Abstract
Interactions of membrane-associated proteins play important roles in many cellular processes. The yeast two-hybrid assay is of limited utility for the analysis of such interactions, due to the need for soluble protein partners, whose interaction is assessed in the nucleus. The advent of the Ras-recruitment system (RRS) has enabled the study of membrane-associated proteins interacting with cytoplasmic proteins fused to Ras. Constitutive membrane association of the Ras fusion protein is expected to complement the growth defect of the yeast strain CDC25-2, assayed in the RRS, independent from the interaction with a membrane-bound partner. We describe the adaptation of the RRS to the analysis of interactions between two membrane-associated proteins using a model system. These results may facilitate the study of protein–protein interactions between membrane-bound proteins and further increase the utility of the RRS.
INTRODUCTION
An estimated 40% of all cellular proteins exert their actions at the cell membrane either as transmembrane proteins or as membrane-associated proteins (1). Membrane association can be conferred by hydrophobic or basic interactions, by protein–lipid interactions or by lipidation such as palmitoylation, farnesoylation or myristoylation. As a starting point of intracellular actions induced by extracellular signals, these membrane-bound proteins participate in numerous interactions with other membrane-bound or transmembrane proteins, often organized in high molecular mass protein complexes. Dissecting these complexes and the diverse downstream signaling pathways is a major goal of molecular biology and pharmaceutical research.
The study of protein–protein interactions has been greatly facilitated by the invention of yeast two-hybrid technology (2). Although there are some reports describing the application of this approach to soluble domains of membrane proteins, such as the tumor necrosis factor and the insulin receptor (3–7), the general utility of the two-hybrid assay for the study of membrane-associated proteins is inherently limited, due to the requirement of solubility and nuclear localization of the interacting proteins. The SOS- or Ras-recruitment system (RRS) allows for the analysis of the interaction between a membrane-bound protein and a soluble partner fused to SOS or Ras (8,9). The RRS exploits the strict requirement of membrane localization of a constitutively active human Ras mutant to complement the temperature-sensitive phenotype of the mutant Saccharomyces cerevisiae strain CDC25-2. Membrane localization is conferred by the interaction of a cytoplasmic ‘bait’ protein fused to the Ras mutant lacking its CAAX box with a membrane-bound ‘prey’, resulting in yeast growth at the non-permissive temperature of 36°C (9). In light of these considerations, constitutive membrane localization of the Ras fusion protein, e.g. by lipid modification, is expected to rescue the mutant yeast phenotype at 36°C (10). Indeed, since the function of Ras can be impaired when fused to a heterologous protein, its activity in a specific ‘bait’ fusion protein is often confirmed by expressing it as fusion to a membrane localization signal (such as the N-terminal signal for myristoylation derived from v-Src) prior to the screening procedure. This suggests that the analysis of interactions between two membrane-associated proteins with the RRS is inherently impossible. However, lipid modification of proteins is a central regulatory mechanism, not only in terms of localization, but also by influencing protein–protein interactions and complex formation between membrane proteins (11). In this study, the interaction between the myristoylated C-terminus of the epidermal growth factor receptor (EGFR) (also named ErbB1) and the adapter protein growth factor receptor binding protein 2 (Grb2), N-terminally fused to a myristoylation signal, is used as a model to show that the RRS can be applied to membrane-bound ‘bait’ proteins. The interaction of Grb2 with the kinase domain of ErbB was chosen since it is well characterized and can serve as a model for EGFR signal transduction. The functional expression of the ErbB kinase domain at the yeast plasma membrane and its activity-dependent interaction with adapter proteins represents a novel approach to the study of receptor tyrosine kinase signaling (F.Köhler, manuscript in preparation). The data presented here strongly suggest that the interaction of membrane-associated proteins in their natural environment at the inner leaflet of the plasma membrane can be analyzed using the RRS.
MATERIALS AND METHODS
Standard methods have been used for yeast maintenance, manipulation and plasmid construction (12,13).
