Abstract
The specific recognition of homopurine–homo pyrimidine regions in duplex DNA by triplex-forming oligonucleotides (TFOs) provides an attractive strategy for genetic manipulation. Alkylation of nucleobases with functionalized TFOs would have the potential for site-directed mutagenesis. Recently, we demonstrated that a TFO bearing 2-amino-6-vinylpurine derivative, 1, achieves triplex-mediated reaction with high selectivity toward the cytosine of the G-C target site. In this report, we have investigated the use of this reagent to target mutations to a specific site in a shuttle vector plasmid, which replicates in mammalian cells. TFOs bearing 1 produced adducts at the complementary position of 1 and thereby introduced mutations at that site during replication/repair of the plasmid in mammalian cells. Reagents that produce covalent cytosine modifications are relatively rare. These TFOs enable the preparation of templates carrying targeted cytosine adducts for in vitro and in vivo studies. The ability to target mutations may prove useful as a tool for studying DNA repair, and as a technique for gene therapy and genetic engineering.
INTRODUCTION
The wealth of sequence information from human and other genome projects highlights the need for facile and efficient strategies for genome manipulation. This requires the selective recognition of duplex DNA, and has stimulated the development of various synthetic candidates for this purpose. Successful reagents would be used for gene knockout for target validation, to facilitate gene knock in, for strain and transgenic animal construction and, perhaps, for gene therapy. One approach that has been of interest for many years is based on the DNA triple helix (1).
A triple helix can form when a triple helix-forming oligonucleotide (TFO) binds in a sequence-specific manner in the major groove of the duplex DNA containing homopurine–homopyrimidine stretches (2,3). Depending on the nature of the target sequences, triplexes can be formed by third strands consisting of pyrimidines (parallel triplex) or purines (antiparallel triplex), and a binding code for both has been described (4,5). Triple helix formation can prevent transcription factor binding to promoter sites and block m-RNA synthesis in vitro and in vivo (6–10). Reactive chemical groups linked to TFOs have been used to stabilize triplex formation via covalent linkage with bases in the target sequence. These conjugates have been applied to the inhibition of gene expression, in what has been termed the ‘antigene strategy’ (11).
Alternatively, TFO conjugates have been used to manipulate the sequence of genes containing target sequences (12). Replication/repair of targeted adducts can result in mutagenesis of the target site (13). This has been shown in recent work with TFOs, linked to psoralen, that direct mutagenesis to specific sites in reporter genes (14–16). In these experiments photoactivation of the targeted psoralen produces a cross-link in the duplex (as well as the linkage of the third strand to the target sequence), which during replication or repair of the plasmid results in mutations. In another approach, psoralen-TFOs designed to bind a target sequence in the chromosomal hypoxanthine phosphoribosyl transferase gene have been introduced into cells and, after photoactivation, colonies with mutations at the target site were recovered (16–18). These results demonstrate proof of principle that TFOs can be developed as reagents for gene targeting and sequence modulation.
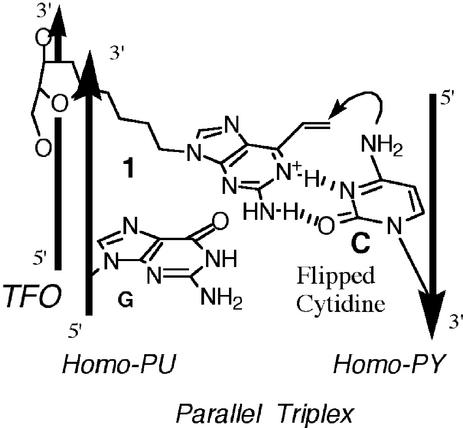
Although psoralen has been a very useful mutagen, it has some serious drawbacks. It is sharply restricted in reactivity to pyrimidines, has a strong bias towards thymines, with cross-link formation limited to 5′-TA or AT sites (19,20). This greatly reduces the target options for gene mutagenesis, and knockout applications. In addition, reactivity requires photoactivation, and only those TFOs bound at the time of UVA exposure will contribute to the mutagenesis. Consequently, there is a need for additional compounds that can be linked to TFOs while retaining efficient DNA reactivity. TFOs have been conjugated with aryl nitrogen mustard (21), aziridine units (22) or a minor groove-reactive compound (23) and used for interstrand cross-linking, but these reactions were not efficient. Furthermore, the biological consequences following reaction with the DNA target sequences, such as mutagenesis, have not been reported. Recently, we demonstrated that the new nucleoside derivative 1 exhibited triplex-mediated reactivity with high selectivity toward the cytosine at a G-C target site (Fig. 1) (24–27). Here, we report that TFOs bearing 1 (2-amino-6-vinylpurine) have been used to achieve site-specific modification at a reaction site in the supF reporter gene. We describe mutagenesis of that site during passage of the plasmid in repair-deficient human cells. Cytosine adducts are relatively rare and the new TFO derivatives make possible the study of targeted cytosine derivatives in both biochemical and biological analyses.
Figure 1.
The reaction of 1 with the flipped cytosine.
