Abstract
Single-nucleotide polymorphism (SNP) analysis is a powerful tool for mapping and diagnosing disease-related alleles. Mutation analysis by polymerase-mediated single-base primer extension (minisequencing) can be massively parallelized using DNA microchips or flow cytometry with microspheres as solid support. By adding a unique oligonucleotide tag to the 5′ end of the minisequencing primer and attaching the complementary antitag to the array or bead surface, the assay can be ‘demultiplexed’. Such high-throughput scoring of SNPs requires a high level of primer multiplexing in order to analyze multiple loci in one assay, thus enabling inexpensive and fast polymorphism scoring. We present a computer program to automate the design process for the assay. Oligonucleotide primers for the reaction are automatically selected by the software, a unique DNA tag/antitag system is generated, and the pairing of primers and DNA tags is automatically done in a way to avoid any crossreactivity. We report results on a 45-plex genotyping assay, indicating that minisequencing can be adapted to be a powerful tool for high-throughput, massively parallel genotyping. The software is available to academic users on request.
INTRODUCTION
Single-nucleotide polymorphisms (SNPs) have been estimated to occur at a rate of about one in every thousand nucleotides in the human genome (1,2). To date, more than 1.4 million SNPs have been identified, comprising a substantial proportion of all common human variation. Tools to routinely analyze a growing number of SNPs will play a key role in medical diagnosis, they will make it possible to perform studies to identify genes that confer risk for common diseases (3), and they will provide new insights into the history of human populations by allowing studies of human genetic diversity (4). Such applications could involve the simultaneous screening of thousands of SNPs, constituting a pressing need for robust, high-throughput and cost efficient SNP scoring methods.
The minisequencing approach to SNP analysis involves the annealing of an oligonucleotide primer directly adjacent to the mutation site, and polymerase-mediated single-base extension using labeled dideoxynucleotide triphosphates (ddNTPs) (4). By combining this technique with the analytical power of flow cytometry using multiplexing microsphere arrays, the technology can be used to simultaneously analyze multiple, potentially hundreds to thousands of sites (5).
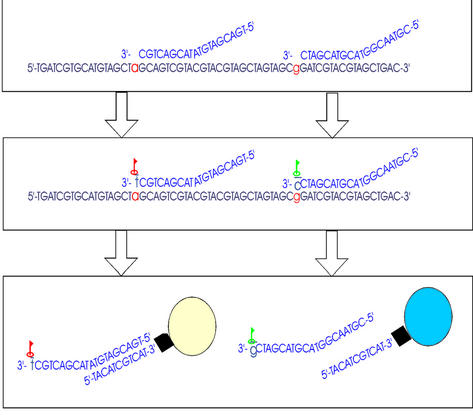
By attaching unique DNA tags to the 5′ end of each minisequencing primer, and by covalently binding the complementary tags (antitags) to microspheres or the chip surface, the primers are sorted, and the assay is thus demultiplexed. The fluorescent label introduced in the single base extension will then reveal the genotype. Figure 1 illustrates the experiment.
Figure 1.
Flow cytometry based minisequencing. Step one involves annealing of the primer–tag oligomers to the template strand, adjacent to the polymorphism sites. In step two, the polymerase extends the primer by fluorescently labeled ddNTPs. Step three involves dissociation of the primer and the template strand, and annealing between the tags and their microsphere or array bound antitags. Finally, microsphere color (or array spot position) and base label reveal genotype.
To permit multiplexed SNP scoring as described above, minisequencing primers must be appropriately chosen. Most importantly, such primers must not false prime, i.e. bind to a different site on the template strand (thus resulting in the incorporation of an arbitrary nucleotide unrelated to the SNP); they must not form homo- or heterodimers; they should not form hairpins; but they must bind immediately adjacent to the 3′ side of the mutation and extend in the polymerase reaction. Furthermore, a tag/antitag oligonucleotide code is required in order to ‘sort’ the extended primers to the corresponding elements on the microarray respectively to the correct microspheres in the flow cytometry assay, and thus ‘demultiplex’ the experiment. These tags must not show any cross-hybridization, they must be carefully chosen to avoid reactions with any of the minisequencing primers, and the pairing of primers and tags poses another challenging combinatorial problem, as, here as well, hairpins and crossreactivity over the joint between primer and tag must be taken into account.
