Abstract
The quadruplex structures of the human telomere sequences AG3(T2AG3)3 I and (T2AG3)4 II were investigated in the presence of Na+ and K+ ions, through the cross-linking of adenines and guanines by the cis- and trans-[Pt(NH3)2(H2O)2](NO3)2 complexes 1 and 2. The bases involved in chelation of the cis- and trans-Pt(NH3)2 moieties were identified by chemical and 3′-exonuclease digestions of the products isolated after denaturing gel electrophoresis. These are the four adenines of each sequence and four out of the 12 guanines. Two largely different structures have been reported for I: A from NMR data in Na+ solution and B from X-ray data of a K+-containing crystal. Structure A alone agrees with our conclusions about the formation of the A1–G10, A13–G22, A1–A13 platinum chelates at the top of the quadruplex and A7–A19, G4–A19 and A7–G20 at the bottom, whether the Na+ or K+ ion is present. At variance with a recent proposal that structures A and B could be the major species in Na+ and K+ solutions, respectively, our results suggest that structure A exists predominantly in the presence of both ions. They also suggest that covalent platinum cross-linking of a human telomere sequence could be used to inhibit telomerase.
INTRODUCTION
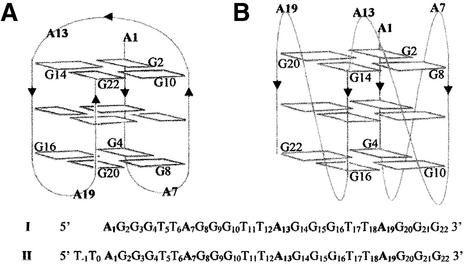
Human telomere DNA, located at the ends of each chromosome, contains a double-stranded sequence formed of (TTAGGG/CCCTAA) repeats prolonged at the 3′ extremity by a (T2AG3)n single strand (1). This strand is able to adopt, in vitro, folded quadruplex structures. Two of them have been described for AG3(T2AG3)3 I. One is the major form in Na+ solution, A, as determined from NMR data (2), and the other, B, was obtained from the X-ray structure of a K+-containing crystal (3) (Fig. 1). Both A and B result from the stacking of three G tetrads stabilized by ‘sandwiched’ monovalent cations. They differ: (i) by the relative orientations of the DNA backbones of the four G3 columns, either antiparallel for the opposing ones in A, or all parallel to each other in B; and (ii) by the positions and shapes of the three T2A connecting loops placing the four adenines at both ends of the quadruplex in A or all of them at one end in B. This introduces quite different distances between these adenines and between them and the guanines of the external tetrads.
Figure 1.
Schematic representations of the folded structures of AG3(T2AG3)3 I derived from those used by Patel (6): (A) from NMR, in Na+ solution, proposed by Wang and Patel (2); (B) in the crystalline state with K+ ions, proposed by Parkinson et al. (3). The sequences of AG3(T2AG3)3 I and (T2AG3)4 II are given with the nucleotide numbering chosen in this paper for the purpose of comparison.
We have shown previously that the cis- and trans-diamminediaquaplatinum(II) complexes (1 and 2) are able to cross-link guanines which are brought close to each other within the quadruplex structure of the Tetrahymena (T2G4)4 sequence (4). In this case, only the four guanines located in the loops were platinated. At variance with (T2G4)4, the folded human sequence AG3(T2AG3)3 presents, from its NMR structure (2), only four adenines N7 and/or N1 directly accessible to the platinum complexes. The N7-binding sites of all the guanines are masked by hydrogen bonding within the G tetrads. Compared with guanines, adenines are about 10 times less nucleophilic (5) but have two potential N1- and N7-binding sites which can participate in cross-linking. It was thus interesting to determine whether the di-functional platinum complexes 1 and 2 could cross-link the quadruplex structures of AG3(T2AG3)3 efficiently in Na+ and K+ solutions and give clues to their differences (6).
From the biological point of view, cisplatin was shown to induce telomere loss in cells (7) and the inhibition of telomerase in human testicular cancer cells (8). It forms 1,2-intrastrand adducts on double-stranded telomere sequences (9), but no evidence for cross-linking of the quadruplex structures was looked for. Such a cross-linking might have a similar effect to those of compounds stabilizing the stacking of G tetrads, and inhibiting telomerase activity (10–14) with subsequent induction of tumor cell death (15,16).
We have studied the two telomere sequences AG3(T2AG3)3 I and (T2AG3)4 II both in the presence of Na+ or K+. The choice of I and II was made to take into account any particular behavior of I which might result from the 5′-terminal position of A1; in II, the latter is flanked on its 5′ side by a T2 dinucleotide. The di-functional complexes cis- and trans-[Pt(NH3)2(H2O)2]2+ gave several cross-linked products of I and II identified by chemical and 3′-exonuclease digestions coupled to gel electrophoresis analyses. Chelates and bis-chelates of the cis- and trans-Pt(NH3)2 moieties were formed between purines remote from each other in the sequence. Only structure A could account for the platinum chelates deduced from our data. They involve four adenines in the stem and loops of the quadruplex structure and four guanines which belong to the external G tetrads. There is no qualitative difference between the cross-linked products from I or II, in either Na+ or K+ solution.
MATERIALS AND METHODS
Materials
AG3(T2AG3)3, (T2AG3)4 and the 8, 11, 16, 20 and 33mer markers were synthesised by Eurogentec, purified by 15% denaturating gel electrophoresis and located by UV shadowing. After elution and ethanol precipitation, they were desalted on a Sephadex G25 column and the 1 mM aqueous solu tion was stored at –20°C. T4 polynucleotide kinase and [γ-32P]ATP were from Pharmacia Biotech, NaCN from Merck, and dimethylsulfate (DMS), diethylpyrocarbonate (DEPC) and piperidine from Sigma. Cis-[Pt(NH3)2(H2O)2](NO3)2 and trans-[Pt(NH3)2(H2O)2](NO3)2 were prepared as previously described (4).
5′-End-labeling of oligonucleotides
AG3(T2AG3)3, (T2AG3)4 and the 8, 11, 16, 20 and 33mer markers were 5′-end-labeled using polynucleotide kinase and [γ-32P]ATP. The reaction products were purified by 20% denaturing gel electrophoresis without EDTA or on a Sephadex G25 column.
