Abstract
Very-short-patch repair (Vsr) enzymes occur in a variety of bacteria, where they initiate nucleotide excision repair of G:T mismatches arising by deamination of 5-methyl-cytosines in specific regulatory sequences. We have now determined the structure of the archetypal dcm-Vsr endonuclease from Escherichia coli bound to the cleaved authentic hemi-deaminated/hemi-methylated dcm sequence 5′-C-OH-3′ 5′-p-T-p-A-p-G-p-G-3′/3′-G-p-G-p-T-pMe5C-p-C formed by self-assembly of a 12mer oligonucleotide into a continuous nicked DNA superhelix. The structure reveals the presence of a Hoogsteen base pair within the deaminated recognition sequence and the substantial distortions of the DNA that accompany Vsr binding to product sites.
INTRODUCTION
Methylation is widely used in prokaryotes and eukaryotes for ‘tagging’ selected regions of DNA, and encoding epigenetic information in the genome. For example, in prokaryotes, methylation is used to indicate the parental strand for post-replicative mismatch repair (1), and to protect cognate sequences against cleavage by the cells own restriction endonucleases (2). In eukaryotes, methylation plays a key role in transcriptional regulation via the recruitment of chromatin (3), and underlies phenomena such as genome imprinting and X-inactivation (4). Elective methylation in bacteria can occur on the exocyclic amino groups of cytosine or adenine that protrude into the major groove, or on the C5 position of the cytosine ring. In eukaryotes, methylation is believed to occur exclusively on C5 of cytosine in the context of 5′-C-p-G-3′ sequences (5).
The mechanism of cytosine methylation common to all DNA cytosine methyltransferases (6) promotes hydrolytic deamination of the N4 exocyclic amino group, and 5meC is more susceptible to spontaneous deamination than the unmethylated base. Unlike deamination of cytosine, which generates the non-standard base uracil, deamination of 5meC generates the normal DNA base thymine. Thus, 5meC:G base pairs become promutagenic G:T mismatches which if not repaired, give rise to C:G→T:A transition mutations. Eukaryotes possess at least two repair enzymes, thymine DNA glycosylase (TDG) (7) and 5meC-binding domain glycosylase 4 (8), which recognise G:T mispairs specifically within the context of 5′-T-p-G-3′/3′-G-p-5meC-5′, and initiate a base-excision repair pathway by hydrolysing the N-glycosidic bond connecting the thymine base to the deoxyribose. While some bacteria possess homologues of TDG (9), these simpler mismatch-specific uracil-DNA glycosylase (MUG) enzymes are primarily active against U:G and ethenoC:G mismatches (10,11), displaying only slight activity to G:T mispairs (12), and are unlikely to provide a major pathway for repair of deaminated 5meC:G.
An alternative nucleotide excision repair pathway for G:T mismatches is provided in bacteria by the very-short-patch-repair (Vsr) endonucleases (13,14). These enzymes initiate repair by cleaving the phosphodiester backbone on the 5′ side of the thymine in a G:T mismatch. Extension of the generated 3′-OH by a repair DNA polymerase such as Escherichia coli polymerase I combined with displacement and excision of the mismatched 5′ ‘flap’ and ligation can then restore the original sequence. The archetypal Vsr endonuclease is the dcm-Vsr which specifically recognises and cleaves hemi-deaminated/hemi-methylated dcm sequence in E.coli and some other enteric bacteria. Vsr-initiated very-short-patch-repair reinstates the hemi-methylated dcm sites which are the substrate for the dcm maintenance methyltransferase (15). Although the role of the dcm sites in E.coli is obscure, the presence of a specific repair mechanism suggests that their maintenance is important to E.coli in a way that is not yet appreciated. Homologues of dcm-Vsr have been identified in a number of bacteria, in association with restriction– modification systems in which cytosine 5-methylation is used to prevent DNA self-cleavage by restriction endonucleases (16). The presumed role of the associated Vsr enzymes in these systems is to initiate repair of G:T mismatches resulting from deamination of the 5meC:G base pairs arising in the methylated restriction target sequences.