MATERIALS AND METHODS
Synthesis of TFOs
5′-O-(4,4′-dimethoxytrityl)-5-methyluridine-2′-O-methyl-3′-O-(β-cyanoethyl-N,N-diisopropyl) phosphoramidite, N4-formamidine(4,4′-dimethoxytrityl)-5-methylcytidine-2′-O-methyl-3′-O-(β-cyanoethyl-N,N-diisopropyl) phosphoramidite, and 5′-O-(4,4′-dimethoxytrityl)-5-methyluridine-2′-O-methyl-3′- O-succinamido-N6-hexanamido-N3-propyl CPG supports were purchased from Chemgenes, Ashland, MA. 5′-O-(4,4′-di methoxytrityl)-2′-deoxy-3′-O-(β-cyanoethyl-N,N-diisopropyl) phosphoramidite, N4-benzoyl(4,4′-dimethoxytrityl)-5-methylcytidine-2′-deoxy-3′-O-(β-cyanoethyl-N,N-diisopropyl) phosphoramidite, and 5′-O-(4,4′-dimethoxytrityl)-5-thymidine-2′- deoxy-3′-O-succinamido-N6-hexanamido-N3-propyl CPG sup ports were purchased from Glen Research. 2-Phenoxy acetylamino-9-[1′-butyl-2′-deoxy-5′-O-dimethoxytrityl-3-O- (N,N-diisopropyl-β-cyanoethylphosphoramidyl)-d-ribofura nosyl]-6-(2-methylthioethyl)purine was synthesized following previous procedures (23–27). The oligonucleotides were synthesized on CPG supports (500 Å) using an Expedite 8909 synthesizer in a 1.0 µmol scale using commercial 2-cyanoethyl phosphoramidites and the amidite precursor described above according to the regular protocol. The 5′-terminal dimethoxytrityl-bearing oligodeoxynucleotides (ODNs) were removed from the solid support by treatment with 28% aqueous ammonia at 55°C for 1 h. Purification was performed by reverse-phase HPLC (nacalai tesque COSMOSIL 5C18-MS, 10 × 250 mm) using a linear gradient of 10–40% acetonitrile in 0.1 M TEA buffer at a flow rate of 4.0 ml/min, then the purified ODNs were passed through a Sep-Pak (ODS) with H2O (10 ml), 2% trifluoroacetic acid (5 ml), H2O (10 ml) and 50% acetonitrile (3 ml). The purified oligonucleotides were characterized by MALDI-TOF mass spectrometry.
Activation of the TFOs
A solution of 5 mM mono magnesium perphthalate (MMPP, 1 µl) in a carbonate buffer, adjusted to pH 10 with aqueous 0.01 M NaOH, was added into a solution of the purified ODN 2a–5a (1 nmol) and each mixture was left at room temperature for 30 min. An aqueous 2 M NaOH solution (3 µl) was added to the above mixture and left for an additional 30 min. Each mixture was adjusted to pH 4.5 with 2 M CH3COOH and used for all experiments.
Thermal denaturation experiments
The target duplexes (5′-GTAGAAGAAAAAAGAGAAA; TTTCTCTTTTTTCTTCTAC) were dissolved in 50 mM Tris, 100 mM NaCl and 2 mM MgCl2 (pH 6.0, 7.0, respectively). These solutions were heated at 80°C for 10 min and allowed to come to room temperature in 30 min. The TFO was added to the mixture and incubated at 4°C overnight. The thermal denaturation experiments were performed in a Cary 3E UV-visible spectrophotometer fitted with a thermostat sample holder and temperature controller. Triplexes were heated from 5 to 80°C at a rate of 0.4°C/min and the absorbance at 260 nm was recorded as a function of the temperature. The data were processed using SigmaPlot 5.0 software to determine the first derivative of the melting curves and the Tm value was obtained.
Binding assay
The pyrimidine strand (5′-ATTTCTCTTTTTTCTTCTA) was 32P-end-labeled using [γ-32P]ATP (NEN) and T4 polynucleotide kinase (New England Biolabs, Beverly, MA) and annealed with the purine strand (5′-TAGAAGAAAAAAGAGAAAT). The 32P-labeled duplex was incubated with varying concentrations of 2b–5b in the binding buffer (40 mM MES, 10 mM MgCl2, pH 4.5) at 32°C. After 17 h, loading dye (0.1% xylene cyanol and 0.1% bromophenol blue, 95% formamide; 4 µl) was added to each mixture and heated at 90°C for 5 min. These mixtures were loaded onto a 15% polyacrylamide gel containing 7 M urea using TBE buffer. Gels were visualized using a Fuji Phosphorimager and quantitated using the ImageQuant software.
Restriction enzyme protection assay
SupF12, digested by EcoN1 and PflM1, was incubated with varying concentrations of 2b–5b in the binding buffer (40 mM MES, 10 mM MgCl2, pH 4.5) at 32°C. After 17 h, each of the plasmid–TFO complexes were separated from the unbound oligonucleotides by ethanol precipitation. This mixture was resuspended in the buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM dithiothreitol) and digested with XbaI (New England Biolabs). The digests were analyzed by 1% agarose gel electrophoresis and the relative amount of DNA in the protected and non-protected bands was quantitated by fluorescence image analysis.
Polymerase stop assay
These assays were carried out using primer 5′-CGGCAGATTTAGAGTCTGCTC corresponding to nucleotides 124–144 of supF12. The primer was 32P-labeled using [γ-32P]ATP (NEN, 3000 mCi/mmol) and T4 polynucleotide kinase (New England Biolabs). Primer extension was carried out in a total volume of 20 µl containing 0.2 µg of modified or unmodified supF12 or unmodified pNS14 digested by BglII (New England Biolabs), 32P-labeled primer, T4 polymerase buffer (New England Biolabs), BSA (2 µg) and 250 µM of each dNTP. The DNA was denatured by heating to 95°C for 5 min, and the primer was annealed during subsequent cooling to 4°C. Extension was initiated by the addition of 3 U T4 DNA polymerase (New England Biolabs) and elongation was allowed to proceed at 37°C. After 1 h, 8 µl of stop solution (95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanole) were added to each mixture and heated at 95°C for 5 min. The DNA fragments were resolved on an 8% polyacrylamide gel containing 7 M urea using the TBE buffer system. Gels were visualized using a phosphorimager or film autoradiography.