Platination of AG3(T2AG3)3 and (T2AG3)4 in the presence of Na+ or K+
5′-End radiolabeled I and II mixed with 100 µM non-radiolabeled material were incubated with 300 µM platinum complex (1:3 molar ratio) in a total volume of 10 µl. This ratio was chosen because it gave optimal platination kinetics. A 10:1 excess of platinum gave the same cross-linked products. For mass spectrometry analysis, 300 µM non-radiolabeled oligonucleotides were reacted with 900 µM platinum complex in a final volume of 16.5 µl. The reactions were run for 2 h at 25°C in 50 mM NaClO4 or KClO4 with cis-[Pt(NH3)2(H2O)2](NO3)2 1 or trans-[Pt(NH3)2(H2O)2](NO3)2 2. With 2, the platination was also run for 15 h.
The platinated oligonucleotides were separated by 20% polyacrylamide denaturing gel electrophoresis and located either by autoradiography when mixed with radiolabeled oligonucleotides or by UV-shadowing in the absence of labeling. They migrated differently according to their masses and charges.
Mass spectrometry
The major platinated products were analyzed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry with a linear time of flight (TOF) analyzer matrix using 2, 4, 6 trihydroxyacetophenone.
Determination and quantification of the platinum-binding sites by chemical probes
The gel migrations of the platinated oligonucleotides depend on the type of adducts. An increasing number of bound complexes slows down the migration, essentially because each of them brings an additional mono or dicationic charge. However, the migration is accelerated when the adducts are platinum chelates forming loops (4). After elution from the gel and ethanol precipitation, the material from each band was treated with DMS/piperidine (probe of guanine N7) or DEPC/piperidine (probe of adenine N7) in Maxam–Gilbert sequencing conditions (4,17). The fragments from digestion of platinated I and II were deplatinated by 1 and 2 M NaCN, respectively, for 18 h at 37°C and precipitated to allow a comparative gel analysis with the fragments obtained from the same treatment of unplatinated I and II. In both cases, the various spots were quantified using a Dynamic Molecular Phosphorimager with the Imagequant software for data processing. The percentage of inhibition of cleavage by DMS/piperidine (or DEPC/piperidine) observed for a guanine (Gi or adenine Ai) corresponds to the percentage of this guanine (or adenine) at position i which was platinated on the isolated oligonucleotide(s). This value is reported as the percentage of platinated guanine (or adenine) in Tables 1–3. These data indicated the following major platination sites: G4, G10, G20 and G22; A1, A7, A13 and A19. In parallel with chemical cleavage, we used 3′-exonuclease digestion to obtain a complementary assignment of the platinated sites, including adenine N1.
Table 1. Identification of the platinum chelates formed by reaction of cis-[Pt(NH3)2(H2O)2]2+ 1 with AG3(T2AG3)3 I and (T2AG3)4 IIa.
| Analysis of the platinated fragments from gel bands 1.1 and 1.2 (Fig. 2) | Platinated sitesb | ||||
|---|---|---|---|---|---|
| A1 | G10 | A13 | G22 | ||
| % Platinated G | 1.1 (I) | 80 | 80 | ||
| (DMS) | 1.2 (I/IIb) | 80 | 80 | ||
| % Platinated A– | 1.1 (I) | <15 | <15 | ||
| N7 (DEPC)c | 1.2 (I/IIb) | <15 | <15 | ||
| 3′-Exonuclease | 1.1 (I) | A1 and G2d | G10 | A13 and G14d | G22 |
| stopd | 1.2 (I/IIb) | A1 and G2d | G10 | A13 and G14d | G22 |
| ‘n-mer like’ migration of the platinated fragment after digestion | 1.1 (I) 1.2 (I/IIb) | 9 9/11 | 9–10e 9–10e/12 | 17 16/18 | 20 21/23 |
| Base position in | 1.1 (I) | 5′ | 3′ | 5′ | 3′ |
| the chelate | 1.2 (I/IIb) | 5′ | 3′ | 5′ | 3′ |
| Conclusion | 1.1 (I) | One bis-chelate A1–G10/A13–G22 | |||
| 1.2 (I/IIb) | Two chelates A1–G10 or A13–G22 | ||||
aSee text for step-by-step analysis.
bII is a 24mer but for the sake of comparison we have adopted the same base numbering as that of the 22mer I (Fig. 1). However, the size of the fragments is the actual one.
cDEPC/piperidine treatment does not detect A–N1 binding. The lower limit of reliable quantification by gel analysis is 15%.
dFor some adducts, the 3′-exonuclease digestion exhibits a 1 nt premature arrest. This is easily detected thanks to the assignment of the platination sites by chemical digestion.
eThe two values result from the non-identity of the migration scales of the free and platinated fragments.
Table 3. Identification of the platinum chelates formed by reaction of trans-[Pt(NH3)2(H2O)2]2+ 2 with (T2AG3)4 IIa.
| Analysis of the platinated fragments from gel bands 2.1, 2.2, 2.3 and 2.4 (Fig. 3) | Platinated sitesb | |||||||
|---|---|---|---|---|---|---|---|---|
| A1 | A7 | G10 | A13 | A19 | G20 | G22 | ||
| % Platinated G | 2.1 and 2.3 | 60 | <15 | 50 | ||||
| (DMS) | 2.2 | 30 | <15 | 30 | ||||
| 2.4 | <15 | 30 | 30 | |||||
| % Platinated | 2.1 and 2.3 | 45 | <15 | 35 | <15 | |||
| A–N7 | 2.2 | 55 | 85 | 60 | 80 | |||
| (DEPC)c | 2.4 | <15 | <15 | <15 | <15 | |||
| 3′-Exonuclease | 2.1 and 2.3 | A1 | G10 | A13 and G14d | G22 | |||
| stopd | 2.2 | A1 | A7 | G10 | A13 | A19 | G22 | |
| 2.4 | A1 | A7 | A13 | A19 | G20 | G22 | ||
| Platinated | 2.1 and 2.3 | 3 | 12 | 15 | 24 | |||
| fragment size | 2.2 | 3 | 9 | 12 | 15 | 21 | 24 | |
| (mer)b | 2.4 | 3 | 9 | 15 | 21 | 22 | 24 | |
| ‘n-mer like’ migration of the platinated fragment after digestion | 2.1 and 2.3 2.2 2.4 | 10 10 10 | 14 13 | 11–12e 16–17e | 14–15e 14–15e 14 and 17i | 19 20 | 21 | 18 (2.1) 20 (2.3) 20 23 |
| Base position | 2.1 and 2.3 | 5′ | 3′ | 5′f | 3′ | |||
| in the chelate | 2.2 | 5′ | 5′ | 3′f | 5′ or 3′f | 3′ | 3′ | |
| 2.4 | 5′ | 5′ | 5′ and 3′i | 3′ | 3′ | 3′ | ||
| Conclusion | 2.1 and 2.3 | One bis-chelate A1–G10/A13–G22g | ||||||
| 2.2 | Three bis-chelates A1–G10/A7–A19; A1–A13/A7–A19 and A13–G22/A7–A19 | |||||||
| 2.4h | Four chelates A1–A13; A13–G22; A7–A19; A7–G20 | |||||||
aSee text for step-by-step analysis.