We have now determined the structure of a complex between the E.coli dcm-Vsr and a continuous ‘nicked’ DNA duplex designed to self-assemble and generate an authentic dcm-Vsr product site at the juxtaposition of adjacent oligonucleotides. The structure explains the high affinity of Vsr enzymes for their nicked product, and identifies the protein–DNA interactions that may play a role in determining specificity for the G:T mismatch and for the dcm context. Structural changes in the DNA itself that result from hemi-deamination may explain the observed preference for the hemi-methylated product. The success of the self-assembling ‘nicked’ duplex in forcing a crystal lattice suggests that this might be a powerful general method for co-crystallisation of protein–DNA complexes.
MATERIALS AND METHODS
Cloning, expression and crystallisation
Escherichia coli Vsr-dcm endonuclease was cloned by PCR amplification from E.coli JM109. Due to uncertainty as to which of two possible methionines provided the authentic start codon, two different clones were constructed; a ‘long’ clone expressing a 156 residue protein from the first candidate ATG, and a ‘short’ clone expressing a 143 residue protein from the second ATG. Both constructs were cloned into pRSETB and expressed in E.coli B834 (DE3). Cells were grown in 1.5× Oxoid Nutrient Broth to an A600 of 0.5–0.9. Following induction with 0.8 mM IPTG the broths were maintained at 37°C overnight prior to harvesting.
Cells were resuspended in buffer containing 20 mM Tris pH 7.5, 2 mM MgCl2, 50 mM NaCl and 1 mM PMSF. The cells were lysed by sonication and were subsequently incubated on ice with 1% streptomycin sulphate for 30 min prior to centrifugation. The clarified lysate was then applied to a Q-Sepharose column equilibrated in 20 mM Tris pH 7.5, 2 mM MgCl2, 50 mM NaCl and 0.1 mM PMSF. Protein was eluted from the column using a linear salt gradient (0.1–1 M NaCl). Fractions containing Vsr were then concentrated prior to size exclusion chromatography (Superdex-75) in buffer containing 50 mM HEPES pH 7.0 and 200 mM NaCl. Vsr-containing fractions were diluted to 100 mM NaCl and were applied to a heparin–Sepharose column equilibrated in 50 mM HEPES pH 7.0 and 100 mM NaCl. Protein was eluted using a linear salt gradient (0.1–1 M NaCl). Purified Vsr was then concentrated by diluting to a salt concentration of 100 mM NaCl followed by loading onto a 1 ml HiTrap SP-Sepharose (Pharmacia) column equilibrated in the same buffer. Protein was eluted in buffer containing 50 mM HEPES pH 7.0 and 1 M NaCl, in which the protein was stored at 4°C prior to crystallisation trials.
Co-crystallisation of the ‘long’ Vsr protein was performed with several oligonucleotides designed to self-anneal as a continuous nicked duplex, generating the dcm-Vsr endonuclease reaction product sequence (5′-C-OH-3′ 5′-p-T-p-A-p-G-p-G-3′/3′-G-p-G-p-T-pMe5C-p-C) at the abutment of consecutive oligonucleotides. There was an absolute requirement for a 5′ phosphate group to achieve crystallisation. The Vsr–DNA complexes were obtained by mixing the protein with the annealed oligonucleotides with a slight excess of DNA (DNA to protein 1.1:1 molar ratio) prior to dilution to obtain a final buffer concentration of 50 mM HEPES pH 7.0 and 150 mM NaCl. The complex was then concentrated using a Vivaspin 500 device (MW cut-off 5000 Da; Vivascience) to the required protein concentration. Small crystals containing ‘long’ Vsr were obtained in microbatch experiments at 16°C with self-annealing oligonucleotides 12 bases in length, in either 15% PEG 4000, 0.1 M sodium citrate pH 5.6, 0.2 M sodium acetate and 25 mM HEPES pH 7.0 or 15% PEG 4000, 0.1 M Tris–HCl pH 8.5, 0.2 M sodium acetate and 25 mM HEPES pH 7.0 at a protein concentration of 4 mg/ml, but only gave weak diffraction to 8 Å. Experiments with the ‘short’ Vsr protein also gave crystals with a self-annealing 12mer (5′-pTpApGpGpCp5meCpTpGpGpApTpC-3′) using the microbatch method at 14°C in 25 mM HEPES pH 7.0, 75 mM NaCl, 15% PEG 8000, 50 mM sodium cacodylate pH 6.5, 75 mM ammonium sulphate and 10% glycerol, with a protein concentration of 2.5 mg/ml. These crystals diffracted to beyond 4 Å on a rotating anode source and had space group P61 or P65 with unit cell dimensions a = 102.81 Å, c = 64.29 Å. Isomorphous crystals were obtained using a second 12mer in which thymine was replaced by 5-iodo-uracil (5′-pTpApGpGpCp5meCp5IUpGpGpApTpC-3′).
