Abstract
Yth1, a subunit of yeast Cleavage Polyadenylation Factor (CPF), contains five CCCH zinc fingers. Yth1 was previously shown to interact with pre-mRNA and with two CPF subunits, Brr5/Ysh1 and the polyadenylation-specific Fip1, and to act in both steps of mRNA 3′ end processing. In the present study, we have identified new domains involved in each interaction and have analyzed the consequences of mutating these regions on Yth1 function in vivo and in vitro. We have found that the essential fourth zinc finger (ZF4) of Yth1 is critical for interaction with Fip1 and RNA, but not for cleavage, and a single point mutation in ZF4 impairs only polyadenylation. Deletion of the essential N-terminal region that includes the ZF1 or deletion of ZF4 weakened the interaction with Brr5 in vitro. In vitro assays showed that the N-terminus is necessary for both processing steps. Of particular importance, we find that the binding of Fip1 to Yth1 blocks the RNA–Yth1 interaction, and that this inhibition requires the Yth1-interacting domain on Fip1. Our results suggest a role for Yth1 not only in the execution of cleavage and poly(A) addition, but also in the transition from one step to the other.
INTRODUCTION
In eukaryotes, accurate processing of the 3′ ends of primary transcripts is an essential step that promotes transcription termination (1–3) and transport of mRNA from the nucleus (4–6). Cells are able to change the type and amount of mRNA derived from many genes by regulation of this step (7,8). The resulting poly(A) tail is also important for optimal translation and for determining mRNA stability (9–12).
The processing of the 3′ end of the primary transcripts consists of a tightly coupled two-step reaction involving site-specific endonucleolytic cleavage and poly(A) addition onto the upstream cleaved product. This reaction requires cis-acting signal sequences on the RNA and trans-acting protein com ponents (13,14). Uncoupling cleavage and polyadenylation in vitro (15,16) led to the biochemical identification of factors involved in either one or both steps of the process. In Saccharomyces cerevisiae, fractionation of whole cell extracts has identified five functionally distinct activities involved in cleavage and polyadenylation (17,18). Cleavage requires Cleavage Factor I (CF I) and Cleavage Factor II (CF II), while tail synthesis requires poly(A) polymerase (Pap1), CF I, CF II, Polyadenylation Factor I (PF I), and poly(A) binding protein (Pab1). CF I consists of five subunits, Rna14, Rna15, Pcf11, Clp1 and Hrp1 (18–21). PF I can also be isolated as part of a larger complex that contains CF II components, and has been designated Cleavage Polyadenylation Factor (CPF) (21,22). CPF includes Pap1, Fip1, Pta1, Mpe1, Pfs2, Cft1/Yhh1, Cft2/Ydh1, Brr5/Ysh1, Yth1 (21,23) and additional uncharacterized proteins (24). A subcomplex consisting of Cft1, Cft2, Brr5 and Pta1 was found to carry out CF II activity (25,26). Homologs for many of the CF I and CPF subunits have been found in higher eukaryotes (21,25,27–29), illustrating the conserved nature of this process.
In yeast, at least four cis-acting signals are important for mRNA 3′ end processing: a UA-rich element found at a variable distance upstream of the poly(A) site (30), an A-rich element located ∼20 nt upstream of the poly(A) site (31,32), U-rich sequences around the poly(A) site (33,34) and the poly(A) site, whose sequence is most frequently Py(A)n (35,36). The 3′ processing complex must contain RNA binding subunits that recognize these individual signals and thus direct the protein complex to a pre-mRNA for cleavage at the poly(A) site. In CF I, these are Hrp1, which interacts with the UA-rich element (37–40), and Rna15, which binds specifically to the A-rich element when Hrp1 and Rna14 are present (40). Among CPF subunits, at least three proteins (Cft1, Cft2 and Yth1) have RNA-binding properties. UV cross-linking experiments with purified CF II showed that Cft2 is cross-linked to GAL7 mRNA only when the UA-rich element is present (25). RNase H protection assays demonstrated that the CPF complex, as well as Cft1, Cft2 and Yth1 individually, interacts with the region around the CYC1 RNA poly(A) site (41–43).
As our knowledge of eukaryotic cleavage/polyadenylation factors increases, further understanding of the molecular mechanism underlying this essential process will require a thorough dissection of how the interactions of these factors with each other and with substrate promote cleavage, the transition from a cleavage to a polyadenylation complex, and processive synthesis of a correctly sized poly(A) tail. In this study, we focus such an analysis on Yth1, a 24 kDa subunit of CPF known to be essential for both cleavage and polyadenylation (29,41). Yth1 and its mammalian homolog CPSF-30 contain a well conserved central section of five CCCH zinc fingers, with regions of divergent sequence on either side (29). Members of the rare CCCH zinc finger family have in common two or more repeats of a motif consisting of Cys and His residues in the form CX8CX5CX3H. Proteins bearing this motif have roles in RNA binding, RNA localization and RNA stability in a wide variety of organisms (44–48). Zinc fingers in general have been implicated not only in nucleic acid binding but also in protein–protein interactions (49,50).
In addition to the interaction of Yth1 and RNA described above, Yth1 interacts with Fip1, whose binding to Pap1 regulates this enzyme’s activity and connects it to the processing machinery (29,51). Yth1 also associates with Brr5 (41), a CPF subunit necessary for both steps of processing (27). The structure of Yth1 suggests that it is organized into seven discrete domains that might contribute in unique ways to Yth1 function. Experimental evidence has begun to support this conclusion. Barabino et al. (29,41) showed that a deletion of the C-terminus that includes most of ZF5 (yth1-1) weakened the interaction with Fip1 and abrogated polyadenylation. However, mutation of a highly conserved residue in ZF2 (yth1-4) demonstrated that Yth1 was important for both cleavage and polyadenylation. This mutation preserved Fip1 binding but interfered with the interaction with RNA and the rest of CPF. In addition, a mutation of H142N in ZF4 (yth1-5) caused shorter poly(A) tails but had no effect on cleavage.
In this study, we create mutations throughout the protein to identify functions for the other Yth1 domains and to clarify the roles of ZF4 and the C-terminus. For example, we have found that ZF2 and ZF4 are both essential for RNA interaction and deletion of either one causes lethality. However, further analysis shows that these two zinc fingers have different functions. Yth1 lacking ZF2 cannot rescue the in vitro cleavage defect of a yth1-4 mutant extract. In contrast, Yth1 without ZF4 can function in cleavage, but shows no interaction with Fip1, a protein essential for poly(A) addition. Moreover, we find that Fip1 inhibits the interaction between Yth1 and RNA. Our results suggest that rearrangement of the post-cleavage complex to one active for polyadenylation might involve a switch in which ZF4 binds Fip1 instead of RNA.
MATERIALS AND METHODS
Strains
The YTH1 knockout strain SB7 carrying the yth1-1 allele was a kind gift from Drs Silvia M. L. Barabino and Walter Keller. SB7 (29) was used to create the strain YT1 with a disruption of the chromosomal copy of the essential YTH1 gene and a centromeric vector containing the wild-type YTH1 gene and a URA3 selectable marker [MATa, ade2, leu2, ura3, trp1Δ, his3, yth1::TRP1, YCplac33-YTH1 (CEN4 URA3 YTH1)]. Plasmids carrying a LEU2 marker and a mutated YTH1 gene were introduced into YT1. These transformants were plated on 5-fluoroorotic acid (5-FOA) medium to force loss of the plasmid carrying the wild-type YTH1 gene. The resulting yeast strains, YT2 [same as YT1 but YCplac11-YTH1 (CEN4 LEU2 YTH1)], YT3 (YCplac11-yth1ΔN1), YT4 (YCplac11-yth1ΔZ3), YT5 (YCplac11-yth1ΔC2), YT6 (YCplac11-yth1ΔC1), YT7 (YCplac11-yth1G130R) and YT8 (YCplac11-GST-YTH1) were used in this study. The mutation G130R was created by error-prone PCR (52).