bII is a 24mer but for the sake of comparison we have adopted the same base numbering as that of the 22mer I. However, the size of the fragments is the actual one.
cDEPC/piperidine treatment does not detect A–N1 binding. The lower limit of reliable quantification by gel analysis is 15%.
dFor some adducts, the 3′-exonuclease exhibits a one nucleotide premature arrest. This is easily detected thanks to the assignment of the platination sites by chemical digestion.
eThe two values result from the non-identity of the migration scales of the free and platinated fragments.
fBecause of the presence of another platinum chelate at the 5′ side of this base, its position in the chelate cannot be deduced from the migration length of the digested platinated fragment. This was done thanks to the measured distances between the purine nitrogens in structure A indicating the geometrically feasible chelates (Table 4).
gThe same bis-chelate is present in products from 2.1 and 2.3. The latter carries a supplementary platinum complex whose binding site has not been identified.
hMass spectrometry data indicate one platinum per oligonucleotide.
iThe two fragments result from the same A13 digestion stop. See text.
Determination of the platinum-binding sites by 3′-exonuclease digestion
This digestion is stopped by platinum mono-adducts and chelates (18). The isolated platinated products were incubated in 10 mM Tris–HCl, pH 8.0 buffer, in the presence of 2 mM MgCl2 and 0.5 mg/ml tRNA with the 3′-exonuclease phosphodiesterase I from Crotalus adamanteus venom 0.23 U/ml (Worthington Biochemical Corporation) diluted 5, 10 or 30 times in a 5 µl total volume for 30 min at 37°C. The digested fragments were purified on a 20% denaturing gel (see Figs 4 and 5). Each of them was eluted from the gel, precipitated and deplatinated by 1 and 2 M NaCN for I and II, respectively, for 18 h at 37°C. After precipitation, the deplatinated fragments were migrated on 20% denaturing gel and their migrations were compared with those of the fragments obtained by partial digestion of the starting oligonucleotide (nuclease diluted 30 times).
Figure 4.
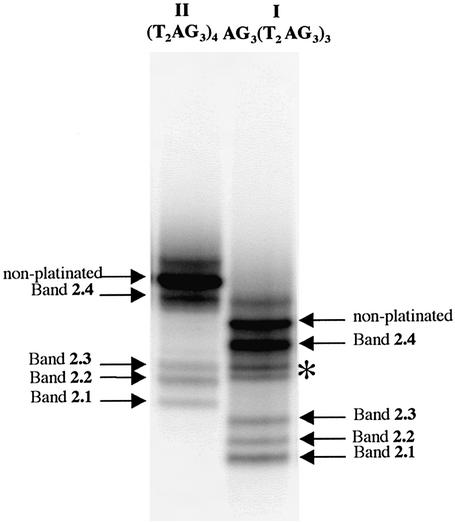
Denaturing gel electrophoresis of the fragments from the 3′-exonuclease digestion of AG3(T2AG3)3 I and of its platinated products from reaction with trans-[Pt(NH3)2(H2O)2]2+ 2 (2.1 to 2.4 from Fig. 3). The digestion reaction was conducted with different enzyme dilutions indicated at the top of each lane (e.g. 1 = none, 5 = five times, 30 = 30 times dilution). The numbered purines correspond to the digestion arrests. They were determined from the migration of the corresponding fragments after deplatination by NaCN. The digestion of I gives the reference scale for the migration of the platinated digested fragments.
Figure 5.
Denaturing gel electrophoresis of the products from the 3′-exonuclease digestion of (T2AG3)4 II and its platinated products from reaction with trans-[Pt(NH3)2(H2O)2]2+ 2 (2.1–2.4 from Fig. 3). The digestion reaction was conducted with different enzyme dilutions indicated at the top of each lane (e.g. 1 = none, 5 = five times, 30 = 30 times dilution). The numbered purines correspond to the digestion arrests. They were determined from the migration of the corresponding fragments after deplatination by NaCN. The digestion of II gives the reference scale for the migration of the platinated digested fragments. For the sake of comparison, we have adopted the same base numbering for the 22mer I and the 24mer II (Fig. 1).
For some adducts, the 3′-exonuclease arrest occurs at the adjacent purine on the 3′-side (i + 1) of the platinated purine. The chemical digestion unambiguously identifies the actual platinated base.
In the case of platinum chelates which cross-link two remote purines, the 3′-exonuclease stops at the first platinated base encountered, i.e. the 3′-base of the chelate (3′-stop). However, it can bypass this nucleotide, with increasing enzyme concentration, and stop at the 5′-base of the chelate (5′-stop). The 3′-stop preserves the platinated loop, which leads to a faster migration of the digestion product than that of the corresponding unplatinated fragment. The 5′-stop, after digestion of the loop between the 3′- and 5′-nucleotides initially chelating the Pt(NH3)2 moiety, leaves a 3′-end platinated fragment. This end binding is through the former 5′-base of the chelate, the former 3′-base being the second purine ligand. Such a linear platinated fragment migrates more slowly than the corresponding unplatinated oligonucleotide (4). Therefore, it is possible to identify the 5′- and 3′-bases of a platinum chelate when the analyzed fragment is mono-platinated. The identification is not straightforward when a second adduct is present on the fragment, either as a mono-coordinated complex or as a second platinum chelate. The uncertainty can be removed by comparing the chelating potential of the identified binding sites with their distances measured on structures A and B, respectively, reported for I from NMR and X-ray data (Table 4).