Plasmids
The plasmid for the expression of the myristoylated C-terminal domain of ErbB1 (pM-ErbBc.t.) was constructed by excising part of the intracellular domain (amino acids 646–1158) of human ErbB1 with XhoI and NarI (NarI cleaves adjacent to the transmembrane domain and a partial digest was followed by treatment with Klenow-polymerase; XhoI cleaves in the poly-linker) from pBKS-ErbB and subcloning into SmaI + XhoI digested pYes2 (Invitrogen) which already contained the v-Src myristoylation signal (8) (kind gift from A. Aronheim). pBKS-ErbB was generated by PCR amplification of a fragment homolog to v-ErbB from a plasmid containing human EGFR (kind gift from U. Kruse) using the forward primer 5′-CCCGGGATGATCCAGTGTGCCCACTACA TTG-3′ and the reverse primer 5′-CCCGGGCTACTTG GCTTCCTTGGG-3′ and subcloning into the EcoRV site of pBKSII (Stratagene). Construction of plasmid pM-Grb2-Ras was a multi-step procedure. Briefly, murine Grb2 was PCR-amplified from a cDNA library and subcloned in frame between the v-Src myristoylation signal and human Ha-Ras devoid of its farnesoylation signal and containing the activating mutation Q61L in pYes2, resulting in pYesM-Grb2-Ras. From this plasmid the myristoylation signal-Grb2-Ras(Q61L)ΔF fusion was excised with HindIII and XhoI and subcloned into the yeast shuttle vectors p425TEF [high copy, TEF promoter: pM-Grb2-Ras(h)], p415GPD [low copy, GPD promoter: pM-Grb2-Ras(m)] and p415TEF [low copy, TEF promoter: pM-Grb2-Ras(l)] opened with HindIII + XhoI (14). The pM-Grb2-Ras plasmids confer complementation of the leu2-negative phenotype and constitutive protein expression. pYes2/pM-ErbBc.t. plasmids confer complementation of the ura3-negative phenotype and expression is under the control of the galactose-inducible and glucose-repressible Gal1 promoter.
Yeast strains and culture
CDC25-2 (Matα, ura3, lys2, leu2, trp1, hisΔ200, ade 2-101, cdc25-2; kind gift from A. Aronheim) were transformed with pM-Grb2-Ras(h), pM-Grb2-Ras(m) and pM-Grb2-Ras(l) together with pYes2 or pM-ErbBc.t., plated on glucose-containing agar plates and incubated at 25°C for 3 days. Five independent colonies of each combination were grown to late log phase in glucose-containing liquid medium at 25°C for 2 days, spotted on a galactose-containing plate and incubated at 36°C for 3 days. Spots were quantified based on grayscale images using the QuantityOne software from BioRad, Hercules, CA, USA. Grayscale density was determined for identical circular areas drawn around each spot. For each expression experiment, five spots were averaged and the background was subtracted. For growth measurements in liquid culture, three independent colonies of CDC25-2 transformed with pM-Grb2-Ras(m) together with pYes2 or pM-ErbBc.t. were grown to late log phase in glucose-containing medium, diluted to optical density (OD) (600) = 0.1 in galactose-containing medium and incubated at 36°C for 2 days followed by OD(600) measurement. All incubations were in synthetic drop-out medium lacking uracil and leucine for maintenance of plasmids (12).
RESULTS
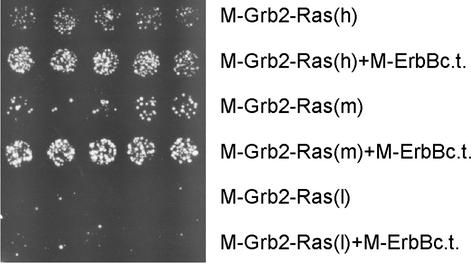
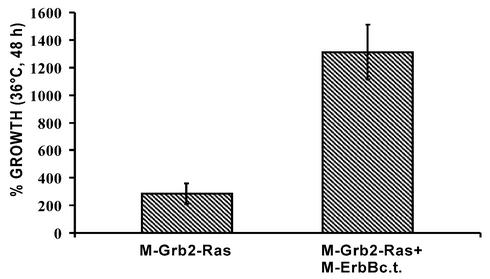
