Abstract
The SOS response, a set of cellular phenomena exhibited by eubacteria, is initiated by various causes that include DNA damage-induced replication arrest, and is positively regulated by the co- protease activity of RecA. Escherichia coli DinI, a LexA-regulated SOS gene product, shuts off the initiation of the SOS response when overexpressed in vivo. Biochemical and genetic studies indicated that DinI physically interacts with RecA to inhibit its co-protease activity. Using nuclear magnetic resonance (NMR) spectroscopy, we show that DinI tightly binds to the central region of RecA (between the N- and C-terminal domains) and that this interaction is enhanced upon the oligomerisation of RecA. On the other hand, DinI did not inhibit the interaction between 4mer single-stranded (ss)DNA and RecA– ATPγS, but had a slight effect on the structure of ssDNA–RecA–ATPγS complexes involving 8mer and 12mer ssDNA. We hypothesise that prevention of repressor binding to the intermolecular cleft region of RecA protomers by DinI, with the possibility of a slight conformational change induced in the DinI-bound ssDNA–RecA–ATPγS complex, together function to inhibit the co-protease activity of RecA.
INTRODUCTION
The SOS response is a set of divergent cellular phenomena induced in eubacterial cells when DNA replication is inhibited by DNA damage or other causes, including activated DNA repair, elevated mutagenesis, prophage induction, inhibition of cell division, cessation of respiration and induction of stable DNA replication, among others (1–4). The discovery of the cleavage of λ CI repressor, and later of LexA repressor, by activated RecA in an ATP- and ssDNA-dependent reaction (5–7) led to an understanding of the initiation of the SOS regulatory system. In brief, stalled DNA replication results in the production of ssDNA regions, to which RecA protein binds and forms nucleoprotein filaments in an ATP-dependent manner. This ssDNA-bound RecA then promotes the self-cleavage of the LexA and λ CI repressors or UmuD (8,9). The cleavage of LexA relieves the repression on SOS genes that were repressed during normal growth. LexA repressor alone controls the expression of more than 30 SOS genes, encoding proteins that function in DNA repair, recombination, mutagenesis and so on (2,10). The cleavage of λ CI repressors induces the temperate phage λ to multiply and eventually lyse the host cells (11), while that of UmuD allows error-prone DNA replication to bypass damaged bases (12).
The ssDNA-bound RecA promotes these cleavage reactions, but RecA does not work as the primary protease, with the actual chemistry of catalysis being carried out by the LexA or λ CI repressors or UmuD. The term ‘co-protease activity’ was put forward to emphasise this function of RecA (11). The nucleoprotein filament constituted by the ATP (or ATPγS)-bound form of RecA and ssDNA interacts with free double-stranded (ds)DNA to promote heteroduplex formation between the ssDNA and the complementary strand of the dsDNA (13–15). Story et al. determined the crystal structure of a helical filament of the ADP-bound state of RecA and proposed that an intermolecular cleft located between two adjacent RecA monomers in the filament acts as the cleavable repressor protein binding site (16,17).
As described, the mechanism of initiation of SOS response has been relatively well studied, but much less is known regarding events leading to the termination of the SOS response. It had been reported that with the repair of damaged DNA, LexA re-accumulates, probably due to a decrease in the co-protease activity of RecA, to repress the SOS genes (2,18). Escherichia coli DinI protein, the expression of which is under the control of the SOS response, shuts off initiation of the SOS response when overexpressed in vivo (19). Moreover, purified DinI was found to inhibit the co-protease activity of RecA in vitro (20). This active ‘shut-off’ mechanism is crucial for survival, since certain SOS functions such as phage induction and mutagenesis are harmful to cells. The recently determined solution structure of DinI indicated that the negatively charged C-terminus, by mimicking the DNA backbone, probably competes with ssDNA for the RecA binding site (21). On the other hand, Yasuda et al. reported that DinI binds to the ssDNA–RecA–ATPγS filament and inhibits the co-protease activity without dissociation of the complex (22). More recently, Voloshin et al. reported that ssDNA was displaced from the ssDNA–RecA–ATPγS filament in the presence of excess amounts of DinI, corroborating the results from their earlier structural studies (23).
Thus, both groups have observed the physical interaction of DinI with RecA, but obtained conflicting results regarding the effects on the DNA binding of RecA. In the present study, we applied NMR spectroscopy to investigate the interaction of DinI and RecA, using DinI labelled with stable isotopes and various RecA fragments. In addition, the effect of DinI binding on the ssDNA–RecA interaction was investigated by the analysis of transferred NOEs (TRNOE) of ssDNA, originating from the RecA-bound state (24). We have also determined the solution structure of recombinant DinI using multidimensional NMR spectroscopy. Based on the results obtained, we present possible mechanisms of interaction between DinI and activated RecA for the inhibition of co-protease activity.
MATERIALS AND METHODS
Protein expression and preparation of E.coli DinI
The dinI gene was cloned from E.coli genomic DNA using the primers 5′-CTGGGATCCATGCGAATTGAA and 3′-CCC GGGAATTCTTATTCGCTGAC by PCR. The amplified PCR product was inserted into the BamHI and EcoRI sites of pGEX-6p expression vector (Amersham Biosciences) and the sequence was confirmed by DNA sequencing. DinI protein was overexpressed as a glutathione S-transferase (GST) fusion protein in E.coli JM109(DE3) cells. Unlabelled DinI was prepared in the following manner. Freshly transformed JM109(DE3) colonies carrying pGEX-6p/DinI from an agar plate (L-broth, 50 µg/ml ampicillin) were directly inoculated into 1 l of L-broth medium. The culture was incubated at 37°C until OD600 = 0.4 and then the temperature was shifted to 30°C. At OD600 = 0.6, 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added and the cells were harvested after fermentation for an additional 6 h. Uniformly 13C/15N- and 15N-labelled proteins were obtained by growing bacteria as mentioned earlier in M9 minimal medium containing 15NH4Cl (1 g/l) either with or without [13C6]d-glucose (2 g/l) as the sole nitrogen and carbon sources, respectively. For the expression of isotope-labelled proteins, the cells were harvested after IPTG induction for an additional 12 h.
