Abstract
The initiation of chromosomal DNA replication in human cell nuclei is not well understood because of its complexity. To allow investigation of this process on a molecular level, we have recently established a cell-free system that initiates chromosomal DNA replication in an origin-specific manner under cell cycle control in isolated human cell nuclei. We have now used fractionation and reconstitution experiments to functionally identify cellular factors present in a human cell extract that trigger initiation of chromosomal DNA replication in this system. Initial fractionation of a cytosolic extract indicates the presence of at least two independent and non-redundant initiation factors. We have purified one of these factors to homogeneity and identified it as the single-stranded DNA binding protein RPA. The prokaryotic single-stranded DNA binding protein SSB cannot substitute for RPA in the initiation of human chromosomal DNA replication. Antibodies specific for human RPA inhibit the initiation step of human chromosomal DNA replication in vitro. RPA is recruited to DNA replication foci and becomes phosphorylated concomitant with the initiation step in vitro. These data establish a direct functional role for RPA as an essential factor for the initiation of human chromosomal DNA replication.
INTRODUCTION
The initiation of chromosomal DNA replication is a key event in the regulation of the cell division cycle in proliferating metazoan cells. Regulatory mechanisms that ensure timely initiation of DNA replication and exact replication of the chromosomal DNA once per cell cycle in eukaryotes have been deduced predominantly from two model systems. Genetic evidence from the yeast Saccharomyces cerevisiae and biochemical data from egg extracts of the amphibian Xenopus laevis have converged into the current regulation model (reviewed in 1–5).
Upon exit from mitosis, chromosomes become licensed for DNA replication by a sequential association of proteins with the chromosomal DNA. The origin recognition complex (ORC) binds to origin DNA and recruits Cdc6 protein, which then, concomitantly with Cdt1 protein, recruits a complex of six minichromosome maintenance proteins (MCM2–MCM7) to form a pre-replicative complex (pre-RC). Initiation of DNA replication, and hence, transition into S phase, is under the control of at least two kinds of protein kinases, cyclin-dependent kinases (Cdks) and the Dbf4-dependent kinase Cdc7. The targets of Dbf4/Cdc7 include the MCM proteins (4,6), which may act as a replicative helicase for unwinding chromosomal DNA (7,8), whereas the physiological targets of Cdks in this transition are less well defined. Later during the multi-step initiation of chromosomal DNA replication, other proteins including MCM10, Cdc45 and replication protein A (RPA) are recruited to the unwound origin DNA, which ultimately leads to the recruitment of DNA primase and DNA polymerases to begin DNA synthesis.
This model most likely reflects the general scheme in eukaryotes because most of its players are evolutionarily conserved. For the study of the regulation of initiation of DNA replication under cell cycle control in human somatic cells, however, a few significant reservations need to be noted that warrant a molecular analysis of this process in systems derived from these cells. As a unicellular organism, yeast is not expected to have all the regulatory pathways conserved that regulate differentiation and proliferation of complex metazoan organisms. The regulation of the early embryonic cell cycles of X.laevis is recapitulated in egg extracts that can replicate any DNA incubated in them (9–11). However, regulation of this embryonic system differs from that of somatic cell cycles, for instance by lacking transcription and site-specificity of initiation of DNA replication (12) and by lacking a G1 phase as found in somatic cycles.
A well-established model system for the enzymology of human somatic DNA replication is the DNA tumour virus SV40. Replication of SV40 DNA depends on the virally encoded protein, large tumour antigen (T-Ag), and on the host cell replication machinery (13). Using biochemical fractionation and reconstitution assays, the host cell proteins required for SV40 replication have been identified and characterised (reviewed in 14–16). The first steps of initiating viral DNA replication depend entirely on the viral control elements. SV40 T-Ag binds DNA sequence-specifically as a double hexamer to the SV40 origin of replication, and its helicase activity mediates bidirectional unwinding of the parental DNA strands (17–20). Possibly the first host cell factor interacting with the activated viral origin is RPA (21), also referred to as human SSB (22) or RF-A (23). RPA is conserved among eukaryotes and is required for many DNA transactions such as replication, repair and recombination (reviewed in 24,25). In the context of SV40 DNA replication initiation, RPA greatly stimulates DNA unwinding by T-Ag at the SV40 origin (21). RPA directly interacts with T-Ag and with the host cell DNA polymerase α (polα)/primase (26). Recruitment of polα/primase to the viral initiating complex by RPA allows primer synthesis (reviewed in 24,25). Downstream of these initiation steps, other host cell proteins are recruited to coordinate the formation of processive DNA replication forks (reviewed in 27).
Despite its success in identifying eukaryotic DNA replication factors, the SV40 system has serious limitations to serve as a model system for the initiation steps of chromosomal DNA replication in human somatic cells. Replication of episomal SV40 in infected host cells evades cell cycle control, employing the virally encoded initiator protein T-Ag and the viral origin. The direct functional identification of host cell proteins required for the initiation of cell cycle controlled chromosomal DNA replication in human somatic cells, therefore, is still of prime interest.
To allow functional identification of these cellular DNA replication initiation factors, we make use of a cell-free system from human cells that initiates semi-conservative chromosomal DNA replication (28,29). Active template nuclei are isolated from cells synchronised in late G1 phase of the cell division cycle. Within these nuclei, DNA replication initiates site-specifically at origins in vitro that also become activated when cells enter S phase in vivo (30). In vitro initiation of semi-conservative DNA replication in mammalian G1 phase nuclei depends on incubation in extracts from S phase cells (28,31) or from asynchronously proliferating human cells (29). Essential initiation factors have been identified in this system as G1/S phase-specific Cdks (28,29,32,33), however, initiation depends on an additional, so far unknown activity present in the cytosolic cell extract (29).
In this paper, we report fractionation and reconstitution experiments to allow separation and functional identification of human cellular initiation factors. Initial fractionation of the cytosolic extract indicates that at least two independent and non-redundant initiation factors are present in the extract. We have purified one of these factors to homogeneity and have identified it as the single-stranded DNA binding protein RPA. Our data establish a functional role for RPA as an essential initiation factor for human chromosomal DNA replication.
