Abstract
We report that HMGN1, a nucleosome binding protein that destabilizes the higher-order chromatin structure, modulates the repair rate of ultraviolet light (UV)-induced DNA lesions in chromatin. Hmgn1–/– mouse embryonic fibroblasts (MEFs) are hypersensitive to UV, and the removal rate of photoproducts from the chromatin of Hmgn1–/– MEFs is decreased as compared with the chromatin of Hmgn1+/+ MEFs; yet, host cell reactivation assays and DNA array analysis indicate that the nucleotide excision repair (NER) pathway in the Hmgn1–/– MEFs remains intact. The UV hypersensitivity of Hmgn1–/– MEFs could be rescued by transfection with plasmids expressing wild-type HMGN1 protein, but not with plasmids expressing HMGN1 mutants that do not bind to nucleosomes or do not unfold chromatin. Transcriptionally active genes, the main target of the NER pathways in mice, contain HMGN1 protein, and loss of HMGN1 protein reduces the accessibility of transcribed genes to nucleases. By reducing the compaction of the higher-order chromatin structure, HMGN1 facilitates access to UV-damaged DNA sites and enhances the rate of DNA repair in chromatin.
Keywords: chromatin/chromosomal proteins/HMGN/knockout mouse/UV repair
Introduction
Efficient and correct repair of the damage induced in DNA by extracellular and intracellular agents is a key factor in maintaining the fidelity of gene expression and preventing mutations leading to disease or death. Inefficient repair of the major DNA ultra violet light (UV) lesions, cyclobutane pyrimidine dimers (CPD) and pyrimidine (6–4) pyrimidone photoproducts (6–4PPs) leads to several pathological conditions such as xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy (de Laat et al., 1999; van Steeg and Kraemer, 1999; Friedberg, 2001; Bootsma et al., 2002). Much of the UV-induced damage in DNA is corrected by nucleotide excision repair (NER), an evolutionarily conserved pathway that must contend with the nucleosomal organization of the nuclear DNA and with the higher-order chromatin structure of the nucleosomal arrays (Smerdon and Conconi, 1999; Thoma, 1999; Green and Almouzni, 2002; Ura and Hayes, 2002).
The rate of repair of UV damage by the NER machinery from nucleosomal DNA is significantly slower than from deproteinized DNA (Wang et al., 1991; Hara et al., 2000; Liu and Smerdon, 2000), and UV damage in the nucleosomal linker regions is repaired faster than in the nucleosomal core itself (Thoma, 1999; Ura et al., 2001). Conversely, activities that disrupt the interaction between histones and DNA such as ATP-dependent remodeling complexes facilitate the removal of the UV damage (Hara and Sancar, 2002). The NER process is affected not only by direct contacts between histones and DNA but also by the structural features of the chromatin fiber. Thus, the linker region in a dinucleosome is repaired significantly slower than that of naked DNA (Ura et al., 2001), and activities that reduce the compaction of the higher-order chromatin structure, such as global histone acetylation (Ramanathan and Smerdon, 1989), enhance the rate of UV repair. Likewise, the increased rate of repair in transcribed chromatin regions correlates not only with the transcription process itself, but also with the decondensation and unfolding of the higher-order chromatin structure in the vicinity of the transcribed regions (Bohr, 1991; Teng et al., 1997). These, and several related findings suggest that the local and higher-order chromatin structures modulate the rate of NER (Hanawalt, 2001a; Green and Almouzni, 2002; Ura and Hayes, 2002); however, direct experimental evidence for a role of the higher-order chromatin structure in DNA repair is still lacking. In chromatin, NER is a complex process involving damage recognition, chromatin remodeling, damage excision, DNA synthesis, DNA ligation and chromatin reassembly (Thoma, 1999; Green and Almouzni, 2002). The various phases of the NER process involve the function of many components, some of which are organized in large multi-protein complexes. Access of these complexes to the damaged site may be impeded by a compact higher-order chromatin, a possibility fully compatible with the suggestion that ability of the various NER components to access the damage site is a key factor regulating the rate of DNA repair in the nucleus (Thoma, 1999; Balajee and Bohr, 2000; Green and Almouzni, 2002).
Nuclear proteins such as the non-histone HMGN proteins (formerly named HMG-14/-17; see Bustin, 2001b) that are known to affect the stability of the higher-order chromatin structure (Bustin, 1999, 2001a) may play a role in regulating access to UV-damaged DNA. Several types of experiments indicated that the binding of HMGN to nucleosomes reduces the compaction of chromatin fibers and enhances transcription from chromatin, but not from deproteinized DNA. HMGN proteins destabilize the higher-order chromatin structure by targeting two main elements known to compact chromatin: histone H1 and the N-terminal tail of histone H3 (reviewed in Bustin, 1999, 2001a).
Here, we demonstrate that chromosomal protein HMGN1, an abundant member of the HMGN family, is part of the cellular mechanism that regulates the rate of UV repair. We find that loss of HMGN1 increases the sensitivity of mice and of mouse embryonic fibroblasts (MEFs) to UV irradiation. We show that a transcribed gene in the chromatin of Hmgn1–/– mice is digested at a slower rate by micrococcal nuclease, implying that it is less accessible. The UV hypersensitivity of the Hmgn1–/– fibroblasts could be rescued by the expression of wild-type HMGN1, but not by mutant HMGN1 proteins that do not bind to chromatin. Our studies indicate that the interaction of HMGN1 with chromatin enhances the rate of removal of UV damage from DNA, and points to an additional cellular component involved in the UV repair process. The present study reports a new, in vivo function for HMGN1, and demonstrates that a chromatin architectural protein is involved in the UV repair process.
Results
Generation of Hmgn1–/– mice
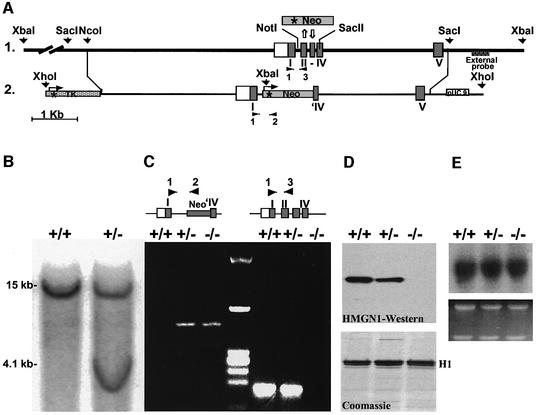
The Hmgn1 gene was inactivated by replacing part of its genomic sequence with a neomycin resistance cassette (Neo; Figure 1A). Southern blot analyses with a 3′ external probe, with an internal probe (from intron IV) and with a Neo probe verified that the vector targeted the correct sequence (Figure 1B). The progeny of male chimera mice were genotyped by PCR with primers that distinguish between the wild-type and the mutated allele (Figure 1C). Western blot analysis indicated that Hmgn1–/– mice do not contain HMGN1 protein, and that in Hmgn1 heterozygotes the amount of protein was approximately half of that detected in the wild-type cells (Figure 1D), supporting our previous observations that the expression of Hmgn1 gene is dosage dependent (Pash et al., 1990). Northern blot analysis (Figure 1E) and western blot analysis (data not shown) indicated that the transcription of the closely related Hmgn2 gene and cellular level of the HMGN2 protein were not affected by the loss of HMGN1 protein.