All purification procedures were performed at 4°C. Harvested cells were suspended in the sonication buffer [50 mM Tris–HCl pH 8.0, 200 mM NaCl, 0.5 mM EDTA, 2 mM mercaptoethanol and 0.1 mM 4-amidinophenylmethane sulfonylfluoride hydrochloride (APMSF)]. The cells were disrupted by sonication and the lysate was cleared by centrifugation at 20 000 g for 1 h. The crude supernatant was loaded onto a glutathione–Sepharose 4B FF column (5 ml) (Amersham Biosciences), pre-equilibrated with the sonication buffer, washed with two volumes of sonication buffer and fractionated with the elution buffer (50 mM Tris–HCl pH 8.0, 200 mM NaCl, 1 mM mercaptoethanol, 10 mM reduced glutathione). The major fractions containing DinI, as confirmed by SDS–PAGE analysis, were collected and dissolved in the digestion buffer (30 mM Tris–HCl pH 8.0, 0.5 mM EDTA, 1 mM mercaptoethanol) using a Centriprep YM-3 cartridge (Millipore). The GST moiety was cleaved off by PreScission Protease (Amersham Biosciences) treatment for 12 h. The cleaved DinI included five additional vector-derived amino acid residues, Gly-Pro-Leu-Gly-Ser, at the N-terminus. The protease-treated solution was loaded onto a ResourceQ column (1 ml) (Amersham Biosciences) pre-equilibrated with the digestion buffer. The protein was eluted using a 0–2 M NaCl linear salt gradient. Subsequently, the fractions containing DinI were concentrated using a Centricon YM-3 cartridge (Millipore) and loaded onto a HiLoad 10/30 Superdex75 column (Amersham Biosciences) pre-equilibrated with the storage buffer (20 mM sodium phosphate pH 6.5, 50 mM NaCl) and eluted with the same buffer. The fractions containing pure DinI were concentrated using a Centricon YM-3 cartridge and stored in the storage buffer containing 50% glycerol.
NMR samples for structure determination (1 mM) and titration experiments (0.1 mM) were concentrated and dissolved in the 1H2O NMR buffer (90% 1H2O and 10% 2H2O containing 20 mM sodium phosphate pH 6.5, 50 mM NaCl and 0.01% NaN3) using a Centricon YM-3 cartridge. NMR samples for measuring TRNOE were concentrated and dissolved in the 2H2O NMR buffer (99.8% 2H2O containing 20 mM [uniform 2H]Tris–2HCl pH 7.1, 6.7 mM MgSO4, 150 mM NaCl). The pH value of the 2H2O NMR buffer was uncorrected for isotope effects.
Protein preparation of full-length and fragment RecA samples
The following proteins, full-length RecA (25,26), N-terminal domain [(RecA1–33) (27)], N-terminally truncated fragment [(RecAΔ1–33) (28)] and the C-terminal domain [(RecA268–330) (29)], were prepared as reported. All samples for NMR titration experiments were concentrated and dissolved in the 1H2O NMR buffer. The full-length RecA sample for TRNOE analysis was concentrated and dissolved in the 2H2O NMR buffer.
Preparation of oligo ssDNA samples
Three oligo ssDNA molecules, 4mer [d(TACG)], 8mer [d(CCTGATAG)] and 12mer [d(ACGACAGGCTAC)], were purchased from Espec Oligo Service Corporation (Ibaraki, Japan). Undesirable organic impurities and metal ions were removed using a succession of cation exchange resins, [AG 50W-X8 resin (Bio-Rad) pre-equilibrated with 10% pyridine, AG 50W-X8 pre-equilibrated with 1 M NaOH and Chelex 100 (Bio-Rad) pre-equilibrated with 50 mM sodium phosphate pH 7.5]. Concentrations of the purified oligo ssDNA molecules were determined by absorbance measurements at 260 nm (the molar extinction coefficient for individual bases A, T, G and C were 15 300, 9300, 11 800 and 7400, respectively) and expressed in moles of entire molecules. Finally, the oligo ssDNA molecules were lyophilised and dissolved in 2H2O before use.
NMR spectroscopy
NMR experiments were performed on a Bruker DRX600 spectrometer. The obtained data were processed on Linux PCs using the AZARA software package (W. Boucher, unpublished work). For the processing of 3-dimensional (3D) NMR data, a 2-dimensional (2D) maximum entropy algorithm was applied for indirect dimensions (30). All of the spectra were analysed on Linux PCs with a combination of customised macroprograms in the ANSIG v.3.3 software (31).
Backbone and side chain resonance assignments for DinI
For the backbone resonance assignment, two types of 3D triple resonance spectra, CBCA(CO)NH (32) and HNCACB (33), were measured. In addition to the above, HNHB (34), CC(CO)NH (35), H(CCCO)NH (36) and HCCH-TOCSY (37) spectra were measured for the side chain resonance assignment. For the collection of NOE-derived distance constraints, 2D 1H-1H NOESY, 3D 15N-separated NOESY and 3D 13C-separated NOESY (38) spectra were measured. All of the above spectra were acquired on 13C/15N-labelled DinI at 20°C.
