Abstract
Errors introduced during PCR amplification set a selectivity limit for microsatellite analysis and molecular mutation detection methods since polymerase misincorporations invariably get confused with genuine mutations. Here we present hairpin-PCR, a new form of PCR that completely separates genuine mutations from polymerase misincorporations. Hairpin-PCR operates by converting a DNA sequence to a hairpin following ligation of oligonucleotide caps to DNA ends. We developed conditions that allow a DNA hairpin to be efficiently PCR-amplified so that, during DNA synthesis, the polymerase copies both DNA strands in a single pass. Consequently, when a misincorporation occurs it forms a mismatch following DNA amplification, and is distinguished from genuine mutations that remain fully matched. Error-free DNA can subsequently be isolated using one of many approaches, such as dHPLC or enzymatic depletion. We present feasibility for the main technical steps involved in this new strategy, conversion of a sequence to a hairpin that can be PCR-amplified from human genomic DNA, exponential amplification from picogram amounts, conversion of misincorporations to mismatches and separation of homoduplex from heteroduplex hairpins using dHPLC. The present hairpin-PCR opens up the possibility for a radical elimination of PCR errors from amplified DNA and a major improvement in mutation detection.
INTRODUCTION
PCR-based amplification is used in almost every aspect of genetic diagnosis, mutation detection and basic research. A major problem with PCR, however, is that all polymerases invariably generate errors during amplification (1–3). Polymerase misincorporations produce slippage errors that complicate microsatellite analysis (4) and set a selectivity limit for molecular mutation detection methods of one mutant sequence in a background of 105–106 wild-type sequences (5). Thus, high selectivity mutation detection, which relies on PCR, often falls short by one to two orders of magnitude of the selectivity required to investigate mechanisms of spontaneous mutagenesis (6), to identify cancer cells at an early stage (7), to detect mutations in single cells (8) or to reliably identify minimal residual disease (9). In all these applications, polymerase misincorporations invariably become disguised as mutations and result to false positives (10).
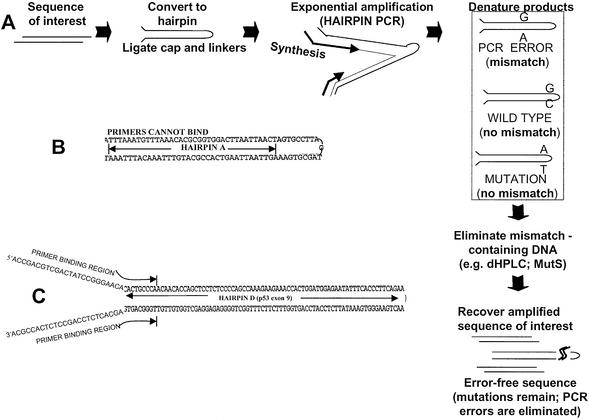
Here we present hairpin-PCR a new form of PCR that completely separates genuine mutations from polymerase misincorporations. This opens up the possibility of eliminating misincorporations and generating ‘error-free’ amplified DNA for mutation detection or other applications (Fig. 1A). Hairpin-PCR is based on the observation that, unlike regular DNA hairpins (Fig. 1B), which tend to suppress PCR (11), a DNA hairpin with non-complementary ends (Fig. 1C) can be efficiently PCR-amplified. A DNA sequence that needs to be PCR-amplified is first converted to a hairpin following ligation of an oligonucleotide ‘cap’ on one end and a pair of non-complementary linkers on the other end (Fig. 1A). Next, primers corresponding to the two non-complementary linkers are used to allow DNA polymerase to displace the opposite strand and amplify the entire complement of the hairpin. These primers can optionally overlap the sequence of interest to confer sequence specificity. Following hairpin amplification, the double-stranded PCR product is heat-denatured and placed rapidly on ice to allow the original hairpins to re-form. By amplifying DNA in a hairpin formation, polymerase errors practically always end up forming a mismatch. Genuine mutations, however, are expected to remain fully matched. Next, the amplified hairpins that contain mismatches can be separated from those that do not, using dHPLC-mediated fraction collection. With subsequent removal of the hairpin caps, the original DNA sequence is recovered. The error-free amplified DNA can then be processed for mutation detection without the limitation of polymerase errors. Below we present feasibility for the main technical steps involved in this strategy, which can lead to a radical elimination of PCR errors from amplified DNA.
Figure 1.
Outline of hairpin-PCR. (A) Scheme for removing PCR errors following amplification of DNA in a hairpin structure. (B) Expected structure and sequence of hairpin A. (C) Expected structure and sequence of hairpin D, an oligonucleotide encompassing both top and bottom strands of p53 exon 9.
