mitochondrial single-stranded DNA-binding protein (mtSSB) and anti-human p32 were as described previously (19,21). Anti-human voltage-dependent anion channel 1 (VDAC) and anti-iron sulfur protein of complex II were produced by immunizing rabbits with C-terminal peptides of these proteins, GHKLGL GLEFQA and TYKEKKASV, respectively. These two antibodies were affinity-purified using the isolated peptides. Anti-60-kDa heat shock protein (HSP60) monoclonal antibodies were from StressGen Biotechnologies Corp. (Victoria, BC, Canada). ExTaq DNA polymerase and DNase I were from Takara (Seta, Japan). Non-ionic detergents, Nonidet P-40 (NP-40) was from Wako (Kyoto, Japan).
For immunoprecipitation, the affinity-purified anti-TFAM antibodies or control IgG were immobilized on magnetic beads (tosylactivated Dynabeads® M-280, Dynal) (0.75 µg of IgG/107 beads) according to the manufacturer’s instructions. Proteins were quantified using a DC protein assay kit (BioRad Laboratories). Bovine serum albumin was used as a standard. Sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and western blotting were performed as described previously (19).
Disruption of mitochondrial membranes
Human placental mitochondria were prepared by a combination of differential and Percoll gradient centrifugation (22). Mitochondria were solubilized essentially according to the method of Miyakawa et al. (13). All solutions contained 1× concentration of a protease inhibitor mix, CompleteMini™ (Roche). Briefly, mitochondria (5 mg protein/ml) were solubilized in 2 ml of TES buffer (0.25 M sucrose, 10 mM Tris–HCl, pH 7.0 and 1 mM EDTA) containing 0.5% NP-40. After incubation for 30 min at 4°C with shaking, the mitochondria were centrifuged for 30 min at 10 000 g at 4°C and separated into a supernatant (S1) and a pellet. The pellet was suspended in 2 ml of the same solubilizing buffer and homogenized with a Potter–Elvehjem homogenizer (P1). The particulate fraction P1 was centrifuged as above. The second supernatant was designated S2. The second pellet was suspended in 2 ml of buffer, homogenized as above, and designated P2. Approximately 20% of mitochondrial proteins were recovered typically from the P2 fraction. Western blotting analysis was performed using 2.5 µl of the preparations.
The P2 fraction (1.0 ml) was mixed with an equal volume of TEN buffer (10 mM Tris–HCl, pH 7.0, 1 mM EDTA and 150 mM NaCl) containing 80% sucrose (w/v) and placed at the tube bottom. Then, stepwise sucrose gradients (5∼30% in TEN buffer, 1 ml each) were layered on top. The gradient steps were serially reduced by 2.5% from the bottom to top. The tube was centrifuged in a Beckman SW41Ti rotor for 15 h at 39 000 r.p.m. (200 000 g) at 4°C. After centrifugation, 1 ml samples were collected from the top down. The pellet at the bottom was homogenized in 1 ml of TES/0.5% NP-40 and designated P3. For western blotting, 5 µl of each fraction was used.
Mitochondria of human U937 cells were prepared by a combination of differential and sucrose gradient centrifugation (23). The mitochondria were sonicated in TES buffer (1 mg/ml) and then centrifuged for 1 h in a Hitachi 120AT rotor at 80 000 r.p.m. (238 000 g) at 4°C. The soluble fraction was recovered. The pellet was homogenized in 1 vol of TES buffer for further analysis. The soluble fraction (200 µl) was applied to a Superose 12HR column (10 × 300 mm, Amersham) and separated using an FPLC system (Amersham) at a flow rate of 0.8 ml/min at 4°C. A fraction was collected every 1 min. For western blotting 20 µl samples were used.