A number of computer programs exist to aid in the primer design process for the polymerase chain reaction (PCR) (6–9 and others). However, some different constraints are required for minisequencing primers, which are not considered by the available software. Only one primer is required, and can be chosen from either the plus or the minus strand, as opposed to primer pairs in the PCR case. That primer must bind immediately adjacent to the 3′ end of the polymorphism for the polymerase to append the next base opposite the SNP. A major complication is the simultaneous extension of all primers in the same reaction. This multiplexing of the minisequencing reaction requires very careful design of the oligonucleotides, as crossreactions between different minisequencing primers will inadvertently cause false results, a problem that is not (and need not be) considered by other primer design software. Furthermore, all primers must work under the same reaction conditions, most importantly, the same temperature, T. It is thus essential to use an accurate model to predict nucleic acid hybridization.
METHODS
Free energy calculation
Given two arbitrary DNA single strands, we now tackle the question of whether they will form a stable duplex at some given temperature. We use the nearest neighbor model (10) for the thermodynamic considerations. However, its application requires prior knowledge about which base pairs form in the annealing reaction. The situation is further complicated as the duplex may contain bulges and loops (11–13).
The Smith–Waterman alignment algorithm (14) is widely used in bioinformatics to align similar subsequences in two given sequences. It calculates a local alignment between the two sequences and returns the optimum alignment found, maximizing or minimizing a score w(x,y) over nucleotide or amino acid pairs formed in the alignment. The algorithm returns the optimum alignment of subsequences, allowing both gaps and mismatches in the two sequences.
We use the Smith–Waterman alignment algorithm to determine the minimum free energy alignment of two DNA strands at a given, fixed temperature. Thus, instead of using a score on amino acid pairs as in the original algorithm, we use the nearest neighbor ΔG parameters. Note that some modifications to the original algorithm are required to account for the nearest neighbors, however, this is relatively straightforward and can be done by storing additional information in the dynamic programming table. Thus, given two single DNA sequences, the algorithm computes the most stable interaction the strands can form, and returns free energy change ΔG for the nucleation reaction. The alignment returned may contain gaps, which correspond to bulges or loops in the DNA duplex. If ΔG < 0, the model predicts favorable energetics for the nucleation reaction, and we assume that the DNA strands interact in the experiment. Note that T is fixed in the calculation and should be set to the temperature at which primer and template annealing is carried out in the experiment, or some lower temperature if an additional ‘margin of safety’ is desired.
Minisequencing primer selection
The SBEprimer program has been implemented to automate the minisequencing primer design process. It designs sets of mutually compatible primers that will minimize the number of experiments required to genotype a given number of polymorphic sites, proceeding through seven iterative steps. (i) Read template sequence and SNP locations. (ii) For each SNP, check if either of the plus-strand or minus-strand primers adjacent to the SNP will false prime. If so, remove the primer. (iii) Generate list of primers adjacent to SNP from both plus and minus strands, requiring the primer length to be within some given limits and the melting temperature to be higher than the reaction annealing temperature. (iv) Check each primer candidate for hairpins and homodimer formation; remove any candidate forming hairpins or homodimer. (v) Calculate all interactions between different primer candidates. (vi) Choose maximal sets of mutually compatible primer candidates, such that each polymorphism has exactly one primer in exactly one primer set. (vii) Output primer sets.
We discuss these steps in more detail in the following.
False priming check (step 2). The minisequencing primer for one polymorphism site can be chosen from either the plus or the minus strand, but it must bind immediately adjacent to the SNP. We speak of the ‘plus’ and ‘minus’ primer in the following; note, however, that for now we make no assumptions regarding the length of the primer, i.e. we do not fix its 5′ end for the time being. By choosing the primer such that its 3′ end binds adjacent to the SNP, the next base added to the primer by the polymerase will oppose the polymorphism and hence allow its typing. However, if the primer binds somewhere else and the polymerase extends there, a false signal is generated. It is thus a requirement to exclude any primer that will false prime.