MATERIALS AND METHODS
Hairpin-forming, long oligonucleotides
Five long oligonucleotides expected to form hairpins were synthesized by Oligos Etc and HPLC-purified (OR, USA). The sequence of hairpins A and D are depicted in Figure 1B–C. Sequences of hairpins B, C and E, which were designed to contain non-complementary ends like hairpin D, were: hairpin B, 5′-ACC GAC GTC GAC TAT CCG GGA ACA CAT GAT TTA AAT GTT TAA ACA CGC GGT GGA CTT AAT TAA CTA GTG CCT TAG GTA GCG TGA AAG TTA ATT AAG TCA CCG CAT GTT TAA ACA TTT AAA TGT ACA GCA CTC TCC AGC CTC TCA CCG CA-3′; hairpin C, 5′-ACC GAC GTC GAC TAT CCG GGA ACA CAA GAT TTA AAT GTT TAA ACA CGC GGT GAC TTA ACA GGC GCG CCT TAA CTA GTG CCT TAG GTA GCG TGA AAG TTA AGG CGC GCC TGT TAA GTC ACC GCG TGT TTA AAC ATT TAA ATC TTG AGC ACT CTC CAG CCT CTC ACC GCA-3′; hairpin E, 5′-ACC GAC GTC GAC TAT CCG GGA ACA GAT CCA TGC ACT GCC CAA CAA CAC CAG CTC CTC TCC CCA GCC AAA GAA GAA ACC ACT GGA TGG AGA ATA TTT CGA CCC TTC AGA AAA CTG AAG GGT CGA AAT ATT CTC CAT CCA GTG GTT TCT TCT TTG GCT GGG GAG AGG AGC TGG TGT TGT TGG GCA GTG CAT GGA TCA GCA CTC TCC AGC CTC TCA CCG CA-3′.
Hairpin-PCR
Designated amounts of hairpins B–D were used in a 25 µl PCR reaction using Titanium AdvantageR polymerase (Clontech, Palo Alto, CA) and the primers 5′-GTG AGA GGC TGG AGA GTG CT-3′, forward; and 5′-ACG TCG ACT ATC CGG GAA CA-3′, reverse. PCR thermo-cycling conditions were: 94°C, 30 s; (94°C, 30 s/68°C, 60 s) × 25 cycles; 68°C, 60 s; 4°C; Hold. The PCR products were then examined via ethidium-stained agarose gel electrophoresis. Alternatively, PCR amplification was conducted using high fidelity Advantage HF-2R polymerase (Clontech) or Pfu Turbo® (Strategene Inc.). In addition, using the same thermocycling conditions, quantitative real-time PCR in the presence of SYBR Green I dye was performed in a Cepheid I SmartCycler™ machine. Primers used for PCR of hairpin A were 5′-TAA ATG TTT AAA CAC GCG GT-3′, forward; and 5′-TAA ATG TTT AAA CAT GCG GT-3′ reverse. To amplify picogram amounts of hairpin D spiked into 100 ng human genomic DNA from HL-60 cells, touchdown PCR was applied: 94°C, 30 s; (94°C, 20 s/65°C, 20 s/68°C, 20 s) × 30 cycles, with annealing temperature decreasing 1°C/cycle; (94°C, 10 s/55°C, 20 s/68°C, 20 s) × 15 cycles; 68°C, 6 min; 4°C; Hold.
dHPLC analysis of oligonucleotide hairpins
To perform separation of mixtures of heteroduplex and homoduplex hairpins, 1 ng hairpins were injected into a WAVE™ dHPLC system (Transgenomics Inc., Cambridge, MA) and run under denaturing conditions at different temperatures, following the company-supplied protocol [Transgenomics Inc. (2002) Single Nucleotide Polymorphism (SNP), Insertion & Deletion on the WAVE® Nucleic Acid Fragment Analysis System; http://www.transgenomic.com/pdf/AN112.pdf]. The dHPLC system was equipped with a fraction collector that allows selection of the elution product according to the DNA retention time on the dHPLC column.