Fig. 1. Targeted disruption of the mouse Hmgn1 gene. (A) 1, outline of the genomic sequence; 2, targeting vector and insertion sites. The targeting vector contained the TK gene with its promoter (asterisk) 5′ to Hmgn1 sequence. The Neo gene linked to HSV TK promoter (asterisk) replaced the Hmgn1 sequence from the middle of intron I to the middle of exon IV. An XbaI site that was introduced in the targeting vector and two genomic XbaI sites flanking the Hmgn1 gene were used to determine the homologous recombination, using the external probe 3′ to exon V. The primers used in PCR for genotyping the mice are indicated by the black arrowheads and numbered 1, 2 and 3. (B) Southern blot of genomic DNA from targeted ES clones digested with XbaI and hybridized with the external probe. The 15 and 4.1 kb fragments correspond to the wild-type and mutated allele, respectively. (C) Genotyping analysis by PCR. DNA samples from Hmgn1+/+, Hmgn1+/– and Hmgn –/– mice were subjected to PCR analysis with primers 1 and 2 to identify the mutated allele, and primers 1 and 3 to identify the wild-type allele. The primer positions are shown in (A). (D) Western blot analysis of 5% PCA protein extracts from Hmgn1+/+, Hmgn1+/– and Hmgn1–/– MEFs. The Coomassie Blue-stained gel below the western blot indicates that the three extracts contained equal amount of histone H1. (E) Northern blot analysis using Hmgn2 cDNA-specific probe shows equal Hmgn2 transcription in the three types of cells. Ethidium bromide staining in the lower panel shows equal amount of RNA was applied.
The appearance of the Hmgn1–/– mice was normal, however, the frequency ratio of the Hmgn1–/– pups from mating of Hmgn1–/+ heterozygotes (>900 pups genotyped) was 0.08 rather than 0.25, as would be expected from a Mendelian distribution of the Hmgn1– allele. Likely, the low number of Hmgn1–/– offspring is due to events occurring in early stages of embryonic development, since the genotype distribution in 11.5 day embryos was the same as that in born pups. Furthermore, in the matings of >150 Hmgn1–/– pairs the average litter size was 7 ± 1.7, while that obtained from Hmgn1+/+ matings was 11 ± 2. The 30% decrease in the litter size from the Hmgn1–/– matings, and the low frequency of Hmgn1–/– pups from mating of Hmgn1 heterozygotes indicate that HMGN1 protein plays a role in embryonic development.
Increased sensitivity to UV-B irradiation in the skin of Hmgn1–/– mice
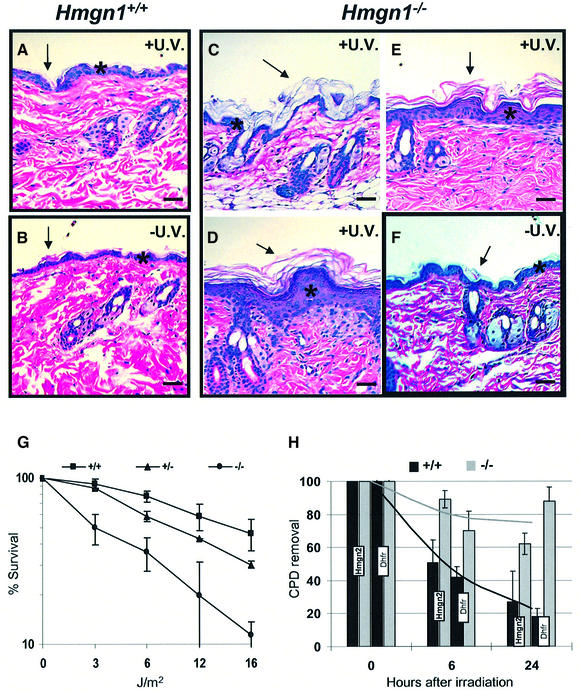
The suggestion that proteins involved in modulating chromatin structure may affect UV repair (Green and Almouzni, 2002; Ura and Hayes, 2002) led us to examine the UV sensitivity of Hmgn1–/– mice. Exposure of the shaved backs of Hmgn1–/– mice and their Hmgn1+/+ littermates to a cumulative UV-B dose of 1200 J/m2, a dose known to produce detectable damage in the skin of XPA–/– mice, (de Vries et al., 1995; de Boer et al., 1999), produced acute alterations in the skin of Hmgn1–/– mice (Figure 2, compare panel F with C and D) but not in the skin of control, wild-type littermates (Figure 2A and B). Histopathological examination of skin samples reveals marked acanthosis and localized hyperkeratosis in the irradiated but not in the non-irradiated areas taken from the Hmgn1–/– mice or in samples taken from irradiated skin of control, Hmgn1+/+ mice (Figure 2). Thus, loss of HMGN1 increased the sensitivity of the skin to the hyperproliferative effects of UV-B irradiation.
Fig. 2. Loss of HMGN1 leads to UV hypersensitivity. (A–F) Increased sensitivity of Hmgn1–/– mice to UV-B. Histology of skin after cumulative irradiation of 1200 J/m2 UV-B. (A and B) Hmgn1+/+ mice, (C–F) Hmgn1–/– mice. Note the increased acanthosis (asterisk; compare E and D with F) and hyperkeratosis (arrows) in the epidermis of the Hmgn1–/– mice. (G and H) Impaired UV repair in MEFs lacking HMGN1. (G) Increased UV sensitivity of Hmgn1–/– fibroblasts. Shown is survival 72 h after irradiation with the indicated doses (see Materials and methods). (H) Decreased rate of gene-specific CPD removal in Hmgn1–/– fibroblasts. Shown is the quantitative analysis of Southern blots with probes specific for the Dhfr and Hmgn2 genes as described in Materials and methods. The 0 h point represents the initial lesion frequency. The bar graphs represent the average of three experiments.
Impaired DNA repair in Hmgn1–/– embryonic fibroblasts
Primary MEFs prepared from 13.5 days old Hmgn1–/– embryos were more sensitive to UV-C irradiation than MEFs prepared from Hmgn1+/+ littermates. The D50 (UV dose resulting in 50% survival) for Hmgn1–/– MEFs was 3 J/m2, a value that is four and a half times lower than the D50 of 13.5 J/m2, observed in the irradiated Hmgn1+/+ MEFs (Figure 2G). The survival of the heterozygote MEFs was intermediate between that of the wild-type and Hmgn1–/– MEF cells, indicating a dose-dependent correlation between loss of HMGN1 protein and the UV sensitivity of the cells.
Cellular survival from UV irradiation is directly linked to the cell’s ability to repair its UV-damaged DNA and remove CPDs from chromatin (Thoma, 1999). In view of the low survival rate of Hmgn1–/– cells after UV irradiation, we tested whether lack of HMGN1 protein impairs the repair of UV damage in cellular chromatin. Since murine cells lack global DNA repair, but have an active transcription-coupled repair (Bohr et al., 1985; Hanawalt, 2001b), we determined the rate of CPD removal from two transcribed genes, Dhfr and Hmgn2. DNA isolated from MEFs at various times following UV-C irradiation was digested with specific restriction enzymes and half of each sample was further treated with T4 endonuclease V, which specifically induces single-strand breaks at each CPD. Therefore, in T4 endonuclease V-treated DNA, the intensity of a restriction fragment is inversely correlated with the number of CPDs present in it (Bill et al., 1991). Quantitative Southern blotting and PCR analysis of the Dhfr and Hmgn2 genes revealed that the rate of CPD removal from the chromatinized DNA in Hmgn1–/– MEFs was significantly impaired. In the Hmgn1–/– cells, over 60% of the UV lesions in the Hmgn2 and 80% of the lesions in the Dhfr genes were still present 24 h after irradiation, while only ∼20% remained in the Hmgn1+/+ (Figure 2H). The inefficient removal of CPDs in Hmgn1–/– MEFs may lead to increased sensitivity to UV-C irradiation.
