Abstract
During eukaryotic translation termination, eRF1 responds to three stop codons. However, in ciliates with variant genetic codes, only one or two codons function as a stop signal. To localize the region of ciliate eRF1 implicated in stop codon discrimination, we have constructed ciliate–human hybrid eRF1s by swapping regions of human eRF1 for the equivalent region of ciliate Euplotes eRF1. We have examined the formation of a cross-link between recombinant eRF1s and mRNA analogs containing the photoactivable 4-thiouridine (s4U) at the first position of stop and control sense codons. With human eRF1, this cross-link can be detected only when either stop or UGG codons are located in the ribosomal A site. Here we show that the cross-link of the Euplotes–human hybrid eRF1 is restricted to mRNAs containing UAG and UAA codons, and that the entire N-terminal domain of Euplotes eRF1 is involved in discriminating against UGA and UGG. On the basis of these results, we discuss the steps of the selection process that determine the accuracy of stop codon recognition in eukaryotes.
Keywords: ciliates/eRF1/stop codon/s4U photocrosslinking/translation termination
Introduction
The presence of a stop codon—UAA, UAG or UGA—in the A site of the ribosome is generally a signal to terminate protein synthesis. This process constitutes the last essential stage of translation, as it ensures the formation of full-sized proteins. Translation termination involves two classes of release factors (RFs). Class 1 RFs recognize stop codons within the ribosomal A site and trigger the hydrolysis of the ester bond connecting the peptide chain and the tRNA at the ribosomal P site. Class 2 RFs are GTPases that stimulate class 1 RF activity and confer GTP dependency upon the termination process. In eukaryotes, a single class 1 RF, eRF1, recognizes all three stop codons. However, in bacteria, a pair of class 1 RFs, namely RF1 and RF2, display an overlapping specificity, decoding either UAA and UAG, or UAA and UGA, respectively. It is now well supported that class 1 RFs bind to the ribosomal A site. They functionally mimic tRNA in that they decode stop codons at the decoding site of the small ribosomal subunit and activate the large ribosomal subunit peptidyl transferase center, which then catalyzes the hydrolysis of the peptidyl-tRNA bond (reviewed in Kisselev and Buckingham, 2000; Ehrenberg and Tenson, 2002). However, the mechanisms by which class 1 RFs interact with the ribosome, discriminate stop codons from sense codons, and trigger peptidyl-tRNA hydrolysis are far from understood. Among the mechanisms that remain to be elucidated, the selection of the stop codon is one of the major questions about the translation termination process.
In bacteria, it has been shown that a tripeptide within the class 1 RF primary sequence shares the ability to determine stop codon specificity (Ito et al., 2000). The Pro-Ala-Thr (PAT) and Ser-Pro-Phe (SPF) tripeptides in RF1 and RF2, respectively, could discriminate between A and G at the second and third positions of stop codon. It was assumed that the ‘tripeptide anticodons’ directly interact with stop codons in the manner of tRNA anticodons. In addition, charge-flip changes at multiple Glu residues located in the sequence adjacent to the ‘tripeptide anticodon’ interfere with the accuracy of stop codon recognition, but do not impair peptidyl-tRNA hydrolysis (Uno et al., 2002). These results strongly support the hypothesis that these regions are positioned at the decoding site of the 30S ribosomal subunit. Surprisingly, in the crystal structure of Escherichia coli RF2, the SPF ‘tripeptide anticodon’ and the universally conserved Gly-Gly-Gln (GGQ) motif, a speculated mimic of the 3′CCA of a tRNA, are ∼23 Å apart from each other (Vestergaard et al., 2001). This distance is much shorter than the expected distance of 75 Å, which separates the anticodon loop and the 3′CCA end of a tRNA. Recent data from cryo-electron microscopy studies reconcile these contradictory results, showing that RF2 adopts a different conformation when bound to the ribosome. In this open conformation, the GGQ motif interacts with the peptidyl transferase center while the SPF tripeptide is situated in the 30S decoding site (Klaholz et al., 2003; Rawat et al., 2003).
In eukaryotes, the tRNA–eRF1 mimicry hypothesis (Ito et al., 1996; Nakamura et al., 2000) was first supported by in vivo and in vitro experiments showing that eRF1 competes with suppressor tRNAs for stop codon recognition (Stansfield et al., 1995; Drugeon et al., 1997; Le Goff et al., 1997). The determination of the crystal structure of human eRF1 revealed a Y-shaped molecule, which roughly resembles a tRNA (Song et al., 2000). It was tentatively concluded that the N-terminal domain (domain 1) forming the stem of the Y was the equivalent of the anticodon arm of tRNA. The middle domain (domain 2) forming one of the arms of the Y corresponded to the acceptor arm of tRNA whereas the other arm of the Y was formed by the C-terminal domain (domain 3) involved in the interaction with eRF3. In addition, it was proposed that the NIKS motif at the tip of domain 1 was the putative anticodon-like site. This domain assignment was based on: (i) the results of random mutagenesis of budding yeast eRF1 showing that mutations which alter stop codon recognition specificity occurred exclusively in the N-terminal domain (Bertram et al., 2000); (ii) mutational analysis of the conserved GGQ motif located at the tip of domain 2 showing that mutation of the glycine residues abolished release activity, but did not affect ribosome binding of human eRF1 (Frolova et al., 1999); and (iii) protein–protein interaction analysis showing that the C-terminal sequence of eRF1 mediates eRF3 binding (Ito et al., 1998; Eurwilaichitr et al., 1999; Merkulova et al., 1999).
An interesting feature of ciliates is their use of alternative nuclear genetic codes. To date, all known changes concern the reassignment of stop codons to sense codons. For example, the stop codons UAA and UAG are translated into glutamine in several species (UGA = stop variant code), whereas in the hypotrich genus Euplotes, UGA is translated into cysteine (Caron and Meyer, 1985; Preer et al., 1985; Harper and Jahn, 1989; for a recent review, see Lozupone et al., 2001). Decoding of reassigned stop codons requires specific cognate tRNAs as shown for Tetrahymena (Kuchino et al., 1985; Hanyu et al., 1986) or a near cognate tRNA acting as natural suppressor as suggested for the tRNACys of Euplotes (Grimm et al., 1998). In addition, it was recently shown that ciliate eRF1s do not respond to the reassigned stop codons in vitro, and thus do not compete with stop codon decoding tRNAs (Kervestin et al., 2001; Ito et al., 2002). It was postulated that the absence of recognition of a reassigned stop codon involves amino acids in ciliate eRF1 sequences that are divergent from those found in eRF1 from organisms using the standard genetic code. Thus, substantial efforts were undertaken to sequence ciliate eRF1 genes (Karamyshev et al., 1999; Inagaki and Doolittle, 2001; Liang et al., 2001; Lozupone et al., 2001; Kervestin et al., 2002). Multiple sequence alignments identified convergent changes in eRF1 from ciliates using the same genetic code deviation. Then, with the help of eRF1 three-dimensional structure, various models for stop codon recognition were designed based on the residues of eRF1 where the convergent changes were observed (Inagaki and Doolittle, 2001; Lozupone et al., 2001; Muramatsu et al., 2001; Inagaki et al., 2002). However, the number of these convergent positions decreases (from 11 to only one for ciliates using UGA = stop variant code) when the set of eRF1 ciliate sequences was expanded (Lozupone et al., 2001; Inagaki et al., 2002; Kervestin et al., 2002), casting doubts on the actual role of these residues in stop codon recognition (Kervestin et al., 2001). Although most of the convergent changes found in ciliate eRF1s were located in the N-terminal domain, none involved the amino acids of the NIKS motif.