RNA processing assays
Yeast cell extracts were prepared as described previously (17). After the ammonium sulfate precipitation, the proteins were resuspended in 100 µl of buffer D (20 mM Tris–HCl, pH 8.0, 50 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol, 2 µM pepstatin A, 0.6 µM leupeptin, 1 mM phenylmethylsulfonyl fluoride) and dialyzed twice against 1 l of the same buffer for 2 h each. Preparation of [α-32P]UTP-labeled full-length GAL7-1 or pre-cleaved GAL7-9 RNAs by run-off transcription and processing assays were carried out as described previously (17). In a total volume of 10 µl, 10 nM RNA (250 000 c.p.m.) was incubated with 1 µl of extract at 30°C for 30 min. The RNA products were resolved on a 5% polyacrylamide–8 M urea gel, and visualized with a PhosphorImager (Storm 860; Molecular Dynamics). For the complementation of cleavage activity in processing-deficient extracts with recombinant proteins, we used 300 ng of GST-Yth1 mutants and substituted dATP for ATP. The strength of the reaction product bands was quantitated by the NIH Image 1.62 program, and divided by number of uridines to normalize the amount of radioactivity in each band.
Recombinant proteins
The coding sequences of Yth1 mutants were cloned into pET-21b(+) (Novagen) in a way that removed the His6-tag, and the recombinant proteins were expressed in Escherichia coli BL21(DE3) pLysS using the T7 expression system (53). The Yth1 proteins were applied onto a DEAE-Sephacel column and the flow-through fractions were collected and used for co-immunoprecipitation as previously described (51). The GST-fused Yth1 deletions were derived from pGEX-6P-2 (Pharmacia). A one step affinity purification on glutathione-Sepharose™ 4B (Pharmacia) was carried out according to the manufacturer’s instructions. Recombinant His6-Fip1 and Pap1 were expressed and purified as described previously (51,54). The yeast plasmid containing the BRR5 gene with the HA3-tag at its C-terminus, pSE360-BRR5(HA)3, was recovered from the yeast strain YOM7 (27) which was a kind gift from Dr Christine Guthrie. The coding sequence of Brr5-HA3 was cloned into pET-16b (Novagen) and His6-Brr5-HA3 was expressed in E.coli BL21-SI (Gibco BRL). The His6-Brr5-HA3 was purified on Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions.
Co-immunoprecipitation
For each immunoprecipitation, 20 µl of a 50% slurry of protein A-agarose (Gibco BRL) was incubated with 10 µl of the monoclonal penta-His antibody (Qiagen) in buffer IP-150 (150 mM KCl, 20 mM Tris–HCl, pH 8.0, 0.1% NP-40) for 2 h at room temperature. The beads were washed with 1 ml of IP-150 three times and resuspended in 100 µl of IP-150 containing 10% fetal calf serum. After incubation for 1 h at 4°C, 500 ng of His6-Fip1 and 6–64 µg of total protein preparation of each recombinant Yth1 were added to the beads. This mixture was incubated for 1 h at 4°C and beads washed with 1 ml of IP-150 three times. The bound proteins were eluted in gel loading buffer containing SDS, and applied on a 12% SDS–polyacrylamide gel. The resolved proteins were immobilized onto a PVDF membrane and detected by immunoblotting with mouse polyclonal antibodies against Fip1 and Yth1 (51). To avoid heavy cross-reaction of the immunoprecipitating antibody with the secondary antibody during the western blotting, the primary and secondary antibodies were preincubated as described previously (55). Reacted His6-Fip1 and Yth1 mutants were visualized by the color reaction using a chromagenic substrate for the enzyme alkaline phosphatase (Gibco BRL) according to the manufacturer’s instructions.
Yth1 and yth1G130R were in vitro translated using TNT® T7 Coupled Reticulocyte Lysate System (Promega) and labeled with [35S]methionine according to the manufacturer’s instructions. Protein A beads were prepared as described above, and 15 µl of each translation reaction and 500 ng of His6-Fip1 were added. Co-immunoprecipitation was performed as described above, and the resolved proteins were visualized by exposing on X-ray films.
GST pull-downs
For each reaction, 20 µl of a 50% slurry of glutathione-Sepharose™ 4B (Pharmacia) was incubated in 100 µl of IP-150 containing 10% fetal calf serum. After incubation for 1 h at 4°C, a mixture of 500 ng of purified GST-Yth1 and 500 ng of His6-Brr5-HA3 was added to the beads. This mixture was incubated for 2 h at 4°C and beads washed with 1 ml of IP-150 three times. The bound proteins were eluted in 50 mM glutathione and applied onto a 10% SDS–polyacrylamide gel. The resolved proteins were immobilized on a PVDF membrane and detected by immunoblotting with rabbit polyclonal antibody against HA (Santa Cruz Biotechnology) and monoclonal antibody against GST (BD Biosciences). Visualization was carried out as described above.
UV cross-linking assays
Yth1 (200 ng), α-32P-labeled GAL7-1 prepared as described above (100 000 c.p.m.), 0.1 µg yeast tRNA, 1 mM magnesium acetate, 75 mM potassium acetate, 10 mM HEPES–KOH, pH 7.0, 2% PEG 8000 and 1 mM DTT were incubated for 10 min at 30°C. After incubation, the mixture was irradiated with 1.2 mJ of energy with a UV Stratalinker model 2400 (Stratagene) on ice. RNA was digested with 4 µg of RNase A for 1 h at 37°C, and then proteins were separated on a 10% SDS–polyacrylamide gel. The gel was stained with Silver Stain Plus (Bio-Rad) and exposed to a PhosphorImager screen (Storm 860; Molecular Dynamics). For RNA binding competition assays, 300 ng of GST-Yth1 was incubated with 66–660 ng of Fip1 or fip1-206 for 10 min at 30°C in 5 µl buffer D followed by the cross-linking assay described above.
RESULTS
Mutation of each Yth1 domain causes a different phenotype in vivo
To investigate whether the multiple zinc finger domains of Yth1 had distinct or overlapping functions, we decided to precisely delete different zinc fingers and test the deletions for viability, for activity in cleavage and polyadenylation, and for interaction with Fip1, Brr5 and RNA. There is a large amount of data indicating that such a deletion analysis of Yth1 would be a valid approach to understand Yth1 function. Numerous structural studies of DNA-binding zinc finger proteins have shown that each zinc finger motif forms a small, functional, independently folded mini-domain, and that the mode of DNA recognition is principally a one-to-one interaction between amino acids of the recognition helix and bases of the DNA (50,56). Deletion analysis of wig-1 (p53-induced mouse zinc finger protein) showed that the first zinc finger is essential for binding to double-stranded RNA, whereas zinc fingers 2 and 3 are dispensable (57). In a similar approach, it was found that zinc fingers 2–4 of MTF-1 (metal response element-binding transcription factor-1) are involved in DNA binding, whereas the first zinc finger appears to function as a metal-sensing domain (58). As the following results demonstrate, each Yth1 zinc finger deletion retained one or more of the known activities of Yth1, suggesting that the mutant proteins are folding properly. In addition to function of the different zinc fingers, we were interested in whether the unique regions of either end of Yth1 were important for Yth1 function.