Conversion of native DNA sequences to a hairpin, and PCR amplification
The primers 5′-AGG CCT TCA TGA CTG ATA CCA-3′ (forward) and 5′-TGA GAT CGA CTG AGA CCC CAA-3′ (reverse) were used to amplify from genomic DNA a 137 bp p53 sequence (nucleotides 2215–2352 of Genbank sequence X54156) flanked by TaqI and AluI restriction sites near each end. Following double digestion of this sequence with TaqI (65°C, 1 h) and AluI (37°C, 1 h) the restricted p53 DNA fragment was purified via QIAquick™ centrifugation columns (Qiagen Inc., Valencia, CA) and then ligated to the hairpin-shaped sequences Cap1, 5′ (phosphate)-CGACGGCGCGCCGCCTTAGGTAGCGTTAGGCGCGCCGT-3′, which ligates TaqI sites and Cap2, 5′ (phosphate)-CTGCCGAGTTCCTGCTTTGAGATGCTGTTGAGUUACGTCGACTATCCTTGAACACCAACTCGGCAG-3′, which ligates AluI (blunt) sites, following the protocol described by Horie and Shimada (12). Briefly, ligation of the two caps to DNA was performed by adding a 100-fold molar excess of each Cap into 10 µM DNA template in the presence of T4 DNA polymerase and incubating the 50 µl reaction volume overnight at 15°C. A 2 µl ligation mixture was then treated with uracil glycosylase (Roche Diagnostics), at 37°C, 30 min, in the company-supplied buffer, 20 µl final volume, in a PCR tube. Upon addition of PCR components and buffer, a reaction was carried out using Titanium polymerase for 35 cycles and the following thermocycling conditions: 94°C, 30 s; (94°C, 30 s/68°C, 60 s) × 25 cycles; 68°C, 60 s; 4°C; Hold. Primers that bind the ligated Cap2 and overlap the target p53 sequence by 12 bases were used in this PCR: 5′-ATGAGATGGGGTCAGCTGCCTTCATCGGCGCGCCCATGATTT 3′ (forward); and CTTCTCCCCCTCCTCTGTTGCTCATCGGCGCGCC-3′ (reverse).
Next, the same p53 sequence flanked by TaqI and AluI sites was converted to a hairpin and amplified from human genomic DNA. Human genomic DNA (1 µg) from an osteosarcoma cell line (ATCC CRL-1543) was digested with TaqI, gel-purified and then digested with AluI. The protocol described above was used to ligate, treat with uracil glycosylase and PCR amplify the target sequence from digested genomic DNA using the same primers and thermocycling conditions. PCR products were examined via ethidium-stained gel electrophoresis. Amplified sequences were then excised from the gel (QIAquick™ gel extraction kit, Qiagen Inc.), and sequenced via dideoxy-sequencing at the Dana Farber Molecular Biology Core Facility. The primers used for sequencing were the same with those used during the hairpin-PCR reaction.
RESULTS AND DISCUSSION
Amplification of DNA hairpins with non-complementary ends
The observation that, if DNA is amplified in a hairpin structure, mismatches should be almost always separated from mutations, urged the development of hairpin-PCR. Indeed, if the polymerase introduces an A>G mutation on the upper DNA strand it is unlikely that, during synthesis of the bottom strand of a single hairpin, it will perform the exact opposite error (T>C mutation) at the same position of the complementary strand. Even for a polymerase with a large error rate of 10–4/base the odds for a double-error event are 10–4 × 10–4 × 0.25 = 2.5 × 10–9, i.e. less than the expected spontaneous mutation rate in somatic tissues (13,14). On the other hand, practically all genuine mutations should remain fully matched following hairpin-PCR, as these reside in both strands from the beginning (Fig. 1A). This complete discrimination of polymerase errors from the mutations should allow subsequent isolation of error-free amplified hairpins by one of many strategies, such as dHPLC (15), CDCE (16), DGGE (17) or enzymatic depletion of mismatches using mismatch recognition proteins, MutS (18), MutY (19) and TDG (20).
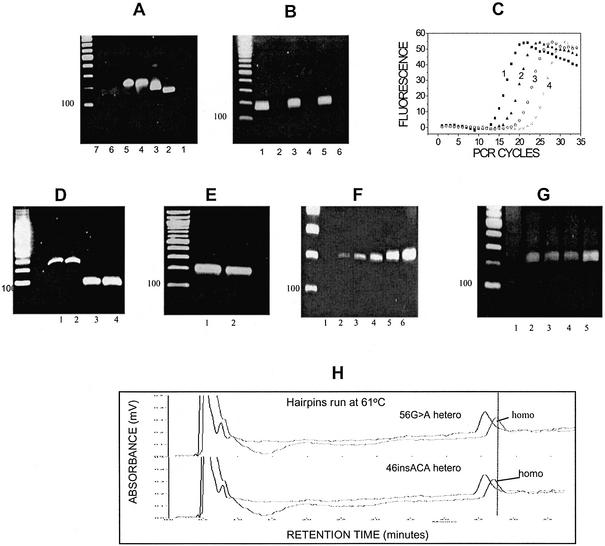