Immunoprecipitation
P2 (240 µg protein) was incubated for 2 h with rotation at 4°C in 1.7 ml of a mixture containing 10 mM Tris–HCl, pH 7.0, 150 mM NaCl, 100 µg/ml bovine serum albumin, 0.5% NP-40, antibody-immobilized magnetic beads (corresponding to 8 µg IgG) and 1× concentration of a protease inhibitor mix, CompleteMini™. The beads were pelleted on the wall of a tube using a magnet. The supernatant was mixed with an equal volume of 2× SDS sample buffer (125 mM Tris–HCl, pH 6.8, 10% glycerol, 4% SDS and 2% 2-mercapthoethanol) and heated for 5 min at 95°C (IP-sup). The beads were washed three times with 1 ml of TBS/NP-40 (10 mM Tris–HCl, pH 7.0, 150 mM NaCl and 0.5% NP-40). Proteins were eluted from the beads in 340 µl of 1× SDS sample buffer without 2-mercapthoethanol by heating for 5 min at 95°C. The beads were pelleted using a magnet and the resulting solution was recovered (IP-pellet). For western blotting, 40 and 4 µl of IP-sup and IP-pellet, respectively, were used.
Immunocytochemistry
HeLa MRV11 cells (23) were cultured on cover slips in 3.5 cm dishes in 2 ml of DMEM (Sigma) containing 10% fetal calf serum. The cells were incubated in the presence of 100 nM Mitotracker Red CMXRos (Molecular Probes) for 20 min. After washing with PBS (10 mM sodium phosphate, pH 7.4 and 150 mM NaCl) three times, the cells were fixed with 2 ml of acetone/methanol (50/50, v/v) for 5 min. After washing with PBS three times, the fixed cells were incubated with 250-fold diluted anti-TFAM affinity-purified antibodies (4 µg/ml) in PBS for 1 h. After washing, the cells were incubated with 250-fold diluted Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes) for 30 min. Fluorescence images were taken using a Radiance 2000 confocal laser microscope (BioRad Laboratories).
Polymerase chain reaction (PCR) detection of mtDNA
Because of difficulties with DNA extraction efficiency, we amplified an mtDNA fragment without extracting DNA. Briefly, 10 µl samples were mixed with 10 µl of 2× digestion buffer (100 mM Tris–HCl, pH 8.0, 2 mM EDTA, 2.0% SDS and 2 mg/ml proteinase K) and incubated for 30 min at 55°C. After the addition of 180 µl of distilled water, the mixtures were further incubated for 10 min at 100°C to inactivate proteinase K. Then the mixtures were cooled down to room temperature, vortexed, and centrifuged for 10 min at 10 000 g. The supernatants were diluted 10-fold with distilled water (thus final samples were 200-fold diluted). One microliter of the DNA samples was used in 20 µl of the reaction mixture for amplification of an mtDNA region (nucleotides 7319–7603). The primers used were: mt3, ttgagaagccttcgcttcgaag; mt4, ttgcgctfcatgtgccattaag. Twenty-five cycles of DNA amplification were performed: DNA denaturing at 94°C for 30 s, annealing at 60°C for 30 s, and DNA synthesis at 72°C for 45 s for each cycle.
DNase I digestion
P2 (36 µg protein) was incubated in 50 µl of a reaction mixture containing 40 mM Tris–HCl, pH 7.5, 8 mM MgCl2, 5 mM dithiothreitol, 0.5% NP-40 and 15 U DNase I for 45 min at 4°C. The reaction mixture was centrifuged for 30 min at 10 000 g at 4°C. The supernatant was mixed with an equal volume of the 2× SDS sample buffer. The pellet was solubilized with 100 µl of the 1× SDS sample buffer. Then, the samples were heated for 5 min at 95°C.