It has been shown that the polymerase reaction requires the terminal 3′ end bases of the primer to form stable base pairs with the template (15). Our procedure to identify potential false priming sites uses a hashtable of all 4mers, for each such 4mer storing a list of the plus and minus primers that contain the reverse Watson–Crick complement of that 4mer at their 3′ end (considering two primers for each polymorphic site, the plus and the minus primer, and leaving their 5′ end open for the time being). This hashtable is then used in a routine that screens the entire template sequence and its complement. For each 4mer in the template sequence, it checks the list of primers contained in the corresponding list in the table, and checks for potential false priming. The program attempts to extend the 4mer duplex, checking if a stable interaction (with negative free energy ΔG) is possible. The extension is done using a variant of the Smith–Waterman alignment algorithm, thus again allowing for bulges and loops in the duplex. The extension is aborted if either a negative free energy interaction has been found (and the primer is consequently removed from the candidate set), or a threshold ‘maximum primer length’ has been reached.
Primer candidate evaluation (steps 3 and 4). The program will then evaluate the remaining candidates further. We will now consider the primer length as well, which has been neglected so far. To do so, SBEprimer will generate all primers from both the plus and minus strand of each SNP (provided they have not been excluded in step 2) of length between two given parameters minlen and maxlen, and with melting temperature Tm above the PCR annealing temperature.
Subsequently, SBEprimer checks for homodimer and hairpin formation. The homodimer check is done by simply calculating the minimum free energy alignment of a primer candidate with itself.
We have tried several different simple heuristics for the hairpin check. Our results indicate that, whenever one of these predicts a hairpin, the homodimer check will also show potential homodimer formation (data not shown). We have thus decided against a dedicated hairpin check module, but consider this to be covered by the homodimer module as well.
Primer multiplexing evaluation (steps 5 to 7). Finally, the primers must be chosen for the multiplexing assay. Given a set S of SNPs and a list Ci, i ∈ {1..|S|} of primer candidates for each s ∈ S, the task is to generate disjoint sets P1 .. Pm of primers that will fulfil the following criteria. (i) For each SNP i, exactly one primer from Ci must be in ∪mj = 1 Pj. This means that each SNP is genotyped by a primer in one set. (ii) Any two primers pa and pb from the same set Pj ‘work together’, i.e. no heterodimer formation occurs between any two primers within the same set. Hence, the primers in one set Pj can be multiplexed. (iii) The number m of different sets is minimized. m corresponds to the number of experiments that have to be run separately in order to genotype all polymorphisms. Ideally, m=1.
The multiplexing module involves the prediction of all pairwise interactions between any of the primer candidates from distinct SNPs. These interactions are calculated using the minimum free energy alignment algorithm, as described above. Again, we assume that two given primers interact if ΔG < 0, and will not interact otherwise.
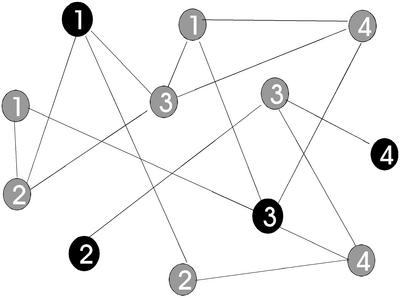
We then use a graph theoretic model to solve the problem. Create an undirected graph G = (V, E) with vertex set V and edge set E as follows: for each primer candidate left after step 5, create one vertex v ∈ V, then label each vertex with the identifier of the polymorphism genotyped by the corresponding primer and finally, create an edge (v1,v2) ∈ E between two vertices v1 and v2, if the minimum free energy alignment for the corresponding primers shows negative ΔG, i.e. if the corresponding primers interact, and if they bear different SNP labels (and hence genotype different polymorphisms). Figure 2 illustrates the construction.
Figure 2.