MATERIALS AND METHODS
Cell culture and synchronisation
Human HeLa S3 and EJ30 cells were cultured as monolayers in Dulbecco’s MEM plus 10% foetal bovine serum, 10 U/ml penicillin and 0.1 mg/ml streptomycin (all from Gibco), and synchronised in early S phase exactly as described (28). Cells were arrested in late G1 phase by treatment with 0.5 mM mimosine (Sigma) for 24 h (34). All cell synchronisations were verified by flow cytometry of isolated nuclei (28).
Preparation of nuclei and cell extracts
Nuclei were prepared by hypotonic treatment of the cells, followed by Dounce homogenisation as described (28). Nuclei were stored in liquid N2 without loss of replication competence (29). Cytosolic and nuclear extracts were prepared as described (28).
Protein purification
All steps were carried out at 4°C. Crude cytosolic HeLa extract was obtained from 4C Biotech (Seneffe, Belgium), thawed on ice and was clarified by ultracentrifugation for 1 h at 100 000 g. This 100 000 g supernatant (S100) was loaded onto a Q Sepharose Hi-Load 26/10 column in an ÄktaFPLC system (Amersham Pharmacia Biotech), pre-equilibrated in buffer A (50 mM Tris–HCl pH 7.8, 1 mM DTT, 1 mM EGTA) containing 200 mM KCl. The protein flowing through the column was collected and the column was eluted sequentially with 280 and 500 mM KCl in buffer A. The resulting protein fractions were termed QFT, QA and QB, respectively. Initially, QA was further purified on Heparin Sepharose and Phenyl Superose (Amersham Pharmacia Biotech), resulting in the isolation of candidate protein bands from acrylamide gels and their subsequent identification as RPA32 by MALDI-TOF (details on request). Subsequently, the purification procedure was modified by replacing heparin with a blue dye as the affinity resin, similarly to Henricksen et al. (35). In this modification, QA was loaded directly onto a 5 ml Blue Sepharose CL-6B column (Amersham Pharmacia Biotech), pre-equilibrated in buffer B (20 mM K-HEPES pH 7.8, 1 mM DTT, 1 mM EGTA) containing 800 mM KCl and extensively washed. This column was then eluted in two steps with 200 mM and 1 M NaSCN in buffer B. The 1 M NaSCN fraction was dialysed overnight into 800 mM (NH4)2SO4 and 500 mM KCl in buffer B, then loaded onto a 0.1 ml Phenyl Superose column pre-equilibrated with the same buffer, using the SMART system (Amersham Pharmacia Biotech). The bound proteins were eluted with a decreasing linear salt gradient to buffer B. The active fractions eluting in the middle of the gradient were pooled and a 60 µl aliquot was layered on top of a 0.6 ml linear 15–40% sucrose gradient in buffer B. The gradient was centrifuged at 40 000 r.p.m. for 24 h in a Beckman SW50.1 rotor and subsequently fractionated into 60 µl fractions.
Recombinant human RPA was expressed in Escherichia coli C41 as a heterotrimer using the p11d-tRPA plasmid (35) and purified using Blue Sepharose CL-6B and Phenyl Superose columns exactly as described above. The endogenous and recombinant human RPA complexes showed identical chromatographic behaviour.
DNA replication reactions and analysis of reaction products
DNA replication reactions contained the following: a buffered mixture of NTPs and dNTPs (elongation buffer; 29), including 150 pmol digoxigenin-dUTP (Roche) as a probe for microscopic detection, 100 µg HeLa unfractionated cytosolic extract or variable amounts of fractions from the protein purification as specified in the figure legends, and 2–5 × 105 late G1 phase nuclei. The reaction volume was adjusted to 50 µl with buffer B containing 100 mM K-acetate (see above). All fractions from the protein purification were first dialysed against this buffer before addition to the reaction. Incubation time was 3 h, unless indicated otherwise.
Detection of DNA replication by confocal microscopy was performed exactly as described (29), except that anti-digoxigenin fluorescein-labelled Fab fragments (Roche) were used at 1:100 dilution for the staining of newly replicated DNA. For staining with RPA antibodies, nuclei were isolated from replication reactions, washed twice in PBS, fixed in 4% paraformaldehyde and transferred to polylysine-coated coverslips exactly as for DNA replication reactions. Primary antibodies were diluted in PBS with 2% dry non-fat milk, 0.1% Triton X-100 and 0.02% SDS. Fluorescein- and Texas Red-linked secondary antibodies were obtained from Amersham Life Science. Nuclear DNA was counterstained with propidium iodide. Stained nuclei were analysed using a Leica confocal laser microscope as specified (30).
For analysis of RPA binding during these rections, the nuclei were washed twice in PBS after the incubation, resuspended in SDS sample buffer, boiled for 10 min and loaded on SDS–polyacrylamide gels. Phosphatase treatments were performed after the PBS wash by resuspending the nuclei in 20 µl of λ phosphatase buffer containing 2 mM MnCl2 and incubating for 30 min at 30°C in the presence of 200 U λ phosphatase (NEB). Western blotting was performed using standard techniques, using the anti-RPA monoclonal antibodies 34A, 70A, 70B and 70C (36) (RMAS of Cancer Research UK) and polyclonal antibody pAb-RPA1. This antibody was raised by Abcam (Cambridge, UK) against the bacterially expressed and purified trimeric human RPA (see above).
RESULTS
The initiation of human chromosomal DNA replication can be studied biochemically in a cell-free system that uses as templates nuclei from human cells synchronised in late G1 phase by the plant compound mimosine (28,29). When these nuclei are incubated in a physiological replication buffer containing nucleoside and deoxynucleoside triphosphates, only ∼5% of nuclei synthesise DNA. They represent a few S phase contaminants in the preparation of nuclei, which continue DNA replication at replication forks that were established in vivo before the preparation (28,30,34). Addition of cytosolic extract from proliferating cells triggers initiation of DNA replication in ∼50% of the late G1 phase nuclei (29,30). Therefore, the cytosolic extract contains soluble activity that is required for the initiation of DNA replication in somatic human cell nuclei. In the first set of experiments, we fractionated the cytosolic initiation extract to separate it into distinct initiation factors, allowing their further purification and identification.