Recently, it has been shown that a combination of four substitutions distributed in two different regions of domain 1 altered the response of human eRF1 to UAA and UAG codons in an in vitro release assay (Seit-Nebi et al., 2002). The implication of NIKS and the conserved surrounding amino acids (i.e. TASNIKS heptapeptide) in the modulation of stop codon discrimination was examined using fusion between fission yeast Schizo saccharomyces pombe eRF1 and Tetrahymena eRF1, which carried KASNIKD in place of the TASNIKS heptapeptide found in eRF1 from eukaryotes with universal genetic code (Ito et al., 2002). As a result, it was shown that the TASNIKS motif alone was not sufficient for stop codon discrimination. Therefore, it was suggested that other regions of the eRF1 N-terminal domain may cooperate to modulate eRF1–stop codon interaction.
For both bacteria and eukaryotes, zero-length photocrosslinking approaches demonstrated that class 1 release factors specifically and tightly contact the invariant uridine in the first position of the stop codons within the ribosome (Brown and Tate, 1994; Chavatte et al., 2001). The synthetic mRNAs contained a close analog of uridine, the 4-thiouridine (s4U), that was able to crossreact with amino acids from ribosomal proteins, residues from ribosomal RNA (rRNA), and release factors when located nearby. In eukaryotes, the formation of the eRF-mRNA cross-link is specific for the presence of a cognate stop codon in the ribosomal A site and correlates with an efficient binding of eRF1 to the A site (Chavatte et al., 2001). In addition, the main cross-linking site was localized to the Lys63 residue of the NIKS motif in human eRF1 (Chavatte et al., 2002). These data confirmed that the N-terminal domain of eRF1 is directly involved in the interaction with stop codons in the 40S subunit decoding center, and pointed the issue of eukaryotic ‘peptide anticodon’ in which the conserved Lys63 would interact with the first base of the termination signal.
In the present work, we focused our efforts on identifying regions in eRF1 that are involved in the discrimination of the second and third bases of stop codons. We used in vitro photocrosslinking, which allowed us to show that under given conditions eRF1 cross-reacts exclusively with in-frame stop and UGG codons (Chavatte et al., 2002). We first ruled out the implication of residues at positions 35, 64, and 126 in the stop codon recognition specificity of eRF1 from ciliates that use the UGA = stop variant code. Then, to question the eukaryotic ‘peptide anticodon’ possibility, we designed a set of human–ciliate hybrid eRF1s that all contained the NIKS motif including the conserved Lys63 from either Tetrahymena thermophila or Euplotes aediculatus. Our data demonstrated that swapping the complete N-terminal domain was necessary for changing the stop codon specificity between omnipotent and ciliate eRF1s. Interestingly, we showed that Euplotes–human hybrid eRF1 still cross-link with the same efficiency to UAA and UAG, but lost their cross-linking ability towards UGA and UGG. These results led us to discuss the mechanisms of stop codon selection and discrimination.
Results
To study eRF1–stop codon interactions within the decoding site of eukaryotic ribosomes, we used the photoaffinity labeling methodology that was applied previously to study the translation termination complex (Chavatte et al., 2001, 2002). Our in vitro assay was composed of 42mer mRNA analogs, high salt-washed 80S ribosomes, in vitro transcribed yeast tRNAAsp and recombinant eRF1 proteins. The nucleobase s4U, an analog of U, was incorporated into synthetic mRNAs in the first position of a stop or control sense codon. When photoactivated, this probe cross-links with protein and nucleic acid residues located nearby. The maximum distance for a cross-link to occur with this zero length probe is estimated to 4 Å (for a review, see Favre et al., 1998). In this reconstituted in vitro translation system, the deacylated tRNAAsp located at the ribosomal P site interacts with the unique GAC codon of the mRNA analogs. The 3′ adjacent triplet is then positioned at the ribosomal A site, allowing the s4U residue to explore its environment. If photocrosslinked, eRF1, rRNA or ribosomal proteins are photoaffinity labeled with [32P]mRNA. The labeled product can be separated by denaturating acrylamide gel electrophoresis and detected by autoradiography. It was shown that the eRF1–mRNA cross-link migrated as a band of 68–70 kDa (53 kDa eRF1 plus 15 kDa mRNA). This cross-linking was dependent on the presence of the phasing tRNAAsp, and a stop or UGG codon within the ribosomal A site. Thus, this in vitro photoaffinity labeling assay should detect modifications in the eRF1–stop codon interaction due to alteration in the decoding capacity of eRF1 variants.