RESULTS
Insolubility of TFAM and mtDNA
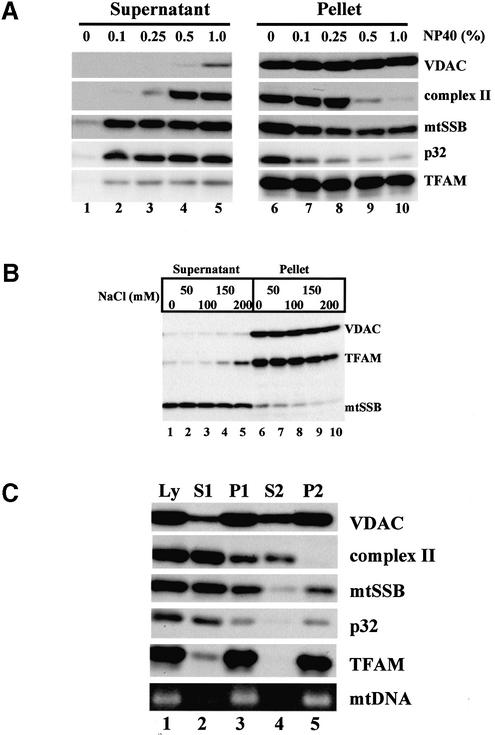
Human placental mitochondria were treated with various concentrations of the non-ionic detergent NP-40, and then separated into supernatants and particulate fractions (Fig. 1A). mtSSB and p32, a matrix protein (21), were largely released into the soluble fraction at 0.1% NP-40 (Fig. 1A, lanes 2 and 7), indicating that the outer and inner membranes were substantially permeabilized. Nevertheless, most TFAM, like the iron sulfur protein of complex II that peripherally attaches to mitochondrial inner membranes, remained in the particulate fraction. At 0.5 and 1.0% NP-40, the iron sulfur protein was released into the supernatants and VDAC, an integral outer membrane protein, was partially released (Fig. 1A, lanes 4, 5, 9 and 10). Thus, at these concentrations, the mitochondrial membranes were substantially, if not completely, solubilized. However, under these conditions, most TFAM molecules were still recovered from the particulate fractions. To examine the effects of ionic strength, we added NaCl to the buffer (Fig. 1B). Because TFAM was not released into the supernatant by the addition of 150 mM NaCl (Fig. 1B, lanes 4 and 9), the association of TFAM with the particulate fractions was not caused by the low ionic strength. Considering that TFAM itself is a soluble protein, mitochondria seem to contain surprisingly few unbound or free TFAM molecules. A small part of mtSSB and p32 remained in the particulate fraction even at 1.0% NP-40 (Fig. 1A, lanes 5 and 10). To examine if the remaining portions were due to artificial entrapment by the pellets, we re-suspended the first 0.5% NP-40 particulate fraction (P1) in the same buffer containing 0.5% NP-40, homogenized, and separated it again into supernatant (S2) and pellet (P2). The iron sulfur protein in P1 was completely removed by this washing from P2, but the levels of mtSSB and p32 remained nearly the same (Fig. 1C, lanes 4 and 5), suggesting that this small portion of mtSSB and p32 is indeed associated with the particulate fraction. Because mtSSB is a soluble protein, the mtSSB in the particulate fractions would be bound to the single-stranded region of mitochondrial D-loops (19).
Figure 1.
Solubilization of human placental mitochondria. (A) Mito chondria were incubated with various concentrations of NP-40 and then separated into supernatants and pellets. The pellets were re-suspended in the original volume of buffer for easily estimating recovery of proteins. Each protein was detected by western blotting. (B) Mitochondria were incubated with 0.5% NP-40 in the presence of various concentrations of NaCl. Then, the mixtures were separated into supernatants and pellets. (C) Mitochondria were incubated with 0.5% NP-40 (Ly), and separated into the first supernatant (S1) and pellet. The first pellet was re-suspended in the original volume of buffer (P1). The P1 was centrifuged and separated into the second supernatant (S2) and pellet. The second pellet was similarly re- suspended in the original volume of buffer (P2). An mtDNA fragment (np 7319–7603) in each sample was amplified by PCR (lowest row).