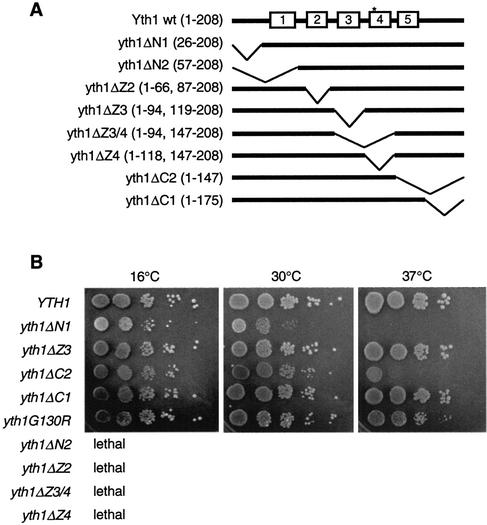
To determine which regions of Yth1 are necessary for cell viability, we replaced the wild-type YTH1 gene with the deletions depicted in Figure 1A using plasmid shuffling and 5-FOA selection as described in Materials and Methods. As shown in Figure 1B, a yeast strain carrying a deletion of the central 24 amino acids of the third zinc finger (ZF3) domain (yth1ΔZ3) shows no growth defect when compared to growth of the wild-type strain. Similarly, normal growth is observed when a yeast strain carries a construct with deletion of the C-terminal 33 amino acids (yth1ΔC1). These results suggest that these regions are dispensable for cell growth.
Figure 1.
(A) A schematic representation of Yth1 mutants used in this study. The five zinc finger motifs are indicated as boxes. The bold line shows the portions remaining in each construct; an asterisk represents the point mutation G130R. Positions of the remaining amino acids are shown in parentheses. (B) Growth behavior of S.cerevisiae carrying mutations in YTH1. Plasmids carrying the mutations were introduced into strain YT1 by plasmid shuffling. The viable strains were grown overnight in liquid culture, and their cell densities normalized to an optical density of 0.5. An equal volume of serially diluted cell suspensions was spotted on solid medium and incubated at the indicated temperature. Pictures were recorded 48 h after plating at 30 and 37°C, and 8 days at 16°C.
However, a strain with a deletion of the N-terminal 25 amino acids (yth1ΔN1) grows slowly at the permissive temperature of 30°C, and cannot grow at 37°C. Furthermore, a longer deletion of the N-terminal 56 amino acids including ZF1 (yth1ΔN2) is lethal. From these results, we conclude that the N-terminal region up to position 56 is essential for Yth1 function. Temperature sensitivity is also observed when a strain carries deletion of the C-terminal 61 amino acids (yth1ΔC2), which includes the fifth zinc finger. This phenotype is similar to the mutant allele yth1-1 that carries a deletion of 50 amino acids at the C-terminus, as shown by Barabino et al. (29). In contrast to yth1ΔN1, the yth1ΔC2 strain grows normally at the permissive temperature and slowly at 37°C. The non-essential nature of the last 33 amino acids suggests that the critical domain removed in yth1ΔC2 may be ZF5. However, we cannot exclude the possibility of an additive effect due to deletion of both regions.
Deletion of ZF2 from amino acids 67 to 86 (yth1ΔZ2) and deletion of the 28 amino acids spanning ZF4 (yth1ΔZ4) are lethal, suggesting that ZF2 and ZF4 are essential. Deletion of the central 52 amino acids including ZF3 and ZF4 (yth1ΔZ3/4) is also lethal, in agreement with the results that ZF4 is essential. A substitution of the conserved glycine residue for arginine at position 130 in ZF4 gives a temperature-sensitive yeast strain (yth1G130R). This result further supports the idea that ZF4 is responsible for an essential function. Finally, none of the viable mutants are cold-sensitive when grown at 16°C.
We next performed western blot analysis to detect Yth1 in cell extracts and determine whether the growth defects might be the consequence of poor expression of the mutant Yth1. As shown in Figure 2A, the temperature-sensitive yth1 mutants were successfully immunodetected in these extracts. In the extract from the strain carrying yth1ΔN1, a degradation product was also detected, although the protein level of yth1ΔN1 was similar to that of wild-type Yth1 (Fig. 2A, lane 2). The protein levels of yth1ΔC2 and yth1G130R were only slightly reduced compared to wild-type Yth1 (Fig. 2A, lanes 3 and 4). We also analyzed the levels of eight other processing components (Cft2, Brr5, Pta1, Rna14, Hrp1, Pap1, Fip1 and Rna15) in these extracts, in order to determine whether depletion of other components contributed to the growth deficiency of the mutant strains. In these extracts, all of the protein levels were constant except for that of Fip1, which showed a very small reduction (Fig. 2A).
Figure 2.
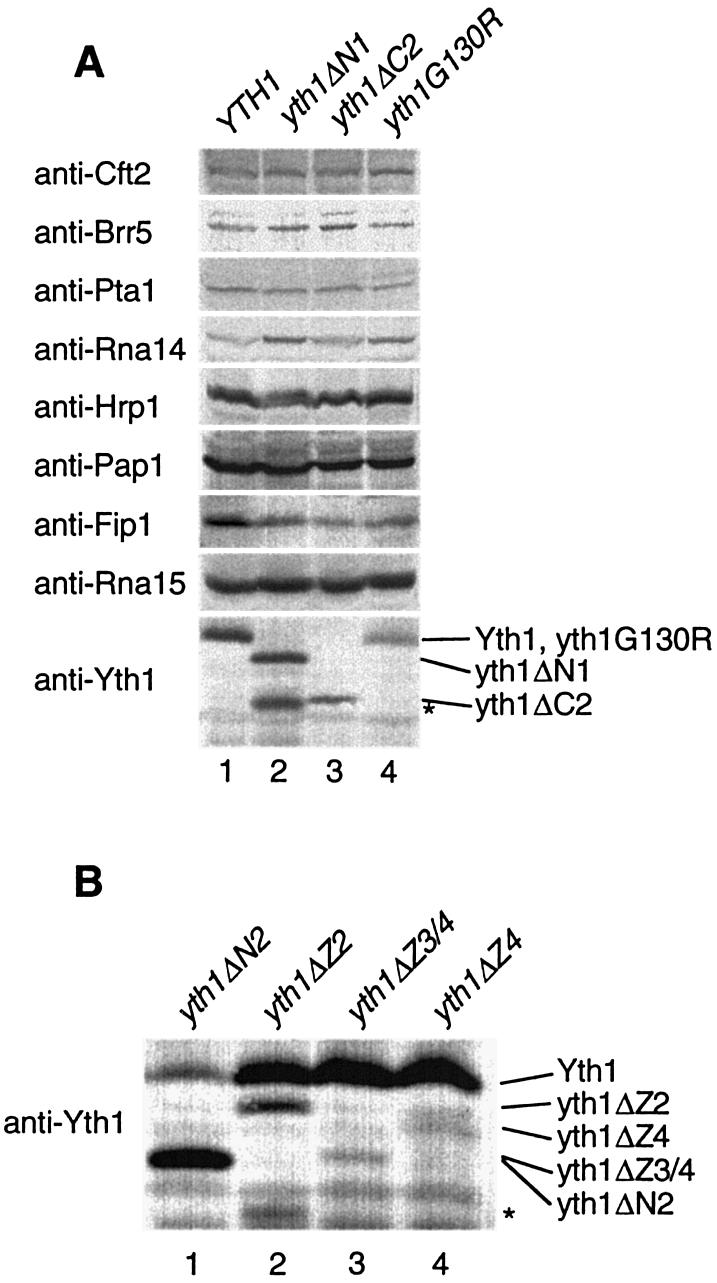
Analysis of the levels of defective yth1 proteins. (A) Immuno detection of Cft2, Brr5, Pta1, Rna14, Hrp1, Pap1, Fip1, Rna15 and Yth1 in mutant yth1 extracts. Thirty micrograms of protein was loaded in each lane. The yth1ΔN1 degradation product is indicated by an asterisk on the right. (B) Immunodetection of Yth1 deletions expressed in a YTH1 wild-type strain. An asterisk indicates truncated yth1ΔZ2.