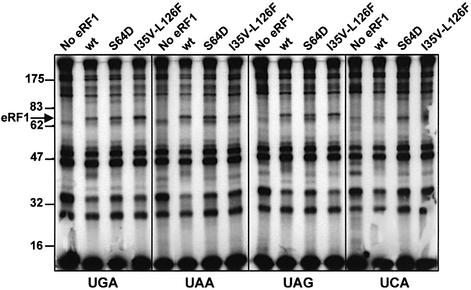
S64D and I35V-L126F substitutions did not alter the binding of human eRF1 to stop codons
It has been proposed that convergent substitutions in the N-terminal domain of eRF1 from ciliates with variant codes are involved in the modification of eRF1 pattern of stop codon recognition (Knight and Landweber, 2000; Lozupone et al., 2001; Inagaki et al., 2002). In the three-dimensional structure of eRF1, amino acid Ser64 (numbering according to human eRF1) is located at the tip of the eRF1 N-terminal domain in the NIKS motif, which was thought to be directly involved in stop codon recognition (Knight and Landweber, 2000; Nakamura et al., 2000; Song et al., 2000). Considering the alignment of all sequences available (Inagaki et al., 2002), this Ser residue is conserved in all eRF1 except in that of T.thermophila and Paramecium tetraurelia, two ciliates using the UGA = stop variant code, which have a Ser to Asp substitution, and in Trichomonas vaginalis, which has a Ser to Asn substitution. The role of Ser64 in eRF1 activity was tested using an in vitro release assay. For the three stop codons, the Ser to Asp substitution (hereafter referred as S64D) decreased eRF1 activity to ∼50% of the wild-type level (Frolova et al., 2002). The comparison of different eRF1 sequences also identified residues Ile35 and Leu126 as sites of convergent substitution (Ile to Val and Leu to Phe, respectively; hereafter referred as I35V and L126F) in eRF1 from ciliates with the UGA = stop variant code (Lozupone et al., 2001). These two residues are close to each other spatially and are located in the β-sheet forming the groove of the N-terminal domain (Song et al., 2000). It has been proposed that these residues play a role in stop codon discrimination in ciliate eRF1, but this hypothesis was not tested experimentally (Lozupone et al., 2001; Inagaki et al., 2002). We have introduced either the S64D substitution or the double I35V-L126F substitution in a C-terminally His-tagged human eRF1, which was overexpressed in E.coli and purified by Ni-agarose column chromatography. The recognition of stop codons by the wild-type and mutant proteins was tested using the cross-linking procedure described above. As shown in Figure 1, both mutant proteins yielded exactly the same pattern of cross-linking as the wild-type eRF1. In the absence of eRF1, the radiolabeled bands correspond to rRNA–mRNA and ribosomal proteins–mRNA cross-links. In the presence of wild-type and mutated eRF1s, an additional band of ∼68 kDa, corresponding to the covalently linked eRF1–mRNA complex, was detected with the three stop codons but not with the sense UCA codon, which was used as a control. Our results suggest that these convergent substitutions in ciliate eRF1 do not play a critical role in the modulation of eRF1–stop codon interaction or in the binding of eRF1 to the ribosome.
Fig. 1. Cross-linking patterns obtained with 42mer mRNA analogs containing stop codons (UAG, UAA or UAG) or a sense codon (UCA) in presence of C-terminally His-tagged human wild-type eRF1 (wt) or mutated human eRF1 containing either S64D or I35V-L126F substitutions. After irradiation, the reaction products were separated onto a 10% SDS–polyacrylamide gel and analyzed by autoradiography. A control reaction without eRF1 (No eRF1) is shown for each mRNA analog. The 68 kDa band corresponding to the eRF1–mRNA cross-link is indicated by an arrow. Molecular mass markers in kDa are indicated on the left.
Swapping of the NIKS motif region between Euplotes, Tetrahymena and human eRF1
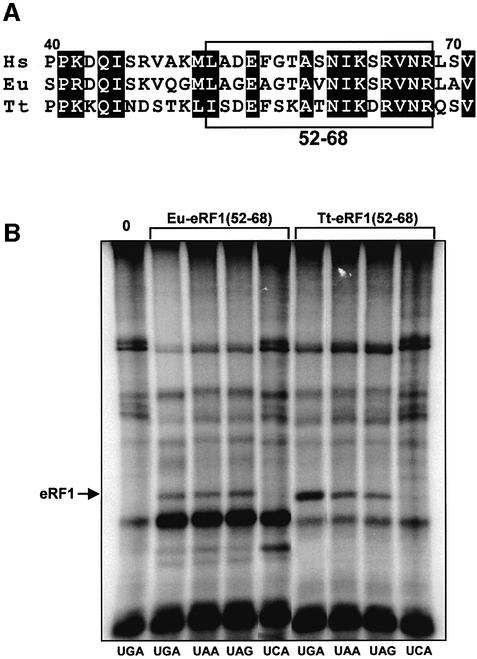
Several arguments support the involvement of the NIKS motif in stop codon discrimination: (i) the motif is located at the tip of the N-terminal domain of eRF1; (ii) it is conserved throughout evolution of eukaryotes; (iii) divergence from the NIKS sequence is found mainly in ciliates with variant genetic codes; and (iv) it has recently been shown that K (Lys63) contacts the U of stop codons. To test this hypothesis, we exchanged amino acids 52–68 of human eRF1 sequence with those from either T.thermophila eRF1 or E.aediculatus eRF1 (boxed in Figure 2A). The recombinant His-tagged eRF1s, Eu-eRF1(52–68) and Tt-eRF1(52–68), were tested for their ability to cross-link to mRNA analogs containing either one of the three stop codons or the sense UCA codon. As shown in Figure 2B, these two recombinant eRF1s cross-react with the three stop codons, but not with the sense UCA codon. For Tt-eRF1(52–68), we noticed that the yield of cross-link was higher with UGA than with UAA and UAG codons. However, the same observation was reported previously for wild-type human eRF1 (Chavatte et al., 2002), suggesting that the variation of the cross-link intensity observed on Figure 2B with Tt-eRF1(52–68) recombinant eRF1 was probably not due to an alteration of the recognition of UAA and UAG stop codons. Taken together, these results suggest that the NIKS motif and the surrounding region are not sufficient for ciliate eRF1 stop codon discrimination. Our results are also consistent with recent data showing that, in vitro, the KATNIKD sequence of T.thermophila eRF1 is not involved in restricting release activity to the UGA codon only (Ito et al., 2002).
Fig. 2. Analysis of the cross-linking patterns obtained in the presence of recombinant human eRF1 containing region 52–68 from either E.aediculatus, Eu-eRF1(52–68) or T.thermophila, Tt-eRF1(52–68). (A) Comparison of eRF1 amino acid sequences from Human (Hs, DDBJ/EMBL/GenBank accession No. P46055), E.aediculatus (Eu, accession No. AAK07830) and T.thermophila (Tt, accession No. BAA85336). The alignment is shown only for the positions 40–71. The swapped region between Euplotes, Tetrahymena and human eRF1, residues 52–68, is boxed. Identical amino acids residues are shaded in black. (B) Cross-linking patterns with 42mer mRNA analogs containing UGA, UAA, UAG, UCA codons (indicated below the autoradiogram) in the presence of recombinant eRF1, Eu-eRF1(52–68) or Tt-eRF1(52–68) as indicated above. The cross-linking pattern of the UGA mRNA analog in the absence of eRF1 is shown in lane 0. The irradiated reactions were separated on a 7.5% SDS–polyacrylamide gel. eRF1–mRNA cross-links are indicated by an arrow.