The NER machinery remains functional in Hmgn1–/– MEFs
Removal of the UV-induced damage from the chromatin by the NER complex involves several steps (Hoeijmakers, 1993; Balajee and Bohr, 2000). The first step requires efficient access of the NER complex to the damaged DNA, the second involves removal of the damage and the third involves restoration of the nucleosome structure at the repaired site. Potentially, HMGN1 may change the repair efficiency by affecting each of these stages either directly or indirectly, or both. By unfolding the chromatin structure at the damaged site, HMGN1 could facilitate access of the NER complex to the damaged site. Alternatively, since HMGN proteins are known to affect transcription from chromatin templates (Bustin, 2001a), HMGN1 may affect the activity of the NER complex indirectly by changing the expression level of one or more of the NER components.
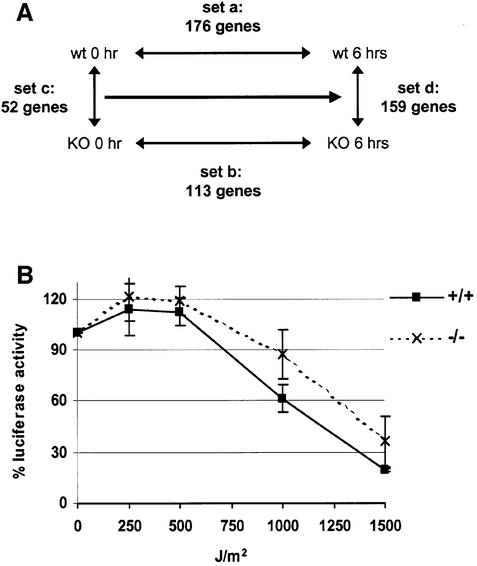
To test whether the expression levels of any known NER components are affected by HMGN1, we used cDNA microarrays to compare four sets of expression profile of Hmgn1–/– and Hmgn1+/+ cells before and 6 h after UV-C irradiation at 3 J/m2 (Figure 3A). In the first set (set a), we compared the expression profile of wild-type Hmgn1+/+ cells before irradiation with that of 6 h after UV-C irradiation. In the second set (set b), we compared the expression profile of Hmgn1–/– cells before irradiation with that of the same cells 6 h after UV-C irradiation. In the third set (set c), we compared the expression profile of wild-type Hmgn1+/+ cells with that of Hmgn1–/– cells before irradiation. In the fourth set (set d), we compared the expression profile of Hmgn1+/+ cells with that of Hmgn1–/– cells 6 h after UV irradiation. The reliability of these microarray results were spot checked by quantitative RT–PCR. The number of genes whose expression changed (either increased or decreased) in the various sets ranged from 52 to 176 (Figure 3A); however, none of these genes are known to be NER components as listed in the data base at http://www.cgal.icnet.uk/DNA_Repair_Genes.html (Wood et al., 2001). We note however that the expression level of a few genes potentially associated with apoptosis and DNA damage, including Gadd45a (Hollander and Fornace, 2002), was induced in Hmgn1–/– cells to a greater extent than in Hmgn1+/+ cells (Table I). Since none of these genes are considered part of the NER machinery, the elevated levels of the transcripts reflect, rather than cause, the decreased ability of the Hmgn1–/– cells to repair UV damage.
Fig. 3. Intact NER in Hmgn1–/– cells. (A) Microarray analysis of gene expression in Hmgn1+/+ and Hmgn1–/– cells after UV-C irradiation at 3 J/m2 UV. The following RNA samples were compared: Array a, Hmgn1+/+ cells, before and 6 h after irradiation; Array b, Hmgn1–/– cells, before and 6 h after irradiation; Array c, non-irradiated Hmgn1+/+ and Hmgn1–/– cells; and Array d, Hmgn1+/+ and Hmgn1–/– cells 6 h after irradiation. The number of genes changed by >1.3-fold in each array experiment is indicated. The arrays contained 15 out of the 27 genes listed as NER components in the database. (B) Host-cell reactivation assay indicates that a UV-damaged luciferase reported plasmid is repaired to a similar extent in Hmgn1–/– and in Hmgn1+/+ cells.
Table I. Genes potentially involved in the response to DNA damage with an altered expression in Hmgn1–/– cells compared with Hmgn1+/+ cells, 6 h after UV irradiation.
| Expression levels KO/WT | Gene name | Full name | Function |
|---|---|---|---|
| 2.1 | Gadd45a | Growth arrest and DNA-damage-inducible 45 α | Induced by stresses. Stimulates DNA repair and inhibits entry into S phase |
| 1.6 | Rad21 | RAD21 homolog (S.pombe) | Involved in repair of ionizing radiation-induced DNA damage in yeast; part of cohesin; cleavage of RAD21 by caspase activates apoptosis |
| 1.4 | Apex1 | Apurinic/apyrimidinic endonuclease 1 | Endonuclease involved in DNA repair |
| 1.3 | Hells | Helicase, lymphoid specific | Helicase; possible role in DNA repair? |
| 1.4 | Smarca5 | SWI/SNF related | Part of chromatin remodelling complex; possible role in DNA repair? |
| 1.7 | Casp12 | Caspase 12 | Protease; induces apoptosis |
| 1.5 | Casp3 | Caspase 3, apoptosis related cysteine protease | Protease; induces apoptosis |
| 1.4 | Tial | Cytotoxic granule-associated RNAbinding protein 1 | Apoptosis; induces DNA fragmentation |
To verify that Hmgn1–/– cells indeed contain a functional NER machinery, we used a ‘host cell reactivation assay’ (Protic-Sabljic and Kraemer, 1985). We tested the ability of Hmgn1–/– and Hmgn1+/+ MEFs to repair the UV-induced damage in luciferase-expressing plasmids, which were irradiated with increasing doses of UV prior to transfection. The luciferase activity recovered from the Hmgn1–/– and Hmgn1+/+ MEFs extracts was very similar (Figure 3B), an indication that the UV-irradiated plasmids were repaired at the same rate in the two cell types. Therefore, we conclude that the UV hypersensitivity observed in the cells lacking HMGN1 protein is not a result of specific changes in the expression of genes coding for the major components of the NER machinery. Taken together, the cDNA microarray and the host cell reactivation results indicate that all the known components necessary to repair the UV-induced damage in cellular DNA are present and functioning in the Hmgn1–/– MEFs.