To examine the expression of the lethal mutants (yth1ΔN2, ΔZ2, ΔZ3/4 and ΔZ4), we asked whether these proteins could be detected in extracts from strains expressing the lethal mutations in a YTH1 wild-type background. All truncations were detected in these extracts; but the protein levels varied (Fig. 2B). yth1ΔN2 was expressed at higher levels than wild-type. The level of yth1ΔZ2 was reduced compared to wild-type and degraded product was also detected (Fig. 2B, lane 2), but the level of full-length protein was similar to that seen for the viable mutants yth1ΔC2 and yth1G130R. The protein levels of yth1ΔZ3/4 and yth1ΔZ4 were significantly decreased when compared to the level of wild-type Yth1 (Fig. 2B, lanes 3 and 4). These results suggest that poor expression may contribute to the lethality of these two deletions. We do not know whether these particular mutations are inherently unstable in vivo, or if poor incorporation into CPF, especially in competition with the endogenous wild-type Yth1, results in free mutant yth1 that is recognized as abnormal and targeted for degradation. As shown below, the fact that recombinant yth1ΔZ4 could rescue the critical function of cleavage in a yth1-defective extract is strong support that the protein is folding properly.
We next examined whether the various Yth1 mutations could function in cleavage and polyadenylation in vitro and if they exhibited wild-type interactions with Fip1, Brr5 and RNA, as described in the following sections.
Effects of mutations on processing in vitro
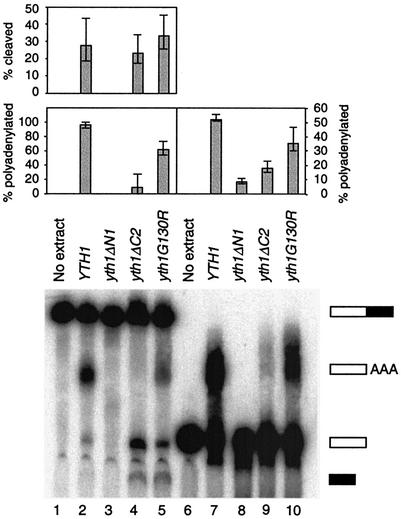
To further define the role of Yth1 in processing, we first prepared extracts from the viable mutant strains grown at permissive temperature and tested them for cleavage in vitro using the radioactive substrate GAL7-1 that contains the GAL7 poly(A) site and flanking sequences. Poly(A) addition was assayed using the radioactive substrate GAL7-9 that ended at the GAL7 poly(A) site. Reactions were performed for 30 min; we have found that very little additional processing occurs after this time period. When the full-length RNA was added to wild-type extract, 27% of the full-length RNA input was cleaved, and 97% of the cleaved RNA was polyadenylated (Fig. 3, lane 2). When the pre-cleaved RNA was added to wild-type extract, 52% of the input RNA was polyadenylated (Fig. 3, lane 7). In extract from the strain carrying yth1ΔN1, cleavage activity was not detectable (Fig. 3, lane 3) and polyadenylation activity was greatly reduced (Fig. 3, lane 8). This result confirms that Yth1 is required in both processing steps, as shown by Barabino et al. (41). Extract prepared from the strain carrying yth1ΔC2 was somewhat reduced for cleavage activity (Fig. 3, lane 4) but much more severely affected for polyadenylation in comparison to YTH1 extract (Fig. 3, lane 9). Extract from the strain carrying yth1G130R showed a similar level of activity as wild-type in cleavage (33%) (Fig. 3, lane 5), but a weakened polyadenylation activity of 62% (compared to 97% for wild-type). The defect in polyadenylation is also evident from the accumulation of cleavage product compared to wild-type extract (Fig. 3, lanes 3 and 5).
Figure 3.
Effects of yth1 mutations on cleavage and polyadenylation in vitro. Radiolabeled full-length GAL7-1 precursor (lanes 1–5) or pre-cleaved GAL7-9 RNA (lanes 6–10) was incubated in yth1 mutant extracts. RNA products were resolved on a 5% polyacrylamide–8 M urea gel and visualized by PhosphorImager scanning. Unreacted precursor (lanes 1 and 6), reactions with wild-type extract (lanes 2 and 7), yth1ΔN1 extract (lanes 3 and 8), yth1ΔC2 extract (lanes 4 and 9) and yth1G130R extract (lanes 5 and 10). Cleavage efficiency shows percentage of [(cleaved product + polyadenylated product)/(full-length RNA + cleaved product + polyadenylated product)]. Polyadenylation efficiency shows percentage of [polyadenylated product/(polyadenylated product + cleaved product)]. The values from three separate experiments for each extract were averaged.
To determine whether in vitro processing can be rescued by exogenous Yth1, we carried out complementation assays. We used extract from the yth1-4 strain, which carries a point mutation in ZF2 (41), and recombinant Yth1 and mutated proteins expressed in E.coli as GST fusions. Reactions were performed in the presence of dATP to block poly(A) addition. None of the recombinant proteins showed any cleavage activity on their own (Fig. 4, lanes 11–16). As reported previously (41), the yth1-4 extract showed very weak processing activity in both steps, but could be rescued for cleavage with wild-type protein. Addition of recombinant GST-Yth1 stimulated cleavage activity, with 27% of the input RNA being processed, compared to 42% for wild-type extract (Fig. 4, lanes 3 and 6). Recombinant GST-yth1ΔZ4 and GST-yth1ΔC2 proteins could also stimulate cleavage, but less efficiently than that of wild-type GST-Yth1 (Fig. 4, lanes 9 and 10). Recombinant GST-yth1ΔN2 and GST-yth1ΔZ2 proteins could not rescue cleavage (Fig. 4, lanes 7 and 8). These results indicate that ZF4 and ZF5 are not required for the cleavage function of Yth1.
Figure 4.
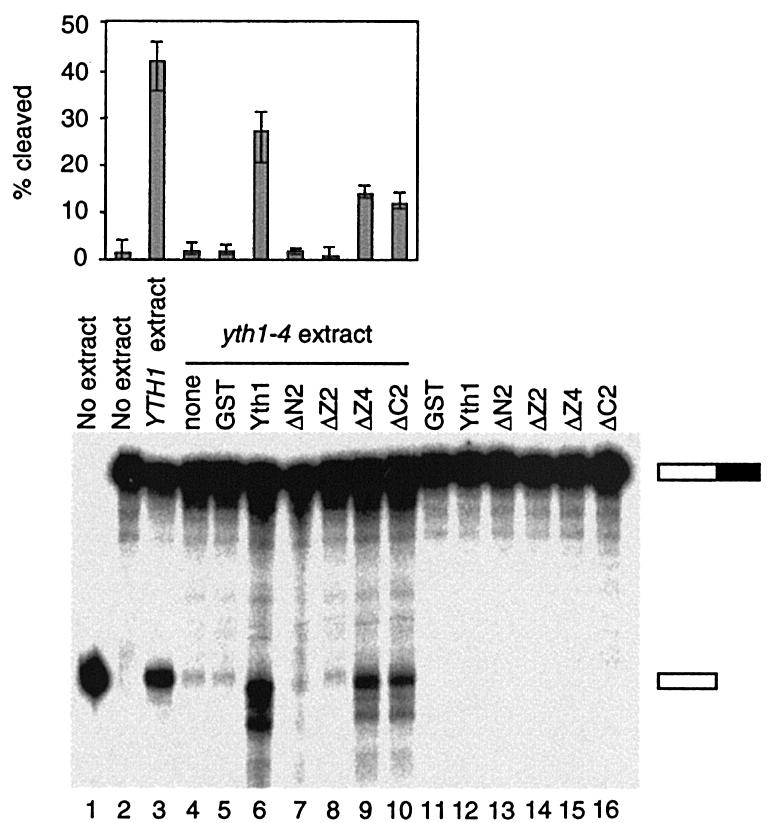
Complementation of cleavage activity in yth1-4 extract by the addition of recombinant GST-Yth1. Lane 1 shows unreacted pre-cleaved GAL7-9 RNA, lane 2 shows unreacted full-length GAL7-1 RNA, and lane 3 shows a control reaction with wild-type extract. The indicated lanes show reactions of yth1-4 extract (lane 4), yth1-4 supplemented with GST (lane 5), GST-Yth1 (lane 6), GST-yth1ΔN2 (lane 7), GST-yth1ΔZ2 (lane 8), GST-yth1ΔZ4 (lane 9) and GST-yth1ΔC2 (lane 10). Lanes 11–16 show GAL7-1 RNA incubated without extract but with GST (lane 11), GST-Yth1 (lane 12), GST-yth1ΔN2 (lane 13), GST-yth1ΔZ2 (lane 14), GST-yth1ΔZ4 (lane 15) and GST-yth1ΔC2 (lane 16). Cleavage efficiency shows percentage of [cleaved product/(full-length RNA + cleaved product)]. The values from three separate experiments were averaged.