Data collection, structure determination and refinement
Native data to 2.75 Å were collected on a CCD detector with 0.933 Å radiation on ID14-2 at the ESRF Synchrotron, Grenoble, France. Diffraction images were processed using MOSFLM (17) and the data merged, scaled and reduced using programs of the CCP4 suite (18). Data to 3.3 Å were collected for a co-crystal with an iodinated oligo. Parameters for the data collection and processing are given in Table 1. The data collection statistics for the native data are acceptable and there is no indication of twinning; however, the data do show an unusually high inherent temperature factor as estimated from Wilson plots (∼105 Å2).
Table 1. Crystallographic statistics.
| Data collection |
All data (outer shell) |
| Resolution range | 50.0–2.75 (2.9–2.75) |
| Rmerge | 0.070 (0.384) |
| I/σ(I) | 6.9 (2.0) |
| Completeness (%) | 99.0 (100) |
| Multiplicity | 4.9 (5.0) |
| No. of unique reflections |
10 084 (1464) |
| Structure refinement |
|
| No. of atoms (protein) | 2942 |
| No. of atoms (all) | 3086 |
| Resolution range | 24.7–2.8 |
| Rcryst | 0.287 |
| Rfree | 0.356 |
| Real-space correlation | 0.90 (protein); 0.87 (DNA) |
| Real-space R-factor | 0.1 (protein); 0.13 (DNA) |
| RMS deviation of bond lengths from ideality (Å) | 0.008 |
| RMS deviation of bond angles from ideality (°) | 1.306 |
The structure of the E.coli Vsr-dcm endonuclease–DNA complex was determined by molecular replacement using AMoRe (19) with the structure of the isolated enzyme (PDB code 1CW0) which was very kindly released ahead of schedule by Dr Kosuke Morikawa. A single solution was obtained in the rotation function, and gave a clear translation function solution in P61 (correlation coefficient = 31.2, 17.5 higher than the next solution), but not in P65 (highest correlation coefficient = 15.3, only 0.2 higher than the next solution) defining the correct enantiomorph of the space group. The native self-rotation functions indicated non-crystallographic 2-fold symmetry perpendicular to the crystallographic 61 axis; however, a second molecular replacement solution for the protein could not be found (best correlation coefficient = 27.1, only 0.2 higher than the next solution). Subsequently, this non-crystallographic 2-fold pseudo-symmetry can be understood to arise from the DNA superhelix.
The protein model transformed by the molecular replacement solution was refined against the observed diffraction data using CNS (20) and difference maps calculated, revealing electron density features consistent with the base planes and phosphate-scattering centres of bound DNA, and indicating the presence of a DNA superhelix. Segments of the DNA indicated by the difference maps were constructed and refined with the protein model in CNS. The staging of the DNA sequence along the continuous nicked DNA superhelix was verified by the positions of >3σ positive peaks in isomorphous difference maps calculated between crystals grown with the native oligonucleotide and crystals grown with an oligonucleotide in which a thymine was substituted with 5-iodo-uracil. Subsequent cycles of manual intervention using ‘O’ and simulated annealing refinement in CNS gave the final model for the asymmetric unit, consisting of 156 protein residues, 24 nucleotides, a Zn2+ ion and 80 solvent molecules. The refined crystallographic R-factor (0.287) and R-free (0.356) are higher than ideal, but difference Fourier maps show no significant positive or negative density features, and the structure is consistent with omit-maps. Real-space R-factors calculated for the final model using CNS show excellent correlation between the model and the electron density (real-space correlation coefficient = 0.90 for protein, 0.87 for DNA; real-space R-factor = 0.10 for protein, 0.13 for DNA). We believe the relatively high reciprocal space R-values result from the very high solvent content, the high thermal parameters particularly of the protein which lacks lattice contacts, and the general weakness and anisotropy of the data. The refined co-ordinates and structure factors have been deposited in the Protein Databank with PDB code 1odg.