Loss of HMGN1 impedes access to UV-damaged sites in cellular chromatin
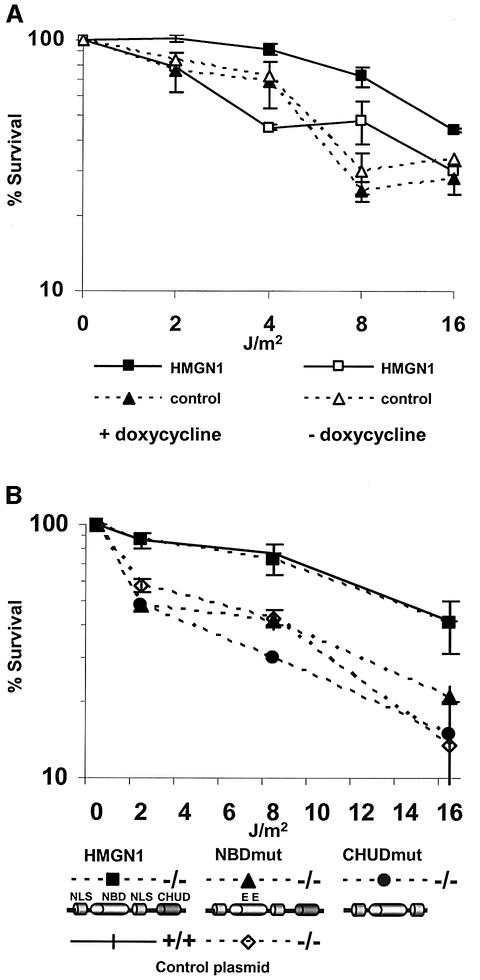
The chromatin structure and transcriptional regulation of transiently transfected plasmids is different from that of the endogeneous cellular chromatin (Archer et al., 1992). Our finding that loss of HMGN1 protein impairs the repair of cellular genes (Figure 2H) but not of transfected plasmids (Figure 3B), raises the possibility that the UV sensitivity of the Hmgn1–/– cells is linked to the ability of HMGN1 to reduce the compaction of the higher-order chromatin structure. To test this possibility, we established revertant Hmgn1–/– MEFs expressing wild-type HMGN1 under the control of the inducible tetracycline response element (TRE) promoter. Induction of the TRE promoter by addition of doxycycline gradually increased the cellular levels of HMGN1 until they were comparable to those in HeLa cells (data not shown). Significantly, induction of HMGN1 expression increased the UV-C survival of the cells (Figure 4A). In the absence of doxycycline, the D50 of the HMGN1 revertant cells remained 3 J/m2 while in cells grown in the presence of the inducer the D50 was 12 J/m2, a level comparable to wild-type MEFs (compare Figure 4A with 2G). The UV sensitivity of control cells transfected with Tet-inducible plasmids that do not express HMGN1 was not affected by addition of doxycycline, and the survival level remains low regardless of whether they were grown in the presence or absence of the inducer (Figure 4A). Thus, the hypersensitivity of the Hmgn1–/– MEFs to UV irradiation is linked directly to the absence of HMGN1 protein.
Fig. 4. Rescue of UV-C sensitivity of Hmgn –/– MEFs. (A) Stable transfection with inducible HMGN1 expressing plasmids. Shown are UV survival curves of cell lines either expressing (filled squares) or not expressing (open squares) HMGN1 in the presence or absence of doxycycline. Note that addition of doxycycline reduced the UV sensitivity of cells expressing HMGN1 protein. (B) Rescue of the UV-C hypersensitivity of Hmgn1–/– cells by transient transfection of fully functional HMGN1 protein. Hmgn1–/– MEFs were transfected with vectors expressing either wild-type or mutated HMGN1 cDNA. The cells were UV irradiated 24 h after transfection and their survival rate determined 72 h later. Note that intact HMGN1, but not mutants that do not bind to nucleosomes (NBDmut) or cannot unfold chromatin (CHUDmut), rescued the UV sensitivity of the Hmgn1–/–. The schemes outline the major functional domains of Hmgn1 protein and the mutants that were transfected. NLS, nuclear localization signal; NBD, nucleosome binding domain; CHUD, chromatin unfolding domain. In the NBD mutant, two Ser residues were mutated to Glu (Prymakowska-Bosak et al., 2001). The CHUD mutant lacks the chromatin unfolding domain (Ding et al., 1997). Control, empty plasmid vector.
To examine the molecular mechanism whereby HMGN1 affects the cellular UV response, we transfected wild-type and Hmgn1–/– MEFs with plasmids express ing either intact HMGN1 protein, the HMGN1S20,24E double point mutant that cannot bind to chromatin (Prymakowska-Bosak et al., 2001), or with the HMGN1-CHUD, a C-terminal deletion mutant that binds to nucleosomes but does not unfold chromatin (Ding et al., 1997). The transfected cells were irradiated with UV-C and the sensitivity to irradiation evaluated 72 h later. The UV hypersensitivity of the Hmgn1–/– MEFs could be rescued by transfection with plasmids coding for intact HMGN1 protein (Figure 4B), a result that is in full agreement with those obtained with the inducible revertant cells expressing HMGN1 (Figure 4A). In contrast, transfection either with plasmids coding for the nucleosomal binding domain (NBD) mutant that cannot bind to nucleosomes, or with the C-terminal deletion mutants that cannot unfold chromatin, did not rescue the UV sensitivity of the Hmgn1–/– MEFs (Figure 4B). Our finding that expression of intact but not of mutant HMGN1 rescues the ability of the cell to survive UV-induced DNA damage suggests that, through a direct interaction with nucleosomes, HMGN1 facilitates alterations in chromatin structure that ultimately enhance the rate of repair of the damaged DNA.
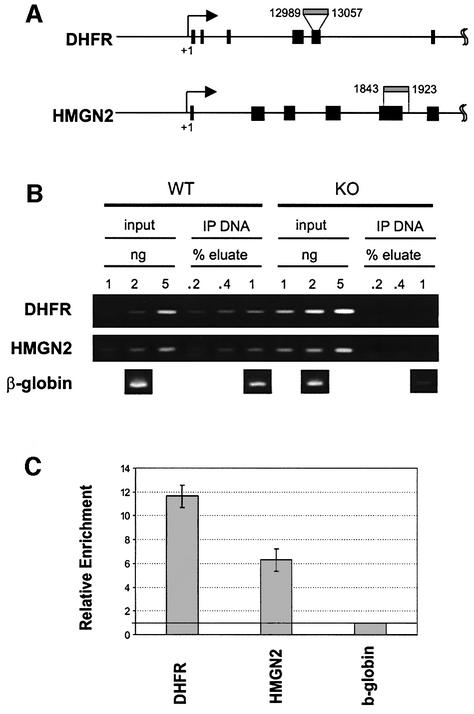
Our finding that HMGN1 rescues the UV hypersensitivity of Hmgn1–/– MEFs, taken together with the decreased rate of CPD removal in these cells (Figure 2H), raises the possibility that HMGN1 protein is at or near the sites of active UV repair. We therefore directly tested whether HMGN1 protein is indeed associated with Dhfr and Hmgn2, the two genes whose rate of UV repair was impaired in Hmgn1–/– MEFs (see Figure 2H). To this end, we isolated chromatin regions containing HMGN1 from Hmgn1+/+ MEFs by chromatin immunoprecipitation assays (ChIP) with affinity purified antibodies to mouse HMGN1. Chromatin isolated from Hmgn1–/– MEFs served as negative controls. As expected, the amount of chromatin DNA recovered from Hmgn1–/– MEFs was negligible (Figure 5B) on the same order as that usually obtained with non-immune IgG. The immunoprecipitated DNA contained both Dhfr and Hmgn2 genes (Figure 5B). To obtain a more accurate indication of the relative amount of HMGN1 in transcribed genes, we analyzed the immunoprecipitated DNA by quantitative RT–PCR. Since in murine cells most of the UV repair is coupled to transcription, we normalized the amounts of the transcribed Dhfr and Hmgn2 genes to β-globin, a gene that is not actively transcribed in MEFs. By quantitative RT–PCR analysis the Dhfr gene was enriched 11-fold and the Hmgn2 gene up to 6-fold over the globin gene (Figure 5C). Thus, HMGN1 is preferentially associated with genes that are actively transcribed.
Fig. 5. Presence of HMGN1 on the chromatin of Hmgn2 and Dhfr genes. (A) Primers used to detect the genes in the ChIP assays. Black boxes indicate exons. (B) PCR analysis of chromatin immunoprecipitations from Hmgn1–/– and Hmgn1+/+ cells with antibodies to HMGN1. Note the lack of signals in the IP DNA from Hmgn1–/– cells. This analysis is not quantitative. (C) Quantification, by real time PCR analysis, of the Hmgn2 and Dhfr genes in IP from Hmgn1+/+ cells. The value for β-globin is set to 1 and the bar graphs represent the average of three experiments.