Barabino et al. (41) found that GST-Yth1 could not rescue the poly(A) addition defect of yth1-4 extract. We obtained a similar result, with no reconstitution of polyadenylation with GST-Yth1 in extracts from the strains yth1-4, yth1ΔN1 and yth1ΔC2, even if the extract was supplemented with Fip1 and Pap1, or pre-treated at 37°C to try to destabilize the heat-labile mutants (data not shown). Moreover, the GST-YTH1 fusion was able to cover a disruption of the wild-type YTH1 gene in vivo, and removal of the N-terminal tag on recombinant Yth1 did not activate polyadenylation activity in vitro. This data supports the previously proposed hypothesis (41) that recombinant Yth1 is unable to displace the endogenous mutant yth1 from interactions with CPF subunits critical for polyadenylation.
Domains of Yth1 involved in protein–protein and protein–RNA interactions
By co-immunoprecipitation assays, Yth1 was shown to interact directly with Fip1 (41,51). We carried out similar assays with the recombinant Yth1 deletion series and His6-tagged Fip1, in order to determine the Fip1 interaction domain of Yth1 (Fig. 5). Two mutants, yth1ΔZ3/4 and yth1ΔZ4, did not co-immunoprecipitate with His6-Fip1 (Fig. 5A, lanes 6 and 7), but all other Yth1 derivatives, including yth1ΔZ3, were immunoprecipitated. Barabino et al. (41) reported that the yth1-1 protein with a deletion of the C-terminal 50 amino acids is impaired in the interaction with Fip1. Similarly, we found that the pull-down of yth1ΔC2 by His6-Fip1 was somewhat less efficient than that of wild-type, but reproducibly detectable, and in striking contrast to the lack of binding of yth1ΔZ4 (Fig. 5A, lane 8). These results indicate that ZF4 is necessary for Fip1 interaction, and that ZF5 probably stabilizes this interaction but is not required. Furthermore, when in vitro translated yth1G130R was used for co-immunoprecipitation instead of proteins expressed in E.coli, yth1G130R showed a weaker interaction with Fip1 compared to Yth1 (Fig. 5C), consistent with the polyadenylation defect caused by this mutation. This result further supports the idea that ZF4 interacts with Fip1.
Figure 5.
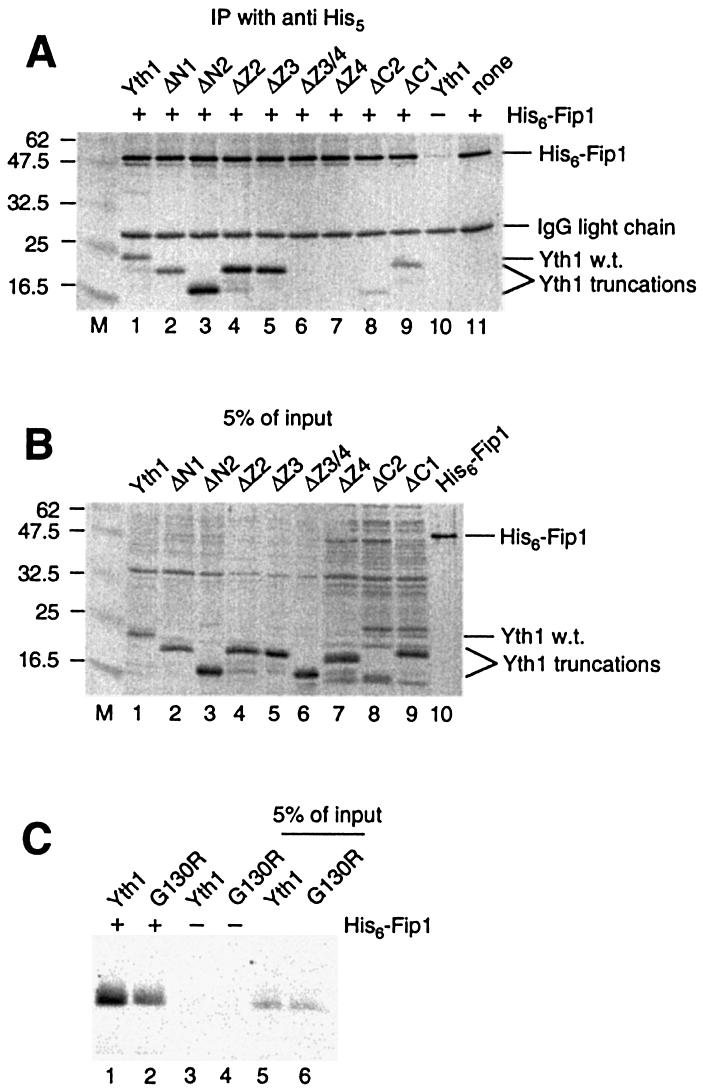
Interactions of yth1 mutants with Fip1. (A) Co-immunoprecipitation of His6-Fip1 and recombinant Yth1 deletions using anti-His5 antibody. (B) Recombinant proteins corresponding to 5% of the input used in (A) loaded directly on the gel. His6-Fip1 and Yth1 mutants were detected by blotting with anti-Fip1 and anti-Yth1 antibodies, and visualized by color reaction as described in Materials and Methods. Molecular weight markers (kDa) are shown in lane M. (C) Co-immunoprecipitation of His6-Fip1 and in vitro translated 35S-labeled Yth1 and yth1G130R using anti-His5 antibody. Lanes 5 and 6 show in vitro translated proteins corresponding to 5% of the input used for immunoprecipitation.
Yth1 also binds to the CPF subunit Brr5 (41), but the interaction domain has not been defined. We next performed a GST pull-down assay of GST-fused Yth1 truncations and recombinant His6-Brr5-HA3, to determine the Brr5 interaction domain of Yth1 (Fig. 6). The wild-type and all of the truncations tested were able to pull down His6-Brr5-HA3, but GST-yth1ΔN2 and GST-yth1ΔZ4 exhibited a weaker interaction (Fig. 6, lanes 4 and 6). Thus, ZF4 and the N-terminus including ZF1 both contribute to efficient binding of Brr5. It would not be surprising that Brr5, a relatively large protein (100 kDa) in comparison to Yth1 (24 kDa), contacted Yth1 in two places. The fact that Yth1 with the ZF4 deletion lost Fip1 binding and had a weakened Brr5 interaction raised the question of whether the Brr5–Yth1 interaction could compete with the Fip1–Yth1 interaction at ZF4. However, in a co-immunoprecipitation assay with all three proteins together, we found that His6-Fip1 and His6-Brr5-HA3 can bind Yth1 at the same time without interference (data not shown).