Initial fractionation of cytosolic extract
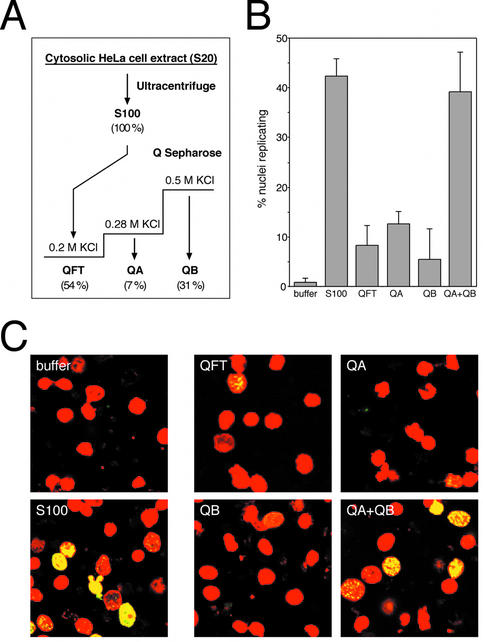
Cytosolic extract from proliferating HeLa cells was separated by anion exchange chromatography into three fractions (Fig. 1A). After clearing by ultracentrifugation the extract was loaded onto Q Sepharose at 200 mM KCl and the protein flowing through (QFT) was collected. Bound protein was eluted in two steps yielding two fractions, termed QA (eluting at 200–280 mM KCl) and QB (eluting at 280–500 mM KCl), respectively. These fractions were tested for DNA replication initiation activity by adding them to late G1 phase template nuclei and DNA replication buffer. When tested separately, the protein flowing through the column (QFT), fraction QA, and fraction QB showed only marginal initiation activity by themselves, sufficient to induce replication in only ∼10% of the nuclei on average (Fig. 1B and C). In contrast, when fractions QA and QB were added together, the initiation activity present in the cytosolic extract was fully reconstituted (Fig. 1B and C).
Figure 1.
Initial fractionation of cytosolic extract. (A) Schematic diagram of fractionation steps. See Materials and Methods for details. The values given in parentheses below each fraction indicate the percentages of total protein recovered in the respective fraction. (B) Quantitative analysis of DNA replication initiated in isolated G1 phase nuclei in vitro. Nuclei from mimosine-arrested HeLa cells were incubated in elongation buffer (buffer), supplemented with 100 µg protein of unfractionated extract (S100), 45 µg of fraction QFT, 15 µg of QA, 35 µg of QB and a combination of 15 µg QA and 35 µg QB as indicated (these masses approximate typical mass percentages of the respective fractions in the S100 extract). Percentages of replicating nuclei were quantitated and mean values and standard deviations of 5–10 independent experiments are shown. The main variance of the percentages of nuclei replicating in the indicated fractions was due to differences between separate preparations of fractions. (C) Representative fields of replicating nuclei. Replication reactions were performed in the presence of the indicated components and nuclei were analysed by confocal fluorescence microscopy. Nuclear DNA is visualised by propidium iodide (red signal) and replicated DNA by fluorescein-conjugated anti-digoxigenin Fab fragments (green signal). Merged images are presented showing sites of replicated nuclear DNA in yellow and non-replicating nuclei in red.
We conclude that there are at least two separate and non-redundant initiation activities present in the cytosolic extract from proliferating human cells, which are both required for triggering DNA replication in late G1 phase nuclei. In the next set of experiments, we purified the initiation factor present in fraction QA to homogeneity.
Purification of the DNA replication initiation factor from fraction QA
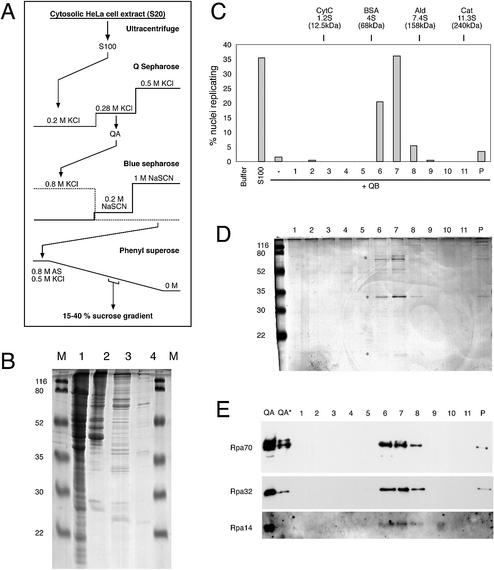
We detected the DNA replication initiation activity present in fraction QA through the subsequent chromatographic purification steps by adding a standard amount of fraction QB to the functional reconstitution assay. The initiation activity of QA was followed as a single peak of activity over two subsequent chromatographic steps, using Blue Sepharose and Phenyl Superose (Fig. 2A and Table 1). During this procedure, proteins with apparent molecular weights of ∼70 and 30–35 kDa were enriched (Fig. 2B). Protein bands in the 30–35 kDa range were excised from a protein gel and subjected to MALDI-TOF mass spectrometry for identification. One of these bands was identified as the 32 kDa subunit of the human single-stranded DNA binding protein RPA (data not shown). To see whether the initiation activity of the highly enriched Phenyl Superose fraction co-purifies further with human RPA, we added a final sucrose sedimentation step to the purification procedure (Fig. 2A and Table 1).
Figure 2.
Purification of the initiation factor present in fraction QA. (A) Schematic diagram of purification procedure. See Materials and Methods for details. (B) Protein analysis of key purification steps. Unfractionated S100 extract (100 µg protein, lane 1), fraction QA (20 µg protein, lane 2), Blue Sepharose eluate (10 µg protein, lane 3) and Phenyl Superose eluate (<1 µg, lane 4) were loaded on a 15% polyacrylamide gel. Proteins were visualised by staining with Coomassie brilliant blue. Molecular masses of marker proteins (M) are indicated. (C–E) Final purification and identification of RPA as the initiation activity present in fraction QA by sucrose gradient centrifugation. (C) Quantitative analysis of initiation of DNA replication. Nuclei from mimosine-arrested EJ30 cells were incubated in 35 µg of fraction QB supplemented with a 20 µl volume of the sucrose gradient fractions (1–11) and the pelleted material (P) as indicated. Incubations in elongation buffer (buffer) or 100 µg of unfractionated extract (S100) were used as controls. Percentages of replicating nuclei of a representative experiment are shown. (D) Protein analysis of fractions of the sucrose gradient step. A 20 µl volume of each sucrose gradient fraction (1–11) and the pelleted material (P) were separated on a 15% polyacrylamide gel and stained with silver salts. Molecular masses of marker proteins (M) are indicated. Asterisks denote the position of Rpa70, 32 and 14 subunits. (E) Identification of RPA by western blot analysis. A 20 µl volume of each sucrose gradient fraction (1–11 and P) and 15 and 5 µg (*) of fraction QA were separated on a 15% polyacrylamide gel and analysed by western blotting. The membrane was sequentially probed with a mixture of monoclonal antibodies 70A, 70B and 70C (Rpa70) (36), monoclonal antibody 34A (Rpa32) (36) and polyclonal antibody pAb-RPA1 (visualising Rpa14; see Fig. 4A for a characterisation of this polyclonal antibody).