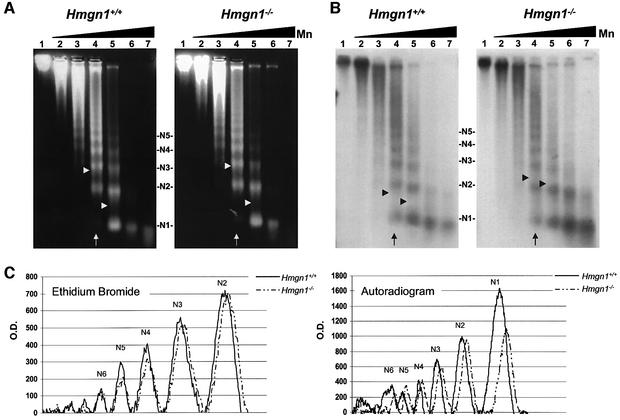
The association of HMGN1 with the Dhfr and Hmgn2 genes may affect their chromatin structure and enhance the NER process by increasing the accessibility of the DNA in these genes. To test this possibility, we digested nuclei isolated from the livers of either Hmgn1–/– or Hmgn1+/+ mice with micrococcal nuclease, a nuclease that has been used previously to asses the relative compactness of chromatin (van Holde, 1988). We then compared the ethidium bromide-stained nucleosomal ladder (N1–N6), which is indicative of the overall rate of digestion of the chromatin, to the ladder resulting from Southern blot analysis with a probe specific for the Hmgn2 gene, which is indicative of the rate of digestion of this specific gene. The rate of chromatin digestion, i.e. the conversion of the chromatin fiber into progressively smaller oligonucleosomal units, is an indication of the accessibility of the linker DNA to the enzyme. In the experiment shown in Figure 6, after 5 min of digestion with 2 U of enzyme (Figure 6, lane 4) the average ethidium bromide-stained nucleosome length in the digest of nuclei isolated from either Hmgn1–/– or Hmgn1+/+ mice was 2.88 and 2.91, respectively (Table II). After 5 min digestion with 11 U (Figure 6, lane 5), the average lengths were 1.42 and 1.46 nucleosomes, respectively. Thus, in both lanes 4 and 5, the ratio of the average nucleosomal lengths (i.e. length in the –/– cells to length in +/+ cells, expressed as Hmgn1–/–/ Hmgn1+/+) is close to 1, an indication that the overall accessibility of the chromatin to micrococcal nuclease is the same in the two cell types. In contrast, in the autoradiogram that measures the accessibility of the Hmgn2 gene, the ratio is close to 1.3, an indication that in the Hmgn1–/– nuclei, the Hmgn2 gene is digested slower than in the wild-type Hmgn1+/+ nuclei. The average Hmgn1–/–/Hmgn1+/+ ratio obtained in three similar experiments, performed with nuclei isolated from the liver of different mice, was 1.02 for the ethidium bromide lanes and 1.48 for the lanes measuring the Hmgn2 organization (Table II). Thus, loss of HMGN1 decreases the rate at which the chromatin region containing the Hmgn2 gene is digested into smaller chromatin fragments, an indication that loss of this protein decreases the accessibility of the Hmgn2 gene to micrococcal nuclease, and by analogy to the NER system. The reduced accessibility of the UV damage to the NER system could account for the reduced removal of the CPDs from the Hmgn1–/– genome and for the lower rate of cell survival.
Fig. 6. Loss of HMGN1 decreases the accessibility of the Hmgn2 gene. Micrococcal nuclease digestions of nuclei isolated from the livers Hmgn1–/– and Hmgn1+/+ mice. (A) Ethidium bromide stain prior to Southern transfer. (B) Southern analysis with Hmgn2 specific probe. (C) Scans of lane 4 (arrow). Arrow heads indicate the length of the average oligonucleosome in the digest calculated as indicated in the methods section. The increased average oligonucleosome length in the autoradiogram of the Hmgn1–/– cells indicates slower rate of digestion of the Hmgn2 chromatin. Lanes 1–7 correspond to 0, 0.05, 0.3, 2, 11, 65 and 400 U micrococcal nuclease in the digest, respectively. N1–N5 denotes the oligonucleosomal size.
Table II. Average nucleosome length (La) following micrococcal nuclease digestion.
| Ethidium bromide |
Autoradiogram |
|||||
|---|---|---|---|---|---|---|
| Hmgn1+/+ | Hmgn1–/– | Hmgn1–/–/Hmgn1+/+ | Hmgn1+/+ | Hmgn1–/– | Hmgn1–/–/Hmgn1+/+ | |
| La – lane 4a | 2.88 | 2.91 | 1.01 | 1.78 | 2.29 | 1.29 |
| La – lane 5a | 1.42 | 1.46 | 1.02 | 1.66 | 2.02 | 1.22 |
| Averageb | 1.02 ± 0.01 | 1.46 ± 0.2 | ||||
aLanes from Figure 6.
bAverage from three different experiments.
La, average nucleosome length was calculated as: La = ΣN1–N6 (PN). Where N1–N6 is the oligonucleosome size (i.e. mono, di) and PN is the fraction of a particular oligonucleosome size out of the total scan (the region covering mono- to hexa-nucleosomes was scanned).
Statistical significance of the two groups was determined by Student’s t-test, p < 0.01.
Discussion
Our main finding is that loss of HMGN1, a nucleosome binding protein that decreases the compaction of the higher-order chromatin structure (Bustin, 2001a), reduces the rate of repair of UV-induced DNA damage. These studies with Hmgn1 null mice demonstrate an in vivo function for HMGN1 and a role for higher-order chromatin structure in DNA repair. We show that the HMGN1-mediated enhancement of the rate of UV repair in chromatin is linked to the ability of HMGN1 to bind to nucleosomes and unfold chromatin. Our data provide experimental evidence that the NER process is affected by the higher-order chromatin structure, and demonstrate that the architectural changes induced in chromatin by structural proteins may ultimately modulate the rate of DNA repair.
Higher-order chromatin structure and DNA repair
Most of the literature on the effect of chromatin structure on repair of UV damage pertains to the role of histone–DNA contacts on this process. The interaction between histone and DNA in the nucleosome reduces the accessibility of the damaged sites and impedes the NER process (Thoma, 1999; Balajee and Bohr, 2000; Hara et al., 2000; Green and Almouzni, 2002). Modification of the histone–DNA interaction by post-translational modification of histones (Ramanathan and Smerdon, 1989), by nucleosome remodeling activities (Hara and Sancar, 2002), or by the transcription process itself enhances the rate of DNA repair (Meijer and Smerdon, 1999; Svejstrup, 2002). It is generally accepted that weakening of the DNA–histone contacts facilitates access of NER components to the damaged DNA; however, recent studies raise the possibility that the repair factors themselves recruit nucleosome remodeling activities to the damaged sites (Hara and Sancar, 2002). An important concept resulting from all these studies is that access to the nucleosomal DNA is an early step in the UV repair process (Meijer and Smerdon, 1999; Thoma, 1999; Green and Almouzni, 2002). Given that access to the damaged DNA is important, it can be expected that the compactness of the higher-order chromatin structure, i.e. the folding of the 10 nm chromatin fiber to form the 30 nm fiber and even more compacted structures, would also impede the action of the NER machinery. Indeed, an additional role for chromatin structure beyond the nucleosome itself in regulating the rate of UV repair has been inferred from several studies on transcription-coupled repair (Meijer and Smerdon, 1999; Svejstrup, 2002). In part, the enhanced rate of repair of transcribed regions can be attributed to the transcription process itself and to the relative ‘decompaction’ of the chromatin structure during transcription. However, a more detailed analysis of the UV repair of the transcribed DHFR and metallothioneine genes (Bohr, 1991), of the yeast MFA2 gene (Teng et al., 1997) and of a promoterless APRT gene (Zheng et al., 2001) suggested that structural features of the chromatin fiber rather than the transcription process itself play a role in the rate of UV repair. An obvious possibility is that structural transitions in the chromatin fiber which affect access to the nucleosome affect the UV repair process. Taken together with previous information on the action of HMGN proteins (Bustin, 1999; 2001a), our finding that HMGN1 enhances the rate of DNA repair strengthens the evidence that the higher-order chromatin structure plays a regulatory role in the UV repair process.