RESULTS AND DISCUSSION
Crystal architecture
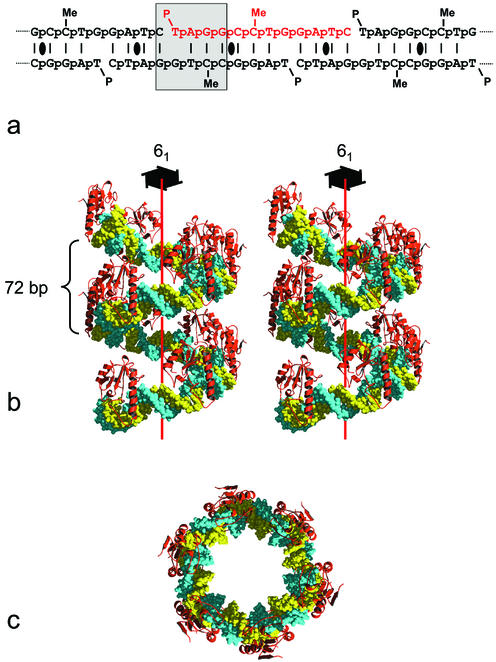
A ‘short’ version of the E.coli dcm-vsr endonuclease consisting of residues 14–156 was expressed and purified (see Materials and Methods) and co-crystallised with an oligonucleotide 5′-p-T-p-A-p-G-p-G-p-C-p-5meC-p-T-p-G-p-G-p-A-p-T-p-C-OH-3′. This oligonucleotide was designed to self-anneal, generating sites corresponding to the product of the cleavage reaction in a regular pattern along the continuous ‘nicked’ duplex (Fig. 1a). As Vsr endonucleases display substantial affinity for the product of their reaction, it was anticipated that this nicked duplex would be periodically adorned by enzyme molecules, generating an inherently ordered array in two dimensions, and would thereby promote formation of diffracting crystals by reducing the dimensionality of the crystallisation process. The Vsr–DNA complex crystallised in space group P61, and ultimate determination of the complex structure (see Materials and Methods) revealed that the oligonucleotide had, as intended, self-assembled into an ‘infinite’ nicked duplex. However, as the cleaved DNA at the dcm-Vsr recognition site was substantially bent by its interaction with the enzyme (see below), the nicked duplex formed a shallow regular superhelix rather than a more-or-less linear B-form structure as we have observed in other DNA repair enzyme complexes using self-assembling oligonucleotides (12,21,22).
Figure 1.
Structure of the dcm-Vsr–DNA superhelix complex. (a) The DNA 12mer self-assembles to generate a continuously nicked DNA duplex. The repeating unit of the superhelix is highlighted in red, and the cleaved hemi-deaminated/hemi-methylated dcm site is boxed. Local dyad symmetries are indicated. (b) Stereo-pair showing two complete unit cells (12 repeats) of the dcm-Vsr–DNA superhelix complex, viewed perpendicularly to the crystallographic 61-screw axis. Each asymmetric unit consists of 12 bp of duplex DNA with a single bound protein molecule. There are no contacts between the protein molecules in different asymmetric units. (c) The dcm-Vsr–DNA superhelix complex viewed down the crystallographic 61-screw axis, showing the large solvent-filled central hole.
The axis of the superhelix formed by the self-assembly of the oligonucleotide is parallel to the crystal c-axis and the DNA obeys the strict 61 screw-symmetry of the space group with a radius of ∼50 Å and a rise of 10.7 Å, giving a repeat of 72 bp, which is comparable to the DNA repeat length observed in the DNA of nucleosome core particles (23) (Fig. 1b and c). The self-assembling oligonucleotide generates two dcm-Vsr product sites in alternating orientations every 12 bp along the continuous nicked duplex DNA. However, only one of these sites is occupied by an enzyme molecule in the crystal. Thus, the asymmetric unit contains one protein molecule, and two copies of the 12mer oligonucleotide, which occupy non-equivalent environments. The dcm-Vsr molecules make no protein–protein contacts with other dcm-Vsr molecules bound to the same ‘infinite’ nicked superhelix, and only a single lattice contact with a protein molecule bound to a different superhelix, defining the two-dimensional packing of the protein–DNA superhelices that completes the formation of the crystal. The crystals thus have a very open structure with a solvent content >70% by volume, and consequently display weak and somewhat anisotropic diffraction.