Table 1. Purification of initiation activity present in fraction QA (RPA).
| Fractionation step | Total protein (mg) | Total activity (106 nuclei initiated) | Specific activity (106 nuclei initiated/mg protein) | Fold purification |
|---|---|---|---|---|
| S100 extract | 224 | 1344 | 6 | 1 |
| Q Sepharose | 12.8 | 1152 | 90 | 15 |
| Blue Sepharose | 0.24 | 240 | 1000 | 167 |
| Phenyl Superose | 0.08 | 75 | 936 | 156 |
| Sucrose gradient | 0.04 | 77 | 1920 | 320 |
For each fractionation step, the total protein mass, the total amount of initiation activity and the specific initiation activity are shown. The increase in purification is calculated as the specific initiation activity of each fractionation step divided by the specific initiation activity of the unfractionated S100 extract.
Initiation activity sedimented with an apparent molecular weight of 100–120 kDa through the sucrose gradient (Fig. 2C), perfectly co-migrating with three polypeptides of apparent molecular masses of 70, 32 and ∼15 kDa (Fig. 2D). The composition of this complex closely resembles that of human RPA, which consists of 70, 32 and 14 kDa subunits (21,25). Western blots using monoclonal antibodies against human RPA (36) clearly identified the large and middle polypeptides of the active fractions as Rpa70 and Rpa32, respectively (Fig. 2E), whereas the smallest subunit was recognised by a polyclonal antibody raised against the trimeric RPA complex (Fig. 2E). These fractionation results strongly suggest that the soluble initiation activity present in fraction QA is human RPA. However, it remains a possibility that different proteins present in minute amounts in the active gradient fractions constitute this activity. Therefore, we sought to consolidate these data by gain-of-function experiments using recombinant human RPA.
RPA is the initiation activity of fraction QA
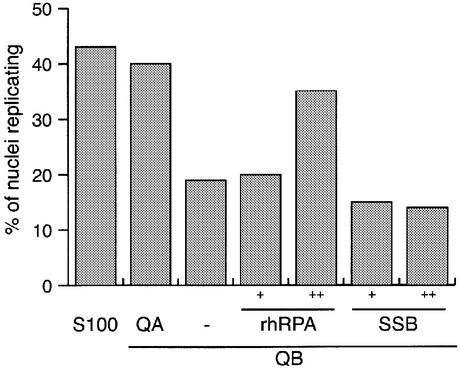
Recombinant trimeric human RPA (rhRPA) was expressed in E.coli and purified to apparent homogeneity (35; see Materials and Methods for details). When tested in the presence of fraction QB, rhRPA clearly triggered initiation of genomic DNA replication in late G1 phase template nuclei (Fig. 3). Next, we tested the specificity of this requirement and asked whether human RPA can be substituted by the heterologous prokaryotic single-stranded DNA binding protein SSB in the initiation reaction. When SSB was added alongside fraction QB, no initiation of genomic DNA replication was observed (Fig. 3). We conclude that initiation of DNA replication in isolated human G1 phase nuclei specifically depends on the human protein RPA, which cannot be substituted by a heterologous prokaryotic single-stranded DNA binding protein.
Figure 3.
Recombinant human RPA, but not prokaryotic SSB can substitute for the initiation activity present in fraction QA. Nuclei from mimosine- arrested HeLa cells were incubated in unfractionated extract (S100) and in 35 µg of fraction QB, supplemented with either 20 µg of fraction QA, replication buffer (–), 12.5 ng (+) or 37.5 ng (++) purified recombinant human RPA (rhRPA) or equimolar amounts of E.coli single-stranded DNA binding protein (SSB), respectively. Percentages of replicating nuclei were quantitated and mean values of two or three independent experiments are shown.
In the next set of experiments, we consolidated the role for RPA in the initiation step of genomic DNA replication by loss-of-function experiments using RPA-specific antibodies.
RPA is required for the initiation step of nuclear DNA replication
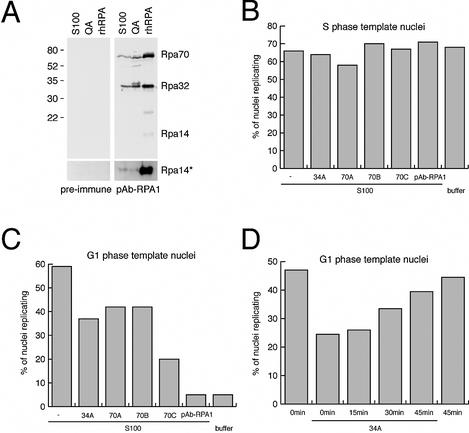
We first used the well-characterised monoclonal antibodies 70A, 70B, 70C and 34A (36), which are specific to their antigen as shown by immunoblotting (data not shown). We added these antibodies to control reactions containing unfractionated cytosolic extract and S phase nuclei as templates for elongation at existing DNA replication forks in vitro. As a further control, S phase nuclei also continued to elongate DNA replication at existing DNA replication forks in the absence of cytosolic extract (Fig. 4B, buffer), consistent with our previous papers (28,29,34). Addition of either monoclonal antibody did not reduce the percentages of S phase nuclei replicating in cytosolic extract (Fig. 4B). We then generated a polyclonal antibody against rhRPA that specifically detects all three subunits of RPA in extracts from human cells and from bacteria expressing rhRPA (Fig. 4A), and termed it pAb-RPA1. Addition of pAb-RPA1 also did not reduce the percentages of S phase nuclei replicating in cytosolic extract (Fig. 4B). To rule out that RPA might be inaccessible to these antibodies in the nuclei, we used fluorescent secondary antibodies and detected a focal RPA-specific pattern by confocal microscopy (data not shown). Therefore, our data indicate that elongation of DNA replication in S phase nuclei is not abolished by any of these RPA-specific antibodies in vitro.