A role for HMGN1 in UV repair
Our experiments both at the whole-animal level and with tissue-culture cells indicate an involvement of HMGN1 protein in the UV response. Low UV-B doses that do not affect the skin of wild-type animals produce significant acanthosis and hyperkeratosis in the skin of Hmgn1–/– mice. These skin lesions are localized to the irradiated areas, an indication that they are a direct effect of UV irradiation rather than general abnormalities in skin proliferation. In tissue culture cells, the increased sensitivity of Hmgn1–/– fibroblasts to UV-C can be linked to a reduced rate of CPD removal from transcribed genes (Figure 2). These cells are under increased stress as indicated by elevated levels of the stress-response gene Gadd45a. The UV-C hypersensitivity of the Hmgn1–/– fibroblasts is directly linked to loss of HMGN1 protein, since controlled induction of HMGN1 expression in Hmgn1–/– fibroblasts reduces their sensitivity to UV irradiation (Figure 4). Significantly, expression of HMGN1 point mutants that do not bind to nucleosomes, or deletion mutants that do not unfold the higher-order chromatin structure failed to rescue the UV hypersensitivity of the Hmgn1–/– fibroblasts. Taken together, the results indicate that the interaction of HMGN1 with nucleosomes and the ability of HMGN1 protein to unfold chromatin affect the cellular response to UV irradiation.
HMGN1 facilitates access to UV-damaged sites
Several different mechanisms could account for an HMGN1 effect on the rate of UV repair. One possibility is that the binding of HMGN1 to nucleosomes weakens the histone–DNA interactions thereby exposing the DNA-damaged sites to the NER complex. This possibility is not compatible with all of the previous information on the interaction of HMGN proteins with nucleosomes demonstrating that HMGN proteins stabilize the structure of nucleosomes (Bustin, 2001a). By this scenario, the absence of HMGN1 should enhance the rate of DNA repair. Our studies with Hmgn1–/– mice and MEFs show reduced DNA repair. A second possibility, that HMGN1 affects the rate of repair by modulating the expression levels of one of the NER components, is not compatible with our analysis of the transcription profiles of the Hmgn1–/– and Hmgn1+/+ fibroblasts, which failed to detect changes in transcription levels of any of the known NER components (Wood et al., 2001). Furthermore, the similar host-cell reactivation results in Hmgn1–/– and Hmgn1+/+ MEFs (Figure 3) also demonstrate that all of the NER components are present and functional in Hmgn1–/– fibroblasts.
We therefore favor a third possibility, that HMGN1 enhances the rate of DNA repair by reducing the compaction of the chromatin fiber and facilitating access of various components involved in the repair of the UV damage (Green and Almouzni, 2002) to the nucleosomes containing the damaged DNA sites. Our suggestion that HMGN1 affects the rate of UV repair by facilitating access to the nucleosomes is supported by our findings that HMGN1 deletion mutants that do not unfold chromatin fail to rescue the UV hypersensitivity of the Hmgn1–/– fibroblasts, and with the reduced rate of micrococcal nuclease digestion of the Hmgn2 gene in these cells.
HMGN may facilitate access to nucleosomes by targeting histone H1 and the amino termini of core histones (Bustin, 2001a; Catez et al,. 2002), two main elements known to stabilize the compacted higher-order chromatin structure. From the UV repair standpoint, perturbation of the interaction of H1 with chromatin has been shown to increase the accessibility of nucleosomes and facilitate the function of histone acetylases (Herrera et al., 2000) and nucleosome remodeling complexes such as SWI/SNF (Hill and Imbalzano, 2000; Horn et al., 2002), activities that could affect the NER process (Green and Almouzni, 2002).
Recent studies reveal that the intranuclear organization of HMGN is dynamic and related to the metabolic state of the cell. The location of the protein is spatially and functionally related to transcription and to stages of the cell cycle (Hock et al., 1998; Phair and Misteli, 2000; Prymakowska-Bosak et al., 2001). The dynamics of the intranuclear movement suggest that HMGN proteins are only transiently associated with any specific chromatin site. Our chromatin immunoprecipitation studies demonstrate that HMGN1 is indeed associated with transcriptionally active regions. Since in murine cells the UV repair is coupled to transcription, the presence of HMGN1 in these genes could account for the effect on the repair process. This possibility is supported by our finding that loss of HMGN1 reduced the accessibility of Hmgn2 to micrococcal nuclease, suggesting perturbation of the chromatin structure in or around this gene. HMGN1 may affect the accessibility of the transcribed chromatin regions to the NER either by facilitating the unfolding of the chromatin fiber, or by binding and stabilizing an already unfolded conformation.
Materials and methods
Generation of Hmgn1–/– mutant mice
Hmgn1-containing clones from an 129 Sv λ EMBL3 phage library were identified by screening with intron-specific probes obtained by PCR of genomic mouse DNA. The 4.5 kbp SacI–NotI and 2.5 kbp SacII–SacI restriction fragments (see Figure 1A) were sequentially subcloned to create the targeting vector containing the NcoI–SacI region of the gene in which the neomycin cassette replaces part of intron 1, exons 2, 3 and part of exon 4. The XhoI-linearized targeting vector was electroporated into ESVJ-1183 cells that were grown in the presence of G418 and gancyclovir. ES cells were genotyped by Southern analysis of Xba1-digested DNA probed with either a 400 bp fragment from intron V DNA, located 3′ to the targeting vector (see ‘external probe’, Figure 1A), or an internal probe spanning a 1 kb region starting from the middle of exon IV (‘internal probe’; not shown in figure), or a Neo probe. The targeted ES cell clones were injected into C57 BL/6 blastocysts and transferred into pseudopregnant NIH black Swiss females. The resulting chimera males were mated with C57BL/6 and 129 Sv females. Genotyping of the tail clip-DNA was carried out by Southern blot analysis using the external probe, and by PCR analysis using the primers outlined in Figure 1A. Gene expression was monitored by northern blot analysis using the mouse Hmgn1 cDNA and by western blot analysis of 5% perchloric acid extracts (PCA) using affinity purified antibodies to mouse HMGN1.
Skin UV-sensitivity measurements
The mice were anaesthetized by IP injection of 2.5% avertin (300 µl per mouse) prior to each treatment. The backs of shaved 8- to 10-week-old Hmgn1+/+ and Hmgn1–/– littermates (nine of each), were irradiated with UV-B (FS20 sunlamp) at a cumulative dose of 1.2 kJ/m2 for 1 week. On the seventh day, 2 h after the last treatment, the animals were euthanized and skin samples were taken from both irradiated and non-irradiated areas and, as additional controls, from non-irradiated animals. The skin specimens were fixed in 10% buffered formalin phosphate, embedded in paraffin, and 6 µm-thick sections were stained with haematoxylin and eosin. The lowest UV-B dose that induced erythema and edema after 24 h on the shaved backs of C57 BL/6 mice was considered as the minimal erythema/edema dose (MED) for this mouse strain.