To develop the basic technical aspects of this approach, we designed long oligonucleotides (B, C, D and E, 149, 168, 200 and 218 nucleotides, respectively) expected to form hairpins with non-complementary ends that do not inhibit primer binding at their ends (Fig. 1C), as well as a regular hairpin A, 131 bp, which lacks the non-complementary ends (Fig. 1B), for comparison. Hairpins D and E encompass the complete sequence of p53 exon 9. One nanogram each hairpin was then used in a 25 µl PCR reaction using Titanium AdvantageR polymerase and primers designed to operate on the non-complementary ends of hairpins B–D, or alternatively on the complementary ends of hairpin A. Hairpins B–D produce a PCR product, while hairpin A does not (Fig. 2A, lanes 1–5). The data indicate that hairpins are readily amplified as long as primers are allowed to bind, and the polymerase is able to synthesize the hairpin complement, presumably by displacing the opposite strand. Omission of either forward or reverse primers abolishes the product (Fig. 2A, lanes 6–7) which indicates that amplification requires both primers and that the full-length hairpin is replicated by the polymerase. Hairpin-PCR was repeated using two proof-reading polymerases, Pfu Turbo™, or Advantage-HF2 and amplification was obtained (Fig. 2B). Figure 2C depicts quantitative real-time hairpin-PCR profiles of hairpin D serial dilutions, using SYBR Green I dye. The exponential nature of amplification is evident. Because of the way hairpin-PCR operates (Fig. 1A), the PCR products are expected to result in double-stranded DNA molecules, each strand of which is a full hairpin. To separate the two strands, and to recover the original hairpins, following purification of the PCR product the samples are denatured at 95°C, 1 min, and rapidly cooled by placing them directly on ice. This procedure does not allow time for substantial cross-hybridization of different DNA strands, while each strand is expected to rapidly form a hairpin due to its self- complementary sequence. Figure 2D demonstrates that rapid cooling converts the hairpin amplification product (lanes 1 and 2) to a band approximately half the size (lanes 3 and 4), which corresponds to the expected monomer hairpin. Next, the forward and reverse primers used for the amplifications in Figure 2A were re-designed to encompass an additional nine nucleotide extension (20 + 9 = 29mers) inside the p53 exon 9 hairpin D sequence. Figure 2E demonstrates that, although the 3′ end of the primers falls within the hairpin portion of the sequence, amplification remains almost unhindered. The data are consistent with the occurrence of primer binding by means of the 20 base overlap with the non-complementary end of the hairpin, and that the 3′ end of the primers temporarily displaces the hairpin sequence. This ‘invasion’ by hybridized oligonucleotides at the DNA ends, also reported by Guilfoyle et al. (21), presumably happens frequently enough to allow polymerase binding and primer extension to occur. Therefore restricting the primers on the non-complementary ends amplifies every hairpin sequence that contains those ends, while using primers with 3′ ends extending into the hairpin sequence renders hairpin-PCR sequence specific.
Figure 2.
PCR amplification and dHPLC separation of hairpin-shaped oligonucleotides. (A) Lanes 1–5, PCR product of hairpins A–D, respectively. Lanes 6 and 7, amplification of hairpin D with only forward or only reverse primer. (B) Amplification of hairpin C using Advantage Titanium® (lane 1), Pfu Turbo® (lane 3) or Advantage HF2® (lane 5) polymerases, respectively. Lanes 2, 4 and 6 are water-controls (no template) in each case. (C) Quantitative real-time PCR of hairpin D. Curves 1–4, starting material of 1 ng, 100, 10 and 1 pg, respectively. (D) Hairpin-PCR (lanes 1 and 2, in duplicate) followed by denaturation and rapid cooling of the product (lanes 3 and 4, in duplicate). (E) Hairpin D amplified with primers that bind the non-complementary ends, and either not extending (lane 1) or extending 9 bases into the hairpin sequence (lane 2). (F) Spiking of p53 exon 9-containing hairpin D into 100 ng p53-negative HL-60 genome, followed by hairpin-PCR using Advantage Titanium® polymerase. Spiking of 0.01 pg hairpin D corresponds to adding a single p53 exon 9 allele in the genome. Lanes 1–6, hairpin D addition of 0, 0.1, 1, 10, 100 and 1000 pg, respectively. (G) Similar to (H), but using Advantage HF2® polymerase. Lanes 1–5, hairpin D addition of 0, 0.01, 0.1, 1 and 10 pg. (H) dHPLC-based separation of 1:1 mixtures of homoduplex and heteroduplex hairpins. The threshold of the fraction collector is set on the trailing (slowest) portion of the homoduplex.