The yeast strain CDC25-2 is unable to grow at the non-permissive temperature of 36°C due to the inability of mutant CDC25 to activate Ras proteins and the resulting failure to activate adenylyl cyclase (15). This growth defect can be complemented by a membrane-associated constitutively active human Ras protein (9). Membrane association can be achieved by fusion to a signal sequence for myristoylation, and complementation depends on the expression level of such a fusion protein. We set out experiments to test whether the interaction of two membrane-associated proteins can be monitored using a modified Ras-recruitment approach as depicted in Figure 1. A fusion protein comprising the mouse Grb2 fused to a human Ras mutant which lacks its farnesoylation signal [hRas(Q61L)ΔF] and contains a myristoylation signal at the N-terminus of Grb2 (M-Grb2-Ras) should lead to weak growth at 36°C when expressed at the appropriate level (Fig. 1A). Co-expression of the intracellular kinase domain of ErbB fused to a signal for myristoylation (M-ErbBc.t.) should result in the interaction of Grb2 with phosphorylated tyrosine residues of M-ErbBc.t. and lead to enhanced yeast growth (Fig. 1B). To test this hypothesis, M-Grb2-Ras was expressed driven either by the TEF promoter from a high copy plasmid (referred to as high expression level), by the GPD promoter from a low copy plasmid (medium expression level) or by the TEF promoter from a low copy plasmid (low expression level). Yeast growth after 72 h at 36°C depended on the expression level of M-Grb2-Ras, with best growth at high expression levels, intermediate growth at medium expression levels and virtually no growth at low expression levels. When M-ErbBc.t. was co-expressed, growth did markedly increase with the high and medium expression levels of M-Grb2-Ras, whereas no difference was observed with low level M-Grb2-Ras expression (Fig. 2). Quantification of the cell density of the shown spots with and without co-expression of M-ErbBc.t. yielded a ratio of 2.5 for the high expression level and 3.3 for the medium expression level. In the case of the low expression level, the cell density was too low for quantification. The medium expression level constructs provided the most evident difference and, thus, were used to assess growth properties in liquid culture. A significant increase in optical density of liquid cultures was observed after 48 h incubation at 36°C when M-ErbBc.t. was co-expressed compared with the membrane localized Ras alone (Fig. 3). The ratio of the cell densities at this time point was 4.6.
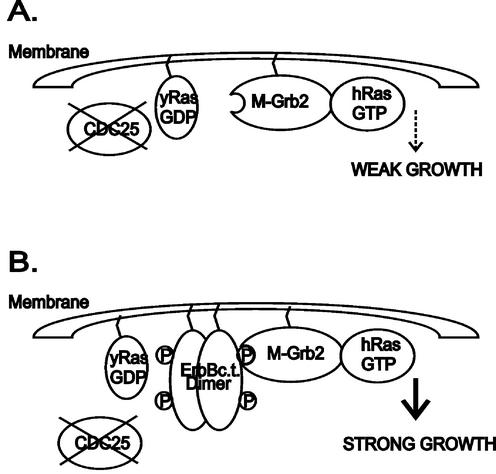
Figure 1.
Schematic representation summarizing the modified Ras recruitment assay. (A) Yeast strain CDC25-2 is unable to grow at 36°C due to a lack of CDC25-activated Ras proteins (yRas GDP). This defect can be overcome by expression of a constitutively active human Ras mutant (RasQ61L). Low level expression of a protein consisting of mouse Grb2 fused to a myristoylation signal and to a constitutively active human Ras mutant lacking its farnesoylation signal (Q61LΔF, hRas GTP) leads to very weak growth at the non-permissive temperature of 36°C. (B) Co-expression of the intracellular kinase domain of ErbB (lacking the trans-membrane domain; ErbBc.t.) fused to a myristoylation signal leads to markedly improved growth at 36°C, most likely by enhancing and stabilizing the membrane localization of M-Grb2-Ras through recruitment to phosphorylated tyrosine residues of the active kinase domain at the cell membrane.
Figure 2.
Differential growth at 36°C of CDC25-2 expressing M-Grb2-Ras or co-expressing M-Grb2-Ras and M-ErbBc.t. at various expression levels. Expression of myristoylated Grb2-Ras(Q61L)ΔF (named M-Grb2-Ras) was driven by the TEF promoter from a high copy plasmid [M-Grb2-Ras(h); high level expression], by the GPD promoter from a low copy plasmid [M-Grb2-Ras(m); medium level expression] or by the TEF promoter from a low copy plasmid [M-Grb2-Ras(l); low level expression]. Expression of myristoylated ErbB C-terminal domain (named M-ErbBc.t.) was driven by the Gal1 promoter from a high copy plasmid. Cells were incubated at 36°C for 72 h.
Figure 3.