Preparation and growth of primary MEFs and MEF cell lines
After removal of the head and viscera, E13.5 embryos were digested in 0.25% trypsin at 37°C, with gentle pipetting for 30 min. The dissociated fibroblasts were allowed to settle and then cultured in 150 cm plates in DMEM with 10% FBS under 5% CO2 at 37°C. Cell lines were established by transforming the primary embryonic fibroblasts with SV40 ts mutant virus (Chou, 1989; Jat and Sharp, 1989).
Generation of Hmgn1 revertant MEFs
SV-40-immortalized Hmgn1–/– embryonic fibroblasts were co-transfected with linearized pTet-On, pTet-tTS (Clontech) and pZeoSV2 (Invitrogen). Cells were plated in 2% methocel (Fluka) containing 50 µg/ml zeocin (Invitrogen) to isolate colonies of stable integrants. Colonies were expanded and screened for the ability to induce doxycycline-dependent expression from a transiently transfected TRE reporter plasmid. The best clone was transfected with pTK-Hyg, and either pBI-G-HMGN1 to generate line 622, or pBI-G-HMGN1S20/24E (Prymakovska-Bosak et al., 2001) to generate line 85, which are derived from pBI-G (Clontech). pBI-G-HMGN1 contains the open reading frame of human HMGN1 inserted in the Sal1–NotI sites. Cells were plated in methocel containing 50 µg/ml zeocin and 100 µg/ml hygromycin (Clontech). Colonies were expanded and screened for doxycycline-induced HMGN expression.
UV survival of MEF cells
All treatments commenced 24 h after plating 5 × 104 cells into 35 mm dishes. The medium was aspirated and replaced with 0.5 ml PBS, the cell plates were chilled and UV-C irradiated (UV Systems, LS-15, 254 nm) at the indicated doses. Fresh medium was added to the plates immediately after irradiation and cell survival was determined 72 h after treatment by counting the number of trypan blue-excluding cells. Survival is expressed as a percentage, using untreated cells as the 100% value. All experiments were conducted in triplicate and were repeated at least twice.
Transfection of MEFs with HMGN1 cDNA expression vectors
Point and deletion mutants of HMGN1 cDNA subcloned into pCI-neo (Promega) mammalian expression vectors were transfected into 5 × 105 MEFs, in 35 mm plates, using Lipofectamine 2000 reagent (Gibco-BRL). Twenty-four hours later, the cells were UV irradiated at the indicated dosages and the cell survival rate was determined 72 h after irradiation, as described above. Survival was expressed as a percentage using transfected non-treated cells as the 100% value. Every experiment was done in triplicate and repeated at least twice. Transfection efficiency for each plasmid in the two cell types was over 60% as determined by cotransfection with GFP HMGN-fusion proteins. Expression levels of the protein from the various transfected plasmids were similar to each other, as determined by immunofluorescence (Prymakowska-Bosak et al., 2001, 2002).
Host-cell reactivation assay
The pGL3 promoter vector (Promega) containing the luciferase gene under the CMV promoter was treated with UV-C at different doses and transfected into 1.5 × 105 MEFs in 35 mm plates using Lipofectamine 2000 reagent (Gibco-BRL). After 48 h, luciferase activity was measured with a luminometer (TD-20/20, Turner Designs) using a luciferase assay system (Promega) as recommended by the manufacturer. Total protein in the cell lysates was measured by Bio-Rad protein assay. The relative luciferase activities done with previously described plasmids were expressed as percentage of expression from non-irradiated control plasmids, which were taken as 100% (Emmert et al, 2002).
Removal rate of CPDs from genomic DNA
Genomic DNA was isolated at various times after UV-C irradiation (30 J/m2) and restricted with either EcoRI (to detect the 15 kb fragment of the Dhfr gene) or BglII (to detect the 10 kb fragment of the Hmgn2 gene), and half of each sample was further digested with T4 Endonuclease V (Epicentre Technologies), which specifically cleaves at CPDs. Quanti tative Southern blot analysis (Image Quant-Molecular Dynamics) of fragments detected with a 2.9 kb EcoRI–EcoRV probe from plasmid pBR327 (a gift from V.Bohr, NIH) containing the Dhfr gene, or a 560 bp BamHI–HindIII probe from plasmid pXT17 containing the Hmgn2 gene, allowed estimation of the rate of removal of the CPDs from the chromatin of the cells. For each time point the intensity of the restriction fragment obtained in DNA that was not digested with T4 endonuclease V was taken as 100%. The results obtained by the Southern blot analysis were independently verified by semi-quantitative PCR, using primers that amplified specific regions of the Dhfr or Hmgn2 genes.
Analysis of gene expression profile
Cells were harvested either before or 6 h after irradiation with 3 J/m2 UV-C. RNA was purified using Trizol (Life Technologies) followed by RNeasy (Qiagen) as recommended by the manufacturers. Fluorescently labeled cDNA was prepared using anchored oligo (dT) primer and the Cyscribe first-strand cDNA labeling kit (AP Biotech). Cy3 and Cy5-labeled samples were combined and hybridized to a glass microarray slide at 65°C overnight. Mouse expression arrays from Advanced Technology Center, NCI/NIH, contained 10 368 cDNA spots from the Incyte mouse GEM2 clone set. Arrays were scanned and quantified using the GenePix 4000A microarray scanner. Two separate reverse-fluor hybridizations were performed for each experiment, and genes were selected that showed >1.3-fold change in both hybridizations. Individual mRNAs were quantified by real-time RT–PCR using SYBR green and an ABI PRISM® 7900HT sequence detection system, as described by the manufacturer (Applied Biosystems). Expression levels were normalized to GAPDH, and differences calculated using the ΔΔCt method (Applied Biosystems).
Analysis of chromatin structure
Nuclei were isolated from mouse livers by ultracentrifugation through layered 1.7 and 2.3 M sucrose solutions (Hewish and Burgoyne, 1973) and digested with micrococcal nuclease (Sigma-Aldrich cat. N5386) for 5 min at 25°C, and the extent of digestion analyzed as described previously (Einck et al., 1986).
Chromatin immunoprecipitation
ChIP experiments were carried out as described previously (Orlando et al., 1997) and modified by Upstate Biotechnology. Formaldehyde (Sigma) was added to a final concentration of 1% directly to the medium of primary Hmgn1+/+ and Hmgn1–/– MEFs grown to 95% confluence in DMEM with 10% (v/v) fetal bovine serum at 37°C. Cells were sonicated to produce ∼200–800 bp DNA fragments. HMGN1-containing fragments were purified with affinity purified rabbit anti-mouse HMGN1 peptide 6 (Bustin et al., 1990) antibodies. Polymerase chain reactions: for semi-quantitative PCR, 30–40 cycle-PCR (95°C for 30 s, 60°C for 30 s and 72°C for 30 s) reactions were performed with 1–5 ng DNA of input samples or with 0.2–1% (v/v) of the IP sample. Real-time PCR were performed using an ABI PRISM 7900HT Sequence Detection System and SYBR Green PCR master mix (Applied Biosystems) with appropriate primers (sequences on request). ChIP values normalized to input.