Sample instance of the generalized graph coloring problem. The vertices correspond to primer candidates, two vertices being connected when the corresponding primers interact and genotype different polymorphisms. Two vertices with equal numbers type the same SNP. The dark black vertices constitute an optimum solution, as one vertex has been chosen for each polymorphism, and the corresponding primers do not interact. In this simple example, all chosen vertices can be used in one single experiment
The multiplexing primer selection problem then transforms to a special version of the graph coloring problem. In the graph coloring problem, the task is to assign a color to each vertex, where no adjacent vertices [vertices v1, v2 connected by an edge (v1,v2) ∈ E] can have the same color, and the number of different colors used over the entire graph is to be minimized. In our case, not all vertices need to be assigned a color, but only one vertex from each group of vertices bearing the same label, which one of them can be freely chosen. This construction ensures that exactly one primer is used for each polymorphic site, and vertices and their respective primers assigned the same color can be multiplexed together in the same experiment.
To solve the problem, the algorithm splits the vertices in |S| distinct groups, according to their label. Then, all vertices v within each group are sorted according to their vertex degree deg(v) := |{(x,y) ∈ E|v = x or v = y}|. Only the vertex n with the lowest degree is kept in each group, all other vertices and the corresponding edges are removed from the graph. Hence, we now have an instance of the standard graph coloring problem, where each vertex has to be assigned a color. A standard algorithm is finally used to color this residual graph.
Running time. It is difficult to provide a theoretical analysis of the running time of the algorithm, as it depends on the number of feasible primer candidates. Clearly, the time required for the Smith–Waterman alignment is O(nm), where n and m are the lengths of the two sequences, respectively. This computation is done for each pair of primer candidates for different SNPs. Similarly, the false priming check has linear time for pre-processing (creating the hashtable), and then time dependent on the number of hits for the extension of the found hits.
In practice, the running time of the entire algorithm is very fast; a 45-plex experiment with a 100 KB false priming check sequence database took 1.15 min; primers for 100 SNPs with a 1 MB database can be computed in ∼33 min with 5 MB memory.
The software is available for different Unix systems and Windows.
Tag generation
A second problem associated with ‘demultiplexing’ the genotyping assay is the need for tag/antitag pairs to be used with the primers. The tags will be conjugated with the minisequencing primer, the resulting oligo having a dual function: the primer part is required for the single base extension reaction, whereas the tag part sorts the extended primer to its corresponding antitag on the bead or chip surface, thus enabling simple readout of the results.
For this to work, there must not be any crossreactivity between the tags and antitags. (i) Each tag must bind to its corresponding antitag. (ii) A tag must not bind to a ‘foreign’ antitag. (iii) No two tags should bind to one another (including the homodimer case). (iv) No two antitags should bind to one another (including the homodimer case).
The problem of designing DNA tag/antitag systems satisfying requirements (i) and (ii) has been previously described. Frutos et al. (16) use a coding theory approach with Hamming distance conditions to avoid crosshybridization. They design octamers with 50% G-C content, differing in at least four bases from each other. The approach followed by Brenner (17) implies the construction of the largest possible λ-free code for a given λ. Morris et al. (18) use De Bruijn sequences of order λ to obtain such λ-free codes. Ben-Dor et al. (19) extend this approach to incorporate a simple thermodynamic model, employing the 2-4-rule: the melting temperature in °C of a duplex is assumed to be equal to twice the number of A-T base pairs plus four times the number of G-C base pairs.
We felt these approaches had two shortcomings. First of all, interactions between different tags (and not involving any antitags) could also interfere with tag/antitag hybridization and thus a clear, strong signal. Secondly, and even more importantly, in order to obtain a high level of multiplexing one would clearly benefit from a more sophisticated thermodynamic model, taking into account, for example, effects of mismatches, bulges and dangling ends.
We have thus implemented the following greedy algorithm to generate such sets:
1 start with empty set T
2 repeat (add tags)
3 repeat (generate sequence)
4 generate a random sequence S and its complement S′
5 until S and S′ form no homodimer
6 if both S and S′ do not interact with any other sequence in T
7 add S, S′ to T
8 until enough tags.