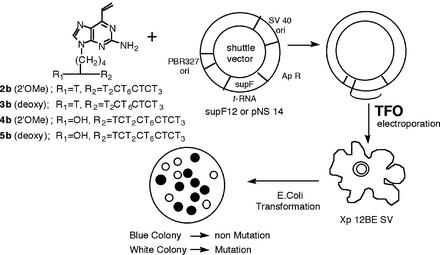
Mutagenesis protocol
Shuttle vector plasmids were constructed as described previously (28). The plasmids contain variant supF genes containing triplex target sequences immediately adjacent to the 5′ end of the mature tRNA sequence. SupF12 and/or pNS 14 were incubated with 2b–5b in the binding buffer (40 mM MES, 10 mM MgCl2, pH 4.5) at 32°C. As control experiments, these plasmids were incubated with only MMPP or H2O in the same condition. After 17 h, each plasmid–TFO complex was separated from the unbound oligonucleotide by ethanol precipitation. The plasmid–triplex complexes were electroporated into SVXP12BE cells. These cells are an immortalized fibroblast line derived from a patient with xeroderma pigmentosum, complementation group A. Nucleotide excision repair activity is extremely low in these cells. The cells were plated for 48 h, during which time the reactive sites were repaired and/or mutagenized, and the plasmid replicated. Progeny plasmids were then harvested, treated with DpnI to remove non-replicated input plasmids (29), and introduced into the Escherichia coli indicator strain MBM 7070 [F–, lacZamCA7020, lacY1, hsdR–, hsdM+, araD139, Δ(araABC-leu)7679, galU, galK, rpsL, thi] (30). The bacteria were spread on indicator plates containing isopropyl-1-thio-β-d-galactopyranoside and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and the frequency of white colonies containing plasmids with mutations in the supF gene was determined. The mutant colonies were purified and the plasmids were isolated for DNA sequence analysis.
DNA sequencing
The purified mutant colonies were picked into proteinase K (Roche)–TE buffer solution (40 ng/µl, 15 µl) and incubated at 65°C for 15 min, 80°C for 15 min. After centrifugation, plasmid DNA was sequenced using the cycle-sequencing kit (Epicentre Technologies), as directed by the manufacturer. The sequencing primer was chosen to bind to the β-lactamase gene just upstream of the supF gene in the vector (31,32).
RESULTS
Reactive TFOs and the targets sequence
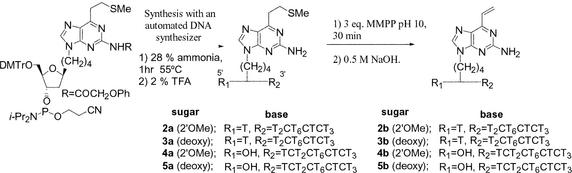
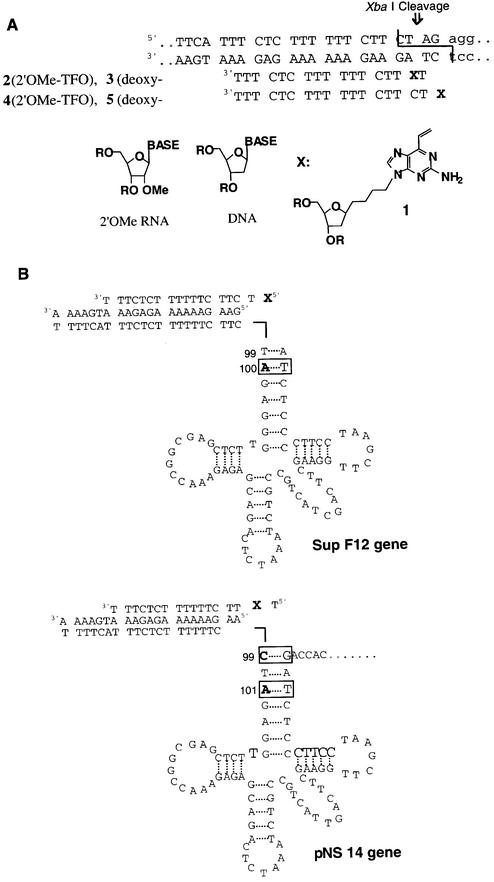
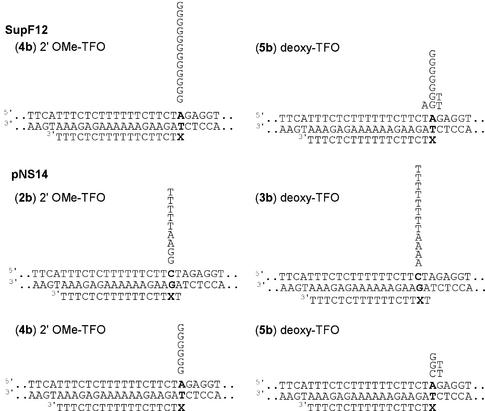
The reactive oligomers were synthesized following published procedures (Scheme 1) (33–35). The target sequences of the reactive oligomers in this study are displayed in Figure 2A. The TFOs were designed to form a triplex with the target sequence embedded within supF mutation marker genes (Fig. 2B) carried in shuttle vector plasmids. In our previous study, a TFO containing the vinylpurine derivative (1) at an internal position reacted selectively with the cytosine (27) within the pyrimidine strand of the duplex. However, such selectivity was not observed when 1 was located at the terminal position (33). In the current experiments, 1 was introduced to internal position of TFO 2 and 3 so as to react with cytidine, and to the terminal position of TFO 4 and 5 such that it would react with the adenosine external to the triplex. In these experiments, 2′-O-methoxy (2′-OMe) (2b, 4b) and deoxy nucleosides (3b, 5b) were used to construct reactive TFOs. The use of the 2′-OMe sugar was expected to enhance pyrimidine triplex stability (34,35).
Scheme 1. Synthesis of reactive TFOs.
Figure 2.
(A) The triplex target sequence and TFOs. (B) Schematic of the supF12 and pNS gene embedded in the pre-tRNA region. The boxed nucleotides are the reactive site of TFOs.