References
- Archer T.K., Lefebvre,P., Wolford,R.G. and Hager,G.L. (1992) Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science, 255, 1573–1576. [DOI] [PubMed] [Google Scholar]
- Balajee A.S. and Bohr,V.A. (2000) Genomic heterogeneity of nucleotide excision repair. Gene, 250, 15–30. [DOI] [PubMed] [Google Scholar]
- Bill C.A., Grochan,B.M., Meyn,R.E., Bohr,V.A. and Tofilon,P.J. (1991) Loss of intragenomic DNA repair heterogeneity with cellular differentiation. J. Biol. Chem., 266, 21821–21826. [PubMed] [Google Scholar]
- Bohr V.A. (1991) Gene specific DNA repair. Carcinogenesis, 12, 1983–1992. [DOI] [PubMed] [Google Scholar]
- Bohr V.A., Smith,C.A., Okumoto,D.S. and Hanawalt,P.C. (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell, 40, 359–369. [DOI] [PubMed] [Google Scholar]
- Bootsma D., Kraemer,K.H., Cleaver,J.E. and Hoeijmakers,J.H.J. (2002) Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. In Vogelstein,B. and Kinzler,K.W. (eds), The Genetic Basis Of Human Cancer. McGraw-Hill, New York, pp. 211–237.
- Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol., 19, 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. (2001a) Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci., 26, 431–437. [DOI] [PubMed] [Google Scholar]
- Bustin M. (2001b) Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci., 26, 152–153. [DOI] [PubMed] [Google Scholar]
- Bustin M., Crippa,M.P. and Pash,J.M. (1990) Immunochemical analysis of the exposure of high mobility group protein 14 and 17 surfaces in chromatin. J. Biol. Chem., 265, 20077–20080. [PubMed] [Google Scholar]
- Catez F., Brown,D.T., Misteli,T. and Bustin,M. (2002) Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO rep., 3, 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J.Y. (1989) Differentiated mammalian cell lines immortalized by temperature-sensitive tumor viruses. Mol. Endocrinol., 3, 1511–1514 [DOI] [PubMed] [Google Scholar]
- de Boer J. et al. (1999) Mouse model for the DNA repair/basal transcription disorder trichothiodystrophy reveals cancer predisposition. Cancer Res., 59, 3489–3494. [PubMed] [Google Scholar]
- de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- de Vries A. et al. (1995) Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature, 377, 169–173. [DOI] [PubMed] [Google Scholar]
- Ding H.F., Bustin,M. and Hansen,U. (1997) Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region of chromosomal protein HMG-14. Mol. Cell. Biol., 17, 5843–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einck L., Fagan,J. and Bustin,M. (1986) Chromatin structure of the cytochrome P-450c gene changes following induction. Biochemistry, 25, 7062–7068. [DOI] [PubMed] [Google Scholar]
- Emmert S. et al. (2002) Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J. Invest. Dermatol., 118, 972–982 [DOI] [PubMed] [Google Scholar]
- Friedberg E.C. (2001) How nucleotide excsion repair protects against cancer. Nat. Rev. Cancer, 1, 22–33. [DOI] [PubMed] [Google Scholar]
- Green C.M. and Almouzni,G. (2002) When repair meets chromatin: first in series on chromatin dynamics. EMBO rep., 3, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P.C. (2001a) Controlling the efficiency of excision repair. Mutat. Res., 485, 3–13. [DOI] [PubMed] [Google Scholar]
- Hanawalt P.C. (2001b) Revisiting the rodent repairadox. Environ. Mol. Mutagen., 38, 89–96. [DOI] [PubMed] [Google Scholar]
- Hara R. and Sancar,A. (2002) The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol., 22, 6779–67787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R., Mo,J. and Sancar,A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol., 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera J.E., Schiltz,R.L. and Bustin,M. (2000) The accessibility of histone H3 tails in chromatin modulates their acetylation by P300/CBP-associated factor. J. Biol. Chem., 275, 12994–12999. [DOI] [PubMed] [Google Scholar]
- Hewish D.R. and Burgoyne,L.A. (1973) Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem. Biophys. Res. Commun., 52, 504–510. [DOI] [PubMed] [Google Scholar]
- Hill D.A. and Imbalzano,A.N. (2000) Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry, 39, 11649–11656. [DOI] [PubMed] [Google Scholar]
- Hock R., Wilde,F., Scheer,U. and Bustin,M. (1998) Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J., 17, 6992–7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers J.H. (1993) Nucleotide excision repair. II: from yeast to mammals. Trends Genet., 9, 211–217. [DOI] [PubMed] [Google Scholar]
- Hollander M.C. and Fornace,A.J.,Jr (2002) Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene, 21, 6228–6233. [DOI] [PubMed] [Google Scholar]
- Horn P.J., Carruthers,L.M., Logie,C., Hill,D.A., Solomon,M.J., Wade,P.A., Imbalzano,A.N., Hansen,J.C. and Peterson,C.L. (2002) Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol., 9, 263–267. [DOI] [PubMed] [Google Scholar]
- Jat P.S. and Sharp,P.A. (1989) Cell lines established by a temperature-sensitive simian virus 40 large-T-antigen gene are growth restricted at the nonpermissive temperature. Mol. Cell. Biol., 9, 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. and Smerdon,M.J. (2000) Nucleotide excision repair of the 5S ribosomal RNA gene assembled into a nucleosome. J. Biol. Chem., 275, 23729–23735. [DOI] [PubMed] [Google Scholar]
- Meijer M. and Smerdon,M.J. (1999) Accessing DNA damage in chromatin: insights from transcription. BioEssays, 21, 596–603. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Pash J., Popescu,N., Matocha,M., Rapoport,S. and Bustin,M. (1990) Chromosomal protein HMG-14 gene maps to the Down syndrome region of human chromosome 21 and is overexpressed in mouse trisomy 16. Proc. Natl Acad. Sci. USA, 87, 3836–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Protic-Sabljic M. and Kraemer,K.H. (1985) One pyrimidine dimer inactivates expression of a transfected gene in xeroderma pigmentosum cells. Proc. Natl Acad. Sci. USA, 82, 6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prymakowska-Bosak M., Misteli,T., Herrera,J.E., Shirakawa,H., Birger,Y., Garfield,S. and Bustin,M. (2001) Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol., 21, 5169–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prymakowska-Bosak M., Hock,R., Catez,F., Lim,J.H., Birger,Y., Shirakawa,H., Lee,K. and Bustin,M. (2002) Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol. Cell. Biol., 22, 6809–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan B. and Smerdon,M.J. (1989) Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J. Biol. Chem., 264, 11026–11034. [PubMed] [Google Scholar]
- Smerdon M.J. and Conconi,A. (1999) Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol., 62, 227–255. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. (2002) Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell. Biol., 3, 21–29. [DOI] [PubMed] [Google Scholar]
- Teng Y., Li,S., Waters,R. and Reed,S.H. (1997) Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial rad16 requisite for repairing upstream control sequences. J. Mol. Biol., 267, 324–337. [DOI] [PubMed] [Google Scholar]
- Thoma F. (1999) Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J., 18, 6585–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura K. and Hayes,J.J. (2002) Nucleotide excision repair and chromatin remodeling. Eur. J. Biochem., 269, 2288–2293. [DOI] [PubMed] [Google Scholar]
- Ura K., Araki,M., Saeki,H., Masutani,C., Ito,T., Iwai,S., Mizukoshi,T., Kaneda,Y. and Hanaoka,F. (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J., 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K.E. (1988) Chromatin. Springer-Verlag, New York, NY.
- van Steeg H. and Kraemer,K.H. (1999) Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol. Med. Today, 5, 86–94. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Wu,X.H. and Friedberg,E.C. (1991) Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem., 266, 22472–22478. [PubMed] [Google Scholar]
- Wood R.D., Mitchell,M., Sgouros,J. and Lindahl,T. (2001) Human DNA repair genes. Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Pao,A., Adair,G.M. and Tang,M. (2001) Cyclobutane pyrimidine dimers and bulky chemical DNA adducts are efficiently repaired in both strands of either a transcriptionally active or promoter-deleted APRT gene. J. Biol. Chem., 276, 16786–16796. [DOI] [PubMed] [Google Scholar]