Where T is the set of tags and antitags generated so far. The generation of a new sequence S in line 4 is done base by base, drawing each base randomly and i.i.d. from the set {A,C,G,T}. New bases are added at the end of S until its melting temperature Tm reaches a predefined bound, say 60°C. This temperature is calculated as
where ΔH and ΔS are enthalpy and entropy changes of duplex formation derived from the nearest neighbor model, R is the gas constant, and CT is the total molar concentration of strands.
Again, we assume that a check for homodimer formation will also catch hairpins, and the interactions in line 6 are calculated using the free energy alignment algorithm described above.
Primer–tag pairing
One central idea of the assay is the independence of the minisequencing primers and the tag/antitag system. In principle, the same set of tags can be used for all experiments, and hence custom microsphere sets or DNA chips can be prefabricated and stored. One problem that needs to be considered is the possibility of interactions between primers and tags/antitags. This may force us to exclude certain tags for a specific assay. Note that we have so far checked for crosshybridization between the tags and antitags, but not considered the case where the primer binds to a tag or the combination between primer and tag leads to additional problems, as some undesired hybridization over the joint of the two may occur or primer foldback becomes a problem. Therefore, we also need to decide which tag to combine with which primer. The following problems need to be addressed. (i) Binding of a tag–primer pair to an undesired antitag on the microspheres or chip surface, leading to wrong signal. (ii) Binding of different primer–tag pairs to one another, leading to false extension in the minisequencing reaction. (iii) Foldback of a primer–tag pair onto itself, causing wrong primer extension and lower signal on the antitags due to competitive reactions.
We have implemented a computer program as part of the SBEprimer package that will, given the set P of primers, a set T of tags with |T| > |P| (i.e. we assume a larger number of tags to be given than primers), choose a subset of the tags and pair them with the primers, ensuring that the above problems are avoided. The process is straightforward: starting with a random pairing, the program will check for hairpins, dimer-formation and crossreactivity between the primer–tag oligonucleotide and the antitags. These checks are performed using the free energy alignment algorithm as in the previous sections. If any problem is encountered, the involved tag/antitag pair is discarded and replaced by a new pair from T. This is iterated until either a feasible combination is found, or no more tags remain in T to exchange.
The program will then output the list of primer–tag pairs and antitags to use in a format that can be used to directly order or assemble the oligonucleotides required.
RESULTS
A set of 45 multiplexed minisequencing primers for scoring 45 SNPs in the human genome was selected using the SBEprimer program. Forty-five tag/antitag pairs were automatically chosen for these primers from a set of 250 generated by the software, and a final list of oligonucleotides that had the sequence of the tag and the minisequencing primer was generated for synthesis.
The MHC complex is located on chromosome 6 in the human genome, and target regions containing the 45 SNP sites were amplified using the PCR. Fifteen amplicons were generated using this technique; the PCR primers are available on our website. The amplicons were pooled together, and treated with shrimp alkaline phosphatase, which removes the excess unused deoxynucleotide triphosphates (dNTPs), and exonuclease I, which removes single-stranded primers used in the PCR.
Single base extension was performed in a 10 µl reaction that consisted of the pooled, clean template, thermosequenase (0.75 U), thermosequenase reaction buffer, biotinylated ddNTPs (7.5 µM), and 45 minisequencing primers (25 nM, each primer). A single reaction was performed for each of the four biotinylated ddNTPs. The cycling conditions used were an initial incubation at 94°C for 10 s, and annealing and extension of the single base at 60°C for 10 s.