Protein–DNA interactions
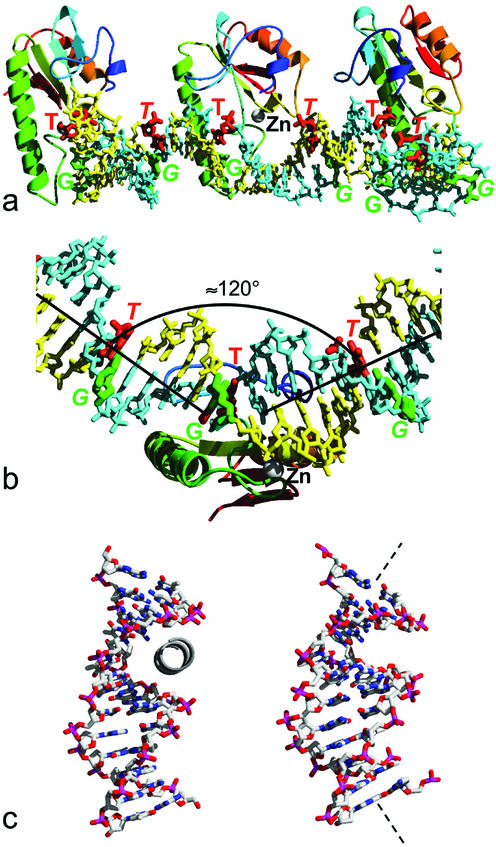
The dcm-Vsr enzyme binds, as intended, to the cleaved recognition sequence (T-strand) 5′-C-OH-3′ 5′-p-T-p-A-p-G-p-G-3′ / 3′-G-p-G-p-T-pMe5C-p-C-5′ (G-strand) formed at the junction of consecutive oligonucleotides along the DNA helix (Fig. 2a). The base-pair stacking in the DNA duplex is disrupted at the site of dcm-Vsr binding, introducing an ∼60° bend in the minor groove face of the duplex (Fig. 2b), whose cumulative effect every 12 bp generates the 72 base-repeat superhelix. The intervening segments of DNA are linear with an essentially canonical B-form conformation.
Figure 2.
Protein-induced DNA bending. (a) Three consecutive repeats generating a half turn around the superhelix axis. The DNA strands are coloured yellow and blue, with the nucleotides in the G:T mismatches coloured green (G) and red (T). The position of the structural zinc atom is indicated in the middle repeat by the silver sphere. (b) The DNA is not uniformly curved, but consists of segments of more-or-less straight B-form DNA kinked by ∼60° where it is bound by the dcm-Vsr enzyme. (c) In the structure of dcm-Vsr bound to a short nicked oligonucleotide (24), an short N-terminal helix binds into the minor groove of the DNA (left). In the structure presented here, the DNA has a greater curvature that closes down the minor groove, preventing binding of the truncated N-terminal segment, which is disordered in this structure.
Overall, the binding of dcm-Vsr with the superhelical DNA duplex is similar to that observed in the structure of dcm-Vsr bound to a short cleaved oligonucleotide (24), with the long helix (82–106) binding in the major groove and the loop at 120–122 interacting with the phosphate backbone on the 5′ side of the mismatched thymine. In the short-oligo dcm-Vsr complex residues 17–22 curl around the sugar–phosphate backbone on the 3′ side of the mismatched thymine, with residues 2–16 inserting into the minor groove of the duplex. Although residues 14–22 are present in our construct, they are entirely disordered, and we observe no protein binding into the much narrower minor groove of the superhelical DNA (Fig. 2c).
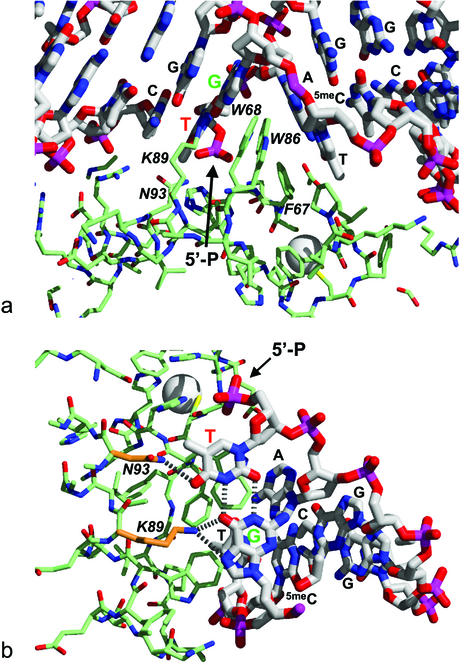
The major disruption to the base stacking and linearity of the DNA results from the insertion of a hydrophobic wedge consisting of the side chains of Phe67, Trp68 and Trp86, between the G:T mismatch and the central A:T base pair of the dcm sequence, as previously described (24) (Fig. 3a). The wedge penetrates the mismatched DNA from the major groove causing a substantial opening of the major groove, but compaction of the minor groove. This asymmetric intercalation is similar to the interaction of the MutS repair protein with DNA mismatches (25,26), but different from the interaction of the MUG in which both grooves are opened up to similar extents (12). Also unlike the MUG enzyme, recognition of the mismatched G:T base pair occurs in situ, with the unique pattern of hydrogen bond acceptors (thymine O4; guanine O6 and N7) presented by the major groove edge of the wobble base pair recognised via interactions with the Nε of Lys89 and the amide nitrogen of Asn93, as previously described (24) (Fig. 3a and b).