Figure 4.
Antibodies specific for human RPA inhibit the initiation of chromosomal DNA replication in vitro. (A) Generation of the anti-RPA polyclonal antibody pAb-RPA1 and its specificity on S100 cytosolic extract, fraction QA and rhRPA. The bottom panel shows an overexposed image of the 14 kDa region of the blot. (B) Effect of anti-RPA antibodies on DNA replication in S phase nuclei. Replication initiation reactions using template nuclei from S phase HeLa cells were supplemented with 5 µg of the indicated monoclonal antibodies or 2 µl of pAb-RPA antiserum. Percentages of nuclei replicating were scored and mean values of two independent experiments are shown. (C) Inhibition of initiation in G1 phase template nuclei by the anti-RPA antibodies. Replication initiation reactions using template nuclei from mimosine-arrested late G1 phase HeLa cells were supplemented with the indicated antibodies as detailed in (B). Percentages of nuclei replicating were scored and mean values of two independent experiments are shown. (D) Late addition of antibody 34A no longer inhibits DNA replication. Replication initiation reactions using late G1 phase template nuclei were supplemented with 5 µg of the monoclonal 34A antibody and dig-dUTP at the indicated times. Percentages of nuclei replicating were scored and mean values of two independent experiments are shown.
We next addressed the requirement for RPA for the initiation step of DNA replication rather than for a general inhibition of DNA chain elongation by using G1 phase nuclei as templates and adding the same antibodies (Fig. 4C). Addition of monoclonal antibodies 34A, 70A, 70B and, to a larger extent, 70C significantly reduced the percentages of G1 phase nuclei initiating DNA replication (Fig. 4C). Furthermore, polyclonal antibody pAb-RPA1 reduced the percentages of nuclei initiating DNA replication in vitro to the background level observed in the absence of cytosolic extract (Fig. 4C), whilst addition of pre-immune serum had no inhibitory effect (data not shown). Therefore, these antibodies are very useful tools for distinguishing between the functions of RPA during initiation versus elongation stages of chromosomal DNA replication. Making use of this observation, we finally added antibody 34A and dig-dUTP after pre-incubation periods to initiation reactions using late G1 phase template nuclei and cytosolic extract. Importantly, addition of the 34A antibody after increasing pre-incubation periods no longer inhibited DNA replication in these nuclei (Fig. 4D). To control for permeability of the nuclear envelope for the 34A antibody during this reaction, we also stained the nuclei with fluorescent secondary antibodies after the incubation. In all cases we could confirm a nuclear localisation of the 34A antibody (data not shown), suggesting that a trivial loss of permeability for the inhibiting antibody by the nuclei is not the reason for the reduced inhibitory action (Fig. 4D), but rather the diminishing need for RPA as an initiation factor. Taken together, these inhibition studies establish a requirement for RPA in the initiation step of chromosomal DNA replication in isolated nuclei.
In the last set of experiments we used the polyclonal antibody pAb-RPA1 to investigate the interaction of RPA with template nuclei during the initiation of DNA replication in vitro.
Recruitment of RPA to nuclei before and during initiation of DNA replication
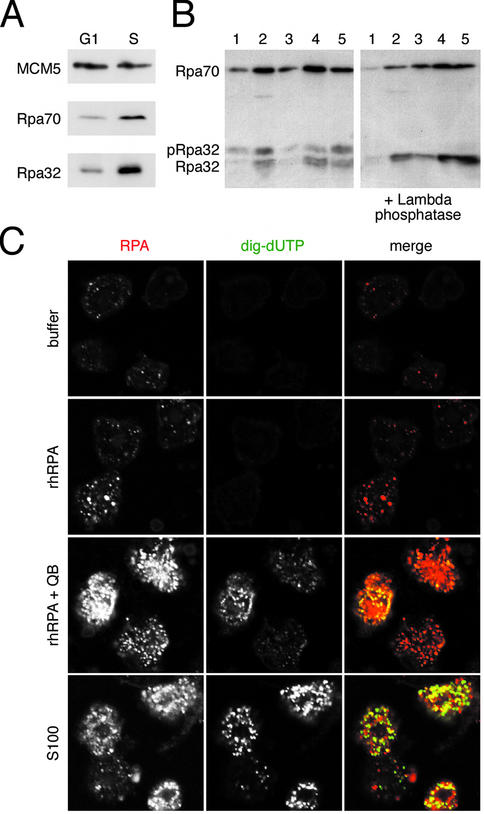
As a reference, we first synchronised human cells in late G1 phase and in early S phase and prepared a high salt nuclear extract to visualise RPA by western blot analysis (Fig. 5A). As control, the pre-RC protein MCM5 is present in similar amounts in these nuclei, as expected (1–5). RPA is bound only in small amounts to late G1 phase nuclei, but significantly increased amounts are bound to S phase nuclei (Fig. 5A), indicating that additional RPA is binding to the nucleus upon a cell’s transgression across the G1/S phase boundary in vivo. We next investigated by western blotting (Fig. 5B) and immunofluorescence microscopy (Fig. 5C) the interaction of RPA with late G1 phase nuclei during the initiation of DNA replication in vitro.
Figure 5.
RPA is recruited to DNA replication foci and becomes phosphorylated in vitro. (A) Comparison of bound RPA in G1 and S phase nuclei in vivo. Nuclear extracts (25 µg protein per lane) from mimosine- arrested late G1 phase and from early S phase cells were analysed by western blot using antibodies specific for MCM5 as a control and polyclonal antibody pAb-RPA1. (B) Nuclear binding and phosphorylation of RPA during initiation of DNA replication in vitro. G1 phase nuclei isolated from in vitro incubations done in replication buffer (lane 1), 100 µg S100 cytosolic extract (lane 2), 30 and 100 ng rhRPA (lanes 3 and 4) and 100 ng rhRPA plus 35 µg of fraction QB (lane 5). The samples shown in the right-hand panel were treated with λ phosphatase before loading onto the same gel. The Rpa70 and Rpa32 subunits were visualised with pAb-RPA1. Phosphorylated Rpa32 is indicated as pRpa32. (C) Confocal microscopy. G1 phase nuclei were incubated in buffer (top row), in 100 ng of rhRPA (second row), in 100 ng rhRPA supplemented with 35 µg QB (third row) and in 100 µg of unfractionated S100 (bottom row). High-resolution micrographs are shown of RPA foci (stained with pAb-RPA1, red), replication foci (dig-UTP, green) and merged images of the same set of nuclei.