Table 4. Cross-linking of AG3(T2AG3)3 I and (T2AG3)4 II by cis- or trans-[Pt(NH3)2(H2O)2]2+ 1 and 2: correlation between the identified platinated purines, their platinum chelation ability and their N–N distances measured on the NMR (A) and X-ray (B) structures of I.
| Theoretical cross-links inferred from the identified platinated sites of I and II by 1 and 2 | Distances (Å) between the purine nitrogens of the theoretical chelates on structures A and B | ||
|---|---|---|---|
| A | B | ||
| Top of A | |||
| A1–G10 | A1N1–G10 | 4.2–6.9 | 25.1 |
| 1 and 2 | A1N7–G10 | 6.4–6.9 | 22.2 |
| A13–G22 | A13N1–G22 | 3.5–5.4 | 21.3 |
| 1 and 2 | A13N7–G22 | 3.9–6.2 | 21.8 |
| A1–A13 | A1N1–A13N1 | 7.8–11.5 | 33.7 |
| 2 | A1N1–A13N7 | 5.1–8.4 | 34.8 |
| A1N7–A13N1 | 6.6–10 | 31.1 | |
| A1N7–A13N7 | 3–6.8 | 32.3 | |
| A1–G22a | A1N1–G22 | 9–12.3 | 16 |
| A1N7–G22 | 8.8–9.6 | 12.6 | |
| Bottom of A | |||
| A7–A19 | A7N1–A19N1 | 4.6–4.8 | 30.8 |
| 2 | A7N1–A19N7 | 6.1–6.4 | 31.4 |
| A7N7–A19N1 | 4.7–7.1 | 31.6 | |
| A7N7–A19N7 | 6.7–7.8 | 32.4 | |
| G4–A19 | G4–A19N1 | 4.2–5.9 | 21.9 |
| 2 | G4–A19N7 | 6.1–8.1 | 21.8 |
| A7–G20 | A7N1–G20 | 5.5–5.9 | 19.7 |
| 2 | A7N7–G20 | 8.1–8.6 | 21.1 |
aA1–G22 platinum chelation is not feasible because of the too long N–N distances between the purines. The two purines are involved in other cross-links.
To exemplify the method used to analyze the adducts from the platination reactions, the case of the products from the reaction between 2 and II, migrating in band 2.4 of Figure 3, is presented in detail. Thirty percent of G22 and G20 were found to be platinated according to their reactivity to DMS, indicating that 30% of the chelates of 2.4 involved G22 or G20. None of the adenines were found to be platinated according to their reactivity to DEPC. It showed that they were either platinated on their N1 or on their N7 position, but to a lower extent than the 15% value which corresponds to the limit of our determinations. Enzymatic digestion was used to determine: (i) the positions of all the platination sites; and (ii) the nucleotides actually chelating the trans-Pt(NH3)2 moiety. (i) The arrests of the 3′-exonucleases gave several fragments still bearing a platinum complex. After cyanide treatment, the migrations of the deplatinated fragments were compared with those of the reference fragments from partial digestion of II. The data allowed the identification of G22, G20, A19, A13, A7 and A1 as the platinated purines in the progression order of the 3′-nuclease (see Fig. 5 and Table 3). (ii) The 5′- and 3′-nucleotides of the chelates were identified thanks to the opposite effects of a platinum monoadduct and a platinum chelate on the gel migration of the digestion fragments, as discussed above. In the case of band 2.4, the fragments resulting from exonuclease arrest at G22 (no digestion at all), G20, A19 and A13 migrated like 23, 21, 20 and 14mers respectively, compared with the 24, 22, 21 and 15mer migrations of the unplatinated oligonucleotides. [It should be remembered that (T2AG3)4 is a 24mer, but that the last nucleotide is denoted G22 because we adopted the AG3(T2AG3)3 numbering for the sake of comparison (Fig. 1)]. These data indicated that G22, G20, A19 and A13 were the 3′-nucleotides of trans-Pt(NH3)2 chelates. Other digestions stopped at A13, A7 and A1, giving fragments migrating like 17, 13 and 10mers, respectively, instead of 15, 9 and 3mers for the corresponding unplatinated fragments. This indicated that there was no more chelate present on these fragments and that the exonuclease had bypassed the 3′-purine of the former chelate and stopped at its 5′-purine. The whole set of data for the digestion of the products of band 2.4 collected in Table 3 led to the conclusion that G22, G20 and A19 were the 3′-bases and A7 and A1 were the 5′-bases of trans-Pt(NH3)2 chelates.
Figure 3.
Denaturing gel electrophoresis (20% acrylamide) of the platination products of AG3(T2AG3)3 I and (T2AG3)4 II by trans-[Pt(NH3)2(H2O)2]2+ 2 in 50 mM K+ solution. Same numbering of the bands as in Figure 2, e.g. 2.1. The products contained in the bands marked by an asterisk were found to be unstable in the digestion conditions and could not be analyzed further.
A13 played both roles in two different products. The analysis of the other adducts was conducted in a similar fashion and is summarized in Tables 1–3.
Determination, on structures A and B, of the distances between the purine nitrogens which appear as potential chelating sites from the digestion data
The N–N distances in Ångstroms were measured: (i) on the six relaxation matrix-refined structures A reported from the NMR study of I (2) available from the Brookhaven Data Base (143D.pdb); for each distance, the interval between the extreme values is given in Table 4; and (ii) on the X-ray crystal structure B recently reported (3) available from the Brookhaven Data Base (1KF1.pdb). When an adenine is considered, the two distances involving N1 and N7 are given, even if the experimental data do not always allow the assignment of the actual coordinated nitrogen.
RESULTS
Assignment of the platinum-binding sites
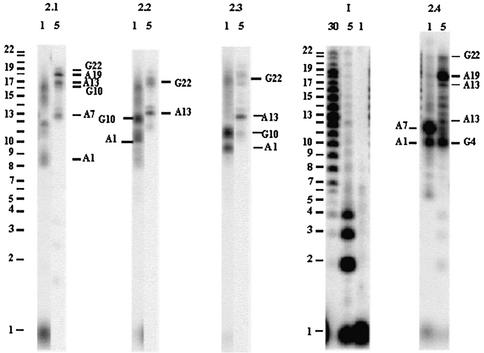
The two telomere sequences AG3(T2AG3)3 I and (T2AG3)4 II were reacted with cis- and trans-diamminediaquaplatinum(II) 1 and 2, in a 1:3 ratio, in the presence of 50 mM Na+ or K+. The platinated products were separated on denaturing gel electrophoresis: bands 1.1 and 1.2 from reactions of 1, and bands 2.1 to 2.4 from those of 2 as shown in Figures 2 and 3. The gel profiles of the reaction mixtures from 1 and 2 were similar for the two cations. Figures 2 and 3 present the data in Na+ and K+ solutions, respectively, for 1 and 2, whereas Figures S1 and S2 (in the Supplementary Material) present the K+ and Na+ data for the same complexes.