Thermal stability of triplexes
The thermal stability of the triplexes was evaluated by measurement of the melting temperature using TFOs containing a stable precursor of 1 in pH 6.0 and 7.0 buffer, shown in Table 1. The analysis, in pH 7.0 buffer, of the triplex formed by the duplex and the deoxy-TFOs showed two transitions with the triplex Tm (3a, 28°C and 5a, 41°C) below the duplex Tm (55°C). The triplex formed by a control TFO without the conjugate had a Tm of 31°C. These results indicated that the position of the vinylpurine influenced triplex stability—when it was in the triplex interior it reduced stability, while when external to the triplex it enhanced stability. As expected from our previous study (16) the triplexes formed by the TFOs containing 2′-OMe sugars showed a single transition with a Tm (2a, 53°C and 4a, 60°C) higher than the duplex Tm. The Tm of the control triplex (TFO with 2′-OMe, no vinylpurine precursor) was 64°C. Thus, in the triplexes formed by the TFOs with the 2′-OMe the vinylpurine residue was destabilizing in both positions, although less so in the external location. At pH 6.0, Tm values with all TFOs were higher than Tm values at pH 7.0. These results indicated that the triple helix between these reactive TFOs and targets would be stable under reaction conditions (pH 4.5 and 32°C).
Table 1. Melting temperature of triple helix using 2a–5a.
| TFO | pH 6.0 | pH 7.0 |
|---|---|---|
| 2a | 60°C | 53°C |
| 3a | 42°C | 28°C |
| 4a | 73°C | 60°C |
| 5a | 55°C | 41°C |
Measured in buffer containing 50 mM Tris, 100 mM NaCl and 2 mM MgCl2. Concentration: [TFO] = [duplex] = 1.0 µM.
Evaluation of reactivity by gel shift assay and restriction enzyme protection assay
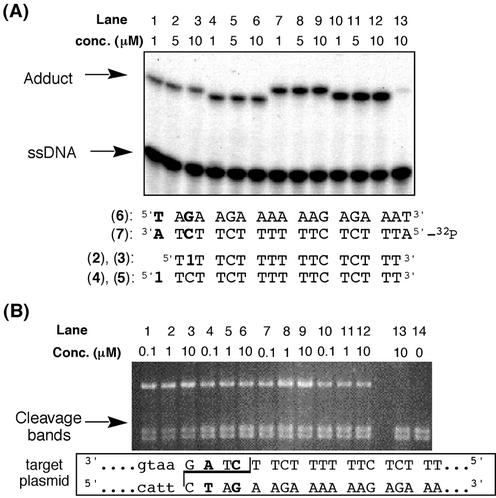
The reaction between the reactive TFOs and the target sequence was estimated by gel shift assay. Triplexes were formed with the TFOs (2–5) and the duplex oligomer with a 5′-32P-labeled pyrimidine strand. The reactions were analyzed by electrophoresis on a 15% denaturing gel (Fig. 3A). The results showed that the 1-bearing TFO (2b–5b) reacted with the target site within the pyrimidine strand. Each TFO (2b–5b) displayed similar levels of reactivity, with ∼25–40% of the duplex target shifted.
Figure 3.
(A) Comparison of the cross-linking reactivity as assayed by gel electrophoresis (15% denaturing gel). Lanes 1–3, the reaction with 2b; lanes 4–6, the reaction with 3b; lanes 7–9, the reaction with 4b; lanes 10–12, the reaction with 5b; lane 13, the reaction with 2a; 1 µM of target duplex 6–7 including 7 labeled with 32P at 5′ end as a tracer in a buffer including 50 mM MES, 10 mM MgCl2. (B) Agarose gel of XbaI protection assay. Lanes 1–3, the reaction with 2b; lanes 4–6, the reaction with 3b; lanes 7–9, the reaction with 4b; lanes 10–12; the reaction with 5b; lane 13, the reaction with 2a; lane 14, the digested marker.
The extent of the reaction of each TFO and the target sequence in the mutation reporter plasmid was also determined. The covalent reaction site was contained in the recognition sequence for the restriction enzyme XbaI (see Fig. 2). Covalent linkage of the TFOs to the target blocked cleavage by the enzyme. The results (Fig. 3B) demonstrated protection with all the TFOs, in the same range as that seen with the band shift analysis. There was little difference in modification levels as a function of TFO concentration because site occupancy was complete at the lowest concentration of TFO. Control experiments showed that, under the conditions of the digestion, the TFO 2a conjugated to the stable precursor did not inhibit enzyme cleavage.
Polymerase stop assay
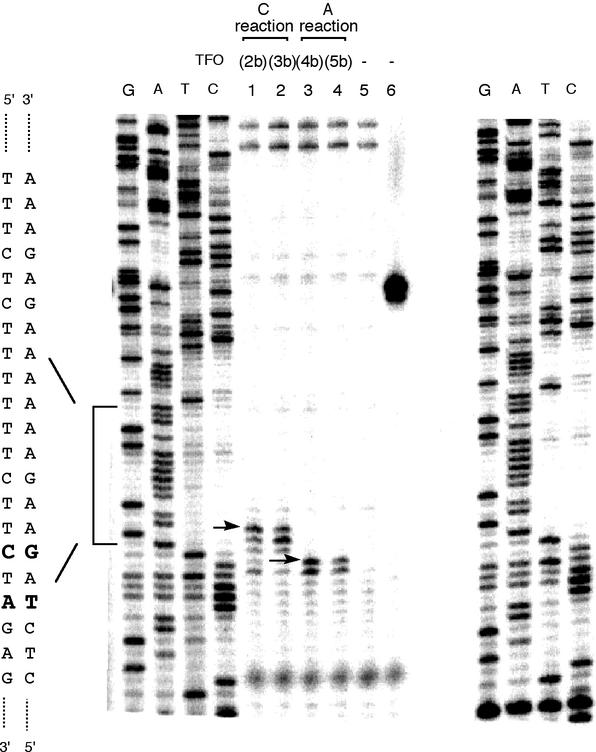
We have used the polymerase stop assay (36) to determine the response to polymerase of adduct formation on the plasmids following the reaction with reactive TFOs (2b–5b). T4 polymerase was used for this assay as a polymerase (Fig. 4). In the reactions of 2b and 3b, designed to react with cytosine, the DNA synthesis paused at one or two bases prior to the adduct, as well as at the adduct site. Stopping one base before, or at the adduct site appeared to occur with similar probability. On the other hand, with 4b and 5b, designed to react with adenine, the stop sites were one or two bases prior to the adduct, and not the adduct site. These results indicated that T4 polymerase responded differently to the two TFO-vinyl adducts, and that the nature of the sugar was not a determinant in the assay.