Figure 6.
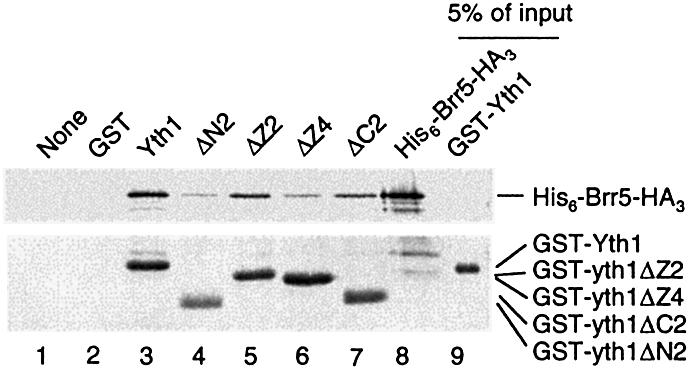
Deletions of the N-terminal region and ZF4 of Yth1 impair the interaction with Brr5. GST-fused Yth1 deletions were tested by a GST pull-down assay with His6- and HA-tagged Brr5 using glutathione-Sepharose beads. His6-Brr5-HA3 is not pulled down when GST-Yth1 is absent (lane 1) or with GST alone (lane 2). Recombinant proteins corresponding to 5% of the input were loaded directly on the gel (lanes 8 and 9). His6-Brr5-HA3 and GST-Yth1 mutants were detected by blotting with anti-HA and anti-GST antibodies, and visualized by color reaction as described in Materials and Methods.
By a primer extension assay, we found that Yth1 binds the U-rich stretches of GAL7 RNA (data not shown), similar to the previous study showing that Yth1 binds RNA with a preference for poly(U) (29). The binding of Yth1 to poly(U) is very weak, and Barabino et al. (41) have used UV cross-linking, a more sensitive assay, to monitor the interaction of Yth1 and RNA. We have also found that this assay corresponds well with the ability of protein to bind RNA (40,54). Thus, to determine the RNA interaction domains of Yth1, we performed UV cross-linking with GST-fused Yth1 derivatives and radiolabeled GAL7-1 RNA. As shown in Figure 7, GST-Yth1 and two truncations, GST-yth1ΔN2 and GST-yth1ΔC2, cross-linked to RNA with comparable efficiencies (Fig. 7A, lanes 1, 2 and 5). In contrast, the cross-linking of GST-yth1ΔZ2 and GST-yth1ΔZ4 to RNA was greatly reduced (Fig. 7A, lanes 3 and 4). This finding suggests that ZF2 and ZF4 must both be present for normal interaction with RNA.
Figure 7.
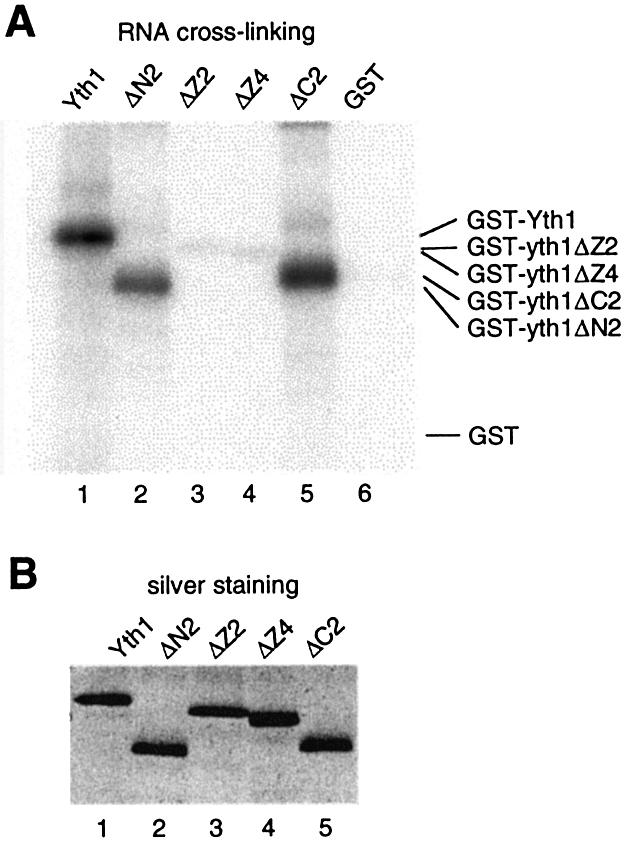
ZF2 and ZF4 are necessary for interaction with RNA. GST-fused Yth1 proteins were UV cross-linked to radiolabeled GAL7-1 pre-mRNA. Proteins were resolved on a 10% SDS–polyacrylamide gel. (A) Phosphor Imager exposure of the silver stained gel of (B). (B) Silver stained picture of the gel.
Fip1 blocks the interaction between Yth1 and RNA
Because ZF4 is needed for interaction with both Fip1 and RNA, we were curious as to whether the presence of Fip1 would interfere with the RNA interaction. To determine this, we performed a UV cross-linking assay using GST-Yth1 and radiolabeled GAL7-1 together with His6-Fip1 as a competitor. We first incubated GST-Yth1 with various amounts of His6-Fip1 or His6-fip1-206, added RNA, and exposed the samples to UV light. The fip1-206 derivative has a deletion of the C-terminal 121 amino acids and does not interact with Yth1 (51). As shown in Figure 8, the cross-linking efficiency was greatly reduced as the amount of His6-Fip1 increased. When the GST-Yth1:His6-Fip1 molar ratio was 1:1, cross-linking efficiency dropped to 21% of that seen without Fip1 (Fig. 8B). However, addition of His6-fip1-206 did not affect cross-linking efficiency. This result suggests that Fip1 occupancy of ZF4 inhibits the interaction of Yth1 with RNA.
Figure 8.
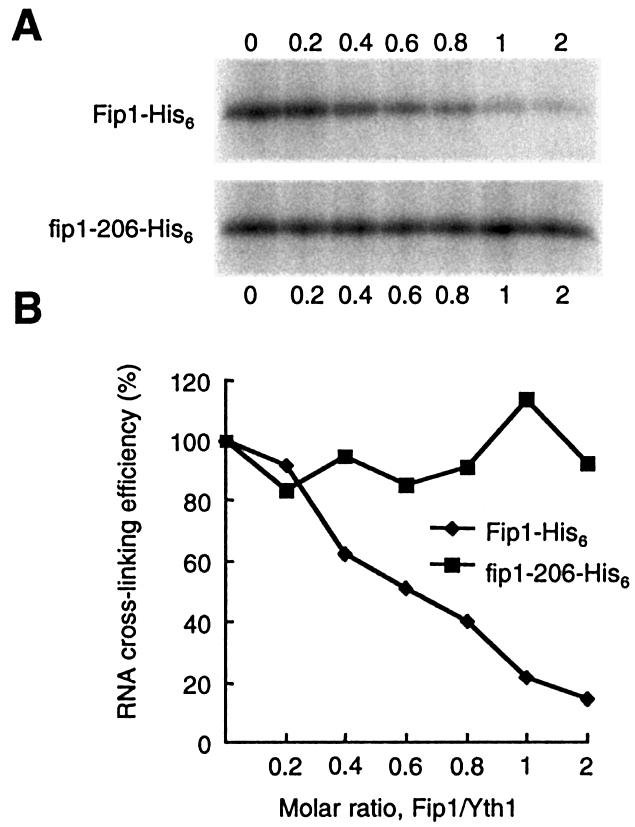
Fip1 interaction with Yth1 blocks interaction between Yth1 and RNA. (A) GST-Yth1 was incubated with various amounts of His6-Fip1 or His6-fip1-206, and cross-linked to radiolabeled GAL7-1 pre-mRNA. Proteins were resolved on a 10% SDS–polyacrylamide gel and visualized by PhosphorImager scanning. The molar ratio of Fip1/Yth1 is given under each lane. (B) Quantitation of results in (A). Incubation with His6-Fip1 (closed diamonds); incubation with His6-fip1-206 (closed squares).