Quantification of growth differences of CDC25-2 growing in liquid culture at 36°C either expressing M-Grb2-Ras or co-expressing M-Grb2-Ras and M-ErbBc.t. The expression of myristoylated M-Grb2-Ras was driven by the GPD promoter from a low copy plasmid and the expression of myristoylated M-ErbBc.t. was driven by the Gal1 promoter from a high copy plasmid. For each growth measurement three individual clones were used. Yeast cells were diluted to a measured OD(600) between 0.045 and 0.051. Percent growth is the OD(600) read after 48 h incubation relative to the starting OD(600). The values shown represent the mean of the measurements of three independent clones.
DISCUSSION
The data shown strongly suggest that the RRS can be applied to the study of interactions between two membrane-associated proteins. By titrating the expression level of the Ras fusion protein, differential growth can be observed depending on whether it interacts with a membrane-bound partner or not. This is most likely due to globally enhanced and stabilized membrane localization of the Ras fusion protein. Alternatively or additionally, a certain clustering and concomitant localized increase in the concentration of activated Ras protein at the plasma membrane might cause a threshold to be crossed that is necessary for efficient Ras signaling. The existence of such a threshold has been observed in other experimental settings (personal observations) and can be deduced from the inability of M-Grb2-Ras at low expression level, even with co-expressed M-ErbBc.t., to complement the temperature sensitive phenotype of CDC25-2.
The observed interaction-dependent growth differences might be further increased by tuning the expression level of the Ras fusion protein. Expression driven by the GPD promoter from a low copy plasmid is approximately 4.5 times higher compared with expression driven by the TEF promoter from a low copy plasmid (14). Using appropriate expression vectors, it should be feasible to adjust expression levels such that, only upon interaction of the proteins of interest, is the threshold for complementation of the mutant CDC25-2 phenotype surpassed.
Grb2 interacts with phosphorylated tyrosine residues on EGFR. Tyrosine phosphorylation is effected by ligand-induced receptor oligomerization and subsequent auto- and cross-phosphorylation of the intracellular domain (16). The oncogene ErbB lacks most of the extracellular ligand-binding domain and part of the extreme C-terminus of EGFR and is active independently of ligand. Thus, a protein comprising the intracellular kinase domain of ErbB tethered at the membrane by a myristoyl moiety resembles the ErbB signaling properties and leads to tyrosine phosphorylation. The efficiency of activation depends on ‘molecular collision’ events and thus on the concentration of the myristoylated ErbB kinase domain at the membrane. Although expressed under control of the strong Gal1 promoter from a high copy plasmid, the amount of phosphorylated M-ErbBc.t. may be rather low, e.g. due to endogenous phosphatases or inefficient auto- and cross-activation. Thus, there are good chances that the interaction-dependent growth differences are more pronounced when studying proteins which are constitutively binding-competent, i.e. in which the interaction does not depend on further modifications, such as phosphorylation as with M-ErbBc.t. In addition, interaction of a protein with a partner which is more tightly associated with the membrane, e.g. by a transmembrane domain, would be expected to result in a further increase of the discriminatory parameter.
However, it remains to be demonstrated if the observed interaction-dependent growth differences will be sufficient for the identification of novel or known interacting proteins in a screening procedure. Nonetheless, our results suggest that known interactions can be verified in an independent assay and that such interactions can be further characterized, e.g. in terms of defining and mapping amino acid residues or protein domains involved in mediating the interaction. Furthermore, our approach may be useful as a tool to screen for compounds or peptides that have the potential to disrupt such identified protein interactions.
Recently, an assay system for the analysis of membrane proteins based on the pheromone response pathway in yeast has been described (17). Interaction of a protein fused to Ste 18 (the Gγ subunit of the yeast pheromone receptor-associated heterotrimeric G-protein) with a transmembrane protein results in disruption of the pheromone pathway and down-regulation of a pheromone-inducible promoter. The feasibility of the assay has been demonstrated with the known binding partners nSec1/Syntaxin1a and SNT-1/FGFR3. Although described as an assay to monitor interactions between a membrane protein and a cytoplasmic protein, it should be noted that at least one of the tested proteins (SNT-1) is membrane-associated via myristoylation as is the Gγ subunit (via prenylation) itself. The underlying mechanism is believed to involve the competition between binding of the dimer consisting of Gβ and the Gγ fusion protein to components of the pheromone response pathway and binding of the heterologous part of the Gγ fusion protein to its transmembrane protein partner. This competitive mechanism may be disadvantageous, and it has been stated by the authors that the assay may be applicable to strongly interacting proteins only. Differences in reporter activity upon co-expression of both interacting proteins were determined to be ∼4-fold, which is comparable to the data presented in this study.