Stop codon recognition by Euplotes–human hybrid eRF1s
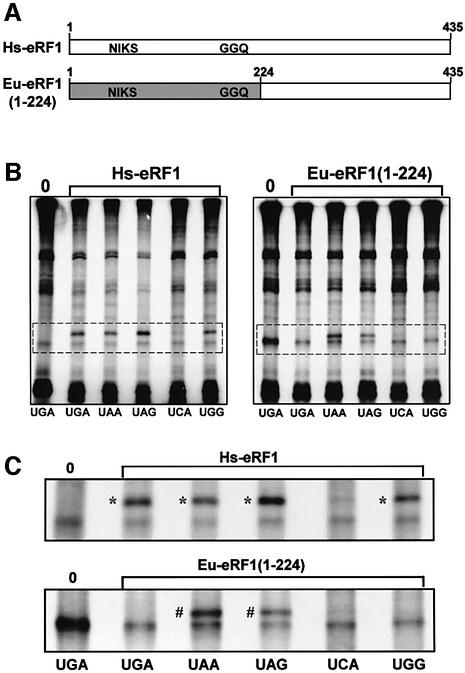
To identify the region of eRF1 involved in stop codon discrimination, we constructed hybrid eRF1s in which regions of human eRF1 were swapped for equivalent regions of Euplotes eRF1. The recombinant genes were constructed using existing restriction sites of either Euplotes or human eRF1 gene so that the encoded sequence at the border of the swapped regions is conserved (see Materials and methods). First, we generated a hybrid eRF1, named Eu-eRF1(1–224), which contains the N-terminal domain and a portion of the middle domain including the GGQ motif from Euplotes eRF1, and the remaining sequence from human eRF1 (Figure 3A). The cross-linking activities of human eRF1 (Hs-eRF1) and Eu-eRF1(1–224) to stop codons and to UGG and UCA sense codons were compared. Figure 3B shows the total cross-linking patterns in these experiments and Figure 3C shows an enlargement of the region of the autoradiogram that contains the mRNA–eRF1 cross-link bands. Confirming our previous results (Chavatte et al., 2002), human eRF1 cross-reacted with all three stop codons, with the UGG codon, but not with the UCA codon (Figure 3B and C). The cross-linking of Hs-eRF1 to UGG suggested that the mechanism of stop codon recognition by eRF1 was less stringent than was previously reported based on the analysis of eRF1 release activity (Frolova et al., 1994). In contrast, the hybrid Eu-eRF1(1–224) cross-linked only with UAA and UAG stop codons (Figure 3C). The absence of a cross-link with UGA was expected as Euplotes uses UGA as a cysteine codon. Interestingly, there was no cross-link with UGG, which suggests that Euplotes eRF1 differs from Hs-eRF1 in its ability to discriminate between A and G at the second positions of the stop codon.
Fig. 3. Comparison of the cross-linking pattern of human eRF1 (Hs-eRF1) with recombinant Eu-eRF1(1–224). Eu-eRF1(1–224) contains residues 1–224 from E.aediculatus eRF1 and residues 225–435 from human eRF1. (A) Schematic representation of the amino acid sequences of Hs-eRF1 and recombinant Eu-eRF1(1–224). The approximate locations of the NIKS (domain 1) and GGQ (domain 2) motifs are indicated. The region of Euplotes eRF1 in Eu-eRF1(1–224) is shaded in light gray. (B) Cross-linking patterns of 42mer mRNA analogs containing UGA, UAA, UAG, UCA or UGG codons (as indicated below the autoradiogram) in the presence of Hs-eRF1 or Eu-eRF1(1–224) as indicated above the autoradiograms. The cross-linking pattern of the UGA mRNA analog in the absence of eRF1 is shown in lane 0. The irradiated reactions were analyzed by 7.5% SDS–PAGE. The region containing the eRF1–mRNA cross-links is boxed with broken line. (C) Enlargement views of regions boxed with broken lines in (B). Cross-links between mRNA analogs containing the canonical stop (UGA, UAA, UAG) or sense (UGG and UCA) codons as indicated below the autoradiograms and Hs-eRF1 (upper panel) or recombinant Eu-eRF1(1–224) (lower panel). The cross-linking pattern of the UGA mRNA analog in the absence of eRF1 is shown in lane 0. An asterisk indicates a Hs-eRF1–mRNA cross-link and a hash symbol indicates a Eu-eRF1(1–224)–mRNA cross-link.
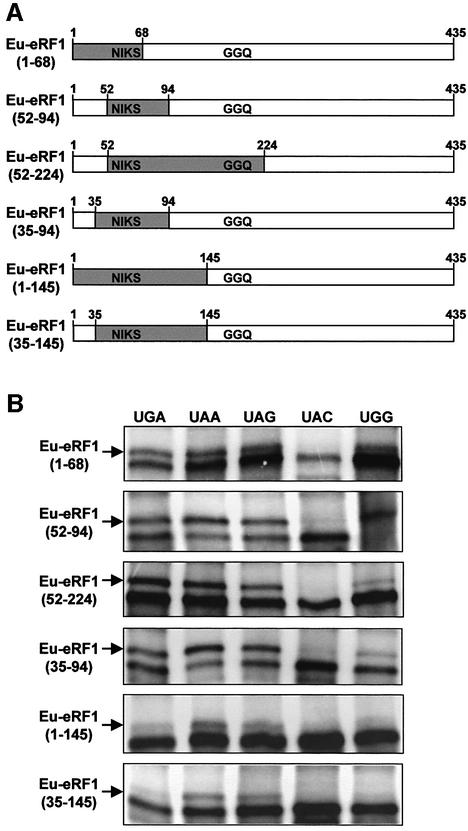
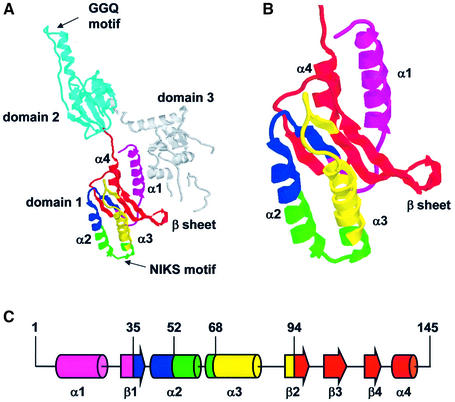
To further define the region of Euplotes eRF1 involved in discrimination, several hybrid eRF1s (Figure 4A) were tested for their ability to interact with the same set of stop and sense codons. All these hybrids had the NIKS motif region (amino acids 52–68) of Euplotes eRF1 in common. As shown in Figure 4B, the hybrid Eu-eRF1(1–68), which contained the N-terminal portion of Euplotes eRF1 extending to the NIKS motif, exhibited the same pattern of cross-link as wild-type human eRF1, i.e. a cross-link with the three stop codons and with UGG. The two hybrids beginning at the NIKS region (position 52) of Euplotes eRF1 and extending either to amino acid 94 or to amino acid 224 (including the GGQ motif), namely Eu-eRF1(52–94) and Eu-eRF1(52–224), also exhibited a wild-type human eRF1 cross-link pattern (Figure 4B). However, these three hybrid eRF1s, Eu-eRF1(1–68), Eu-eRF1(52–94) and Eu-eRF1(52–224), contained only a part of the α2-helix–loop–α3-helix (hereafter referred as α2–loop–α3) structure of Euplotes eRF1 (Figure 5). Since the α2–loop–α3 of eRF1 was proposed to mimic the anticodon arm of tRNA (Knight and Landweber, 2000; Nakamura et al., 2000; Song et al., 2000), we constructed two hybrids that contained the sequence of Euplotes eRF1 extending to the extreme N-terminus or to the amino acid at position 35, namely Eu-eRF1(1–94) and Eu-eRF1(35–94). The sequence from position 1 to 35 contains the first α-helix of eRF1 domain 1. In the three-dimensional structure of eRF1, this helix α1 is located at the interface with the C-terminal domain (domain 3). The helix α1 does not participate in the formation of the ‘pseudo anticodon arm’ or in the groove of the domain 1, which is composed of a