Figure 4.
Analysis of polymerase stop sites on SupF12 after adduct formation in vitro by reactive TFO (2b–5b). The primer was situated between bases 124 and 144 of the SupF12 gene. Lanes 1–6, T4 polymerase primer extension: lane 1, TFO 2b; lane 2, TFO 3b; lane 3, TFO 4b; lane 4, TFO 5b; lane 5, no TFO; lane 6, no TFO and supF12 digested by BglII. Arrows indicate polymerase STOP sites with adducts marked. The square brackets show the triplex target sites.
Mutagenesis of TFO-vinylpurine adducts in repair-deficient cells
TFO-targeted mutagenesis in human cells was studied using two shuttle vectors. These contain versions of the supF gene, an amber suppressor tyrosine tRNA gene of E.coli, which serves as a mutation reporter (28). The plasmids also contain the SV40 and pBR327 origins of replication, and the β-lactamase gene. pSupF12 and pNS14 were constructed with variant, but functional, supF mutation marker genes containing the triple helix region placed in the 5′ pre-tRNA region of the gene, and the covalent reaction sites embedded in the 5′ end of the mature tRNA gene (see Fig. 2B). Figure 5 shows the experimental strategy and the schematic of the variant supF genes. After the reaction between the plasmids and the reactive TFOs the covalent complexes were transfected into the SVXP12BE cells, which have a severe defect in nucleotide excision repair due to inactivating mutations in the XPA gene (37). Mutation frequencies are higher in these cells than in wild-type cells because of the repair defect. Recovery of mutant plasmids is more efficient and experiments can be done on a smaller scale than with wild-type host cells. After time for replication and mutagenesis, the progeny plasmids were recovered from the cells and introduced into the bacterial tester strain, MBM7070. Mutant colonies containing inactivated supF genes were detected as white colonies among the wild-type blue colonies. The mutation frequencies of the individual preparations ranged from 0.19 to 0.35% (Table 2). The white colonies were purified and the sequences of the mutant supF genes determined.
Figure 5.
Experimental strategy for targeted mutagenesis of supF gene mediated by triple helix formation in vitro.
Table 2. Mutation frequency of reactive TFO-treated plasmid in SVXP12BE cells.
| TFO | Plasmid | No. of colonies | Mutation frequency (%) | |
|---|---|---|---|---|
| Mutant | Total | |||
| 4b | supF12 | 15 | 4381 | 0.34 |
| 5b | supF12 | 11 | 3139 | 0.35 |
| 2b | pNS14 | 10 | 3052 | 0.33 |
| 3b | pNS14 | 14 | 4503 | 0.29 |
| 4b | pNS14 | 8 | 3926 | 0.20 |
| 5b | pNS14 | 5 | 2672 | 0.19 |
| 2a | pNS14 | 0 | 5816 | <0.02 |
| Control | pNS14 | 0 | 6665 | <0.02 |
Numbers represent the frequency of mutations seen in supF gene.
The supF gene is sensitive to mutations in the sequence of the mature tRNA gene (nucleotides 99–183). Mutations outside this region are generally not scored. Thus, supF12 can report mutations at the adenine positioned two bases into the mature tRNA gene. However, mutations at the cytosine in the triplex region, expected from triplexes formed by TFOs 2b and 3b, would not be scored by supF12. Consequently, after the demonstration of mutagenesis in supF12 by TFOs 4b and 5b, we constructed pNS14 in which the triplex region was advanced into the mature gene region so as to report mutations at both the A at 101 and the C at 99 (38).
The mutation spectra indicated that almost all mutations were at the A or C targeted by the appropriate TFO conjugate (Fig. 6). However, there were differences in the kinds of mutations as a function of the TFO and the target base. The 2′-OMe-TFO 4b, designed to react with A, showed exclusively A→G mutations in both supF12 and supNS14. When the deoxy-TFO 5b (A reactive) was used, more diverse kinds of mutations were observed and they appeared at positions adjacent to the target A. When pNS14 was modified with TFOs 2b and 3b, the mutations were of diverse type but only at the target C.
Figure 6.
Spectra of supF gene mutations produced in SVXP12BE cell. Point mutations are indicated above each base pair, with the listed base representing a change from the sequence in the top strand.
DISCUSSION
The purpose of these studies was to investigate the feasibility of targeted mutagenesis by reactive TFOs bearing 1. The TFOs were designed to direct reaction of 1 with either C or A, within or immediately outside the triplex. In addition, the oligonucleotides were constructed with either 2′-OMe or 2′-deoxyribonucleotides. Thus, we were able to assess the contribution of TFO composition on mutagenesis by the vinylpurine adducts.
The thermal stability experiments of the triplexes revealed that the nature of the sugar and the position of the vinylpurine influenced triplex stability. As expected from previous studies (39), the triplexes formed by the 2′-OMe-TFOs were more stable than those formed by the deoxy-TFOs. However, the vinylpurine introduced at the penultimate location within the triplex destabilized the triplex relative to controls regardless of sugar structure. This is probably due to the loss of hydrogen bonding at the derivative site. In contrast, when the vinylpurine was positioned at the terminus of the TFO so as to react with the adenine external to the triplex, triplex stability was enhanced with the deoxy-TFOs, but was decreased with the 2′-OMe-TFOs. This dichotomy presumably reflects the differences in structure of triplexes formed by the two kinds of TFOs. Triplex formation by 2′-OMe-TFOs imposes relatively little distortion on the underlying duplex, while the corresponding deoxy-TFO requires more extensive conformational changes (40). In addition, deoxy oligomers as the third strand have been shown to produce strong binding sites for intercalators at the 5′ junctions between the duplex and the triple helix (41). Thus, the vinylpurine moiety might provide intercalative and/or stacking interactions for stabilization of the triplex with the deoxy-TFO at such a 5′ junction, whereas it might perturb the otherwise stable triplex with the 2′-OMe-TFO.