Incubation of template nuclei in replication buffer without RPA in vitro showed the presence of a small amount of RPA (Fig. 5B, lane 1), present at faint intranuclear sites in a significant proportion of the nuclei, but these nuclei did not detectably incorporate dig-dUTP into the genomic DNA (Fig. 5C, top row). When this incubation reaction was supplemented with increasing amounts of soluble rhRPA, increasing amounts of RPA were bound to the nuclei (Fig. 5B, lanes 3 and 4). This RPA was visible at discrete intranuclear sites, which again did not detectably incorporate dig-dUTP (Fig. 5C, second row). Therefore, RPA can associate with late G1 phase nuclei at discrete sites in the absence of genomic DNA replication, but these sites are of as yet unknown function.
Importantly, incubation of these nuclei in the reconstituted initiation system containing both rhRPA and fraction QB or in unfractionated S100 initiation extract also leads to an increased association of RPA with the template nuclei in a discrete focal pattern (Fig. 5B, lanes 2 and 5, and C, third and fourth rows). The majority of these intranuclear RPA sites also incorporated dig-dUTP into the chromosomal DNA (Fig. 5C, third and fourth rows).
During incubation of nuclei under all of these in vitro conditions, we observed a mobility shift of a significant fraction of the nuclear bound Rpa32 subunit (Fig. 5B, left panel), suggesting its phosphorylation (reviewed in 24,25). Treatment of the nuclei with λ phosphatase after the in vitro incubation entirely removed the slower migrating form of Rpa32 (Fig. 5B, right panel), indicating that nuclear bound Rpa32 becomes phosphorylated in this in vitro system.
Taken together, these results show that the initiation of genomic DNA replication coincides with a phosphorylation of Rpa32 and the binding of RPA in a focal pattern to the template nuclei. However, these events can both also occur under conditions where DNA synthesis is not initiated. Importantly, when DNA synthesis is triggered by the additional presence of factors present in QB, the intranuclear sites where human chromosomal DNA replication is initiated, and further elongated, do co-localise with RPA.
DISCUSSION
In this paper, we used fractionation and reconstitution experiments to functionally identify human cellular factors that trigger initiation of chromosomal DNA replication in isolated cell nuclei. Initial fractionation of a cytosolic extract containing initiation activity indicated that at least two independent and non-redundant initiation factors are present in the extract. One of these factors was purified to homogeneity and functionally identified as the single-stranded DNA binding protein RPA. These data establish a functional role for RPA as an essential initiation factor for human chromosomal DNA replication.
In this study, we used as templates nuclei isolated from cells synchronised in late G1 phase by a treatment with the plant compound mimosine (34). We have shown before that these nuclei initiate semi-conservative DNA replication very efficiently in human cell extracts in an origin-dependent manner (29,30). Using these nuclei, we have shown in this paper that soluble initiation factors can be fractionated into at least two separate activities, one of which we have identified as RPA. It is a formal possibility that these activities would only be required to initiate DNA replication in nuclei from mimosine-arrested cells, and not necessarily in nuclei isolated from cells synchronised in an initiation-competent state by other means. We have addressed this possibility and naturally synchronised human EJ30 cells in late G1 phase by serum stimulation of quiescent cells. When incubated in unfractionated extract from asynchronously proliferating HeLa cells, or in the reconstituted system of fractions QA and QB, identical percentages of these G1 phase nuclei initiated DNA replication over and above the background of true S phase nuclei (data not shown). We have shown before that such ‘naturally’ synchronised nuclei prepared from rodent cells initiate DNA replication in extracts from S phase human cells (31,32), and a recent report showed similar data for such nuclei from primary human Wi38 cells (37). We therefore propose as a working hypothesis here that the activities of fractions QA (RPA) and QB are required generally to initiate DNA replication in nuclei from human late G1 phase cells, and do not represent a special case specific for nuclei prepared from cells ‘chemically’ synchronised by mimosine.
Initiation of chromosomal DNA replication in late G1 phase nuclei depends on incubation in a cytosolic extract from proliferating cells (29). In the absence of this extract, or the factors contained in it, no new DNA replication foci are initiated in vitro but DNA replication can proceed further at replication forks that were established in vivo and that are clustered into foci prior to the isolation of the nuclei (28–30,34). By fractionating the initiating cytosolic extract, we have now identified RPA as one of the essential factors required to trigger initiation of new DNA replication foci in late G1 phase nuclei. This inductive functional assay provides strong direct evidence that soluble RPA originally present in the cytosolic extract is required to trigger replication in vitro and is recruited to forming replication foci within the nuclear templates. Our observations have now added the context of chromosomal DNA replication in nuclei of human somatic cells to the list of systems that require RPA as a replication initiation factor. Apart from the strict requirement to initiate DNA replication, the precise molecular mechanism of RPA involvement in the initiation of chromosomal DNA replication is not yet defined. However, in comparison with other initiation systems, some conclusions about its mode of action can be deduced.
In the SV40 replication initiation system, the molecular roles for human RPA are well defined (24,25). After parental DNA strand separation by the unwinding activity of the virally encoded initiator protein T-Ag (17–19), RPA binds to the single-stranded DNA and recruits DNA polα/primase to the unwound origin resulting in the synthesis of RNA primers and short DNA strands. Apart from binding single-stranded DNA, RPA physically interacts with T-Ag and with polα/primase. Under defined biochemical experimental conditions, the heterologous single-stranded DNA binding protein SSB of the prokaryote E.coli can substitute for human RPA in the stimulation of T-Ag-dependent origin unwinding (38,39), suggesting that the function of RPA in this unwinding reaction may be the protection and stabilisation of emanating single-stranded DNA. We found that SSB cannot substitute for RPA in the initiation of genomic DNA replication (Fig. 3), suggesting that the single-strand protection function is not the exclusive activity of RPA in the genomic context. Importantly, the recruitment of polα/primase by RPA cannot be substituted by prokaryotic SSB, indicating that direct protein–protein interactions between RPA and the initiating polα/primase are required (39). In support of this, we found that monoclonal antibodies 34A, 70A and 70B inhibit initiation in our system (Fig. 4). These antibodies were shown before to inhibit SV40 replication and, more specifically, inhibit stimulation of polα/primase, but not DNA unwinding (36). Furthermore, we found that antibody 70C, inhibiting SV40 replication and, more specifically, DNA polymerase δ and T-Ag-dependent DNA unwinding of the SV40 origin (36), also inhibited initiation in our system (Fig. 4). These data support the notion that recruitment of replicative DNA polymerases, and stimulation of their activities, may be a key function of RPA during the initiation of chromosomal DNA replication in human cell nuclei.