Solution structure calculation
Cross-peaks from 3D 15N-separated and 13C-separated NOESY spectra were picked and assigned unambiguously wherever possible. For each spectra, two separate lists, one of unambiguous NOE restraints, the other of ambiguous restraints, were produced from the normalised cross-peak list by the ‘connect’ program within AZARA, allowing 0.04 and 0.3 p.p.m. tolerance for errors in the 1H and 15N/13C dimensions, respectively. Hydrogen bond derived distance constraints were obtained from the analysis of slowly exchanging amide protons in the 3D 15N-separated NOESY spectrum. Dihedral angle restraints for backbone φ and ψ angles were generated from the calculation of the Chemical Shift Index (CSI) (39) using 1Hα, 13Cα and 13Cβ chemical shifts. These restraints files were input into CNS (40) based protocols and subjected to a simulated annealing protocol. Manual iterative assignment processes were performed on the ambiguous NOEs by considering the contributions from possible assignment candidates. In all, 80 structures were calculated, and 20 selected on the basis of having lowest NOE-derived energies.
NMR titration of 15N-labelled DinI with full-length and fragment RecA samples
NMR titration experiments were performed on 15N-labelled DinI (0.1 mM) by measuring a series of 2D 1H-15N HSQC (41,42) spectra in the presence of equimolar amounts of unlabelled full-length RecA or at various concentrations of RecA fragments. For the titration experiments with RecA1–33 and RecA268–330, the protein concentrations of RecA fragments were 0, 0.01, 0.02, 0.05, 0.075, 0.1, 0.15 and 0.2 mM. The experiments with RecAΔ1–33 were performed at concentrations of 0, 0.01, 0.02, 0.05, 0.1, 0.15 and 0.2 mM. All NMR data were acquired at 20°C.
Transferred NOESY experiments of ssDNA in the presence of RecA and/or DinI
For the measurements of transferred NOESY spectra of 4mer, 8mer and 12mer ssDNAs in the presence of RecA and/or DinI, four types of samples, (i) ssDNA and ATPγS, (ii) ssDNA, ATPγS and RecA, (iii) ssDNA, ATPγS, RecA and DinI and (iv) ssDNA, ATPγS and DinI, were prepared. For sample (iii), RecA was mixed and incubated with ssDNA and ATPγS for 5 min at 25°C prior to the addition of DinI. The final concentrations of ssDNA and ATPγS were 1.0 mM each, while those of RecA and DinI were 0.1 mM each. Samples containing the above mixtures were lyophilised and dissolved in 2H2O (99.96%) (Euriso-top, France) before measurements. The transferred NOESY spectra were acquired at 25°C under essentially identical conditions as described earlier (24).
RESULTS
The NMR resonance assignment and structure determination
Although DinI is a relatively small protein (81 amino acids), it behaved like a high molecular weight protein at higher concentrations (∼2 mM), as deduced from preliminary NMR relaxation measurements (data not shown). Consistent with this observation, NMR signals of concentrated samples were broader than those expected for a 9 kDa protein, and thus the assignment of side chain resonances benefited from the HCCH-TOCSY spectrum. Indeed, DinI behaved as dimers during purification, for which DinI fractions were highly concentrated before application onto the column. To confirm the dimeric behavior of DinI, the molecular weight was analysed using Superdex 200 HR 10/30 (Amersham Biosciences) gel filtration column chromatography containing appropriate molecular weight markers and was estimated to be ∼16 kDa (data not shown). Voloshin et al. also reported a similar behaviour of DinI in their study (23). However, we found that the NMR signals were broadened in a concentration-dependent manner, with a significant improvement in line width upon reducing the concentration below 1.0 mM (data not shown), suggesting that DinI does not form a stable homodimer. This conclusion was also supported by the absence of intermolecular NOEs measured on a 1:1 mixture (0.5 mM each) of unlabelled and 13C/15N-labelled proteins (data not shown). We therefore concluded that the NMR signal contribution from monomeric species should be predominant at protein concentrations around 1.0 mM (broader line shapes are observed if the predominant species is a dimer) and performed measurements for resonance assignments and structural analysis at this concentration.
Backbone 1H, 13C and 15N resonances were assigned (including the extra five amino acid residues at the N-terminus) by analysis of spin–spin connectivities in 3D CBCA(CO)NNH and HNCACB spectra. Side chain 1H and 13C resonance assignments, excluding aromatics, were obtained from the analysis of 3D HNHB, CC(CO)NH, H(CCCO)NH and HCCH-TOCSY spectra. Further, side chain NH2 (Asn/Gln), NHε (Arg) and NHε1 (Trp) resonances and other aromatic side chain resonances were assigned by the analysis of intraresidue NOEs from 3D 15N-separated and 13C-separated NOESY spectra, respectively. Chemical shift assignments are listed in Supplementary Material (Table S1).
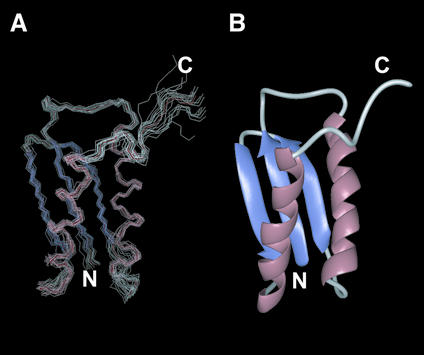
Figure 1 shows the solution structures of DinI calculated from a total of 2617 unambiguous and 572 ambiguous NOEs, 74 hydrogen bond-derived distance restraints and 61 φ/ψ dihedral angle restraints. The mean root mean square deviation (rmsd) from the average for backbone Cα, N and C′ atoms and all non-proton atoms in residues 1–78 were 0.56 and 1.01 Å, respectively. Structural statistics of the final ensemble of 20 refined structures are presented in Supplementary Material (Table S2). The structure consists of two α-helices and a three-stranded β-sheet, as shown in Figure 1B, and is almost identical to the recently published solution structure of DinI (PDB accession code 1GGH, previously 1F0A) (21). The rmsd between the average structures from these two different solution structure ensembles was as low as 1.31 Å for backbone Cα atoms in residues 1–78, in spite of the slightly different structure calculation approaches employed (while Ramirez et al. used a limited NOE supplemented with residual dipolar coupling based strategy, we relied on a solely NOE based approach that included use of ambiguous NOE restraints). The regions with slightly larger rmsd were localised to the N-terminus of the second β-strand, the N-terminus of the second α-helix and the C-terminus of the protein. The C-terminal region of the protein is primarily not well defined in our ensemble of structures. The differences in the other two regions can be attributed to their proximity to the slightly longer non-structured N-terminal extension present in our construct, in addition to the slightly different buffer and experimental conditions employed. The differences in Cα position between the average structures of DinI from the two NMR studies are presented as Supplementary Material (Figure S1).