The gel shift and the restriction protection assays showed that 1 reacts with the complementary position in the pyrimidine strand in a reasonable yield. However, we were unable to devise conditions such that 100% of the target reacted with the TFO, regardless of TFO concentration (Fig. 3). This was probably because the vinyl derivatives can react with nucleophiles such as water in the reaction mixture. To avoid these side reactions, we have developed a new strategy for reaction in which a reactive species is auto-generated within a duplex (42,43). In addition, we have demonstrated that oligonucleotides bearing the 2-amino-6-(1-ethylsulfoxy) vinylpurine derivative achieved efficient reaction with cytosine under neutral conditions (44). Application of these strategies to triplex-mediated modification and mutagenesis is now underway.
The data with the T4 polymerase stop assay indicate that the vinyl adducts at cytosine block replication at one and two bases prior to, as well as at, the adduct site. Some incorporation did occur at the modification site. On the other hand, replication of adenine–vinyl cross-linked templates was blocked at one and two bases prior to the reacted site, with no incorporation at the adduct site. These results were independent of the sugar composition of the TFO. Thus, the polymerase stop sites depended only on the nature of the adducted base. It has been reported that the adenosine-(+) CC-1065 adduct terminates replication by T4 polymerase at one and two bases prior to the modified site (45). Our adenine– vinyl adducts appear to have a similar effect.
In the mutagenesis experiments, the targeted adducts formed by 2′-OMe-TFO 4b led to only A:T to G:C transitions. These mutations, induced by the adenine-vinyl adducts, are in agreement with the mutation study using benzo[a]pyrene-adenine adducts in repair-deficient ‘E.coli’ cells (36), in which it was suggested that the A to G transitions could be a consequence of the structural distortion of the reacted base. The adenine-only mutations with 2′-OMe-TFO 4b, in both SupF12 and pNS14, suggest that the modification reaction was specific to the target A. In contrast to 4b, the deoxy-TFO 5b showed less restricted mutagenesis in both the kinds and location of the mutations. We have previously reported that the deoxy-TFOs containing the vinyl derivative 1 at the terminal position showed reactivity, in addition to the adenine, to the guanine adjacent to the complementary position within the pyrimidine strand (43). This reaction might be attributable to the duplex conformation in the presence of the TFO, and also would be responsible for the mutation at the guanine by the deoxy-TFO 5b. In the presence of 2′-OMe-TFO, the underlying duplex is relatively undistorted, while the deoxy-TFO induces distortion of the duplex to some extent (40). It has also been shown that the duplex is bent by a deoxy-TFO at the junction between the double and the triple helix (46). We suggest that the distortion of the duplex at the triplex:duplex junction by the deoxy-TFO 5b is the basis for the reaction with the adjacent G, as well as the A.
Adducts between the cytosine and the vinylpurine nucleoside with TFO 2b and 3b generated diverse mutations. The reaction with 3b caused mutations C to A and C to T, and mutation C to G was additionally observed with 2b. Apparently mutagenic incorporation at the site of TFO-cytosine adducts was much more relaxed than with the adenine adducts. Recently, error prone DNA polymerases have been described (47,48). Adducts of deoxycytidine with glycidaldehyde have been reported to be miscoding in replication by some of these polymerases, in in vitro experiments. Bypass capacity and base incorporation specificity differed with the different polymerases (49). These polymerases are involved in bypass replication and mutagenesis of DNA adducts, and probably are responsible for mutagenesis of the vinyl adducts studied here.
The data presented here suggest that a TFO with 2′-OMe sugars, targeting a specific base for reaction, can provoke a highly selective mutational response. This will serve as the basis for the development of new more reactive, and selective reagents for targeted mutagenesis. These reagents will have an advantage over the psoralen-TFOs in that the covalent base modifications can accumulate during the incubations, rather than only at the time of photoactivation.