The reactions were then incubated with 2 µl of microsphere mix that contained 45 microspheres conjugated to 45 antitags, which were complementary to the tags associated with each of the 45 minisequencing primers. The incubation allowed for the hybridization of tag/antitag pairs, and the capture (and thus demultiplexing) of the minisequencing primers that had now been extended by a single biotinylated ddNTP, onto microspheres. This hybridization was performed in a binding buffer that contained 100 mM Tris–HCl, 1 mM EDTA and 800 mM NaCl. The hybridization cycle consisted of an initial increase in temperature to 80°C, to allow for the denaturation of all DNA single strand interactions, followed by a stepwise decrease in temperature to 25°C, by holding at 70, 60, 50, 40 and 35°C for 1 min. This decrease in temperature allowed for the gradual annealing of the specific tag with its complementary antitag on the microsphere. Following hybridization, the microspheres were washed two times with the same buffer that also contained 0.02% Tween 20, to prevent the microspheres from sticking to each other, as well as the tubes they were contained in. The wash step was performed to remove all excess, unextended biotinylated ddNTPs that would bind non-specifically to the fluorescent stain. The hybridized minisequencing primers were then resuspended in 35 µl of buffer that contained streptavidin-conjugated phycoerythrin (23 nM), and incubated for 15 min at room temperature. The biotin-ddNTP extended minisequencing primers are thus stained with the fluorescent dye, which will be detected on the LUMINEX Flow-Cytometer. All reactions were transferred to 96 well plates to enable analysis by the LUMINEX Flow-Cytometer. Data were analyzed using Microsoft Excel.
The 45 SNPs were genotyped in a collection of 84 samples from the NIH Polymorphism Discovery Resource Set (Coriel Cell Repository, Camden, NJ). The background signal from a no template control sample was subtracted from each sample, and the nucleotide bases present at the SNP sites were identified using thresholds that considered both the absolute signal intensities from the incorporated ddNTPs and the relative signals from the two possible alleles. The genotypes for 35 of these SNPs were determined by conventional sequencing followed by analysis with Phrap and PolyPhred (20).
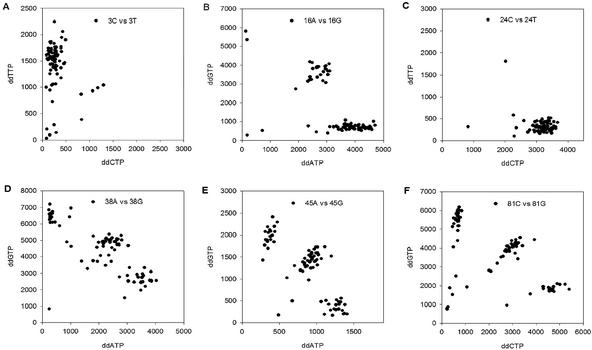
The results of multiplexed genotyping are presented in Table 1. Overall, the genotyping success rate was ∼90% for the more than 3500 SNPs genotyped. Data for six representative SNPs are presented in Figure 3. In general, failed PCR is the single biggest cause of a no result. A small number of SNPs (14,33,40) had high background signals in unknown samples as well as in no template control reactions. This background, presumably due to some undesired interactions between primers, necessitated the use of higher threshold levels. For a large number of SNPs, we genotyped by con ventional sequencing the same NIH sample set. Concordance between the two methods for the majority of SNPs was 95–100%. Some SNPs showed lower concordance, including the three SNPs with high background signals. For other SNPs, the discrepancy centered on the identification of heterozygotes. For some samples, the flow cytometry data led us to review the sequencing data and identify a number of occasions where the automated sequence analysis software missed a heterozygote. In other cases, no obvious cause for discrepancy for specific samples could be identified and these are possibly due to allelic bias introduced by PCR or the sequencing reactions.
Table 1. Multiplexed minisequencing performance.