DISCUSSION
While most of the components of the eukaryotic polyadenylation machinery have been identified, little is known about the architecture of the complex or the function of many of the subunits. In this study, we have identified domains in Yth1 important for the protein’s activity and its interactions with RNA and the CPF subunits Fip1 and Brr5. The main conclusions from our study are: (i) ZF4 is important for all three interactions and for polyadenylation, but is dispensable for the function of Yth1 in cleavage; (ii) the unique N-terminus of Yth1 (before the five zinc fingers) is essential for both cleavage and poly(A) addition; (iii) only four of the five zinc fingers are necessary; (iv) Fip1 prevents the interaction of Yth1 and RNA. The results of this study are summarized in Figure 9. This new data and the earlier studies with Yth1 (29,41) support a functional organization of Yth1 in which the C-terminal part is dedicated to polyadenylation, while the N-terminal half is required for both processing steps.
Figure 9.

Schematic representation of Yth1 function. (A) Protein–protein and protein–RNA interacting domains of Yth1. The zinc finger motifs are indicated as boxes. The N-terminal region including ZF1 and ZF2 is required for cleavage, the N-terminus including ZF1 and ZF2, as well as ZF4 and ZF5, are important for polyadenylation. ZF2 and ZF4 are involved in interaction with RNA. ZF4 and the C-terminus, probably through ZF5, is involved in interaction with Fip1. The N-terminus and ZF4 are important for interaction with Brr5. (B) Model of how Yth1 might play an important role for transition from cleavage to polyadenylation. During cleavage, Yth1, as part of the cleavage/polyadenylation complex, is associated with RNA. When cleavage is completed, Yth1 switches from RNA to Fip1, and polyadenylation is initiated. Only the Yth1, Fip1 and Pap1 subunits of CPF are shown.
Our deletion analysis indicates that the N-terminal half can be further subdivided into distinct functional domains. For example, removal of sequence before ZF1 causes a temperature-sensitive growth phenotype and gives extracts impaired for both cleavage and polyadenylation. Further truncation to eliminate ZF1 causes lethality and this mutant Yth1 cannot restore cleavage in vitro. However, there are no defects in the interaction with Fip1 or RNA, but there is reduced binding to Brr5. It is not clear at this time whether the functional deficit due to loss of the N-terminus is related to the poor interaction with Brr5, or instead with another factor, which like Brr5, is essential for both steps of 3′ end formation.
We have found that cells with a deletion of ZF2, also in the N-terminal half of Yth1, are not viable. An analysis by Barabino et al. (41) had demonstrated ZF2 to be critical for Yth1 function. In their temperature-sensitive yth1-4 allele, mutation of a conserved residue in this region (W70A) weakened interaction with the rest of CPF, but not with Fip1 or Brr5, and caused defects in both the cleavage and poly(A) addition steps. Recombinant yth1-4 protein also showed reduced cross-linking to RNA. Consistent with this study, we show that the yth1 protein completely lacking ZF2 could no longer interact well with RNA or rescue cleavage in vitro, but retained full interaction with Fip1 and Brr5. It is not known which CPF subunit contacts ZF2 of Yth1 and whether RNA binding and CPF interaction contribute equally to ZF2 function.
The central zinc finger, ZF3, can be eliminated without any consequences to cell survival. This result was surprising given that all of the Yth1 homologs from other species have five zinc fingers (29). In contrast, deletion of the next zinc finger, ZF4, located in the C-terminal half, was lethal. However, unlike yth1ΔZ2, yth1ΔZ4 can rescue cleavage in vitro. Interestingly, ZF4 is also important for the cross-linking of Yth1 to RNA. These two findings suggest that contact with the RNA precursor through ZF4 must not be important for cleavage. In addition to the RNA binding defect, a ZF4 deletion results in complete loss of Fip1 binding and a weakened Brr5 interaction. Moreover, the fact that the G130R point mutation in ZF4 reduces the efficiency of Fip1 binding and impairs polyadenylation but not cleavage confirms that Fip1 interaction is an important function of ZF4.
The ZF4 data is consistent with our previous work showing that extracts from cells with a disruption of the Yth1 binding domain on Fip1 (fip1-206) are inactive for polyadenylation (51). This fip1 mutant has completely lost Yth1 interaction, but is not affected in the ability to bind to Pap1 and to inhibit Pap1 activity. However, unlike the ZF4 deletion, a strain with this fip1 mutation is viable unless grown at 37°C, suggesting that contacts made between the Fip1/Pap1 heterodimer and components of the polyadenylation complex other than Yth1 are sufficient to incorporate these two proteins into the holoenzyme. This result, together with our new data, suggests that the essential nature of ZF4 in Yth1 is due to at least two functions: the interaction with Fip1 and the interaction with RNA.
The final zinc finger, ZF5, is an important but non-essential domain of Yth1 that contributes to the efficiency of Fip1 binding. A deletion from the C-terminus which includes ZF5 (yth1ΔC2) causes slower growth at 37°C and extracts deficient for poly(A) addition but not cleavage. Surprisingly, deletion of the C-terminus beyond the zinc fingers causes no effect on cell growth, even though the interaction with Fip1 may be somewhat diminished. Thus, interactions of Fip1 with ZF4 and ZF5 are sufficient for normal polyadenylation.
By RNA cross-linking assays, we have found that Yth1 does not bind to RNA and Fip1 at the same time. One interpretation of this result is that ZF4 helps to mediate the switch from cleavage to polyadenylation. A model for this additional function of Yth1 is illustrated in Figure 9B. Fip1 binds Pap1 through a domain different from the one responsible for Yth1 interaction. At the cleavage step, Yth1, along with other CPF subunits, binds to RNA at the U-rich regions immediately flanking the poly(A) site, but perhaps not tightly associated with Fip1. Following cleavage, Yth1 continues to bind RNA just upstream of the newly created 3′ end. At this point, it may be important for Fip1 to displace Yth1 from RNA, so that Pap1 can access the 3′ end and initiate poly(A) synthesis. Additional research is needed to understand what changes in the configuration of the complex occur following cleavage and how these changes promote polyadenylation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Silvia M. L. Barabino and Walter Keller for the yeast strain SB7, and Christine Guthrie for the yeast strain YOM7. We are grateful to Kimberly Sparks, Shipra Agrawal and Alexander Zhelkovsky for critically reading the manuscript and all members of the Moore laboratory for helpful discussions. This work was supported by NIH grants RO1 GM57218-01A2, GM41752.