REFERENCES
- 1.Felsenfeld G., Davies,D.R. and Rich,A. (1957) Formation of a three stranded polynucleotide molecule. J. Am. Chem. Soc., 79, 2023–2024. [Google Scholar]
- 2.Thuong N.T. and Helene,C. (1993) Sequence specific recognition and modification of double helical DNA by oligonucleotides. Angew. Chem. Intl. Ed. Eng., 32, 666–690. [Google Scholar]
- 3.Frank-Kamenetskii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 4.Letai A.G., Palladino,M.A., Fromm,E., Rizzo,V. and Fresco,J.R. (1988) Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry, 27, 9108–9112. [DOI] [PubMed] [Google Scholar]
- 5.Fossella J.A., Kim,Y.J., Shih,H., Richards,E.G. and Fresco,J.R. (1993) Relative specificities in binding of Watson–Crick base pairs by third strand residues in a DNA pyrimidine triplex motif. Nucleic Acids Res., 21, 4511–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasquez K.M. and Wilson,J.H. (1998) Triplex-directed modification of genes and gene activity. Trends Biochem. Sci., 23, 4–9. [DOI] [PubMed] [Google Scholar]
- 7.Bailey C. and Weeks,D.L. (2000) Understanding oligonucleotide-mediated inhibition of gene expression in Xenopus laevis oocytes. Nucleic Acids Res., 28, 1154–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aich P., Ritchie,S., Bonham,K. and Lee,J.S. (1998) Thermodynamic and kinetic studies of the formation of triple helices between purine-rich deoxyribo-oligonucleotides and the promoter region of the human c-src proto-oncogene. Nucleic Acids Res., 26, 4173–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapozzi V., Cogoi,S., Spessotto,P., Risso,A., Bonora,G.M., Quadrifoglio,F. and Xodo,L.E. (2002) Antigene effect in K562 cells of a PEG-conjugated triplex-forming oligonucleotide targeted to the bcr/abl oncogene. Biochemistry, 41, 502–510. [DOI] [PubMed] [Google Scholar]
- 10.Faria M., Wood,C.D., Perrouault,L., Nelson,J.S., Winter,A., White,M.R., Helene,C. and Giovannangeli,C. (2000) Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc. Natl Acad. Sci. USA, 97, 3862–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helene C., Thuong,N.T. and Harel-Bellan,A. (1992) Control of gene expression by triple helix-forming oligonucleotides. The antigene strategy. Ann. N. Y. Acad. Sci., 660, 27–36. [DOI] [PubMed] [Google Scholar]
- 12.Vasquez K.M., Wang,G., Havre,P.A. and Glazer,P.M. (1999) Chromosomal mutations induced by triplex-forming oligonucleotides in mammalian cells. Nucleic Acids Res., 27, 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins B.D., Wilson,J.H., Wensel,T.G. and Vasquez,K.M. (1998) Triplex targets in the human rhodopsin gene. Biochemistry, 37, 11315–11322. [DOI] [PubMed] [Google Scholar]
- 14.Havre P.A. and Glazer,P.M. (1993) Targeted mutagenesis of simian virus 40 DNA mediated by a triple helix-forming oligonucleotide. J. Virol., 67, 7324–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., Levy,D.D., Seidman,M.M. and Glazer,P.M. (1995) Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell. Biol., 15, 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puri N., Majumdar,A., Cuenoud,B., Natt,F., Martin,P., Boyd,A., Miller,P.S. and Seidman,M.M. (2002) Minimum number of 2′-O-(2-aminoethyl) residues required for gene knockout activity by triple helix forming oligonucleotides. Biochemistry, 41, 7716–7724. [DOI] [PubMed] [Google Scholar]
- 17.Majumdar A., Khorlin,A., Dyatkina,N., Lin,F.L., Powell,J., Liu,J., Fei,Z., Khripine,Y., Watanabe,K.A., George,J. et al. (1998) Targeted gene knockout mediated by triple helix forming oligonucleotides. Nature Genet., 20, 212–214. [DOI] [PubMed] [Google Scholar]
- 18.Puri N., Majumdar,A., Cuenoud,B., Natt,F., Martin,P., Boyd,A., Miller,P.S. and Seidman,M.M. (2001) Targeted gene knockout by 2′-O-aminoethyl modified triplex forming oligonucleotides. J. Biol. Chem., 276, 28991–28998. [DOI] [PubMed] [Google Scholar]
- 19.Kean J.M. and Miller,P.S. (1993) Detection of psoralen cross-link sites in DNA modified by psoralen-conjugated oligodeoxyribonucleoside methylphosphonates. Bioconjug. Chem., 4, 184–187. [DOI] [PubMed] [Google Scholar]
- 20.Hearst J.E. (1989) Photochemistry of the psoralens. Chem. Res. Toxicol., 2, 69–75. [DOI] [PubMed] [Google Scholar]
- 21.Reed M.W., Lukhtanov,E.A., Gorn,V., Kutyavin,I., Gall,A., Wald,A. and Meyer,R.B. (1998) Synthesis and reactivity of aryl nitrogen mustard-oligodeoxyribonucleotide conjugates. Bioconjug. Chem., 9, 64–71. [DOI] [PubMed] [Google Scholar]
- 22.Shaw J.-P., Milligan,J.F., Krawczyk,S.H. and Matteucci,M. (1991) Specific, high efficiency, triple helix-mediated cross-linking to duplex DNA. J. Am. Chem. Soc., 113, 7765–7766. [Google Scholar]
- 23.Lukhtanov E.A., Mills,A.G., Kutyavin,I.V., Gorn,V.V., Reed,M.W. and Meyer,R.B. (1997) Minor groove DNA alkylation directed by major groove triplex forming oligodeoxyribonucleotides. Nucleic Acids Res., 25, 5077–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagatsugi F., Uemura,K., Nakashima,S., Maeda,M. and Sasaki,S. (1995) Design and synthesis of new cross-linking agents. Nucleic Acids Symp. Ser., 171–172. [PubMed] [Google Scholar]
- 25.Nagatsugi F., Usui,D., Kawasaki,T., Maeda,M. and Sasaki,S. (2001) Selective reaction to a flipping cytidine of the duplex DNA mediated by triple helix formation. Bioorg. Med. Chem. Lett., 11, 343–345. [DOI] [PubMed] [Google Scholar]