Another yeast-based assay for protein–protein interactions which can be applied to membrane proteins depends on the interaction-mediated reconstitution of split ubiquitin (18). It has been successfully used for the study of proteins of the endoplasmic reticulum membrane but there are no reports describing the application to proteins of the plasma membrane (19,20).
The results of the study presented here show that the RRS can be applied to membrane proteins. This conceptually rather surprising finding further expands the utility of the system and may contribute to the dissection of biologically and pharmaceutically relevant signaling complexes at the plasma membrane.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank A. Aronheim for the gift of plasmids and the CDC25-2 yeast strain and M. Hager and A. Zimmermann for helpful discussions. This work was supported by a grant of the state Baden-Württemberg, Germany to F.K.
REFERENCES
- 1.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M., Louis,E.J., Mewes,H.W., Murakami,Y., Philippsen,P., Tettelin,H. and Oliver,S.G. (1996) Life with 6000 genes. Science, 274, 563–567. [DOI] [PubMed] [Google Scholar]
- 2.Fields S. and Song,O.K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 3.Chinnaiyan A.M., O’Rourke,K., Tewari,M. and Dixit,V.M. (1995) FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell, 81, 505–512. [DOI] [PubMed] [Google Scholar]
- 4.Wallach D., Boldin,M.P., Kovalenko,A.V., Malinin,N.L., Mett,I.L. and Camonis,J.H. (1998) The yeast two-hybrid screening technique and its use in the study of protein–protein interactions in apoptosis. Curr. Opin. Immunol., 10, 131–136. [DOI] [PubMed] [Google Scholar]
- 5.Furlanetto R.W., Dey,B.R., Lopaczynski,W. and Nissley,S.P. (1997) 14-3-3 proteins interact with the insulin-like growth factor receptor but not the insulin receptor. Biochem. J., 327, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey B.R., Furlanetto,R.W. and Nissley,S.P. (1998) Cloning of human p55 gamma, a regulatory subunit of phosphatidylinositol 3-kinase, by a yeast two-hybrid library screen with the insulin-like growth factor-I receptor. Gene, 209, 175–183. [DOI] [PubMed] [Google Scholar]
- 7.Xu P., Jacobs,A.R. and Taylor,S.I. (1999) Interaction of insulin receptor substrate 3 with insulin receptor, insulin receptor-related receptor, insulin-like growth factor-1 receptor, and downstream signaling proteins. J. Biol. Chem., 274, 15262–15270. [DOI] [PubMed] [Google Scholar]
- 8.Aronheim A., Zandi,E., Hennemann,H., Elledge,S.J. and Karin,M. (1997) Isolation of an AP-1 repressor by a novel method for detecting protein–protein interactions. Mol. Cell. Biol., 17, 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broder Y.C., Katz,S. and Aronheim,A. (1998) The ras recruitment system, a novel approach to the study of protein–protein interactions. Curr. Biol., 8, 1121–1124. [DOI] [PubMed] [Google Scholar]
- 10.Hubsman M., Yudkovsky,G. and Aronheim,A. (2001) A novel approach for the identification of protein–protein interaction with integral membrane proteins. Nucleic Acids Res., 29, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casey P.J. (1995) Protein lipidation in cell signaling. Science, 268, 221–225. [DOI] [PubMed] [Google Scholar]
- 12.Rose M., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Mumberg D., Müller,R. and Funk,M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- 15.Petitjean A., Higler,F. and Tatchell,K. (1990) Comparison of thermosensitive alleles of the CDC25 gene involved in the cAMP metabolism of Saccharomyces cerevisiae. Genetics, 124, 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olayioye M.A., Neve,R.M., Lane,H.A. and Hynes,N.E. (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J., 19, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrhard K.N., Jacoby,J.J., Fu,X.-Y., Jahn,R. and Dohlman,H.G. (2000) Use of G-protein fusions to monitor integral membrane protein–protein interactions in yeast. Nat. Biotechnol., 18, 1075–1079. [DOI] [PubMed] [Google Scholar]
- 18.Johnsson N. and Varshavsky,A. (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA, 91, 10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stagljar I., Korostensky,C., Johnsson,N. and Te Heesen,S. (1998) A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl Acad. Sci. USA, 95, 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dünnwald M., Varshavsky,A. and Johnsson,N. (1999) Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol. Biol. Cell, 10, 329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]