| SNP | Genotypes called (%) | Concordance (%) |
|---|---|---|
| 1 | 89.3 | – |
| 2 | 94.0 | 96.2 |
| 3 | 91.7 | 98.7 |
| 4 | 86.9 | 97.3 |
| 5 | 92.9 | 100 |
| 6 | 97.6 | 91.4 |
| 7 | 92.9 | 98.7 |
| 8 | 79.8 | 98.5 |
| 9 | 72.6 | 98.4 |
| 10 | 77.4 | 100 |
| 11 | 82.1 | – |
| 12 | 95.2 | 95.0 |
| 13 | 92.9 | 91.0 |
| 14 | 86.9 | 98.6 |
| 15 | 96.5 | 98.8 |
| 16 | 96.4 | 98.8 |
| 17 | 91.7 | 94.8 |
| 18 | 91.7 | – |
| 19 | 72.6 | – |
| 20 | 94.0 | 100 |
| 21 | 94.0 | 100 |
| 22 | 94.0 | 97.5 |
| 23 | 97.7 | 97.6 |
| 24 | 95.2 | 95.0 |
| 25 | 97.6 | 97.6 |
| 26 | 96.5 | 97.5 |
| 27 | 97.6 | 100 |
| 28 | 97.6 | 100 |
| 29 | 96.5 | 97.5 |
| 30 | 94.1 | – |
| 31 | 66.7 | – |
| 32 | 96.4 | 90.1 |
| 33 | 50.0 | 76.2 |
| 34 | 91.7 | 100 |
| 35 | 91.7 | 100 |
| 36 | 91.7 | 100 |
| 37 | 91.7 | 100 |
| 38 | 91.7 | 96.1 |
| 39 | 91.7 | – |
| 40 | 60.7 | – |
| 41 | 94.0 | – |
| 42 | 94.0 | – |
| 43 | 89.2 | – |
| 44 | 90.5 | 97.4 |
| 45 | 86.9 | 97.2 |
Forty-five SNPs were genotyped in 83 samples. Presented are the percentages of samples genotyped for each SNP in a single experiment, as well as the concordance with genotypes determined by conventional DNA sequencing.
Figure 3.
Multiplexed minisequencing results. Data from six representative SNPs from the 45-plex assay. The fluorescence intensities after subtraction of the microsphere autofluorescence for each of the two possible bases occurring at a given site are shown. Genotypes were assigned after setting thresholds on both the absolute intensities as well as the relative intensities from the two bases.
DISCUSSION
The results presented here show that automated primer design can significantly enhance multiplexed minisequencing as a tool for high-throughput genotyping. We have previously demonstrated multiplexed minisequencing using microsphere arrays and flow cytometry (5), however a key to success is the careful design of minisequencing primers to avoid undesirable homo- or heterodimer interactions. Commercially available oligonucleotide design software has minimal multiplex primer design capability for PCR primers, none has multiplex primer design capability for minisequencing primers. The design of highly multiplexed minisequencing assays therefore involves extensive manual primer design, and is a rate limiting step in the development of new assays.
High throughput and low cost assays are not feasible without high levels of multiplexing; currently available genotyping tools need to be highly parallelized to satisfy the requirements of pharmacogenomics and of routine SNP analysis in medical institutions. Such applications might involve the scoring of millions of SNPs per day.
Such multiplexing is not possible without a way to ‘demultiplex’ the experiment. We have demonstrated that this can be done using a tag/antitag system, which sorts the signals from the minisequencing reaction to the corresponding beads. The higher the level of multiplexing desired, the more complex becomes the problem of designing such a crosshybridization-free tag/antitag code, and the more relevant becomes a profound thermodynamic algorithm to predict interactions. We have generated a set of 250 such tags using the SBEprimer software and demonstrated its quality.
The selection of appropriate primers for the multiplexing assay is a second crucial requirement. Such primers must not false prime, and they must work together in the same assay. The manual design of minisequencing primers quickly becomes impossible if more than just a few primers are pooled together. The SBEprimer software can automatically design appropriate primer sets, and thus enables high levels of multiplexing.
One important feature of the SNP assay presented is its applicability to heterozygote detection. If two different alleles are present, the experiment will show high signal for both bases, and will make two base-calls. This is a must for any useful genotyping technology, and easily done with the assay presented here.
The quality of the results obtained on the 45-plex experiment presented with its high ratio of true to false signal indicates that much higher levels of parallel genotyping can be achieved using the flow cytometry-based technology with the SBEprimer software package; it is likely that much higher levels of multiplexing are possible.
CONCLUSION
Our results show that minisequencing can be adapted to be a powerful tool for high-throughput, cost-efficient genotyping. The simultaneous screening of 45 polymorphic sites has been demonstrated, with basecalls confirmed by independent sequencing. The manual design of such multiplexed genotyping assays is a laborious process, but can be highly automated using the SBEprimer package presented in this work.