Figure 1.
Solution structure of DinI. (A) The backbone traces of an ensemble of 20 refined structures superimposed on the backbone Cα, N and C′ atoms. (B) Ribbon representation of the structure closest to that of the mean of the ensemble. Molecular graphics images were produced using the MidasPlus software from the Computer Graphics Laboratory (University of California, San Francisco).
Interaction between DinI and full-length RecA
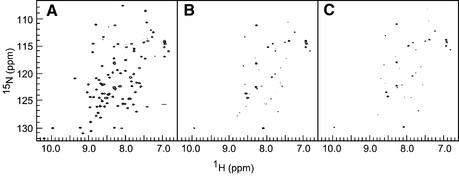
Both Yasuda et al. and Voloshin et al. have reported that DinI associates with RecA directly, based on biochemical and genetic studies (22,23). We therefore measured 1H-15N HSQC spectra of DinI with and without RecA in order to detect this interaction (Fig. 2). Upon the addition of a half-molar equivalent of RecA to 15N-labelled DinI, the signal intensity of DinI was approximately halved (data not shown). No change was observed in the chemical shifts or the line shapes of the remaining signals. When the concentration of RecA was increased to equimolar amounts, the NMR signals of DinI were almost completely suppressed (Fig. 2B) except for some sharp signals from the side chain NH2 groups and backbone amide groups of the N-terminal vector-derived residues. From the knowledge that RecA polymerises and forms a large protein complex even in the absence of DNA (16,43), the loss in the signal intensity (attenuation of the signal) can be explained by the extreme line broadening of NMR signals induced by rapid transverse relaxation of the RecA-bound fraction of DinI. Therefore, the result of this NMR titration experiment confirmed that DinI binds tightly to RecA.
Figure 2.
Physical interaction between fragments of RecA and DinI. Contour plots from 1H-15N HSQC spectra of 15N-labelled DinI (A) in the absence of RecA, (B) in the presence of 0.1 mM RecA and (C) in the presence of 0.2 mM RecAΔ1–33, showing the line broadening of DinI signals in the presence of RecA. All spectra were measured on 0.1 mM 15N-labelled DinI in 20 mM sodium phosphate, pH 6.5, containing 50 mM NaCl and 0.01% NaN3 (90% H2O:10% 2H2O) at 20°C.
When the NaCl concentration in the 1:1 mixture of 15N-labelled DinI and RecA was increased from 50 to 250 mM, the signal intensity over the entire region was slightly restored (data not shown). This indicates that a fraction of the RecA-bound DinI was released with the increase in NaCl concentration, since the 1H-15N HSQC spectrum of DinI alone does not change significantly between 50 and 250 mM NaCl (data not shown). The reduced population of RecA-bound DinI at higher salt concentrations probably reflects the importance of the electrostatic contribution to the DinI–RecA interaction, although we cannot completely exclude the indirect effects of increased salt concentration.
Interaction between DinI and RecA fragments
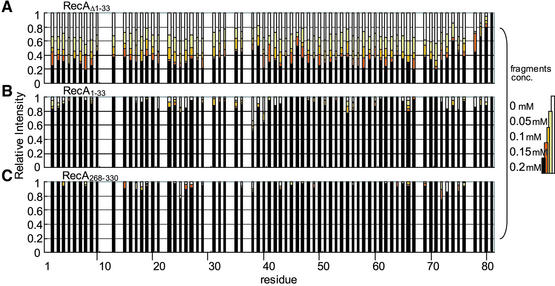
In order to obtain a more detailed picture on the interaction of DinI with RecA, we prepared three RecA fragments, RecA1–33, RecA268–330 and RecAΔ1–33 (corresponding to the N- and C-terminal domains and an N-terminally truncated fragment, respectively), according to the crystal structure of RecA (16). It has been reported that deletion of RecA1–33 leads to a decrease in the self-assembly of RecA (27), whereas RecA268–330 plays an important role for dsDNA binding (29,44). RecAΔ1–33 was used for NMR experiments, instead of the fragments encompassing the central region between the N- and C-terminal domains of RecA (referred to as the ‘central region’), which could not be prepared in a soluble form (unpublished observations). RecAΔ1–33 exists in an almost monomeric state at low concentrations (28). The interactions of DinI with the three different RecA fragments were then examined by NMR titration experiments. In the case of RecA1–33 and RecA268–330, no significant changes in cross-peak intensities and chemical shifts were observed. In contrast, addition of RecAΔ1–33 to 15N-labelled DinI showed a decrease in cross-peak intensities but no change in chemical shifts or line shapes (Fig. 2C), resembling the results for full-length RecA. Figure 3 summarises the changes in cross-peak intensity during the titration experiments with RecA1–33, RecA268–330 and RecAΔ1–33. It is obvious that neither RecA1–33 nor RecA268–330 interacts with DinI, whereas RecAΔ1–33 tightly binds to DinI. However, RecAΔ1–33 showed less cross-peak intensity attenuation compared to full-length RecA (see Fig. 2C).
Figure 3.