- 26.Nagatsugi F., Kawasaki,T., Tokuda,N., Maeda,M. and Sasaki,S. (2001) Site-directed alkylation to cytidine within duplex by the oligonucleotides containing functional nucleobases. Nucl. Nucl., 20, 915–919. [DOI] [PubMed] [Google Scholar]
- 27.Nagatsugi F., Matsuyama,Y., Maeda,M. and Sasaki,S. (2002) Selective cross-linking to the adenine of the TA interrupting site within the triple helix. Bioorg. Med. Chem. Lett., 12, 487–489. [DOI] [PubMed] [Google Scholar]
- 28.Levy D.D., Magee,A.D., Namiki,C. and Seidman,M.M. (1996) The influence of single base changes on UV mutational activity at two translocated hotspots. J. Mol. Biol., 255, 435–445. [DOI] [PubMed] [Google Scholar]
- 29.Peden K.W., Pipas,J.M., Pearson-White,S. and Nathans,D. (1980) Isolation of mutants of an animal virus in bacteria. Science, 209, 1392–1396. [DOI] [PubMed] [Google Scholar]
- 30.Parris C.N., Levy,D.D., Jessee,J. and Seidman,M.M. (1994) Proximal and distal effects of sequence context on ultraviolet mutational hotspots in a shuttle vector replicated in xeroderma cells. J. Mol. Biol., 236, 491–502. [DOI] [PubMed] [Google Scholar]
- 31.Seidman M.M., Dixon,K., Razzaque,A., Zagursky,R.J. and Berman,M.L. (1985) A shuttle vector plasmid for studying carcinogen-induced point mutations in mammalian cells. Gene, 38, 233–237. [DOI] [PubMed] [Google Scholar]
- 32.Parris C.N. and Seidman,M.M. (1992) A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene, 117, 1–5. [DOI] [PubMed] [Google Scholar]
- 33.Nagatsugi F., Kawasaki,T., Usui,D., Maeda,M. and Sasaki,S. (2000) Design and synthesis of novel cross-linking reagents triggered by triple helix formation. Nucleic Acids Symp. Ser., 44, 39–40. [DOI] [PubMed] [Google Scholar]
- 34.Torigoe H., Hari,Y., Sekiguchi,M., Obika,S. and Imanishi,T. (2001) 2′-O,4′-C-methylene bridged nucleic acid modification promotes pyrimidine motif triplex DNA formation at physiological pH: thermodynamic and kinetic studies. J. Biol. Chem., 276, 2354–2360. [DOI] [PubMed] [Google Scholar]
- 35.Marquez V.E., Siddiqui,M.A., Ezzitouni,A., Russ,P., Wang,J., Wagner,R.W. and Matteucci,M.D. (1996) Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? J. Med. Chem., 39, 3739–3747. [DOI] [PubMed] [Google Scholar]
- 36.Chary P., Latham,G.J., Robberson,D.L., Kim,S.J., Han,S., Harris,C.M., Harris,T.M. and Lloyd,R.S. (1995) In vivo and in vitro replication consequences of stereoisomeric benzo[a]pyrene-7,8-dihydrodiol 9,10-epoxide adducts on adenine N6 at the second position of N-ras codon 61. J. Biol. Chem., 270, 4990–5000. [DOI] [PubMed] [Google Scholar]
- 37.Jones C.J. and Wood,R.D. (1993) Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry, 32, 12096–12104. [DOI] [PubMed] [Google Scholar]
- 38.Canella K.A. and Seidman,M.M. (2000) Mutation spectra in supF: approaches to elucidating sequence context effects. Mutat. Res., 450, 61–73. [DOI] [PubMed] [Google Scholar]
- 39.Escude C., Sun,J.S., Rougee,M., Garestier,T. and Helene,C. (1992) Stable triple helices are formed upon binding of RNA oligonucleotides and their 2′-O-methyl derivatives to double-helical DNA. C. R. Acad. Sci. III, 315, 521–525. [PubMed] [Google Scholar]
- 40.Asensio J.L., Carr,R., Brown,T. and Lane,A.N. (1999) Conformational and thermodynamic properties of parallel intramolecular triple helixes containing a DNA, RNA, or 2′-OMeDNA third strand. J. Am. Chem. Soc., 121, 11063–11070. [Google Scholar]
- 41.Perrouault L., Asseline,U., Rivalle,C., Thuong,N.T., Bisagni,E., Giovannangeli,C., Le Doan,T. and Helene,C. (1990) Sequence-specific artificial photo-induced endonucleases based on triple helix-forming oligonucleotides. Nature, 344, 358–360. [DOI] [PubMed] [Google Scholar]
- 42.Nagatsugi F., Kawasaki,T., Usui,D., Maeda,M. and Sasaki,S. (1999) Highly selective and efficient cross-linking to cytidine based on a new strategy for autoactivation within a duplex. J. Am. Chem. Soc., 121, 6753–6754. [Google Scholar]
- 43.Kawasaki T., Nagatsugi,F., Usui,D., Maeda,M. and Sasaki,S. (2000) Efficient crosslinking to cytidine using substituted phenylsulfide derivatives of 2-amino-6-vinylpurine nucleoside via synchronous activation within a duplex. Nucleic Acids Symp. Ser., 44, 129–130. [DOI] [PubMed] [Google Scholar]
- 44.Nagatsugi F., Tokuda,N., Maeda,M. and Sasaki,S. (2001) A new reactive nucleoside analogue for highly reactive and selective cross-linking reaction to cytidine under neutral conditions. Bioorg. Med. Chem. Lett., 11, 2577–2579. [DOI] [PubMed] [Google Scholar]
- 45.Sun D. and Hurley,L.H. (1992) Effect of the (+)-CC-1065-(N3-adenine)DNA adduct on in vitro DNA synthesis mediated by Escherichia coli DNA polymerase. Biochemistry, 31, 2822–2829. [DOI] [PubMed] [Google Scholar]
- 46.Chomilier J., Sun,J.S., Collier,D.A., Garestier,T., Helene,C. and Lavery,R. (1992) A computational and experimental study of the bending induced at a double-triple helix junction. Biophys. Chem., 45, 143–152. [DOI] [PubMed] [Google Scholar]
- 47.Ohmori H., Friedberg,E.C., Fuchs,R.P., Goodman,M.F., Hanaoka,F., Hinkle,D., Kunkel,T.A., Lawrence,C.W., Livneh,Z., Nohmi,T. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- 48.Ling H., Boudsocq,F., Woodgate,R. and Yang,W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell, 107, 91–102. [DOI] [PubMed] [Google Scholar]
- 49.Singer B., Medina,M., Zhang,Y., Wang,Z., Guliaev,A.B. and Hang,B. (2002) 8-(Hydroxymethyl)-3,N(4)-etheno-C, a potential carcinogenic glycidaldehyde product, miscodes in vitro using mammalian DNA polymerases. Biochemistry, 41, 1778–1785. [DOI] [PubMed] [Google Scholar]