four-stranded β-sheet (β1-β2-β3-β4) surrounded on both sides by helices α2 and α3 (Figure 5). Extension of the Euplotes eRF1 sequence to the N-terminal region in hybrids Eu-eRF1(1–94) and Eu-eRF1(35–94) did not restore the discriminating potential observed with Eu-eRF1(1–224)—only shown for Eu-eRF1(35–94) in Figure 4B. Two hybrids were constructed that contained either the entire domain 1 of Euplotes eRF1, i.e. from the N-terminus to the hinge connecting domain 1 to domain 2, or the domain 1 lacking the 35 first amino acids, Eu-eRF1(1–145) and Eu-eRF1(35–145) respectively (Figure 4A). As shown in Figure 4B, hybrids Eu-eRF1(1–145) and Eu-eRF1(35–145) efficiently cross-linked to UAA and UAG stop codons. However, for both hybrids, a faint cross-link was reproducibly observed with UGA, suggesting the existence of a weak interaction with this codon. Interestingly, these two hybrids did not cross-react with a UGG codon. Taken together, these observations suggest that: (i) the discriminating potential of Euplotes eRF1 was restored by the addition of its entire N-terminal domain to the hybrid; and (ii) the discrimination towards UGA was linked to the discrimination towards UGG.
Fig. 4. Localization of the Euplotes eRF1 region implicated in stop codon discrimination. (A) Schematic representation of the human– Euplotes hybrid eRF1s constructed. The regions of Euplotes eRF1 are shaded in light gray. The approximate locations of the NIKS and GGQ motifs are indicated. Numbering is according to human eRF1 amino acids sequence. (B) mRNA–eRF1 cross-links obtained for the recombinant eRF1s are indicated on the left. The 42mer mRNA analogs containing the canonical stop (UGA, UAA, UAG) or sense (UAC, UGG) codons are indicated. The irradiated reactions were analyzed by 7.5% SDS–PAGE. Only the mRNA–eRF1 cross-linking regions of the autoradiograms are shown (as in Figure 3C), and the eRF1–mRNA cross-links are marked by an arrow.
Fig. 5. The structure of eRF1. (A) Crystal structure of human eRF1 with the ribbon representation of the secondary structure. Major domains and secondary structure elements are labeled. Functionally important motifs are indicated by an arrow with the one letter amino acid code (GGQ and NIKS). In domain 1, the different colors indicate the regions of human eRF1 substituted by regions from Euplotes eRF1: amino acids 1–34 are colored pink, amino acids 35–51 are in dark blue, amino acids 52–68 are in green, amino acids 69–94 are in yellow and amino acids 95–145 are in red. Domain 2 is colored light blue and domain 3 is in gray. The coordinate data were obtained from the Protein Data Bank (accession code ccss 1TD9). (B) Enlargement of domain 1. (C) Linear representation of domain 1 (residues 1–145). The regions of human eRF1 swapped for Euplotes eRF1 are represented in cylinders (α-helices) and large arrows (β-strands) using the same colors as in (A). The positions of the junctions (35, 52, 68, 94) are indicated.
Discussion
One of the major questions regarding the mechanism of translation termination is how RFs recognize stop codons and how they discriminate stop codons from sense codons. This question was investigated in eubacteria by switching the recognition specificity of RF1 and RF2 (Ito et al., 2000; Nakamura and Ito, 2002). Using domain swapping and mutagenesis studies, it was shown that a tripeptide conserved among bacterial RFs determines release factor specificity. Based on these results, it was proposed that the discriminator tripeptides are functionally equivalent to the anticodons of tRNAs. In eukaryotes, eRF1 from ciliates with variant genetic codes provides an interesting tool to solve this problem. Indeed, it has been shown using an in vitro release assay based on mammalian ribosomes that eRF1s from Euplotes and Tetrahymena do not respond to stop codons when they are used as sense codons by these organisms (Kervestin et al., 2001; Ito et al., 2002). Furthermore, genetic approaches and site-directed mutagenesis experiments have localized the recognition site to the N-terminal domain of eRF1 (Bertram et al., 2000; Frolova et al., 2002; Ito et al., 2002; Seit-Nebi et al., 2002), thus confirming the analysis of the crystal structure of eRF1 by Song et al. (2000). However, at the molecular level, complementation tests in yeast and in vitro release activity assays cannot distinguish between different steps in termination, including binding of eRF1 to the ribosome, activation of the peptidyl transferase center or interaction of eRF1 with the stop codons. In fact, these kind of studies gave contradictory results in the case of Tetrahymena eRF1 (Ito et al., 2002). As a first step towards understanding stop codon selection in eukaryotes, we recently mapped the region of human eRF1 interacting with the U of the stop codons using a photocrosslinking assay (Chavatte et al., 2002). In this study, we used this photoaffinity method to localize the regions of ciliate eRF1s that are involved in stop codon discrimination.