Differential effect of RecA fragments on signal intensities of 15N-labelled DinI. Changes in 1H-15N cross-peak intensity for observable residues of [15N]DinI upon titration with increasing concentrations of (A) RecAΔ1–33, (B) RecA1–33 or (C) RecA268–330 are plotted against the residue number. The intensity of each cross-peak was normalized relative to the intensity of the C-terminal residue 81. Data for overlapping cross-peaks and proline residues were excluded from these plots.
Effect of DinI on the interaction of RecA–ATPγS with 4mer ssDNA
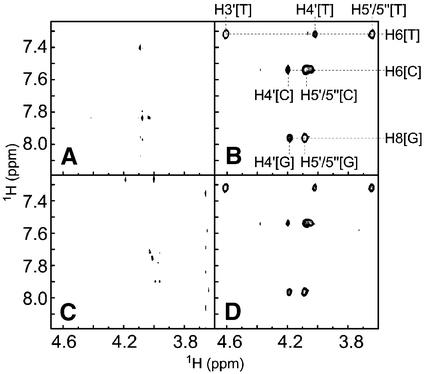
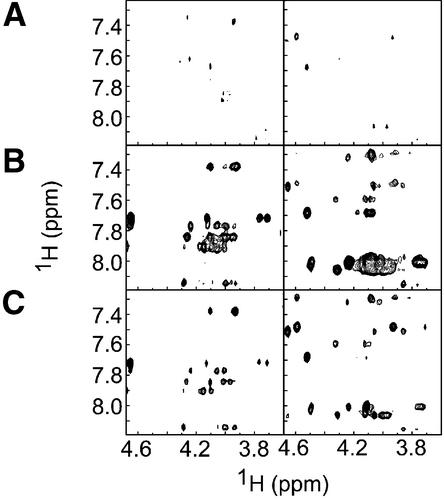
Next, we proceeded to determine the effect of DinI binding on the integrity of the ssDNA–RecA–ATPγS complex. We had previously reported that TRNOEs were observable for oligo ssDNA in the presence of RecA and proposed an unusual extended DNA structure through deoxyribose–base stacking induced by RecA (24). To investigate the influence of DinI on the interaction between ssDNA and RecA, we measured 2D transferred NOESY spectra of a 4mer ssDNA, ATPγS and RecA mixture with and without DinI and probed for changes in the patterns of TRNOE-derived cross-peaks. The sample containing 4mer ssDNA and ATPγS showed only few positive NOE cross-peaks in the 2D transferred NOESY spectrum (Fig. 4A), as described by Nishinaka et al. (24). TRNOEs due to the RecA-bound state of 4mer ssDNA were observed in the 2D transferred NOESY spectrum in the presence of ATPγS and RecA (Fig. 4B), whereas no TRNOEs were found for the sample containing 4mer ssDNA, ATPγS and DinI, suggesting that DinI failed to bind ssDNA (Fig. 4C). Finally, the addition of DinI into the mixture of 4mer ssDNA, ATPγS and RecA caused no significant change in TRNOE-derived cross-peaks (Fig. 4D). These results not only eliminate the idea that DinI alters the 4mer ssDNA binding activity of RecA, but also discount the possibility that DinI inhibits the interaction between 4mer ssDNA and RecA–ATPγS. Since it was already reported that one RecA molecule binds to three bases of ssDNA (43), we further analysed the interactions with longer ssDNA (8mer and 12mer).
Figure 4.
Interaction of DinI with 4mer ssDNA–RecA–ATPγS complex. Contour plots of NOE patterns of ssDNA from 2D transferred NOE spectra on the following samples: (A) ssDNA, (B) ssDNA with RecA, (C) ssDNA with DinI and (D) ssDNA with RecA and DinI are shown. The experiments were performed as described in Materials and Methods. The assignment of TRNOE cross-peaks was done according to Nishinaka et al. (24).
Effect of DinI on the interaction of RecA–ATPγS with 8mer and 12mer ssDNA
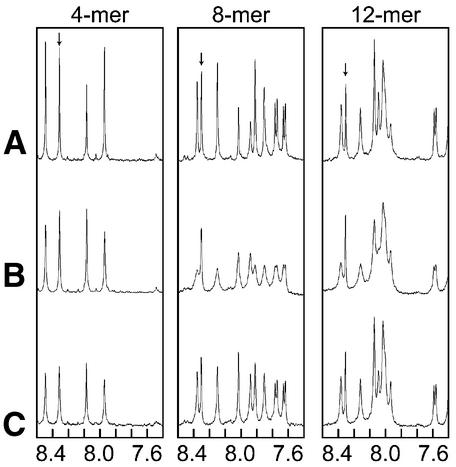
In the presence of RecA, the signals from ssDNA were broadened due to the fast exchange between the free and RecA-bound states (Fig. 5B) (24). For the interaction of 4mer ssDNA and RecA–ATPγS, addition of DinI caused negligible changes in the DNA line shape (Fig. 5, left column). In contrast, addition of DinI sharpened the signals from both 8mer and 12mer ssDNA in the presence of RecA and ATPγS (Fig. 5, centre and right columns). The narrowing of line shape specifically occurred for the signals from ssDNA, but not for ATPγS, which also showed line broadening because of fast exchange between the free and RecA-bound states. This result indicates the possibility that DinI alters the exchange regime of ssDNA for RecA–ATPγS. One possible cause of this altered exchange of RecA-bound ssDNA is a change in the affinity of ssDNA for the RecA–ATPγS in the presence of DinI. On the other hand, changes in TRNOE patterns caused by the addition of DinI were observed in the 2D transferred NOESY spectra of 8mer and 12mer ssDNA in the presence of RecA and ATPγS (Fig. 6). Most of the TRNOE cross-peaks induced by the interaction with RecA were still observed at a similar position, but had attenuated intensities. In addition, for some cross-peaks, increased intensities were observed upon the addition of DinI.
Figure 5.