Association of TFAM with mtDNA
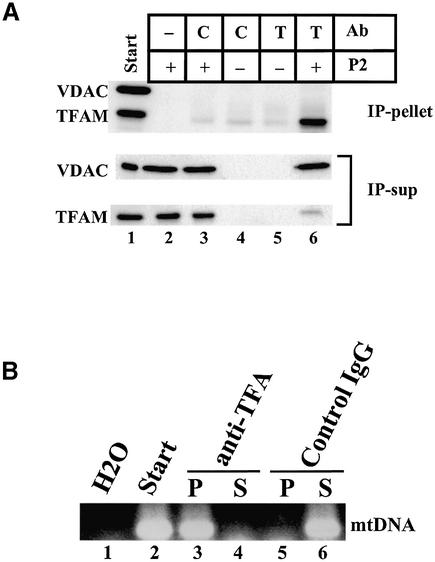
TFAM is a member of the HMG protein family and is a non-specific DNA-binding protein. Therefore, TFAM is likely to interact with mtDNA. We detected mtDNA by a PCR method. As shown in Figure 1C, most mtDNA was recovered from the particulate fractions (lanes 3 and 5 in the bottom row). Even after we further separated the NP-40 insoluble fraction by stepwise sucrose-density gradient centrifugation, TFAM and mtDNA were recovered from the same pellet fraction in 45% sucrose but not in the other fractions (data not shown). As long as we use centrifugation methods, we cannot rule out the simple co-sedimentation of TFAM and mtDNA. To confirm that TFAM and mtDNA are associated with each other, we immunoprecipitated the NP-40 insoluble fraction (P2) using anti-TFAM antibodies. Almost all TFAM was immunoprecipitated by the antibodies (Fig. 2A, lane 6). Non-specific signals were observed in lanes 3–5. Because those signals were observed in the absence of P2 (lanes 4 and 5) but not in the absence of IgG (lane 1), it is likely that IgG light chains detached from the beads migrated to the same position as TFAM and were detected by the secondary anti-IgG antibody. Under these conditions, most mtDNA (Fig. 2B) was co-immunoprecipitated by anti-TFAM, suggesting that most mtDNA is associated with TFAM. No immunoprecipitation of VDAC (Fig. 2A, lane 6) neglects that outer membranes were non-specifically immunoprecipitated. Digestion of mtDNA with DNase I in the particulate fraction released most TFAM into the supernatant (Fig. 3), suggesting that essentially all TFAM is associated with mtDNA.
Figure 2.
Immunoprecipitation by anti-TFAM. P2 samples were incubated with rotation in the presence of antibody-immobilized magnetic beads. The beads were pelleted using a magnet. The resulting supernatants (IP-sup) were removed. After washing three times, proteins were eluted from the beads by heating (IP-pellet). C, control IgG; T, anti-TFAM. (A) VDAC and TFAM were detected by western blotting. In lane 2, unconjugated magnetic beads were used. (B) mtDNA was detected by PCR. P, IP-pellet; S, IP-sup.
Figure 3.
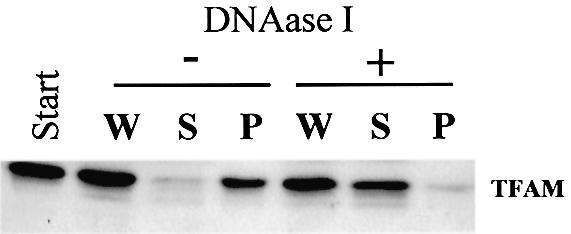
DNase I digestion of P2. P2 was digested with DNase I and centrifuged. TFAM was detected by western blotting. W, before centrifugation; S, supernatant; P, pellet.
Immunolabeling of TFAM
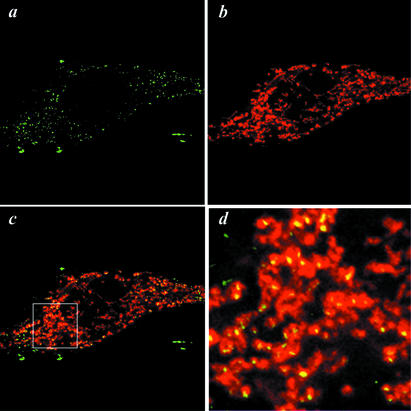
Immunolabeling of HeLa MRV11 cells using anti-TFAM showed a granular pattern in cytoplasm (Fig. 4a). Mitochondria were stained in a tubular form with a fluorescent dye, Mitotracker Red (Fig. 4b). The merged image shows that a large part of the dots were located within mitochondria (Fig. 4c and d). This pattern indicates that TFAM does not exist diffusely in the mitochondrial matrix. The number of dots is approximately a few hundred in one HeLa MRV11 cell (Fig. 4), and the mtDNA copy number per cell is estimated by Southern blotting to be ∼1000 (19). Given that each dot may represent one TFAM/mtDNA complex, such a complex would contain a few copies of mtDNA on average.
Figure 4.
Immunolabeling of HeLa cells. TFAM in HeLa MRV11 cells was identified using affinity-purified anti-TFAM antibodies and Alexa Fluor 488- conjugated goat anti-rabbit IgG antibodies (a). Mitochondria were stained with Mitotracker Red (b). (c) A merged image. (d) A magnified image of the square outlined region in (c).