REFERENCES
- 1.Hirose Y. and Manley,J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- 2.Cramer P., Srebrow,A., Kadener,S., Werbajh,S., de la Mata,M., Melen,G., Nogues,G. and Kornblihtt,A.R. (2001) Coordination between transcription and pre-mRNA processing. FEBS Lett., 498, 179–182. [DOI] [PubMed] [Google Scholar]
- 3.Proudfoot N.J., Furger,A. and Dye,M.J. (2002) Integrating mRNA processing with transcription. Cell, 108, 501–512. [DOI] [PubMed] [Google Scholar]
- 4.Long R.M., Elliott,D.J., Stutz,F., Rosbash,M. and Singer,R.H. (1995) Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA, 1, 1071–1078. [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y. and Carmichael,G.C. (1996) Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol., 16, 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z. and Krug,R.M. (2000) Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol., 8, 376–383. [DOI] [PubMed] [Google Scholar]
- 7.Proudfoot N. (1996) Ending the message is not so simple. Cell, 87, 779–781. [DOI] [PubMed] [Google Scholar]
- 8.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs A.B. and Varani,G. (2000) Eukaryotic translation initiation: there are (at least) two sides to every story. Nature Struct. Biol., 7, 356–361. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell P. and Tollervey,D. (2001) mRNA turnover. Curr. Opin. Cell Biol., 13, 320–325. [DOI] [PubMed] [Google Scholar]
- 11.Tucker M. and Parker,R. (2000) Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem., 69, 571–595. [DOI] [PubMed] [Google Scholar]
- 12.Wilusz C.J., Wormington,M. and Peltz,S.W. (2001) The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell Biol., 2, 237–246. [DOI] [PubMed] [Google Scholar]
- 13.Wahle E. and Ruegsegger,U. (1999) 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore C.L. and Sharp,P.A. (1985) Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell, 41, 845–855. [DOI] [PubMed] [Google Scholar]
- 16.Butler J.S. and Platt,T. (1988) RNA processing generates the mature 3′ end of yeast CYC1 messenger RNA in vitro. Science, 242, 1270–1274. [DOI] [PubMed] [Google Scholar]
- 17.Chen J. and Moore,C. (1992) Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol., 12, 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kessler M.M., Zhao,J. and Moore,C.L. (1996) Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J. Biol. Chem., 271, 27167–27175. [DOI] [PubMed] [Google Scholar]
- 19.Minvielle-Sebastia L., Preker,P.J. and Keller,W. (1994) RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science, 266, 1702–1705. [DOI] [PubMed] [Google Scholar]
- 20.Amrani N., Minet,M., Wyers,F., Dufour,M.E., Aggerbeck,L.P. and Lacroute,F. (1997) PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol., 17, 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preker P.J., Ohnacker,M., Minvielle-Sebastia,L. and Keller,W. (1997) A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J., 16, 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohnacker M., Barabino,S.M., Preker,P.J. and Keller,W. (2000) The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J., 19, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vo L.T., Minet,M., Schmitter,J.M., Lacroute,F. and Wyers,F. (2001) Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol. Cell. Biol., 21, 8346–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M., Remor,M., Hofert,C., Schelder,M., Brajenovic,M., Ruffner,H., Merino,A., Klein,K., Hudak,M., Dickson,D., Rudi,T., Gnau,V., Bauch,A., Bastuck,S., Huhse,B., Leutwein,C., Heurtier,M.A., Copley,R.R., Edelmann,A., Querfurth,E., Rybin,V., Drewes,G., Raida,M., Bouwmeester,T., Bork,P., Seraphin,B., Kuster,B., Neubauer,G. and Superti-Furga,G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J., Kessler,M.M. and Moore,C.L. (1997) Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem., 272, 10831–10838. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J., Kessler,M., Helmling,S., O’Connor,J.P. and Moore,C.L. (1999) Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol., 19, 7733–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chanfreau G., Noble,S.M. and Guthrie,C. (1996) Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF). Science, 274, 1511–1514. [DOI] [PubMed] [Google Scholar]
- 28.Stumpf G. and Domdey,H. (1996) Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor. Science, 274, 1517–1520. [DOI] [PubMed] [Google Scholar]
- 29.Barabino S.M., Hubner,W., Jenny,A., Minvielle-Sebastia,L. and Keller,W. (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev., 11, 1703–1716. [DOI] [PubMed] [Google Scholar]
- 30.Guo Z., Russo,P., Yun,D.F., Butler,J.S. and Sherman,F. (1995) Redundant 3′ end-forming signals for the yeast CYC1 mRNA. Proc. Natl Acad. Sci. USA, 92, 4211–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo P., Li,W.Z., Hampsey,D.M., Zaret,K.S. and Sherman,F. (1991) Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J., 10, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo Z. and Sherman,F. (1995) 3′-end-forming signals of yeast mRNA. Mol. Cell. Biol., 15, 5983–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Helden J., del Olmo,M. and Perez-Ortin,J.E. (2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res., 28, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heidmann S., Obermaier,B., Vogel,K. and Domdey,H. (1992) Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 4215–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo P., Li,W.Z., Guo,Z. and Sherman,F. (1993) Signals that produce 3′ termini in CYC1 mRNA of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7836–7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessler M.M., Henry,M.F., Shen,E., Zhao,J., Gross,S., Silver,P.A. and Moore,C.L. (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev., 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S. and Hyman,L.E. (1998) A specific RNA-protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res., 26, 4965–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentini S.R., Weiss,V.H. and Silver,P.A. (1999) Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA, 5, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross S. and Moore,C.L. (2001) Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol., 21, 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barabino S.M., Ohnacker,M. and Keller,W. (2000) Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J., 19, 3778–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dichtl B. and Keller,W. (2001) Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J., 20, 3197–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dichtl B., Blank,D., Sadowski,M., Hubner,W., Weiser,S. and Keller,W. (2002) Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J., 21, 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabara H., Hill,R.J., Mello,C.C., Priess,J.R. and Kohara,Y. (1999) pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development, 126, 1–11. [DOI] [PubMed] [Google Scholar]
- 45.Reese K.J., Dunn,M.A., Waddle,J.A. and Seydoux,G. (2000) Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell, 6, 445–455. [DOI] [PubMed] [Google Scholar]
- 46.Hendriks E.F., Robinson,D.R., Hinkins,M. and Matthews,K.R. (2001) A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J., 20, 6700–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai W.S., Carballo,E., Thorn,J.M., Kennington,E.A. and Blackshear,P.J. (2000) Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem., 275, 17827–17837. [DOI] [PubMed] [Google Scholar]
- 48.Lai W.S., Kennington,E.A. and Blackshear,P.J. (2002) Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem., 277, 9606–9613. [DOI] [PubMed] [Google Scholar]
- 49.Mackay J.P. and Crossley,M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- 50.Laity J.H., Lee,B.M. and Wright,P.E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol., 11, 39–46. [DOI] [PubMed] [Google Scholar]
- 51.Helmling S., Zhelkovsky,A. and Moore,C.L. (2001) Fip1 regulates the activity of Poly(A) polymerase through multiple interactions. Mol. Cell. Biol., 21, 2026–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- 53.Studier F.W. (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol., 219, 37–44. [DOI] [PubMed] [Google Scholar]
- 54.Zhelkovsky A.M., Kessler,M.M. and Moore,C.L. (1995) Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s). J. Biol. Chem., 270, 26715–26720. [DOI] [PubMed] [Google Scholar]
- 55.Langstein J. and Schwarz,H. (1997) Suppression of irrelevant signals in immunoblots by preconjugation of primary antibodies. Biotechniques, 23, 1006–1008, 1010. [DOI] [PubMed] [Google Scholar]
- 56.Klug A. (1999) Zinc finger peptides for the regulation of gene expression. J. Mol. Biol., 293, 215–218. [DOI] [PubMed] [Google Scholar]
- 57.Mendez-Vidal C., Wilhelm,M.T., Hellborg,F., Qian,W. and Wiman,K.G. (2002) The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res., 30, 1991–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bittel D.C., Smirnova,I.V. and Andrews,G.K. (2000) Functional heterogeneity in the zinc fingers of metalloregulatory protein metal response element-binding transcription factor-1. J. Biol. Chem., 275, 37194–37201. [DOI] [PubMed] [Google Scholar]