Influence of ssDNA length on the binding of DinI to ssDNA–RecA–ATPγS complex. 1D 1H spectra of ssDNA (4mer, 8mer and 12mer) (A) in the absence of RecA, (B) in the presence of RecA and (C) in the presence of both RecA and DinI, indicating the narrowing of signals from DNA bases (ATPγS signals are indicated by arrows). Signals from RecA or DinI are too broad to be observed under this condition.
Figure 6.
DinI influences the integrity of ssDNA–RecA–ATPγS complex. Contour plots of a similar region as in Figure 4, from TRNOE spectra of (A) ssDNA alone, (B) ssDNA in the presence of RecA and (C) ssDNA in the presence of both RecA and DinI. The changes with 8mer and 12mer ssDNA are represented on the left and right columns, respectively.
DISCUSSION
Using NMR spectroscopy we showed that DinI binds to RecA in a 1:1 stoichiometry. This result is consistent with the observations of Yasuda et al. (22). NMR titration experiments of DinI with three RecA fragments showed that the DinI-binding interface of RecA is not located in the N- or C-terminal domains, suggesting that (i) the central region of RecA or (ii) the region between the central region and N- or C-terminal domains or (iii) the intermolecular clefts between neighbouring RecA in the filament may be responsible for DinI-binding activity. If the region between the central region and N- or C-terminal domains in fact binds to DinI, it should be manifested in the NMR spectra upon titrating DinI with the individual domains. The lack of changes in the HSQC spectra of DinI argues against a role for these regions in DinI binding. Even though both RecAΔ1–33 and full-length RecA caused similar attenuation for almost the entire region of DinI, the level of attenuation by RecAΔ1–33 was lower. Supposing that the DinI-binding site is located in the intermolecular cleft rather than the central region, this difference in affinity for DinI between the full-length RecA and RecAΔ1–33 may be explained by considering the altered oligomerisation properties of the two RecA samples. Although RecAΔ1–33 reportedly retained most of the activities of full-length RecA, it was however defective in self-assembly at lower concentrations, ∼0.05 mM (28). In the range of concentrations used for the titration experiments, the majority of RecAΔ1–33 is probably in the monomeric state (T. Mikawa and S. Kuramitsu, personal communication), whereas under similar conditions, a large population of the full-length RecA is oligomerised. At lower concentrations, RecAΔ1–33 did bind to DinI as opposed to the N- and C-terminal regions, whereas it was weaker than that of full-length RecA at the same concentration. However, at higher concentrations (0.2 mM) RecAΔ1–33 showed comparable attenuation of DinI signals to full-length RecA (Fig. 2C). Hence, the similarity in DinI binding to concentrated RecAΔ1–33 and full-length RecA could be attributed to their similar oligomeric natures. This leaves us with the possibility that the DinI-binding surface may be located in the intermolecular clefts and the decreased affinity of DinI for low concentrations of RecAΔ1–33 could be explained by the fewer intermolecular clefts available for binding.
In addition to the above possibility, we investigated the influence of DinI on the integrity of the activated nucleoprotein filament of RecA (ssDNA–RecA–ATPγS). Our transferred NOESY experiments did not show any significant effect of DinI on the interaction of 4mer ssDNA with RecA–ATPγS, suggesting that DinI does not induce the release of 4mer ssDNA from RecA. In contrast, changes in the line shapes as well as NOE cross-peak patterns (increase or decrease in cross-peaks intensity) were observed in experiments with 8mer and 12mer ssDNA. This observation cannot be explained by simple attenuation in the affinity of ssDNA for RecA– ATPγS. Slight conformational distortion of oligo ssDNA in the RecA-bound state or exchange between multiple conformers of RecA-bound ssDNA in the presence of DinI are some of the additional possibilities that can be considered. These contradictory results with 4mer as against 8mer and 12mer oligo ssDNA need to be addressed carefully based on the knowledge that one RecA molecule binds to three bases of ssDNA, as mentioned earlier (43). In our experiments, the 8mer or 12mer ssDNA molecule presumably binds to the ssDNA-binding site presented by two or more adjacent RecA molecules, whereby cooperative effects between neighbouring molecules come into the picture. Thus, there is a possibility that though DinI binding did not affect the ssDNA-binding site of a single RecA molecule, yet the putative cooperative interactions of RecA in the nucleoprotein filament were somehow altered.
Our transferred NOE results were inconsistent with the results of Voloshin et al., where they showed that DinI displaced ssDNA from the RecA–ssDNA complex. However, in their case, the displacement of ssDNA from the complex was hardly detectable when ssDNA–RecA–ATPγS complex was mixed with DinI at near equimolar amounts (the conditions employed in our study), but was significant at higher DinI concentration (15-fold or more compared to RecA) (23). On the other hand, Yasuda et al. reported that DinI neither prevented the formation of ssDNA–RecA– ATPγS complex (over 30-fold DinI to RecA condition) nor drastically altered the structure of the ssDNA–RecA–ATPγS complex (2-fold DinI to RecA) (22). This latter result of Yasuda et al. is consistent with the results from TRNOE experiments, performed under equimolar DinI to RecA conditions. However, limitations of the TRNOE method (use of longer oligo ssDNA, higher concentrations of each component, etc.) prevented us from performing experiments involving higher ratios of DinI to RecA, as reported by these two groups. Nevertheless, as mentioned above, our transferred NOESY experiments, performed at 1:1 DinI to RecA stoichiometry, provide enough evidence to suggest the possibility that DinI alters the nature of the ssDNA–RecA– ATPγS complex, without affecting the release of bound ssDNA.