Figure 2.
Denaturing gel electrophoresis (20% acrylamide) of the platination products of AG3(T2AG3)3 I and (T2AG3)4 II by cis-[Pt(NH3)2(H2O)2]2+ 1 in 50 mM Na+. The numbering of the bands, e.g. 1.1, indicates the complex (here 1) and the migration order on the gel.
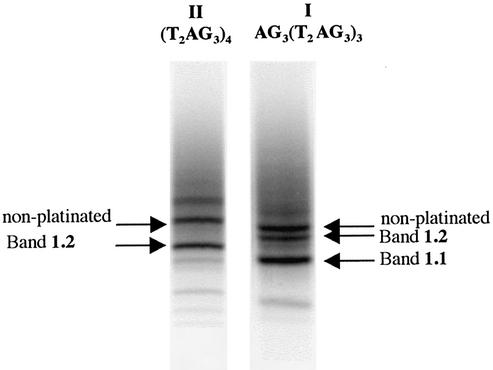
Mass spectrometry (MALDI-TOF) indicated that products from 1.2 and 2.4 bore one platinum adduct. This, together with the faster gel migration of 1.2 and 2.4 compared with that of the non-platinated oligonucleotide, led to the conclusion that these products contained one platinum per oligonucleotide chelated by two remote purines in the sequence. A platinum chelate between adjacent purines or a mono-adduct bound to only one purine should have retarded the migration (4). The faster gel migration of the products from 1.1 and 2.1–2.3 compared with that of the products from 1.2 and 2.4 suggested that the former contained two chelates, as already found in the case of (T2G4)4 (4). Bis-cross-linking results in accelerated migrations despite the addition of a cationic charge.
The assignments of the platinated sites were based on the data from the DMS/piperidine treatment which identifies the N7-platinated guanines and from DEPC/piperidine treatment which identifies the N7-platinated adenines; N1-bound adenines are not detected. 3′-Exonuclease digestions also allowed the identification of the platinated bases thanks to enzyme arrest at their 3′-side. The use of increasing enzyme concentrations permitted the assignment of the 5′- and 3′-bases of a chelate thanks to the typically different gel migrations of the digested fragments. In the case of an oligonucleotide bearing a single platinum chelate, it was therefore possible to identify it unambiguously. In the case of a mixture of mono-platinated oligonucleotides or of multiplatinated oligonucleotides, the identification of the platinated bases had to be completed by an evaluation of their geometrical aptitude to be involved in platinum chelation. This was done from the measure of the distances between their nitrogen ligands on both the NMR (2) and X-ray structures (3) A and B reported for I (Table 4).
To appreciate the relevance of the N–N distance to the chelating ability of these two atoms, we referred to four examples known in the DNA series: the 2.75 Å value between the two GN7 of a cis-Pt(NH3)2 interstrand cross-link (19), the 4.1 Å value between the GN7 and CN3 of a trans-Pt(NH3)2 interstrand cross-link (20) and the 3.3 and 4.5 Å values calculated for the pentacoordinated intermediates of a GN7 coordination to the platinum atom present in cis- and trans-[Pt(NH3)2(GN7)(H2O)] mono-adducts, respectively (4). None of these distances exceeds 5 Å. Their comparison with those calculated for structures A and B (Table 4) shows that no platinum chelate involving the identified purine ligands can be formed on structure B.
The whole set of data supporting the identification of the various chelates formed is presented in Tables 1–3.
Platination of AG3(T2AG3)3 and (T2AG3)4 by cis-diamminediaquaplatinum(II)
After 2 h of reaction with 1, the amounts of conversion of I and II as a function of the monovalent cation were 60 and 20%, respectively, in the presence of K+, and 60% for both in the presence of Na+. The bis-chelate of bands 1.1 and the chelates of bands 1.2 accounted for 40% of the converted material for both I and II (Fig. 2). The analysis of the platination sites identified A1, G10, A13 and G22, with G10 and G22 being the 3′-bases of the chelates (Table 1). As G10 was never found as the 5′-base of any chelate, the G10–A13 and G10–G22 cross-links could be excluded. In both cases, the nitrogen distances on structure A would have exceeded 11 Å. Similarly, A13 was never identified as the 3′-base of any chelate, eliminating the A1–A13 cross-link (N–N distances 3–11.5 Å). Table 4 indicates that, for structure A, only the A1–G10 and A13–G22 chelates are geometrically possible. For structure B, none of the identified platinated sites are closer than 12.6 Å to each other, which excludes any chelation possibility on this structure. These data support the formation of two cis-Pt(NH3)2 chelates at A1N1–G10 and A13N1/N7–G22 present in band 1.2, and the formation of one bis-chelate A1–G10/A13–G22 found in band 1.1.
Platination of AG3(T2AG3)3 and (T2AG3)4 by trans-diamminediaquaplatinum(II)
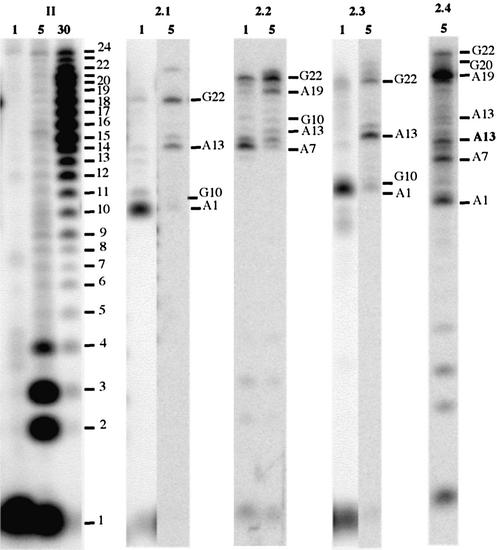
After 15 h of reaction with 2, the amounts of conversion of I and II were 70 and 45%, respectively, in the presence of either K+ or Na+. Compared with 1, the trans isomer 2 gave more platinum chelates migrating faster than the starting oligonucleotide on the gel (2.1 to 2.4). They accounted for 65% of the converted material for both I and II (Fig. 3).