Figure 3.
Protein–DNA interactions. (a) The ∼60° bend in the DNA is the result of asymmetric intercalation from the major groove, of a hydrophobic wedge formed by Phe67, Trp68 and Trp86, between the G:T mismatch and the central A:T base pair of the hemi-deaminated/hemi-methylated dcm sequence. The positions of the terminal thymidine 5′ phosphate at the nick site is indicated. (b) The unique pattern of hydrogen bond accepting groups presented by the major groove edge of the ‘wobble’ conformation G:T mismatch is recognised by hydrogen bonds from the ε-amino of Lys89 and the amide-NH2 of Asn93.
While the presence of a G:T mispair is the overwhelming requirement, the nature of the flanking base pairs exerts a substantial influence on the efficiency of DNA backbone cleavage by dcm-vsr (14,27–30). Thus, replacement of the 5-methyl-cytosine present in the authentic hemi-methylated/hemi-deaminated dcm sequence with cytosine reduces kcat/Km to ∼10% that for the native oligonucleotide sequence (30), while replacement of the central A-T base pair by G-C decreases hydrolysis rate to ∼5%. (28). Surprisingly, apart from the interactions with the G:T mismatch itself (see above), there are few direct interactions in this or the previous dcm-Vsr structures between the enzyme and the other bases within the pentanucleotide dcm sequence that would mediate this extended sequence specificity. In particular, there are no interactions that explain the requirement for a central A:T or T:A base pair with only slight preference for its orientation.
Base pair rearrangements
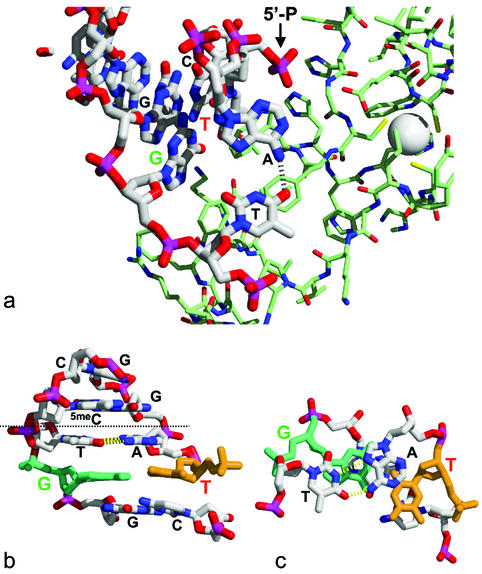
In the structure presented here, the central A:T base pair in the enzyme-bound dcm sequence does not form a normal Watson–Crick base pair. Instead, the adenine glycosidic bond has a syn rather than anti conformation and forms a Hoogsteen base pair with the opposing thymine (Fig. 4a). Hoogsteen A:T base pairs are believed to exist at a low level in bulk DNA in equilibrium with the Watson–Crick form, but had not been observed in a protein–DNA complex until recently (31). In the previously reported structure of dcm-Vsr bound to a short oligonucleotide (24), the central A:T base pair was in the normal Watson–Crick conformation. At first sight the reason for the difference between these two structures is not obvious. Both structures have an A:T rather than T:A base pair so that the adenine is on the same strand as the thymine of the G:T mismatch, and in both structures the DNA is nicked on the 5′ side of the G:T mismatch. As the mismatch substrate for this enzyme occurs in a fully methylated dcm sequence, the cytosine in the G:C base pair on the other side of the central A:T would be 5-methylated in vivo, as it is in this present structure. However, in the previous dcm-Vsr complex structure an unmodified cytosine was included at this position, so that the difference in the observed conformations of the central A:T base pairs in the two structures could be due to the presence or absence of this 5-methyl group.