Yasuda et al. reported that DinI fails to release ssDNA from the ssDNA–RecA–ATPγS complex, by examining the ssDNA-dependent ATPase activity of a poly(dT)–RecA– ATPγS complex at various DinI concentrations, keeping the DNA concentration sufficiently high (3 µM) (22). Under such high DNA conditions it is difficult to observe minor alterations in ATPase activity upon the addition of DinI. We therefore performed a similar set of experiments by varying the DNA concentrations (0–16 µM) (45) at a fixed DinI concentration, thereby ensuring increased sensitivity of the assay to low level changes in ATPase activity that may be induced by DinI. The results showed that DinI did not affect the ATPase activity even under extremely low ssDNA concentrations (data not shown), consistent with the results of Yasuda et al. (22). If DinI did drastically alter the structure of the ssDNA–RecA– ATPγS complex to release the ssDNA, then it would be reflected in a decreased or abrogated ATPase activity. The fact that there is no change in ATPase activity supports our discussion on the TRNOE experiments that DinI binds to the oligo ssDNA–RecA–ATPγS complex without disassociating the complex.
The present study suggests possible mechanisms by which DinI suppresses the co-protease activity of RecA against self-cleavable proteins. The region where the central region and the intermolecular clefts are exposed on the outer surface of the RecA filament has been proposed by Story et al. to serve as the hypothetical repressor-binding site (16). They also discussed that this region presents an optimal binding surface to accommodate smaller sized proteins such as LexA, λ CI, UmuD and so on. Harmon et al. have reported that LexA binds within a deep helical groove on the active RecA filaments (46). Based on the above discussions, we can interpret our results as suggesting that the DinI-binding region on RecA overlaps with those of the repressors. Thus, in line with the prevailing hypothesis, prevention of RecA–repressor interaction by DinI binding to the intermolecular clefts probably inhibits the co-protease activity of RecA, leading to the termination of the SOS reaction. On the other hand, Mustard and Little suggested that λ CI and UmuD bind to the intermolecular clefts in RecA, whereas LexA does not bind in this region (9). This indicates that the binding sites on RecA for different repressors might not be localised to the cleft region. Hence, simple competition for repressor-binding sites by DinI may not be sufficient to completely suppress the co-protease activity of RecA. Thus, in addition to the above, DinI-induced minor conformational changes in the ssDNA– RecA–ATPγS complex also probably function to inhibit the RecA co-protease activity.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Ad Bax and Ben Ramirez for kindly providing structural coordinates prior to publication. The authors also thank Dr Brian O. Smith for valuable suggestions during structure calculations and Dr Takeshi Yasuda for a critical reading of the manuscript. Dr Koji Takio is acknowledged for his support of the measurements on the DRX600 spectrometer at the Division of Biomolecular Characterization, RIKEN. This work was supported in part by Grants-in-Aid for Encouragement of Young Scientists from the Ministry of Education, Science, Sports and Culture (11740419) and the Japan Society for Promotion of Science (13740432) to Y.I., grants from the MR Science Program (RIKEN) to T.S. and Y.I. and CREST, Japan Science and Technology (JST) to T.S.
REFERENCES
- 1.Witkin E.M. (1976) Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol. Rev., 40, 869–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Little J.W. and Mount,D.W. (1982) The SOS regulatory system of Escherichia coli. Cell, 29, 11–22. [DOI] [PubMed] [Google Scholar]
- 3.Walker G.C. (1994) Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev., 48, 60–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedberg E.G., Walker,G.C. and Siede,W. (1995) SOS responses and DNA damage tolerance in Prokaryotes. In DNARepair and Mutagenesis. ASM Press, Washington, DC, pp. 407–464.
- 5.Roberts J.W., Roberts,C.W. and Craig,N.L. (1978) Escherichia coli recA gene product inactivates phage λ repressor. Proc. Natl Acad. Sci. USA, 75, 4714–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig N.L. and Roberts,J.W. (1980) E. coli recA protein-directed cleavage of phage λ repressor requires polynucleotide. Nature, 283, 26–30. [DOI] [PubMed] [Google Scholar]
- 7.Little J.W., Edmiston,S.H., Pacelli,L.Z. and Mount,D.W. (1980) Cleavage of the Escherichia coli lexA protein by recA protease. Proc. Natl Acad. Sci. USA, 77, 3225–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev., 63, 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mustard J.A. and Little,J.W. (2000) Analysis of Escherichia coli RecA interactions with LexA, lambda CI and UmuD by site-directed mutagenesis of recA. J. Bacteriol., 182, 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez de Henestrosa A.R., Ogi,T., Aoyagi,S., Chafin,D., Hayes,J.J., Ohmori,H. and Woodgate,R. (2000) Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol., 35, 1560–1572. [DOI] [PubMed] [Google Scholar]
- 11.Little J.W. (1984) Autodigestion of lexA and phage lambda repressors. Proc. Natl Acad. Sci. USA, 81, 1375–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang M., Shen,X., Frank,E.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (1999) UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA, 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata T., Cunningham,R.P., DasGupta,C. and Radding,C.M. (1979) Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc. Natl Acad. Sci. USA, 76, 5100–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEntee K., Weinstock,G.M. and Lehman,I.R. (1979) Initiation of general recombination catalysed in vitro by the recA protein of Escherichia coli. Proc. Natl Acad. Sci. USA, 76, 2615–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard-Flanders P., West,S.C. and Stasiak,A. (1984) Role of RecA protein spiral filaments in genetic recombination. Nature, 309, 215–220. [DOI] [PubMed] [Google Scholar]