The analysis of the mixture of chelates of band 2.4 from the reaction of 2 with II has been described in Materials and Methods (Figs 4 and 5, and Table 3). For the products from the reactions of 2 with both I and II, the same procedure identified the following platination sites: A1, G4 (for I), A7, A13, A19, G20 (for II) and G22. Among these, G4, A7 and A13 were identified as 5′-bases and A13, A19, G20 and G22 as 3′-bases of the chelates. Table 4 indicates that only the five chelates A1N7–A13N7, A13N1/N7–G22 and A7N1/N7–A19N1, together with G4–A19N1 for I and A7N1–G20 for II, are compatible with structure A.
The analysis of the digestion data from the products of bands 2.1–2.3 (Figs 4 and 5, and Tables 2 and 3) concluded that several bis-chelates were formed. It revealed G10 as the only additional platination site compared with those of the adducts with a single chelate. The corresponding chelate A1N1–G10 can be formed from structure A (Table 4).
Table 2. Identification of the platinum chelates formed by reaction of trans-[Pt(NH3)2(H2O)2]2+ 2 with AG3(T2AG3)3 Ia.
| Analysis of the platinated fragments from gel bands 2.1, 2.2, 2.3 and 2.4 (Fig. 3) | Platinated sites | |||||||
|---|---|---|---|---|---|---|---|---|
| A1 | G4 | A7 | G10 | A13 | A19 | G22 | ||
| % Platinated G | 2.1 | <15 | <15 | <15 | ||||
| (DMS) | 2.2 and 2.3 | <15 | 60 | 80 | ||||
| 2.4 | <15 | <15 | <15 | |||||
| % Platinated A–N7 | 2.1 | <15 | 60 | <15 | 60 | |||
| (DEPC)b | 2.2 and 2.3 | <15 | <15 | 35 | <15 | |||
| 2.4 | <15 | 30 | 45 | 35 | ||||
| 3′-Exonuclease | 2.1 | A1 and G2c | A7 | G10 | A13 | A19 | G22 | |
| stopc | 2.2 and 2.3 | A1 | G10 | A13 | G22 | |||
| 2.4 | A1 | G4 | A7 | A13 | A19 | G22 | ||
| ‘n-mer like’ migration of the platinated fragment after digestion | 2.1 2.2 and 2.32.4 | 9 99 | 10 | 1313 | 16 9–10d | 16–17d 1216 and 12e | 18 18 | 18–19d 19 (2.2) 20 (2.3) 21 |
| Base position | 2.1 | 5′ | 5′ | 3′e | 5′ and 3′f | 3′ | 3′ | |
| in the chelate | 2.2 and 2.3 | 5′ | 3′ | 5′f | 3′ | |||
| 2.4 | 5′ | 5′ | 5′ | 5′ and 3′e | 3′ | 3′ | ||
| Conclusion | 2.1 | Three bis-chelates A1–G10/A7–A19; A1–A13/A7–A19 and A13–G22/A7–A19 | ||||||
| 2.2 and 2.3 | One bis-chelate A1–G10/A13–G22g | |||||||
| 2.4h | Four chelates A1–A13; A13–G22; A7–A19; G4–A19 | |||||||
aSee text for step-by-step analysis.
bDEPC/piperidine treatment does not detect A–N1 binding. The lower limit of reliable quantification by gel analysis is 15%.
cFor some adducts, the 3′-exonuclease exhibits a 1 nt premature arrest. This is easily detected thanks to the assignment of the platination sites by chemical digestion.
dThe two values result from the non-identity of the migration scales of the free and platinated fragments.
eThe two fragments result from the same A13 digestion stop. See text.
fBecause of the presence of another platinum chelate at the 5′ side of this base, its position in the chelate cannot be deduced from the migration length of the digested platinated fragment. This was done thanks to the measured distances between the purine nitrogens in structure A, indicating the geometrically feasible chelates (Table 4).
gThe same bis-chelate is present in the products from 2.2 and 2.3. The latter carries a supplementary platinum complex whose binding site has not been identified.
hMass spectrometry data indicate one platinum per oligonucleotide.
For the products of bands 2.1 (I) and 2.2 (II), the digestions data and the N–N distances (Figs 4 and 5 and Tables 2–4) suggested that I and II gave three bis-cross-linked compounds, with the same A7–A19 chelate at the bottom of structure A and three different ones at the top: A1–A13, A1–G10 and A13–G22.
For the products from bands 2.2 (I) and 2.1 (II), the same analysis indicated that both oligonucleotides gave only one bis-chelate A1–G10/A13–G22 at the top of A.
For the products from band 2.3, the data suggested that for I and II the bis-chelate A1–G10/A13–G22 was formed, together with several mono-adducts which could not be identified.
As for the platination products formed with 1, Table 4 shows that structure B cannot account for any of the chelates of the trans-Pt(NH3)2 moiety, compatible with the identified platinated sites from the reaction with 2.
DISCUSSION
We have shown that the difunctional platinum complexes cis- and trans-[Pt(NH3)2(H2O)2]2+ 1 and 2 can be used to cross-link the quadruplex structures adopted by the oligonucleotides AG3(T2AG3)3 I and (T2AG3)4 II in Na+ and K+ solutions. The characterized cross-linked adducts accounted for 40 and 65% of converted I and II after reaction with 1 or 2. We had already used this method successfully with (T2G4)4 (4).
The identification of the platinated purines was based on DMS/piperidine detection of platinated GN7 and DEPC/piperidine detection of platinated AN7, both completed by the analysis of the 3′-exonuclease-digested platinated fragments. This allowed the unambiguous characterization of any single platinum chelate formed on an oligonucleotide. When a mixture of single platinum chelates from the same oligonucleotide was analyzed, or when two chelates were present on the same oligonucleotide, the purine ligands were assigned thanks to the distances between their potentially chelating nitrogens measured on the two structures A (2) and B (3). For the nucleic bases chelating the cis- or trans-Pt(NH3)2 moieties within intra- or interstrand cross-links, no N–N distance longer than 5 Å has been reported (4,19,20). On structure B, none of the potentially chelating nitrogens are closer than 12.6 Å, excluding cross-linking by 1 or 2.