To investigate the amplification efficiency of hairpin-PCR, 100 ng purified human genomic DNA from a cell line that lacks the p53 gene (HL-60 cells), was mixed with decreasing amounts of the p53 exon 9-containing hairpin D. One human genome (∼3 × 109 bp) is ∼1.5 × 107 times the size of hairpin D, therefore spiking 10–2 pg hairpin D into 100 ng genomic DNA is approximately equivalent to adding a single copy p53 exon 9 in a hairpin formation in the genome. Figure 2F and G demonstrate hairpin-PCR amplification of p53 exon 9 using two different polymerases. Amplification from 0.01–0.1 pg hairpin D in the presence of genomic DNA is obtained. The amplification efficiency of hairpin-PCR appears comparable with that of regular PCR.
dHPLC separation of homoduplex from heteroduplex hairpins
To demonstrate that hairpins containing a single base mismatch, such as those expected to result from polymerase misincorporations, can be distinguished from fully matched hairpins via dHPLC, we injected homoduplex hairpin into a WAVE™ dHPLC system equipped with a fraction collector. Two more hairpins were synthesized. These were identical to the homoduplex hairpin except that they were synthesized to contain sequence changes, 56G>A or 46insACA, respectively. Upon folding, these hairpins form mismatches which simulate a potential misincorporation by Taq polymerase (18) and a Taq slippage error (4), respectively. One nanogram of each heteroduplex and homoduplex hairpin was injected separately into dHPLC, or, alternatively, mixed (1:1) and injected as a mixture. At a partially denaturing temperature of 61°C, the peaks from the heteroduplex hairpins could be distinguished from the fully matched, homoduplex hairpin, Figure 2H. Setting the threshold of the fraction collector on the trailing part of the homoduplex peak allows the collection of mainly (70–80%) homoduplex hairpin out of this mixture. This example simulated a worse case scenario, where the heteroduplex DNA was 50% of the overall sample. Normally, however, the heteroduplex peak resulting from PCR errors will be a smaller fraction (∼1–10%) of the homoduplex peak (2). From the data in Figure 2H it can be estimated that if PCR errors are confined to 10 or 1% of the sequences, one would collect >95 and >99% homoduplex DNA, respectively, resulting in a radical elimination of heteroduplex hairpins from the mixtures. In dHPLC chromatography almost all possible base changes and PCR errors are detectable [Transgenomics Inc. (2002) Transgenomic Optimase™ Polymerase Delivers Highest Fidelity in PCR for WAVE® System Analysis (US); http://www.transgenomic.com/pdf/AN119u.pdf], however individual base changes can result to varying degrees of separation of heteroduplexes from the homoduplex peak (15). Nevertheless, homoduplex DNA tends to have the longest retention time on the column (15). By re-cycling the collected homoduplex through the dHPLC for a second time and by collecting the trailing portion of the homoduplex each time should practically filter-out the misincorporations.
Conversion of native DNA sequences to hairpins and PCR amplification
To enable the scheme in Figure 1A, conversion of a native DNA fragment to a hairpin that can be amplified directly from human genomic DNA is required. To convert a sequence to a hairpin with non-complementary ends we performed ligation of two different oligonucleotide ‘caps’, Cap1 and Cap2, at the positions of two restriction sites encompassing the sequence (Fig. 3A). Cap1 and Cap2 are small oligonucleotides designed to form a hairpin that ligates both top and bottom strands at the respective DNA restriction site (12). In addition, Cap2 contains two centrally located uracils. Following the simultaneous ligation of both caps at the two DNA ends, a treatment with uracil glycosylase removes the uracils and generates abasic sites at the center of Cap2. During the heating step of the subsequent PCR reaction the glycosylase is inactivated and a strand break is expected to form via beta elimination at the abasic sites (22), which allows the hairpin to obtain a structure that can be PCR-amplified. To demonstrate the application, a 91 bp p53 sequence flanked by AluI and NlaIII restriction sites was generated following a double digestion of a larger DNA fragment, which had been first amplified from genomic DNA using regular PCR. Following ligation of caps 1 and 2, the resulting 145 bp fragment was amplified using primers overlapping the non-complementary linkers and the p53 sequence itself. A ∼290 bp double-stranded product was observed (Fig. 3B, lane 1).
Figure 3.