In Xenopus egg extracts, RPA is essential for DNA replication (40,41). RPA is recruited to chromatin in this system in a focal pattern by the ‘focus forming activity’ of another protein, FFA-1 (42), which is the orthologue protein of the Werner syndrome gene product helicase WRN in humans (43). FFA-1 itself is also required for DNA replication in Xenopus eggs and co-localises with RPA at replication foci (44). In a modified nucleus-free system from Xenopus eggs, supercoiled plasmid DNA can be replicated by subsequent incubations in membrane-free egg cytosol followed by a nucleoplasmic extract (45). Similar to the steps observed in the initiation of SV40 replication, plasmid DNA in this system was first unwound in the extracts in a manner that depended either on RPA or bacterial SSB, but recruitment of polα/primase and DNA synthesis constitute a separate step that required RPA and could not be substituted by SSB (46). Upstream of this reaction, generation of unwound DNA and hence recruitment of RPA and DNA polymerases required ORC, Cdc6, MCM2–MCM7, MCM10 and Cdc45, and depended on the protein kinase activities of Cdk2 and Cdc7 (46,47), consistent with other reports from Xenopus and the yeast S.cerevisiae (48–51).
The underlying principle that can be deduced from these systems seems thus to be that origin DNA is unwound by different molecular machines ranging from viral T-Ag to activated pre-RC complexes in a manner stimulated by either RPA or SSB. The common RPA-specific step, which cannot be substituted by SSB, is the recruitment of DNA polα/primase to the unwound origins. We therefore conclude from our data and these published results that this recruitment step is most likely a key step mediated by soluble RPA in the context of initiation of chromosomal DNA replication in human cell nuclei.
We found that Rpa32 becomes phosphorylated in vitro under all experimental conditions tested and thus coincides with, but does not strictly correlate with the initiation of chromosomal DNA replication. Rpa32 phosphorylation is not required for DNA replication in the SV40 system, but it is stimulated upon binding to single-stranded DNA and it occurs in a cell cycle-specific manner and in response to DNA damage (reviewed in 24,25). Our data cannot rule out a function of Rpa32 phosphorylation for the initiation of chromosomal DNA replication. However, because Rpa32 phosphorylation also occurs under conditions where initiation is not triggered, it is not the limiting step leading to initiation, which is induced by the cytosolic extract or its active fractions. Our data are thus consistent with two scenarios leading to a recruitment of RPA into intranuclear foci and its phosphorylation in the absence of efficient initiation of chromosomal DNA replication. Firstly, some DNA in our template nuclei might be damaged, and secondly, a limited unwinding of DNA might have already occurred.
The molecules that catalyse origin unwinding in human cell nuclei, and thus act as host cell counterparts of viral T-Ag, have not been identified. Possible candidates include the MCM protein-associated helicase activity (7,8) or that of the FFA-1 orthologue Werner syndrome gene product WRN helicase (43). However, our fractionation and reconstitution approach provides an inductional direct screen for initiation factors, which might include these relatively elusive unwinding factors. In support of this, preliminary order-of-addition experiments with fractions QA (RPA) and QB already suggest that RPA can be added after addition of QB, but not vice versa, to initiate genomic DNA replication in late G1 phase nuclei (data not shown), suggesting that RPA may be the last acting factor in a multistep cascade of initiation reactions. We are currently fractionating fraction QB further to purify the initiation factors contained in it. Their identity will shed more light on the molecular mechanism of the initiation of genomic DNA replication in human cells.
Acknowledgments
ACKNOWLEDGEMENTS
We thank David Santamaria for critically reading this manuscript, Alla Zaltsman, Anne Jones, Gerry O’Beirne and Ron Laskey for discussions, Marc Wold for the expression plasmid p11d-tRPA and Ron Laskey for the MCM5 antibody. This work was supported by a project grant from Cancer Research UK (C1471/A2612, old CRC reference SP2546/0101). C.C. and A.B were supported by Amersham Pharmacia Biotech and D.S. was supported initially by Peterhouse, Cambridge.