Sonication of mitochondria
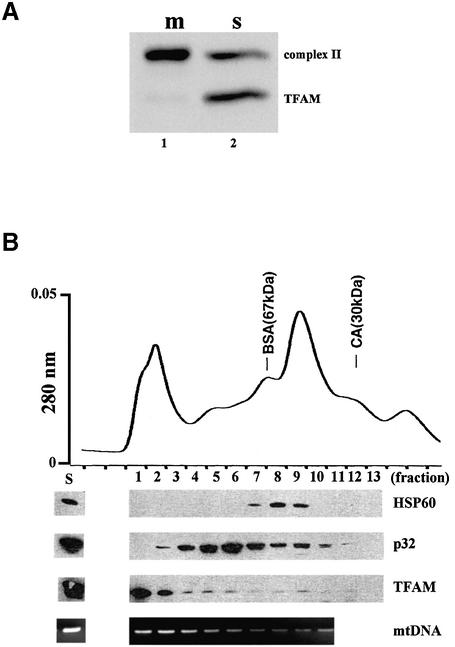
We disrupted mitochondria by sonication without detergent. The sonication procedure, in contrast to detergent solubilization, released TFAM into the soluble fraction (200 000 g supernatant) but little of the iron sulfur protein (Fig. 5A), being consistent with the idea that the NP-40 insolubility of the TFAM/mtDNA would be via macromolecular substances that can be readily disrupted by sonication, such as DNA, cytoskeleton and membranes. The supernatant was applied to size exclusion chromatography. p32, which is known to exist as a homotrimer (24), and HSP60, a matrix protein, were detected at the positions corresponding to their expected molecular weights (Fig. 5B). Although purified TFAM exists as a monomer in solution (1), TFAM appeared mainly in the void volume (>500 kDa), but also was distributed in many fractions associated with molecular weights larger than the TFAM monomer. mtDNA fragments eluted almost in parallel with TFAM (Fig. 5B), and mtDNA in the soluble fraction was immunoprecipitated by anti-TFAM (data not shown). These results suggest that TFAM still maintains a complex with mtDNA after the disruption of mtDNA molecules by sonication.
Figure 5.
Sonication of mitochondria. Mitochondria of U937 cells were sonicated. After centrifugation, soluble (s) and particulate (m) fractions were prepared. The particulate fraction was re-suspended to the original volume. (A) Iron sulfur protein of complex II and TFAM were detected by western blotting. (B) The soluble fraction was separated by size exclusion chromatography. The topmost panel is a profile of UV absorbance. HSP60, p32 and TFAM were detected by western blotting. mtDNA was detected by PCR. BSA, bovine serum albumin; CA, carbonic anhydrase.
DISCUSSION
The results in this study are the first to clearly indicate that most TFAM molecules associate with mtDNA and vice versa. The TFAM/mtDNA ratio is ∼900:1 in the NP-40-insoluble fraction of human placental mitochondria (19). Taken together, these results suggest that ∼900 molecules of TFAM are bound to one molecule of mtDNA on average. This amount of TFAM could cover the mtDNA entirely. Hence, we can no longer say that human mtDNA is naked. LSP, a promoter for transcription of the light strand (25), would be bound first and left last by TFAM, because TFAM has higher affinity to LSP than to the rest of the mtDNA molecule (2,20,26) and fully activates the LSP-specific transcription at a level where a ratio of TFAM/DNA template is ∼10 in the presence of mitochondrial transcription factor B (3). There fore, under the conditions where ∼900 molecules of TFAM are bound to mtDNA, LSP is likely to be persistently occupied by TFAM. Given that binding of TFAM to LSP initiates transcription of mtDNA, TFAM may be (at least potentially) abundant enough to constitutively and fully activate the mitochondrial transcription in a normal state where mitochondrial transcription factors B are present. This is compatible with the recent finding that TFAM levels can be substantially reduced without significant inhibition of mtDNA transcription in insect cells (27). Consistent with this, mitochondrial gene expression largely depends on the mtDNA copy number (28).