Because multiple sequence alignment studies have pointed out the potential role of conserved substitutions of ciliate eRF1 sequences in stop codon recognition (Inagaki and Doolittle, 2001; Lozupone et al., 2001; Inagaki et al., 2002), we first analyzed the binding capacities of human eRF1 mutants carrying either the S64D or the I35V-L126F substitutions. We found that these substitutions do not alter the interaction of eRF1 with any of the three stop codons. This finding strongly suggests that these substitutions are not directly responsible for the changes in ciliate eRF1s stop codon specificity, and hence, that residues Ile35, Ser64 and Leu126 are probably not the major determinants in the interaction between eRF1 and the second and third bases of the stop codon. However, for the S64D mutated eRF1, there is a discrepancy between the presence of a cross-link with the three stop codons (Figure 1) and the alteration of release activity in the in vitro release assay (Frolova et al., 2002). The same discrepancy was reported for the I62M mutation (Chavatte et al., 2002). Taking into account that the Ile62 and Ser64 residues of human eRF1 are adjacent to the conserved Lys63 that contacts the U of the stop codons, we can hypothesize, following other lines of arguments based on structural data (Inagaki et al., 2002), that residues at positions 62 and 64 could be involved in interacting with ribosomal components of the decoding site rather than with the bases of the stop codon. These interactions could be important to relay signals which activate the peptidyl transferase center of the ribosome. In this case, the decrease of eRF1 activity when these residues are modified could be the result of the loss of the activation signal.
The amino acids Ile62 and Lys63 are among the most conserved residues in eRF1. In contrast, the surrounding region is well conserved in eukaryotes with the standard genetic code, but highly divergent in ciliate with variant codes. In the structure of eRF1, this region constitutes the α2–loop–α3 structural motif speculated to mimic the anticodon loop of tRNA (Figure 5). The pseudo anticodon loop model suggests that residues within the α2–loop–α3 region are directly involved in stop codon recognition and function as discriminator residues in variant code eRF1s. Several lines of experimental evidences cast doubt on this model. For example, swapping of this region with that of either Tetrahymena or Euplotes eRF1 does not modify the stop codon specificity of human eRF1 (Figure 3). Similarly, it has been shown by Ito et al. (2002) that, in vitro, replacement of the Tetrahymena KADNIKD sequence with the standard code eRF1 TASNIKS sequence did not alter the UGA-only release activity of Tetrahymena–Schizosaccharomyces pombe hybrid eRF1. Thus, we agree with these authors that the specificity of stop codon recognition may require other structural elements of eRF1. The analysis of the binding capacities of Euplotes–human hybrid eRF1s also confirms this hypothesis. Indeed, the restriction of stop codon recognition to UAA and UAG, a characteristic of Euplotes eRF1 supported by in vitro release assays (Kervestin et al., 2001), was obtained only when the entire N-terminal domain of Euplotes eRF1 was present (Figure 4). One possibility is that, in addition to the conserved residues of the NIKS motif which contact the U of the stop codon, the discriminating potential of Euplotes eRF1 requires amino acid residues located near the beginning of domain 1 (in region 35–52), as well as other residues near the end (in region 94–145). As shown in Figure 5, these two linear sequences are distant in primary structure but close in the tertiary structure. The fact that the region from residue 35 to 52 is important for discrimination (Figure 4) is intriguing because this region is not conserved among all organisms, particularly in eRF1 from protists with the standard code (for an extensive eRF1 sequence alignment, see Inagaki et al., 2002). These observations argue against the simple explanation that the small number of conserved residues that are sufficient for the interaction with stop codons are also sufficient for discrimination. Furthermore, with the exception of the very N-terminal region that forms an α helix at the interface with the C-terminal domain, the integrity of all other structures, i.e. the two helices α2 and α3 packing the four-stranded antiparallel β-sheet (Figure 5), seems to be absolutely required to restore Euplotes eRF1 specificity. Therefore, we assume that the discriminating potential of Euplotes eRF1 is determined by the structure itself rather than discrete amino acid residues distributed along the N-terminal domain. The preservation of a discriminating structure could involve the residues that interact with the bases of the stop codons as well as residues implicated in domain conformation.
In this report, we attempted to further analyze our puzzling finding that human eRF1 interacts with UGG codon (Chavatte et al., 2002). In examining the cross-linking activity of Euplotes–human eRF1 hybrids, we observed that the absence of interaction with UGG seems to correlate with the absence of interaction with UGA. To interpret these data, we assumed, following the hypothesis proposed by Yoshimura et al. (1999) for translation termination in bacteria, that the mechanism of stop codon selection by eRF1 parallel the sequence of interactions leading to the selection of the cognate aminoacyl–tRNA during the elongation cycle. In this case, the ternary complex EF-Tu·GTP·aa-tRNA binds first to the ribosomal A site, irrespective of the codon located in this site. This is followed by a reversible step of codon recognition (first step of selection), which induces conformational changes in the ternary complex and the irreversible step of GTP hydrolysis. Subsequently, the aminoacyl-tRNA is accomodated in the A site (second step of selection or proofreading step) and takes part in the peptidyl-transferase reaction (Pape et al., 1998). In the sequence of interactions between eRF1 and the ribosome, the initial binding of eRF1, which is independent of the nature of the codon in the A site, is followed by codon recognition, which supposes an interaction between eRF1 and the A site codon. This selection step is essential for the accuracy of translation termination. The data presented here, together with the data of a previous photocrosslinking study (Chavatte et al., 2002), show that during this step eRF1 accurately discriminates between U-purine-purine codons and other sense codons. Our experiments with Euplotes–human hybrid eRF1s suggest that the discriminating potential of Euplotes eRF1 is: (i) restricted to UA-purine codons; and (ii) requires more than the N-terminal domain since a faint band of eRF1-UGA cross-link is reproducibly observed with Eu-eRF1(1–145) and Eu-eRF1(35–145) hybrids (Figure 4B), but not with the Eu-eRF1(1–224) hybrid (Figure 3C). Thus, an appropriate eRF1 conformation is of paramount importance for the accuracy of discrimination. Does the selection step induces conformational changes in the interacting components? The modification of the mRNA–rRNA cross-linking pattern when eRF1 is added to the reaction strongly argues for rearrangements within the ribosome decoding site (Chavatte et al., 2001). Furthermore, conformational changes in eRF1 have been speculated (Chavatte et al., 2002; Ehrenberg and Tenson, 2002), because the distance between the tip of the N-terminal domain and the GGQ motif, which is thought to interact with the peptidyl tranferase center, is larger than the distance between the decoding site of the small ribosomal subunit and the peptidyl tranferase center (100 Å versus 75 Å). Thus, we assume that interactions with U-purine-purine codons stabilize eRF1 in a conformation that allows it to proceed to the next step, which likely increases selectivity allowing the discrimination of UGG from stop codons. Given that eRF3 strongly interacts with eRF1 and that its GTPase activity increases the sensitivity of the in vitro release assay by a factor of 10 (Zhouravleva et al., 1995), eRF3 is a good candidate to promote the transition towards the next step of selection. However, very little is known about the role of eRF3 GTPase activity. Understanding the whole translation termination process will require additional information on the complex set of interactions connecting the participants.