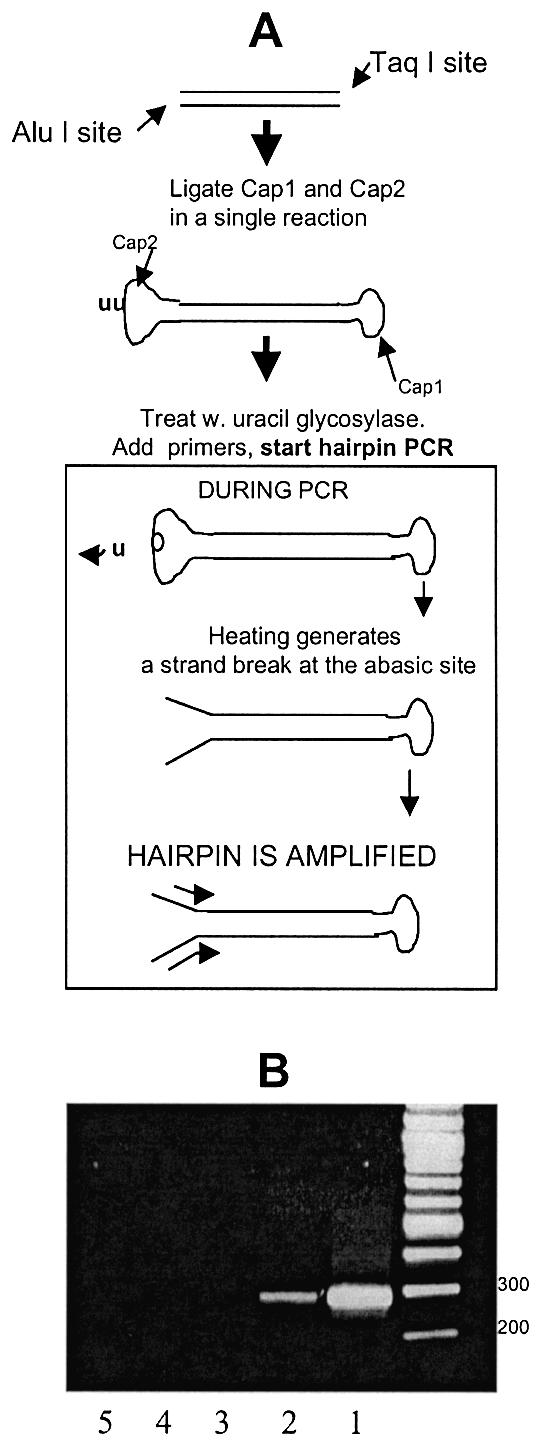
Conversion of a DNA sequence to a hairpin and PCR amplification. (A) Procedure used to convert a native DNA sequence, flanked by two different restriction sites, into a hairpin with non-complementary ends that can be amplified. The hairpin-shaped oligonucleotides Cap1 and Cap2 are ligated to the 5′- and 3′-ends of both sequences. During hairpin-PCR, primers extending into the sequence are used to confer sequence specificity. (B) Conversion of a native p53 sequence flanked by TaqI/AluI sites to a hairpin, followed by hairpin-PCR. (Lane 1) Hairpin-PCR product obtained by applying the scheme in Figure 2A for an isolated p53 sequence. (Lane 2) Hairpin-PCR product obtained by applying the scheme in Figure 2A to human genomic DNA, in order to directly amplify the same AluI/TaqI target sequence depicted in Lane 1. (Lane 3) As in lane 2, but omitting the addition of ligase from the scheme in Figure 2A. (Lanes 4 and 5) As in lane 2, but omitting the forward or reverse primer, respectively, from PCR.
Next, human genomic DNA expected to generate the same AluI/TaqI-flanked p53 fragment following a double enzymatic digestion was subjected to the same procedure. A ∼290 bp was generated when the full scheme of Figure 3A was applied (lane 2) but not when DNA ligase was omitted (lane 3) or when a single primer was used in the hairpin-PCR reaction (lanes 4 and 5). The DNA fragment was then excised from the gel and sequenced. Sequencing verified that the correct sequence had been amplified and that the expected hairpin structure of the amplified sequence had formed.
Both unknown and known mutation detection methods are affected by PCR errors and the most selective methods are those that are affected most. The principal limitation for mutation scanning via constant denaturant capillary electrophoresis (CDCE) is the fidelity of the polymerase used (3,23). High selectivity mutation scanning via DGGE and dHPLC is ultimately limited by polymerase error rate (3,24) [Transgenomics Inc. (2002) Transgenomic Optimase™ Polymerase Delivers Highest Fidelity in PCR for WAVE® System Analysis (US); http://www.transgenomic.com/pdf/AN119u.pdf]. Some of the high selectivity assays for RFLP-based known mutation detection [PCR/RE/LCR (25); Radioactivity-based PCR–RFLP (26); RSM (27,28); APRIL-ATM (29) and others reviewed in (5)] utilize PCR in at least one stage prior to RFLP-selection. Therefore these are also limited by PCR errors (30). The ability to amplify DNA without being limited by polymerase-introduced errors would significantly impact mutation detection and cancer diagnosis. A mismatch-binding protein, MutS, was used previously to deplete mismatches caused by PCR errors, in order to improve DNA synthesis fidelity (18). However, low frequency genuine mutations are also converted to mismatches and eliminated in this process, thus there is no benefit to mutation detection. In contrast, hairpin-PCR converts polymerase errors to mismatches while it also retains mutations in the homoduplex DNA. Forcing the polymerase to keep a double record of the sequence effectively boosts the DNA replication fidelity, as it is unlikely that a misincorporation will happen at the same position in both DNA strands simultaneously. We demonstrated amplification of small (75–145 bp) sequences in hairpin formation. However, polymerases can displace much longer (>1 kb) DNA stretches during synthesis (31) thus it is possible that DNA amplification in a hairpin structure will also occur for larger sequences. On the other hand, if dHPLC-based separation of mismatch-containing hairpins from homoduplexes is applied, an upper DNA size limit of 450–500 bp is imposed, as dHPLC separation is inefficient beyond this (15). Most genotyping/mutation detection methods operate on DNA sizes <500 bp (32), therefore the DNA size limitation should not hinder application of hairpin-PCR in the area of diagnostics. Non-dHPLC-based methods based on enzymatic degradation of heteroduplex hairpins [e.g. via MutS (18); MutY (19); TDG (20,33) or other mismatch-binding proteins] may have to be developed in order to apply this approach to larger DNA fragments.