REFERENCES
- 1.Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 2.Blow J.J. and Hodgson,B. (2002) Replication licensing—origin licensing: defining the proliferative state? Trends Cell Biol., 12, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diffley J.F. and Labib,K. (2002) The chromosome replication cycle. J. Cell Sci., 115, 869–872. [DOI] [PubMed] [Google Scholar]
- 4.Lei M. and Tye,B.K. (2001) Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci., 114, 1447–1454. [DOI] [PubMed] [Google Scholar]
- 5.Kelly T.J. and Brown,G.W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem., 69, 829–880. [DOI] [PubMed] [Google Scholar]
- 6.Masai H. and Arai,K. (2002) Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell Physiol., 190, 287–296. [DOI] [PubMed] [Google Scholar]
- 7.Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 8.Labib K. and Diffley,J.F. (2001) Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- 9.Lohka M.J. and Masui,Y. (1983) Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science, 220, 719–721. [DOI] [PubMed] [Google Scholar]
- 10.Blow J.J. and Laskey,R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- 11.Newport J. (1987) Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell, 48, 205–217. [DOI] [PubMed] [Google Scholar]
- 12.Hyrien O., Maric,C. and Mechali,M. (1995) Transition in specification of embryonic metazoan DNA replication origins. Science, 270, 994–997. [DOI] [PubMed] [Google Scholar]
- 13.Li J.J. and Kelly,T.J. (1984) Simian virus 40 DNA replication in vitro. Proc. Natl Acad. Sci. USA, 81, 6973–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challberg M.D. and Kelly,T.J. (1989) Animal virus DNA replication. Annu. Rev. Biochem., 58, 671–717. [DOI] [PubMed] [Google Scholar]
- 15.Hurwitz J., Dean,F.B., Kwong,A.D. and Lee,S.H. (1990) The in vitro replication of DNA containing the SV40 origin. J. Biol. Chem., 265, 18043–18046. [PubMed] [Google Scholar]
- 16.Stillman B. (1989) Initiation of eukaryotic DNA replication in vitro. Annu. Rev. Cell. Biol., 5, 197–245. [DOI] [PubMed] [Google Scholar]
- 17.Stahl H., Droge,P. and Knippers,R. (1986) DNA helicase activity of SV40 large tumor antigen. EMBO J., 5, 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean F.B., Bullock,P., Murakami,Y., Wobbe,C.R., Weissbach,L. and Hurwitz,J. (1987) Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc. Natl Acad. Sci. USA, 84, 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wold M.S., Li,J.J. and Kelly,T.J. (1987) Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc. Natl Acad. Sci. USA, 84, 3643–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mastrangelo I.A., Hough,P.V., Wall,J.S., Dodson,M., Dean,F.B. and Hurwitz,J. (1989) ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature, 338, 658–662. [DOI] [PubMed] [Google Scholar]
- 21.Wold M.S. and Kelly,T. (1988) Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl Acad. Sci. USA, 85, 2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wobbe C.R., Weissbach,L., Borowiec,J.A., Dean,F.B., Murakami,Y., Bullock,P. and Hurwitz,J. (1987) Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc. Natl Acad. Sci. USA, 84, 1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairman M.P. and Stillman,B. (1988) Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J., 7, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iftode C., Daniely,Y. and Borowiec,J.A. (1999) Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol., 34, 141–180. [DOI] [PubMed] [Google Scholar]
- 25.Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- 26.Dornreiter I., Erdile,L.F., Gilbert,I.U., von Winkler,D., Kelly,T.J. and Fanning,E. (1992) Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J., 11, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stillman B. (1994) Smart machines at the DNA replication fork. Cell, 78, 725–728. [DOI] [PubMed] [Google Scholar]
- 28.Krude T., Jackman,M., Pines,J. and Laskey,R.A. (1997) Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell, 88, 109–119. [DOI] [PubMed] [Google Scholar]
- 29.Krude T. (2000) Initiation of human DNA replication in vitro using nuclei from cells arrested at an initiation-competent state. J. Biol. Chem., 275, 13699–13707. [DOI] [PubMed] [Google Scholar]
- 30.Keller C., Hyrien,O., Knippers,R. and Krude,T. (2002) Site-specific and temporally controlled initiation of DNA replication in a human cell-free system. Nucleic Acids Res., 30, 2114–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoeber K., Mills A.D., Kubota,Y., Marheineke,K., Krude,T., Romanowski,P., Laskey,R.A. and Williams,G.H. (1998) Cdc6 causes premature entry into S phase in a mammalian cell-free system. EMBO J., 17, 7219–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laman H., Coverley,D., Krude,T., Laskey,R. and Jones,N. (2001) Viral cyclin-cyclin-dependent kinase 6 complexes initiate nuclear DNA replication. Mol. Cell. Biol., 21, 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coverley D., Laman,H. and Laskey,R.A. (2002) Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nature Cell Biol., 4, 523–528. [DOI] [PubMed] [Google Scholar]
- 34.Krude T. (1999) Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res., 247, 148–159. [DOI] [PubMed] [Google Scholar]
- 35.Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) Recombinant replication protein A: expression, complex formation and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- 36.Kenny M.K., Schlegel,U., Furneaux,H. and Hurwitz,J. (1990) The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem., 265, 7693–7700. [PubMed] [Google Scholar]
- 37.Stoeber K., Tlsty,T.D., Happerfield,L., Thomas,G.A., Romanov,S., Bobrow,L., Williams,E.D. and Williams,G.H. (2001) DNA replication licensing and human cell proliferation. J. Cell Sci., 114, 2027–2041. [DOI] [PubMed] [Google Scholar]
- 38.Dodson M., Dean,F.B., Bullock,P., Echols,H. and Hurwitz,J. (1987) Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science, 238, 964–967. [DOI] [PubMed] [Google Scholar]
- 39.Kenny M.K., Lee,S.H. and Hurwitz,J. (1989) Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerases alpha and delta. Proc. Natl Acad. Sci. USA, 86, 9757–9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang F. and Newport,J.W. (1993) Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J. Cell Sci., 106, 983–994. [DOI] [PubMed] [Google Scholar]
- 41.Adachi Y. and Laemmli,U.K. (1994) Study of the cell cycle-dependent assembly of the DNA pre-replication centres in Xenopus egg extracts. EMBO J., 13, 4153–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan H. and Newport,J. (1995) FFA-1, a protein that promotes the formation of replication centers within nuclei. Science, 269, 1883–1885. [DOI] [PubMed] [Google Scholar]
- 43.Yan H., Chen,C.Y., Kobayashi,R. and Newport,J. (1998) Replication focus-forming activity 1 and the Werner syndrome gene product. Nature Genet., 19, 375–378. [DOI] [PubMed] [Google Scholar]
- 44.Chen C.Y., Graham,J. and Yan,H. (2001) Evidence for a replication function of FFA-1, the Xenopus orthologue of Werner syndrome protein. J. Cell Biol., 152, 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter J., Sun,L. and Newport,J. (1998) Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell, 1, 519–529. [DOI] [PubMed] [Google Scholar]
- 46.Walter J. and Newport,J. (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA and DNA polymerase alpha. Mol. Cell, 5, 617–627. [DOI] [PubMed] [Google Scholar]
- 47.Wohlschlegel J.A., Dhar,S.K., Prokhorova,T.A., Dutta,A. and Walter,J.C. (2002) Xenopus mcm10 binds to origins of DNA replication after mcm2-7 and stimulates origin binding of cdc45. Mol. Cell, 9, 233–240. [DOI] [PubMed] [Google Scholar]
- 48.Mimura S. and Takisawa,H. (1998) Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J., 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mimura S., Masuda,T., Matsui,T. and Takisawa,H. (2000) Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells, 5, 439–452. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka T. and Nasmyth,K. (1998) Association of RPA with chromosomal replication origins requires an Mcm protein and is regulated by Rad53 and cyclin- and Dbf4-dependent kinases. EMBO J., 17, 5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou L. and Stillman,B. (1998) Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]