- 16.Story R.M., Weber,I. and Steitz,T.A. (1992) The structure of the E. coli recA protein monomer and polymer. Nature, 355, 318–325. [DOI] [PubMed] [Google Scholar]
- 17.Story R.M. and Steitz,T.A. (1992) Structure of the recA protein-ADP complex. Nature, 355, 374–376. [DOI] [PubMed] [Google Scholar]
- 18.Little J.W. (1983) The SOS regulatory system: control of its state by the level of RecA protease. J. Mol. Biol., 167, 791–808. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda T., Nagata,T and Ohmori,H. (1996) Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J. Bacteriol., 178, 3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasuda T., Morimatsu,K., Horii,T., Nagata,T. and Ohmori,H. (1998) Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J., 17, 3207–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez B.E., Voloshin,O.N., Camerini-Otero,R.D. and Bax,A. (2000) Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Sci., 11, 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasuda T., Morimatsu,K., Kato,R., Usukura,J., Takahashi,M. and Ohmori,H. (2001) Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenisis. EMBO J., 20, 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voloshin O.N., Ramirez,B.E., Bax,A. and Camerini-Otero,R.D. (2001) A model for the abrogation of the SOS response by an SOS protein: a negatively charged helix in DinI mimics DNA in its interaction with RecA. Genes Dev., 15, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishinaka T., Ito,Y., Yokoyama,S. and Shibata,T. (1997) An extended DNA structure through deoxyribose-base stacking induced by RecA protein. Proc. Natl Acad. Sci. USA, 94, 6623–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata T., Cunningham,R.P. and Radding,C.M. (1981) Homologous pairing in genetic recombination. Purification and characterization of Escherichia coli recA protein. J. Biol. Chem., 256, 7557–7564. [PubMed] [Google Scholar]
- 26.Shibata T., Osber,L. and Radding,C.M. (1983) Purification of recA protein from Escherichia coli. Methods Enzymol., 100, 197–209. [DOI] [PubMed] [Google Scholar]
- 27.Masui R., Mikawa,T. and Kuramitsu,S. (1997) Local folding of the N-terminal domain of Escherichia coli RecA controls protein-protein interaction. J. Biol. Chem., 272, 27707–27715. [DOI] [PubMed] [Google Scholar]
- 28.Mikawa T., Masui,R., Ogawa,T., Ogawa,H. and Kuramitsu,S. (1995) N-terminal 33 amino acid residues of Escherichia coli RecA protein contribute to its self-assembly. J. Mol. Biol., 250, 471–483. [DOI] [PubMed] [Google Scholar]
- 29.Aihara H., Ito,Y., Kurumizaka,H., Terada,T., Yokohaya,S. and Shibata,T. (1997) An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing. J. Mol. Biol., 274, 213–221. [DOI] [PubMed] [Google Scholar]
- 30.Laue E.D., Mayger,M.R., Skilling,J. and Staunton,J. (1986) Reconstruction of phase sensitive 2D NMR spectra by maximum entropy. J. Magn. Reson., 68, 14–29. [Google Scholar]
- 31.Kraulis P.J. (1989) ANSIG: a program for the assignment of protein 1H 2D NMR spectra by interactive computer graphics. J. Magn. Reson., 84, 627–633. [Google Scholar]
- 32.Grzesiek S. and Bax,A. (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc., 114, 6291–6293. [Google Scholar]
- 33.Wittekind M. and Mueller,L. (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. Ser. B, 101, 201–205. [Google Scholar]
- 34.Archer S.J., Ikura,M., Torchia,D.A. and Bax,A. (1991) An alternative 3D NMR technique for correlating backbone 15N with side-chain Hβ resonances in larger proteins. J. Magn. Reson., 95, 636–641. [Google Scholar]
- 35.Grzesiek S., Anglister,J. and Bax,A. (1993) Correlation of backbone and amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. Ser. B, 101, 114–119. [Google Scholar]
- 36.Clowes R.T., Boucher,W., Hardman,C.H., Domaille,P.J. and Laue,E.D. (1993) A 4D HCC(CO)NNH experiment for the correlation of aliphatic side-chain and backbone resonances in 13C/15N-labelled proteins. J. Biomol. NMR, 3, 349–354. [Google Scholar]
- 37.Bax A., Clore,G.M. and Gronenborn,A.M. (1990) 1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson., 88, 425–431. [Google Scholar]
- 38.Marion D., Kay,L.E., Sparks,S.W., Torchia,D.A. and Bax,A. (1989) Three-dimensional heteronuclear NMR of 15N-labelled proteins. J. Am. Chem. Soc., 111, 1515–1517. [Google Scholar]
- 39.Wishart D.S. and Sykes,B.D. (1994) The 13C Chemical-Shift Index: a simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR, 4, 171–180. [DOI] [PubMed] [Google Scholar]
- 40.Brünger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.-S., Kuszewski,J., Nilges,M., Pannu,N.S. et al. (1998) Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Crystallogr., D54, 905–921. [DOI] [PubMed] [Google Scholar]
- 41.Bodenhausen G. and Ruben,D.J. (1980) Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett., 69, 185–189. [Google Scholar]
- 42.Grzesiek S. and Bax,A. (1993) The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc., 115, 12593–12594. [Google Scholar]
- 43.Roca A.I. and Cox,M.M. (1990) The recA protein: structure and function. Crit. Rev. Biochem. Mol. Biol., 25, 415–456. [DOI] [PubMed] [Google Scholar]
- 44.Kurumizaka H., Aihara,H., Ikawa,S., Kashima,T., Bazemore,L.R., Kawasaki,K., Sarai,A., Radding,C.M. and Shibata,T. (1996) A possible role of the C-terminal domain of the RecA protein. A gateway model for double-stranded DNA binding. J. Biol. Chem., 271, 33515–33524. [DOI] [PubMed] [Google Scholar]
- 45.Mikawa T., Masui,R. and Kuramitsu,S. (1998) RecA protein has extremely high cooperativity for substrate in its ATPase activity. J. Biochem., 123, 450–457. [DOI] [PubMed] [Google Scholar]
- 46.Harmon F.G., Rehrauer,W.M. and Kowalczykowski,S.C. (1996) Interaction of Escherichia coli RecA protein with LexA repressor. II. Inhibition of DNA strand exchange by the uncleavable LexA S119A repressor argues that recombination and SOS induction are competitive processes. J. Biol. Chem., 271, 23874–23883. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.