In summary, we demonstrated that DNA hairpins designed to have non-complementary ends are efficiently PCR-amplified and that dHPLC can discriminate among homoduplex and heteroduplex hairpins. Native DNA sequences can be converted to a hairpin structure and amplified from human genomic DNA. The present hairpin-PCR leads to a strategy for a radical elimination of PCR errors that would allow a major boost to the detection of mutations and STR repeats in human tissue.
REFERENCES
- 1.Fujimura F.K., Northrup,H., Beaudet,A.L. and O’Brien,W.E. (1990) Genotyping errors with the polymerase chain reaction. N. Engl. J. Med., 322, 61. [DOI] [PubMed] [Google Scholar]
- 2.Wright P.A. and Wynford-Thomas,D. (1990) The polymerase chain reaction: miracle or mirage? A critical review of its uses and limitations in diagnosis and research. J. Pathol., 162, 99–117. [DOI] [PubMed] [Google Scholar]
- 3.Keohavong P. and Thilly,W.G. (1989) Fidelity of DNA polymerases in DNA amplification. Proc. Natl Acad. Sci. USA, 86, 9253–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perlin M.W., Lancia,G. and Ng,S.K. (1995) Toward fully automated genotyping: genotyping microsatellite markers by deconvolution. Am. J. Hum. Genet., 57, 1199–1210. [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons B.L. and Heflich,R.H. (1997) Genotypic selection methods for the direct analysis of point mutations. Mutat. Res., 387, 97–121. [DOI] [PubMed] [Google Scholar]
- 6.Khrapko K., Coller,H. Andre,P., Li,X.C., Foret,F., Belenky,A., Karger,B.L. and Thilly,W.G. (1997) Mutational spectrometry without phenotypic selection: human mitochondrial DNA. Nucleic Acids Res., 25, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidransky D. (1997) Nucleic acid-based methods for the detection of cancer. Science, 278, 1054–1059. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q., Swiderski,P. and Sommer,S.S. (2002) Truncated amplification: a method for high-fidelity template-driven nucleic acid amplification. Biotechniques, 33, 129–132, 126,–134, 138. [DOI] [PubMed] [Google Scholar]
- 9.Bartram C.R., Yokota,S., Hansen-Hagge,T.E. and Janssen,J.W. (1990) Detection of minimal residual leukemia by polymerase chain reactions. Bone Marrow Transplant, 6 (Suppl. 1), 4–8. [PubMed] [Google Scholar]
- 10.Reiss J., Krawczak,M., Schloesser,M., Wagner,M. and Cooper,D.N. (1990) The effect of replication errors on the mismatch analysis of PCR-amplified DNA. Nucleic Acids Res., 18, 973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diatchenko L., Lau,Y.F., Campbell,A.P., Chenchik,A., Moqadam,F., Huang,B., Lukyanov,S., Lukyanov,K., Gurskaya,N., Sverdlov,E.D. et al. (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. USA, 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horie K. and Shimada,K. (1994) Gene targeting by a vector with hairpin-shaped oligonucleotide caps. Biochem. Mol. Biol. Int., 32, 1041–1048. [PubMed] [Google Scholar]
- 13.Khrapko K., Andre,P., Cha,R., Hu,G. and Thilly,W.G. (1994) Mutational spectrometry: means and ends. Prog. Nucleic Acid Res. Mol. Biol., 49, 285–312. [DOI] [PubMed] [Google Scholar]
- 14.Li-Sucholeiki X.C. and Thilly,W.G. (2000) A sensitive scanning technology for low frequency nuclear point mutations in human genomic DNA. Nucleic Acids Res., 28, E44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao W. and Oefner,P.J. (2001) Denaturing high-performance liquid chromatography: a review. Hum. Mutat., 17, 439–474. [DOI] [PubMed] [Google Scholar]
- 16.Khrapko K., Hanekamp,J.S., Thilly,W.G., Belenkii,A., Foret,F. and Karger,B.L. (1994) Constant denaturant capillary electrophoresis (CDCE): a high resolution approach to mutational analysis. Nucleic Acids Res., 22, 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cariello N.F., Swenberg,J.A., De Bellis,A. and Skopek,T.R. (1991) Analysis of mutations using PCR and denaturing gradient gel electrophoresis. Environ. Mol. Mutagen., 18, 249–254. [DOI] [PubMed] [Google Scholar]