Figure 4.
DNA conformation. (a) Hoogsteen conformation of the central A:T base pair in the protein-bound dcm site. (b) Hoogsteen conformation of the central A:T base pair in the unbound copy of the dcm site, viewed perpendicularly to the DNA helix axis. (c) The unbound dcm site viewed along the DNA helix axis, clipped at the dotted line in (b).
Within the present structure, only every other hemi-deaminated/hemi-methylated dcm site is actually bound by a dcm-Vsr molecule, providing a view of the DNA sequence in both bound and unbound states (Fig. 4b and c). Remarkably, the central A:T base pair even in the unbound dcm sequence is also in a Hoogsteen conformation, showing that this is an inherent property of the sequence and not due to the large bend introduced into the DNA helix by binding of the enzyme. In the structure of the unbound sequence, it is possible to understand the formation of the Hoogsteen base pair as a response to the unusual properties of the base pairs on either side of the central A:T. On one side, the hydrophobic surface of the central adenine in a Watson–Crick conformation becomes exposed by displacement of the mismatched thymine into the major groove in the ‘wobble’ conformation G:T mismatch base pair. On the other side, the thymine 5-methyl of the central thymine in a Watson–Crick conformation would be sterically crowded by the cytosine 5-methyl of the 5meC:G base pair. Flipping the conformation of the central A:T from Watson–Crick to Hoogsteen, pushes the central thymine towards the minor groove, relieving the methyl–methyl clash with the adjacent 5-methyl-cytosine, and rotates the adenine so that its six-membered ring stacks over the ‘wobbled’ mismatched thymine in the major groove. As G:T mismatches or 5meC:G base pairs individually are not known to promote Hoogsteen conformations in adjacent A:T base pairs, it is the unusual combination of the two found in the dcm-Vsr site that appears to be required. As both these factors would be present in vivo then it is likely that the formation of the Hoogsteen base pair is not simply an in vitro effect. The central base pair in the dcm sequence can be in the A:T orientation as here, or in the inverse T:A orientation, so that the central thymine would be adjacent to the thymine of the G:T mismatch in the deaminated sequence. In that orientation, the central thymine would remain well stacked with the guanine of the mismatch, while the potential methyl–methyl steric clash of the central thymine with the mismatch thymine would be relieved by the wobble conformation, so that a Hoogsteen conformation would not be favoured.
Recognition of the hemi-deaminated/hemi-methylated dcm site by the dcm-Vsr endonuclease involves a very substantial distortion of the DNA by the asymmetric intercalation of a protein wedge between the G:T mismatch and the central A:T base pair. Insertion of Trp86 in the hydrophobic wedge is required to bring the side chains Lys89 and Asn93 into position to hydrogen-bond to the edge of the G:T mismatch and stabilise the complex to allow hydrolysis of the phosphodiester bond. Thus, distortion of the DNA adjacent to the mismatch is a key component in Vsr recognition of the hemi-deaminated/hemi-methylated dcm site, and depends on a degree of inherent softness and plasticity of the DNA sequence, evinced by the ability of the central A:T base pair to re-orientate from a Watson–Crick to a Hoogsteen conformation.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Bernard Connolly, David Turner and Rebecca Gonzalez-Nicieza for useful discussion, Tracey Barrett for assistance with data collection and processing, and the ESRF Grenoble for access to synchrotron beam-time. This work was supported by a BBSRC Project Grant.