Materials and methods
Gene manipulation, site-directed mutagenesis and DNA fragment amplification
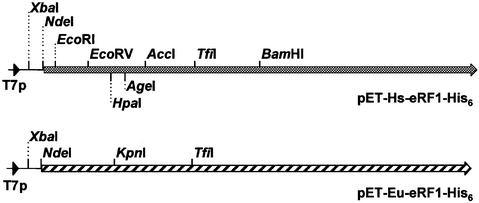
All DNA engineering was carried out using standard protocols (Sambrook et al., 1989). Site-directed mutagenesis was performed using the ExSite PCR-based site-directed mutagenesis kit (Stratagene). The presence of the correct mutation was verified by DNA sequencing. The plasmid pET-Eu-eRF1-His6 (Figure 6) containing the E.aediculatus eRF1 gene in pET21b was described previously (Kervestin et al., 2001). The human eRF1 ORF (from the second GCG codon to the last TAC codon) was amplified by PCR using plasmid pCMVhRF1 (Le Goff et al., 1997) as a template and a pair of appropriate oligonucleotides containing EcoRI or HindIII restriction sites at the 5′ end. The resulting PCR fragment was digested by EcoRI and HindIII and cloned into the EcoRI–HindIII sites of pET21b (Novagen). The final construct, named pET-Hs-eRF1-His6 (Figure 6), contains the human eRF1 ORF followed by a His tag sequence under control of T7 promoter. Note that in this contruct, the human eRF1 ORF is preceded by 16 codons resulting from the fusion with pET21b sequence. Mutations changing Ser64 to Asp (S64D; numbering is according to the human eRF1 protein sequence), Ile35 to Val (I35V) and Leu126 to Phe (L126F) were introduced in plasmid pET-Hs-eRF1-His6 using site-directed mutagenesis and the appropriate oligonucleotides. The plasmid containing the double mutation I35V-L126F was constructed by inserting the XbaI–AccI fragment of the I35V mutant of pET-Hs-eRF1-His6 in place of the corresponding XbaI–AccI fragment in the L126F mutant of pET-Hs-eRF1-His6. Plasmids expressing a recombinant human eRF1 containing amino acids 52–68 of either T.thermophila or E.aediculatus eRF1 (Figure 2A) were constructed in a two step procedure. First, HpaI and AgeI restriction sites were introduced by site-directed mutagenesis at the positions of plasmid pET-Hs-eRF1-His6 encoding amino acids 51 and 67 of eRF1, respectively (Figure 6). Then, two pairs of 50mer oligonucleotides encoding amino acids 52–68 of either T.thermophila or E.aediculatus were synthesized. After annealing the complementary oligonucleotides, the short DNA fragment, having a blunt end at one extremity and an AgeI cohesive end at the other extremity, was ligated to the modified pET-Hs-eRF1-His6 digested with HpaI and AgeI. The resulting plasmids were named pET-Tt-eRF1(52–68) and pET-Eu-eRF1(52–68). Plasmid pET-Eu-eRF1(1–224) expressing a chimeric eRF1 protein containing the N-terminal part of pET-Eu-eRF1(amino acids 1–224) and the C-terminal part of human eRF1 (amino acids 225–435) was constructed using a PCR strategy. The E.aediculatus eRF1 sequence was amplified by PCR using pET-Eu-eRF1-His6 as a template and a pair of appropriate oligonucleotides, with either XbaI or BamHI sites at the 5′ end. The PCR fragment was XbaI–BamHI digested and introduced into XbaI–BamHI-digested pET-Hs-eRF1-His6. Plasmids in which regions of human eRF1 ORF were swapped for the equivalent regions of E.aediculatus eRF1 gene were constructed using a three-fragment-ligation strategy and unique restriction sites present in E.aediculatus eRF1 ORF (KpnI and TfiI) and in human eRF1 ORF (EcoRV, AccI, TfiI and BamHI), as shown in Figure 6. Swapped fragments were introduced into the XbaI–BamHI digested parent plasmid, pET-Hs-eRF1-His6. Note that plasmid pET-Eu-eRF1(1–224) contains the unique BamHI site of human eRF1 ORF, and plasmid pET-Eu-eRF1(52–68) contains the unique KpnI site of the E.aediculatus eRF1 gene. Plasmids containing swapped regions were named taking into account the amino acids of E.aediculatus eRF1 inserted in human eRF1. Plasmid pET-Eu-eRF1(1–68) was constructed with the XbaI–KpnI fragment of pET-Eu-eRF1(1–224) and the KpnI–BamHI fragment of pET-Eu-eRF1(52–68). Plasmid pET-Eu-eRF1(52–224) was generated with the XbaI–KpnI fragment of pET-Eu-eRF1(52–68) and the KpnI–BamHI fragment of pET-Eu-eRF1(1–224). Plasmid pET-Eu-eRF1(1–145) was constructed with the XbaI–TfiI fragment of pET-Eu-eRF1(1–224) and the TfiI–BamHI fragment of pET-Hs-eRF1-His6. Plasmid pET-Eu-eRF1(35–145) was generated using a PCR strategy. The region of the E.aediculatus eRF1 gene encoding amino acids 35–145 was amplified by PCR using pET-Eu-eRF1(1–145) as a template and a pair of appropriate oligonucleotides, with either EcoRV or BamHI sites at the 5′ end. The PCR fragment was EcoRV–BamHI digested and introduced with the XbaI–EcoRV short fragment of pET-Hs-eRF1-His6 into XbaI–BamHI-digested pET-Hs-eRF1-His6. Plasmid pET-Eu-eRF1(1–94) was constructed using a PCR fragment amplified with pET-Eu-eRF1(1–224) as the template and a pair of appropriate oligonucleotides, with either XbaI or AccI sites at the 5′ end and the AccI–BamHI 400 bp fragment of pET-Hs-eRF1-His6, which were inserted into XbaI–BamHI-digested pET-Hs-eRF1-His6. Plasmid pET-Eu-eRF1(35–94) was constructed by inserting the XbaI–KpnI fragment of pET-Eu-eRF1(35–145) and the KpnI–BamHI fragment of pET-Eu-eRF1(1–94) into XbaI–BamHI-digested pET-Hs-eRF1-His6. Plasmid pET-Eu-eRF1(52–94) was constructed by inserting the XbaI–KpnI fragment of pET-Eu-eRF1(52–68) and the KpnI–BamHI fragment of pET-Eu-eRF1(1–94) into XbaI–BamHI-digested pET-Hs-eRF1-His6.