In the presence of Na+ or K+, the same platinum chelates were formed from both I and II (Fig. 6) at the top of structure A for the cis-isomer 1 and at the top and bottom for the trans-isomer 2: (i) at the top, the three chelates characterized were A1–A13, A1–G10 and A13–G22 involving two adenines or one adenine and one guanine from the external top G tetrad (G10 or G22); (ii) at the bottom, the three platinum chelates characterized were A7–A19, G4–A19 or A7–G20 involving two adenines or one adenine and one guanine from the external bottom G tetrad (G4 or G20).
Figure 6.
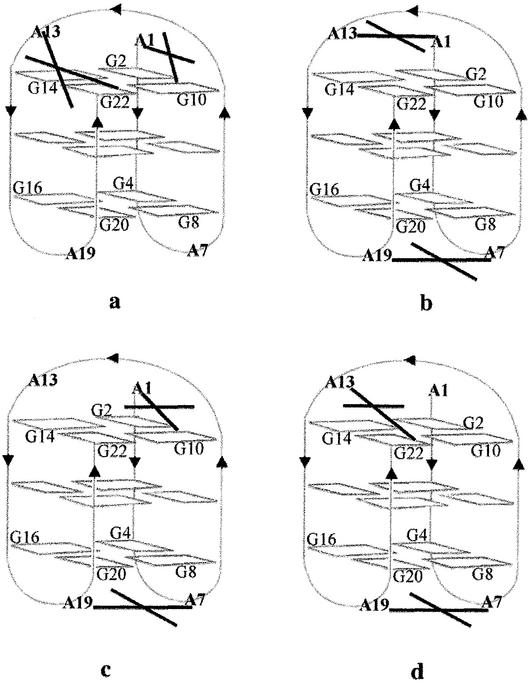
Schematic representation of the various platinum bis-chelates formed upon reaction of AG3(T2AG3)3 I with cis- and trans-[Pt(NH3)2(H2O)2]2+ 1 and 2, respectively (a), (b), (c) and (d). The cross mimics the four bonds of the square planar complex. Both I and II gave the same cross-links. Only structure A is used because structure B (Fig. 1) cannot account for any of the chelates compatible with the sites actually platinated (see text). Only bis-chelates are represented in order to limit the number of figures. They include the cross-links of the single chelates. It is noteworthy that the latter are the major products formed with 2 (bands 2.4, Fig. 3). (a) The A1–G10 and A13–G22 chelates of cis-Pt(NH3)2 were found as single chelates in band 1.2 and as a bis-chelate in band 1.1 (Fig. 2). The same cross-link was also formed by 2 on II (band 2.1, Fig. 3). (b–d) The three bis-chelates formed upon reaction of 2 with I have the same A7–A19 cross-link at the bottom of the quadruplex structure and different chelates at the top: respectively A1–A13, A1–G10 and A13–G22 (band 2.2, Fig. 3).
It is noteworthy that several platinum chelates involve both adenines and guanines (Fig. 6). Whereas the former have accessible N1 and/or N7 atoms in the stem and loops of the quadruplex (Fig. 1), the guanines of the external G tetrads have their N7 site engaged in hydrogen bonding to a neighboring G6NH2. In (T2G4)4, we had found that only the guanines not included in the tetrads were cross-linked (4). For I and II, the results suggest that the first platination occurs on an adenine and is followed by a chelation step involving a close adenine or a more nucleophilic guanine from the adjacent tetrad, provided that its hydrogen bonding is sufficiently disrupted. It is also to be noted that both 1 and 2 gave the same A1–G10 and A13–G22 chelates on I and II (Fig. 6a), revealing a sufficient conformational mobility at the top of the quadruplex structure to accommodate the different geometrical constraints of the two isomers. These depend on the 30° difference for the angle between the two N7–Pt bonds present in the pentacoordinated intermediate of the chelation step (4). Structure 2 gives more single chelates than 1, as already found for (T2G4)4 (4). This could reflect both the slower platination by 2 (21) and a slower chelation step (22,23) on the aqua trans-monoadduct compared with the cis, favoring the spanning of more potential binding sites.
That the cross-linking reactions actually occurred on folded structures of I and II is supported (i) by the remote positions of the bridged purines in the oligonucleotide sequence; (ii) by the fact that the NMR structure A proposed for I (2) accounts for all the platinum chelates compatible with the identified platinated sites; and (iii) by the fact that for a structure even partially unfolded before or after the first platination step, the presence of a single-stranded fragment would have led preferentially to the platination of more than four out of the 12 guanines, which would become more nucleophilic than the adenines once out of the tetrads. In the presence of the platinum excess, they would have given with a high preference 1,2-GG-cis-Pt(NH3)2 chelates with 1 and 1,3-GNG-trans-Pt(NH3)2 chelates with 2, as well documented in the literature (5). Such adducts would have exhibited retarded migration on gel electrophoresis (24,25).
The fact that the two complexes 1 or 2 reacting with I and II gave the same cross-linked adducts shows first that the two thymines at the 5′ end of II do not largely perturb the access to and the type of structure at the top of the quadruplex. Secondly, the fact that the same cross-links were formed either in Na+ or K+ solution suggests that the two oligonucleotides adopt a similar type of folded structure in the presence of the two ions. Previous work with (T2AG3)4 had concluded that there was a major antiparallel quadruplex structure in both Na+ and K+ solutions, from CD data; KMnO4 probing experiments indicated thymine stacking (T6 and T18 with our numbering Fig. 1) and interloop interactions at the bottom of the structure in the presence of Na+ but not K+ (26). Such a difference does not change the antiparallel type of folding. Examination of structure A (2) does not suggest that a T6–T18 stacking should impede A7 platination. Whereas structure A, calculated from NMR data in solution, accounts for all the platinum cross-links determined in this work, none of them is compatible with the N–N distances measured on structure B deduced from X-ray diffraction data on a K+-binding crystal.
Our results show that complexes 1 and 2 have cross-linked efficiently with a 40–65% yield, the adenines and guanines sufficiently close to each other on the quadruplex structure of type A, adopted by I and II in both Na+ and K+ solutions. This suggests that structure A is the major one in the presence of the two cations and does not agree with the proposal that crystal structure B might be the major form in K+ solution.