PDB accession no. 1ogd
REFERENCES
- 1.Modrich P. (1989) Methyl-directed DNA mismatch correction. J. Biol. Chem., 264, 6597–6600. [PubMed] [Google Scholar]
- 2.Bickle T.A. and Kruger,D.H. (1993) Biology of DNA restriction. Microbiol. Rev., 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression—belts, braces and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R. (1997) DNA methylation and imprinting: Why bother? Trends Genet., 13, 323–329. [DOI] [PubMed] [Google Scholar]
- 5.Gruenbaum Y., Stein,R., Cedar,H. and Razin,A. (1981) Methylation of CpG sequences in eukaryotic DNA. FEBS Lett., 124, 67–71. [DOI] [PubMed] [Google Scholar]
- 6.Wu J.C. and Santi,D.V. (1985) On the mechanism and inhibition of DNA cytosine methyltransferases. In Cantoni,G.I. and Razin,A. (eds), Biochemistry and Biology of DNA Methylation. Alan R. Liss, Inc., New York, pp. 119–129. [PubMed]
- 7.Nedderman P. and Jiricny,J. (1993) The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem., 268, 21218–21224. [PubMed] [Google Scholar]
- 8.Hendrich B., Hardeland,U., Ng,H.-H., Jiricny,J. and Bird,A. (1999) The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature, 401, 301–304. [DOI] [PubMed] [Google Scholar]
- 9.Gallinari P. and Jiricny,J. (1996) A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature, 383, 735–738. [DOI] [PubMed] [Google Scholar]
- 10.Saparbaev M. and Laval,J. (1998) 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 95, 8508–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutsenko E. and Bhagwat,A.S. (1999) The role of the Escherichia coli mug protein in the removal of uracil and 3,N-4-ethenocytosine from DNA. J. Biol. Chem., 274, 31034–31038. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- 13.Sohail A., Lieb,M., Dar,M. and Bhagwat,A.S. (1990) A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J. Bacteriol., 172, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennecke F., Kolmar,H., Brundl,K. and Fritz,H.J. (1991) The vsr gene product of E.coli K-12 is a strand and sequence-specific DNA mismatch endonuclease. Nature, 353, 776–778. [DOI] [PubMed] [Google Scholar]
- 15.Lieb M. and Bhagwat,A.S. (1996) Very short patch repair: reducing the cost of cytosine methylation. Mol. Microbiol., 20, 467–473. [DOI] [PubMed] [Google Scholar]
- 16.Kulakauskas S., Barsomian,J.M., Lubys,A., Roberts,R.J. and Wilson,G.G. (1994) Organization and sequence of the Hpaii restriction–modification system and adjacent genes. Gene, 142, 9–15. [DOI] [PubMed] [Google Scholar]
- 17.Leslie A.G.W. (1995) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography, No. 26. MRC Laboratory of Molecular Biology. Cambridge, UK.
- 18.CCP4 (1994) Programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 19.Navaza J. (1994) AMoRE—an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- 20.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 21.Pearl L.H. (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res., 460, 165–181. [DOI] [PubMed] [Google Scholar]
- 22.Barrett T.E., Schärer,O.D., Savva,R., Brown,T., Jiricny,J., Verdine,G.L. and Pearl,L.H. (1999) Crystal structure of a thwarted mismatch DNA glycosylase DNA repair complex. EMBO J., 18, 6599–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White C.L., Suto,R.K. and Luger,K. (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J., 20, 5207–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsutakawa S.E., Jingami,H. and Morikawa,K. (1999) Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell, 99, 615–623. [DOI] [PubMed] [Google Scholar]
- 25.Lamers M.H., Perrakis,A., Enzlin,J.H., Winterwerp,H.H.K., de Wind,N. and Sixma,T.K. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G center dot T mismatch. Nature, 407, 711–717. [DOI] [PubMed] [Google Scholar]
- 26.Obmolova G., Ban,C., Hsieh,P. and Yang,W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature, 407, 703–710. [DOI] [PubMed] [Google Scholar]
- 27.Fox K.R., Allinson,S.L., Sahagun-Krause,H. and Brown,T. (2000) Recognition of GT mismatches by Vsr mismatch endonuclease. Nucleic Acids Res., 28, 2535–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glasner W., Merkl,R., Schellenberger,V. and Fritz,H.J. (1995) Substrate preferences of VSR DNA mismatch endonuclease and their conseqeunces for the evolution of the Escherichia coli K-12 genome. J. Mol. Biol., 245, 1–7. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Nicieza R., Turner,D.P. and Connolly,B.A. (2001) DNA binding and cleavage selectivity of the Escherichia coli DNA G:T-mismatch endonuclease (vsr protein). J. Mol. Biol., 310, 501–508. [DOI] [PubMed] [Google Scholar]
- 30.Turner D.P. and Connolly,B.A. (2000) Interaction of the E.coli DNA G:T-mismatch endonuclease (vsr protein) with oligonucleotides containing its target sequence. J. Mol. Biol., 304, 765–778. [DOI] [PubMed] [Google Scholar]
- 31.Aishima J., Gitti,R.K., Noah,J.E., Gan,H.H., Schlick,T. and Wolberger,C. (2002) A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res., 30, 5244–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]