Human mtDNA is speculated to take on a nucleoid structure (17,18). However, this contention depends almost entirely on morphological observations that mtDNA is stained simply in a punctuate pattern in cells. Structural and biochemical evidence for a mammalian mtDNA nucleoid has been scarcely provided to date. In this study, we biochemically demonstrated that mtDNA associates with TFAM. The packaging of human mtDNA by TFAM may be critical for maintaining mtDNA as is packaging of yeast mtDNA by Abf2p, based on the following: (i) TFAM is able to substitute for Abf2p, a TFAM homolog of yeast, which is not supposed to significantly affect the transcription role for the maintenance of mtDNA (2); (ii) TFAM is abundant enough to cover mtDNA (19); (iii) most mtDNA molecules are associated with TFAM, and vice versa (Figs 2 and 3); (iv) the 50% reduction in TFAM in TFAM+/– mice decreases mtDNA by ∼50% (6). mtDNA depletion accompanies low levels of TFAM without a decrease in its mRNA level (29). Both mtDNA and TFAM could be unstable in a free form.
Both mtDNA and TFAM were recovered from the 10 000 g pellets after NP-40 solubilization (Fig. 1) and from the pellets in 45% sucrose by sucrose-density gradient centrifugation (data not shown), while the yeast mtDNA–protein complexes are recovered from the 16 000 g supernatants and from the boundary between 20 and 40% sucrose (13). This raises the possibility that TFAM/mtDNA complexes are associated with the other macromolecules under our experimental conditions. It is assumed that nuclear DNA is looped into domains by attachment to some underlying skeleton (e.g. a matrix or scaffold; for reviews, see 30). Human mtDNA is supposed to associate with mitochondrial inner membranes (31,32), though the association itself has never been convincingly evidenced. Our 0.5% NP-40 solubilization is incomplete as a large part of VDAC was recovered from the particulate fraction (Fig. 1). Under these conditions, mitochondrial outer and inner membrane proteins, monoamine oxidase and adenine nucleotide translocator, were substantially co-immunoprecipitated by anti-TFAM (identified by mass spectrometry, data not shown), suggesting that the TFAM/mtDNA complex associates with the membranes. The association with membranes may contribute to the sedimentation of the TFAM/mtDNA complex by the lower speed centrifugation. Because VDAC was not co-immunoprecipitated, it is unlikely the mitochondrial membrane fragments were non-specifically co-immunoprecipitated. The TFAM/mtDNA complex might associate with a special mitochondrial membrane domain that is resistant to NP-40 solubilization.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by Uehara Memorial Foundation and Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Technology, Sports, and Culture of Japan.
REFERENCES
- 1.Fisher R.P. and Clayton,D.A. (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol., 8, 3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parisi M.A., Xu,B. and Clayton,D.A. (1993) A human mitochondrial transcription activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol., 13, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falkenberg M., Gaspari,M., Rantanen,A., Trifunovic,A., Larsson,N.G. and Gustafsson,C.M. (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature Genet., 31, 289–294. [DOI] [PubMed] [Google Scholar]
- 4.McCulloch V., Seidel-Rogol,B.L. and Shadel,G.S. (2002) A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol., 22, 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- 6.Larsson N.G., Wang,J., Wilhelmsson,H., Oldfors,A., Rustin,P., Lewandoski,M., Barsh,G.S. and Clayton,D.A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet., 18, 231–236. [DOI] [PubMed] [Google Scholar]
- 7.Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol., 19, 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolffe A.P. (1994) Architectural transcription factors. Science, 264, 1100–1103. [DOI] [PubMed] [Google Scholar]
- 9.Wolffe A.P. (1999) Architectural regulations and Hmg1. Nature Genet., 22, 215–217. [DOI] [PubMed] [Google Scholar]
- 10.Diffley J.F.X. and Stillman,B. (1992) DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem., 267, 3368–3374. [PubMed] [Google Scholar]