Finally, cross-linking of the purines of the quadruplex structure A of I and II by 1 or 2 suggests that it should prevent structured single-stranded telomere sequences from unfolding and might inhibit telomerase activity.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Dr J. C. Blais at Laboratoire de Chimie Organique Structurale, Université Pierre et Marie Curie, France, for mass spectrometry analyses. We thank Johnson-Matthey, Inc. for a generous loan of platinum complexes. This work has been supported by La Ligue Nationale Contre le Cancer.
REFERENCES
- 1.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y. and Patel,D.J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson G.N., Lee,M.P. and Neidle,S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- 4.Redon S., Bombard,S., Elizondo-Riojas,M.A. and Chottard,J.C. (2001) Platination of the (T2G4)4 telomeric sequence: a structural and cross-linking study. Biochemistry, 40, 8463–8470. [DOI] [PubMed] [Google Scholar]
- 5.Martin R.B. (1999) Platinum complexes: hydrolysis and binding to N(7) and N(1) of purines. In Lippert,B. (ed.), Cisplatin. Chemistry and Biochemistry of a Leading Anticancer Drug. Wiley-VCH, Zürick, pp. 183–206.
- 6.Patel D.J. (2002) Structural biology: a molecular propeller. Nature, 417, 807–808. [DOI] [PubMed] [Google Scholar]
- 7.Ishibashi T. and Lippard,S.J. (1998) Telomere loss in cells treated with cisplatin. Proc. Natl Acad. Sci. USA, 95, 4219–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger A.M., Double,J.A. and Newell,D.R. (1997) Inhibition of telomerase activity by cisplatin in human testicular cancer cells. Eur. J. Cancer, 33, 638–644. [DOI] [PubMed] [Google Scholar]
- 9.Burstyn J.N., Heiger-Bernays,W.J., Cohen,S.M. and Lippard,S.J. (2000) Formation of cis-diamminedichloroplatinum(II) 1,2-intrastrand cross-links on DNA is flanking-sequence independent. Nucleic Acids Res., 28, 4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedoroff O.Y., Salazar,M., Han,H., Chemeris,V.V., Kerwin,S.M. and Hurley,L.H. (1998) NMR-based model of a telomerase-inhibiting compound bound to G-quadruplex DNA. Biochemistry, 37, 12367–12374. [DOI] [PubMed] [Google Scholar]
- 11.Perry P.J., Gowan,S.M., Reszka,A.P., Polucci,P., Jenkins,T.C., Kelland,L.R. and Neidle,S. (1998) 1,4- and 2,6-disubstituted amidoanthracene-9,10-dione derivatives as inhibitors of human telomerase. J. Med. Chem., 41, 3253–3260. [DOI] [PubMed] [Google Scholar]
- 12.Perry P.J., Read,M.A., Davies,R.T., Gowan,S.M., Reszka,A.P., Wood,A.A., Kelland,L.R. and Neidle,S. (1999) 2,7-Disubstituted amidofluorenone derivatives as inhibitors of human telomerase. J. Med. Chem., 42, 2679–2684. [DOI] [PubMed] [Google Scholar]
- 13.Han F.X.G., Wheelhouse,R.T. and Hurley,L.H. (1999) Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc., 121, 3561–3570. [Google Scholar]
- 14.Mergny J.L., Lacroix,L., Teulade-Fichou,M.P., Hounsou,C., Guittat,L., Hoarau,M., Arimondo,P.B., Vigneron,J.P., Lehn,J.M., Riou,J.F., Garestier,T. and Hélène,C. (2001) Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc. Natl Acad. Sci. USA, 98, 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izbicka E., Wheelhouse,R.T., Raymond,E., Davidson,K.K., Lawrence,R.A., Sun,D.Y., Windle,B.E., Hurley,L.H. and Von Hoff,D.D. (1999) Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res., 59, 639–644. [PubMed] [Google Scholar]
- 16.Riou J.F., Guittat,L., Mailliet,P., Laoui,A., Renou,E., Petitgenet,O., Megnin-Chanet,F., Helene,C. and Mergny,J.L. (2002) Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl Acad. Sci. USA, 99, 2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen P.E. (1990) Chemical and photochemical probing of DNA complexes. J. Mol. Recognit., 3, 1–25. [DOI] [PubMed] [Google Scholar]
- 18.Gonnet F., Kocher,F., Blais,J.C., Bolbach,G. and Tabet,J.C. (1996) Kinetic analysis of the reaction between d(TTGGCCAA) and [Pt(NH3)3(H2O)]2+ by enzymatic degradation of the products and ESI and MALDI mass spectrometries. J. Mass Spectrom., 31, 802–809. [DOI] [PubMed] [Google Scholar]
- 19.Coste F., Malinge,J.M., Serre,L., Shepard,W., Roth,M., Leng,M. and Zelwer,C. (1999) Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 Å resolution: hydration at the platinated site. Nucleic Acids Res., 27, 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paquet F., Boudvillain,M., Lancelot,G. and Leng,M. (1999) NMR solution structure of a DNA dodecamer containing a transplatin interstrand GN7–CN3 cross-link. Nucleic Acids Res., 27, 4261–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikola M. and Arpalahti,J. (1994) Kinetics and mechanism of the complexation of trans-diamminedichloroplatinum(II) with the purine nucleoside inosine in aqueous solution. Inorg. Chem., 43, 4439–4445. [Google Scholar]
- 22.Brabec V. and Leng,M. (1993) DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues. Proc. Natl Acad. Sci. USA, 90, 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernal-Mendez E., Boudvillain,M., Gonzalez-Vilchez,F. and Leng,M. (1997) Chemical versatility of transplatin monofunctional adducts within multiple site-specifically platinated DNA. Biochemistry, 36, 7281–7287. [DOI] [PubMed] [Google Scholar]
- 24.Bombard S., Kozelka,J., Favre,A. and Chottard,J.C. (1998) Probing the mechanism of an Mn2+-dependent ribozyme by means of platinum complexes. Eur. J. Biochem., 252, 25–35. [DOI] [PubMed] [Google Scholar]
- 25.Escaffre M., Favre,A., Chottard,J.C. and Bombard,S. (2002) Determination of platinated purines in oligoribonucleotides by limited digestion with ribonucleases T1 and U2. Anal. Biochem., 310, 42–49. [DOI] [PubMed] [Google Scholar]
- 26.Balagurumoorthy P. and Brahmachari,S.K. (1994) Structure and stability of human telomeric sequence. J. Biol. Chem., 269, 21858–21869. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.