- 18.Smith J. and Modrich,P. (1997) Removal of polymerase-produced mutant sequences from PCR products. Proc. Natl Acad. Sci. USA, 94, 6847–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakrabarti S., Price,B.D., Tetradis,S., Fox,E.A., Zhang,Y., Maulik,G. and Makrigiorgos,G.M. (2000) Highly selective isolation of unknown mutations in diverse DNA fragments: toward new multiplex screening in cancer. Cancer Res., 60, 3732–3737. [PubMed] [Google Scholar]
- 20.Pan X. and Weissman,S.M. (2002) An approach for global scanning of single nucleotide variations. Proc. Natl Acad. Sci. USA, 99, 9346–9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guilfoyle R.A., Leeck,C.L., Kroening,K.D., Smith,L.M. and Guo,Z. (1997) Ligation-mediated PCR amplification of specific fragments from a class-II restriction endonuclease total digest. Nucleic Acids Res., 25, 1854–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longo M.C., Berninger,M.S. and Hartley,J.L. (1990) Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene, 93, 125–128. [DOI] [PubMed] [Google Scholar]
- 23.Andre P., Kim,A., Khrapko,K. and Thilly,W.G. (1997) Fidelity and mutational spectrum of Pfu DNA polymerase on a human mitochondrial DNA sequence. Genome Res., 7, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cariello N.F., Swenberg,J.A. and Skopek,T.R. (1991) Fidelity of Thermococcus litoralis DNA polymerase (Vent) in PCR determined by denaturing gradient gel electrophoresis. Nucleic Acids Res., 19, 4193–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson V.L., Yin,X., Thompson,B., Wade,K.R., Watkins,J.P., Wei,Q. and Lee,W.R. (2000) Oncogenic base substitution mutations in circulating leukocytes of normal individuals. Cancer Res., 60, 1830–1834. [PubMed] [Google Scholar]
- 26.Nakazawa H., Aguelon,A.M. and Yamasaki,H. (1990) Relationship between chemically induced Ha-ras mutation and transformation of BALB/c 3T3 cells: evidence for chemical-specific activation and cell type-specific recruitment of oncogene in transformation. Mol. Carcinogen., 3, 202–209. [DOI] [PubMed] [Google Scholar]
- 27.Steingrimsdottir H., Beare,D., Cole,J., Leal,J.F., Kostic,T., Lopez-Barea,J., Dorado,G. and Lehmann,A.R. (1996) Development of new molecular procedures for the detection of genetic alterations in man. Mutat. Res., 353, 109–121. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins G.J., Chaleshtori,M.H., Song,H. and Parry,J.M. (1998) Mutation analysis using the restriction site mutation (RSM) assay. Mutat. Res., 405, 209–220. [DOI] [PubMed] [Google Scholar]
- 29.Kaur M., Zhang,Y., Liu,W.H., Tetradis,S., Price,B.D. and Makrigiorgos,G.M. (2002) Ligation of a primer at a mutation: a method to detect low level mutations in DNA. Mutagenesis, 17, 365–374. [DOI] [PubMed] [Google Scholar]
- 30.McKinzie P.B., Delongchamp,R.R., Heflich,R.H. and Parsons,B.L. (2001) Prospects for applying genotypic selection of somatic oncomutation to chemical risk assessment. Mutat. Res., 489, 47–78. [DOI] [PubMed] [Google Scholar]
- 31.Lizardi P.M., Huang,X., Zhu,Z., Bray-Ward,P., Thomas,D.C. and Ward,D.C. (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet., 19, 225–232. [DOI] [PubMed] [Google Scholar]
- 32.Nollau P. and Wagener,C. (1997) Methods for detection of point mutations: performance and quality assessment. IFCC Scientific Division, Committee on Molecular Biology Techniques. Clin. Chem., 43, 1114–1128. [PubMed] [Google Scholar]
- 33.Zhang Y., Kaur,M., Price,B.D., Tetradis,S. and Makrigiorgos,G.M. (2002) An amplification and ligation-based method to scan for unknown mutations in DNA. Hum. Mutat., 20, 139–147. [DOI] [PubMed] [Google Scholar]