- 11.Diffley J.F.X. and Stillman,B. (1991) A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA, 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megraw T.L. and Chae,C.B. (1993) Functional complementarity between the HMG-like yeast mitochondrial histone HM and bacterial histon-like protein HU. J. Biol. Chem., 268, 12758–12763. [PubMed] [Google Scholar]
- 13.Miyakawa I., Sando,N., Kawano,S., Nakamura,S. and Kuroiwa,T. (1987) Isolation of morphorogically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J. Cell Sci., 88, 431–439. [DOI] [PubMed] [Google Scholar]
- 14.Newman S.M., Zelenaya-Toroitskaya,O., Perlman,P.S. and Butow,R.A. (1996) Analysis of mitochondrial DNA nucleoid in wild-type and mutant strain of Saccharomyces cervisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res., 24, 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman B.A., Newman,S.M., Hallberg,R.L., Slaughter,C.A., Perlman,P.S. and Butow,R.A. (2000) In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl Acad. Sci. USA, 97, 7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki N., Sakai,A., Kawano,S., Kuroiwa,H. and Kuroiwa,T. (1998) DNA synthesis in isolated mitochondrial nucleoids from plasmodia of Physarum polycephalum. Protoplasma, 203, 221–231. [Google Scholar]
- 17.Satoh M. and Kuroiwa,T. (1991) Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res., 196, 137–140. [DOI] [PubMed] [Google Scholar]
- 18.Spelbrink J.N., Li,F.Y., Tiranti,V., Nikali,K., Yuan,Q.P., Tariq,M., Wanrooij,S., Garrido,N., Comi,G., Morandi,L., Santoro,L., Toscano,A., Fabrizi,G.M., Somer,H., Croxen,R., Beeson,D., Poulton,J., Suomalainen,A., Jacobs,H.T., Zeviani,M. and Larsson,C. (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature Genet., 28, 223–231. [DOI] [PubMed] [Google Scholar]
- 19.Takamatsu C., Umeda,S., Ohsato,T., Ohno,T., Abe,Y., Fukuoh,A., Shinagawa,H., Hamasaki,N. and Kang,D. (2002) Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep., 3, 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohno T., Umeda,S., Hamasaki,N. and Kang,D. (2000) Binding of human mitochondrial transcription factor A, an HMG box protein, to a four-way DNA junction. Biochem. Biophys. Res. Commun., 271, 492–498. [DOI] [PubMed] [Google Scholar]
- 21.Muta T., Kang,D., Kitajima,S. and Hamasaki,N. (1997) p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem., 272, 24363–24370. [DOI] [PubMed] [Google Scholar]
- 22.Gasnier F., Rousson,R., Lerme,F., Vaganay,E., Louisot,P. and Gateau-Roesch,O. (1993) Use of Percoll gradients for isolation of human placenta mitochondria suitable for investigating outer membrane proteins. Anal. Biochem., 212, 173–178. [DOI] [PubMed] [Google Scholar]
- 23.Kang D., Nishida,J., Iyama,A., Nakabeppu,Y., Furuichi,M., Fujiwara,T., Sekiguchi,M. and Takeshige,K. (1995) Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J. Biol. Chem., 270, 14659–14665. [DOI] [PubMed] [Google Scholar]
- 24.Jiang J., Zhang,Y., Krainer,A.R. and Xu,R.M. (1999) Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl Acad. Sci. USA, 96, 3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clayton D.A. (1991) Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell. Biol., 7, 453–478. [DOI] [PubMed] [Google Scholar]
- 26.Fisher R.P., Lisowsky,T. and Breen,G.A. (1991) A rapid, efficient method for purifying DNA-binding proteins. J. Biol. Chem., 266, 9153–9160. [PubMed] [Google Scholar]
- 27.Goto A., Matsushima,Y., Kadowaki,T. and Kitagawa,Y. (2001) Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem. J., 354, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams R.S. (1986) Mitochondrial gene expression in mammalian striated muscle. J. Biol. Chem., 261, 12390–12394. [PubMed] [Google Scholar]
- 29.Larsson N., Oldfors,A., Holme,E. and Clayton,D.A. (1994) Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun., 200, 1374–1381. [DOI] [PubMed] [Google Scholar]
- 30.Getzenberg R.H., Pienta,K.J., Ward,W.S. and Coffey,D.S. (1991) Nuclear structure and the three-dimensional organization of DNA. J. Cell. Biochem., 47, 289–99. [DOI] [PubMed] [Google Scholar]
- 31.Albring M., Griffith,J. and Attardi,G. (1977) Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl Acad. Sci. USA, 74, 1348–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson D.A., Bartlett,J. and Cook,P.R. (1996) Sequences attaching loops of nuclear and mitochondrial DNA to underlying structures in human cells: the role of transcription units. Nucleic Acids Res., 24, 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]