The thermodynamic alignment algorithm used in the SBEprimer program calculates very accurate interaction profiles for oligonucleotides, and the experiments show how careful design of an assay with computer support enabled complex reactions to be carried out with the desired results. We intend to use a similar method to design primer pairs for multiplexed PCRs. Clearly, this is presently the bottleneck of all genotyping methods, and a higher lever of PCR multiplexing would be highly desirable. We are confident that such experiments will benefit from more accurately determined interactions and automated multiplexing primer design software. SBEprimer can be adapted for such purposes.
Supplementary information is available from our website at http://www.zaik.uni-koeln.de/bioinformatik/sbeprimer.html.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported in part by grants RR01315 and RR14101 from the National Institutes of Health.
REFERENCES
- 1.Cooper D., Smith,B., Cooke,H., Niemann,S. and Schmidtke,J. (1985) An estimate of unique DNA sequence heterozygosity in the human genome. Hum. Genet., 69, 201–205. [DOI] [PubMed] [Google Scholar]
- 2.Venter J., Adams,M., Myers,E., Li,P., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Schafer A. and Hawkins,J. (1998) DNA variation and the future of human genetics. Nat. Biotechnol., 16, 33–39. [DOI] [PubMed] [Google Scholar]
- 4.Syvänen A.-C. (1999) From gels to chips: ‘minisequencing’ primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mutat., 13, 1–10. [DOI] [PubMed] [Google Scholar]
- 5.Cai H., White,P., Torney,D., Deshpande,A., Wang,Z., Marrone,B. and Nolan,J. (2000) Flow cytometry-based minisequencing: a new platform for high-throughput single-nucleotide polymorphism scoring. Genomics, 66, 135–143. [DOI] [PubMed] [Google Scholar]
- 6.Dopazo J. and Sobrino,F. (1993) A computer program for the design of PCR primers for diagnosis of highly variable genomes. J. Virol. Methods, 41, 157–166. [DOI] [PubMed] [Google Scholar]
- 7.Rychlik W. and Rhoads,R. (1989) A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res., 17, 8543–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas K., Busch,M., Mössinger,S. and Thompson,J. (1991) An improved microcomputer program for finding gene- or family-specific oligonucleotides suitable as primers for polymerase chain reactions or as probes. Comput. Appl. Biosci., 7, 525–529. [DOI] [PubMed] [Google Scholar]
- 9.Giegerich R., Meyer,F. and Schleiermacher,C. (1996) In Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, pp. 68–77. [PubMed]
- 10.Breslauer K., Frank,R., Blöocker,H. and Marky,L. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke S. and Wartell,R. (1995) Influence of neighboring base pairs on the stability of single base bulges and base pairs in a DNA fragment. Biochemistry, 34, 4593–4600. [DOI] [PubMed] [Google Scholar]
- 12.LeBlanc D. and Morden,K. (1991) Thermodynamic characterization of deoxyribooligonucleotide duplexes containing bulges. Biochemistry, 30, 4042–4047. [DOI] [PubMed] [Google Scholar]
- 13.Turner D. (1992) Bulges in nucleic acids. Curr. Opin. Struct. Biol., 2, 334–337. [Google Scholar]
- 14.Smith T. and Waterman,M. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 15.Sommer R. and Tautz,D. (1989) Minimal homology requirements for PCR primers. Nucleic Acids Res., 17, 6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frutos A., Liu,Q., Thiel,A., Sanner,A., Condon,A., Smith,L. and Corn,R. (1997) Demonstration of a word design strategy for DNA computing on surfaces. Nucleic Acids Res., 25, 4748–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner S. Methods for sorting polynucleotides using oligonucleotide tags. US Patent 5,604,097 (1997).
- 18.Morris M., Shoemaker,D., Davis,R. and Mittmann,M. Methods and compositions for selecting tag nucleic acids and probe arrays. European Patent application, 97302313 (1997).
- 19.Ben-Dor A., Karp,R., Schwikowski,B. and Yakhini,Z. (2000) Universal DNA tag systems: a combinatorial design scheme. J. Comput. Biol., 7, 503–519. [DOI] [PubMed] [Google Scholar]
- 20.Nickerson D.A., Tobe,V.O. and Taylor,S.L. (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res., 25, 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]