Fig. 6. Schematic representation of plasmid pET21b derivatives expressing C-terminally His-tagged eRF1 from human (pET-Hs-eRF1-His6) or Euplotes (pET-Eu-eRF1-His6) under the control of the T7 promoter (T7p). The restrictions sites used for the construction of the recombinant eRF1s are indicated. The restriction sites indicated under pET-Hs-eRF1-His6 were introduced by site-directed mutagenesis.
Expression and purification of eRF1s
The full-length human eRF1, human eRF1 mutants and chimeric proteins carrying portions of E.aediculatus eRF1 were expressed in E.coli strain BL21(DE3), and purified using Ni-NTA resin, Superflow (Qiagen), as described previously (Kervestin et al., 2001).
Synthesis of s4U-modified RNAs containing oligonucleotides used as mRNAs
Oligonucleotides used as mRNAs were synthesized by in vitro transcription of synthetic DNA templates (Genset) with T7 RNA polymerase using the procedure described in Chavatte et al. (2001). All 42mer mRNAs contained a GAC triplet (encoding for Asp) followed by either a stop (UGA, UAA or UAG) or a sense (UCA, UAC or UGG) codon. They were constructed on the following sequence: 5′-GGGAGAAAAAAGAAAGAAGACs4UNNAAGAAAAAAAAGAAAAA-3′ and named regarding the s4UNN codon (N stands for G, A or C) located in the A site. The nucleotide triphosphate mixture was composed of ATP, GTP, CTP and s4UTP (Amersham Pharmacia Biotech). Replacement of UTP by s4UTP allowed incorporation of the s4U probe in the single position available. These mRNAs were then 5′ end labeled with [γ-32P]ATP (ICN) by T4 polynucleotide kinase, separated by electrophoresis on a 15% polyacrylamide gel containing 7 M urea, and purified by elution and ethanol precipitation.
Ribosomes and yeast tRNAAsp
Isolation of 80S ribosomes from rabbit reticulocyte lysate was performed as reported previously (Chavatte et al., 2001). Standard 80S ribosomes were washed through a 20% sucrose cushion containing 0.5 M KCl (‘high salt-washed’ ribosomes). Yeast tRNAAsp was obtained by in vitro transcription (16 h at 37°C) of a BstNI linearized pUC119 derivative containing the yeast tRNAAsp gene immediately downstream to the T7 RNA polymerase promoter (Frugier et al., 1993).
Cross-linking procedures
Our standard in vitro system was composed of 0.1 µM of 32P-labeled mRNA, 2 µM tRNAAsp, 0.2 µM of high salt-washed 80S ribosomes and 6 µM of eRF1 in a final volume of 10 µl. The reaction mixture contained 50 mM Tris–HCl pH 7.4, 100 mM KCl, 10 mM MgCl2 and 1 mM dithiothreitol. Each sample was incubated for 1 h at 37°C, then placed into a siliconized glass capillary and irradiated for 30 min at 4°C. The light source was a HBO 150 W superpressure mercury lamp placed at 5 cm distance from the sample, providing near ultraviolet light (>320 nm) since shorter wavelengths were removed using a MTO J320A filter. These conditions allowed completion of the cross-linking reactions (t1/2 = 6 min). The irradiated sample was diluted in dye buffer and resolved by electrophoresis on 10 or 7.5% SDS–polyacrylamide gel. The gel was dried and analyzed using a PhosphorImager (Molecular Dynamics). Prestained molecular weight markers were run in parallel.
Acknowledgments
Acknowledgements
We thank R.Giégé and A.Theobald-Dietrich for the kind gift of the plasmid used to obtain the yeast tRNAAsp, and W.C.Merrick and Ludmila Frolova for the generous gift of rabbit ribosomes. We thank D.M.Driscoll, who allowed the completion of this work in her laboratory and who carefully read the manuscript. O.J.-J. was supported by the Association Française contre les Myopathies and by the Association pour la Recherche sur le Cancer (grant no. 5511). L.C. and S.K. hold fellowships from the French Ministère de la Recherche et de l’Enseignement Supérieur (MENESR) and from the Fondation pour la Recherche Médicale.
References
- Bertram G., Bell,H.A., Ritchie,D.W., Fullerton,G. and Stansfield,I. (2000) Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA, 6, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.M. and Tate,W.P. (1994) Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J. Biol. Chem., 269, 33164–33170. [PubMed] [Google Scholar]
- Caron F. and Meyer,E. (1985) Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature, 314, 185–188. [DOI] [PubMed] [Google Scholar]
- Chavatte L., Frolova,L., Kisselev,L. and Favre,A. (2001) The polypeptide chain release factor eRF1 specifically contacts the s4UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem., 268, 2896–2904. [DOI] [PubMed] [Google Scholar]
- Chavatte L., Seit-Nebi,A., Dubovaya,V. and Favre,A. (2002) The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J., 21, 5302–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugeon G., Jean-Jean,O., Frolova,L., Le Goff,X., Philippe,M., Kisselev,L. and Haenni,A.L. (1997) Eukaryotic release factor 1 (eRF1) abolishes readthrough and competes with suppressor tRNAs at all three termination codons in messenger RNA. Nucleic Acids Res., 25, 2254–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M. and Tenson,T. (2002) A new beginning of the end of translation. Nat. Struct. Biol., 9, 85–87. [DOI] [PubMed] [Google Scholar]
- Eurwilaichitr L., Graves,F.M., Stansfield,I. and Tuite,M.F. (1999) The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol., 32, 485–496. [DOI] [PubMed] [Google Scholar]
- Favre A., Saintome,C., Fourrey,J.L., Clivio,P. and Laugaa,P. (1998) Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid–protein interactions. J. Photochem. Photobiol. B, 42, 109–124. [DOI] [PubMed] [Google Scholar]
- Frolova L. et al. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature, 372, 701–703. [DOI] [PubMed] [Google Scholar]
- Frolova L.Y., Tsivkovskii,R.Y., Sivolobova,G.F., Oparina,N.Y., Serpinsky,O.I., Blinov,V.M., Tatkov,S.I. and Kisselev,L.L. (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA, 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L., Seit-Nebi,A. and Kisselev,L. (2002) Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA, 8, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier M., Florentz,C., Schimmel,P. and Giege,R. (1993) Triple aminoacylation specificity of a chimerized transfer RNA. Biochemistry, 32, 14053–14061. [DOI] [PubMed] [Google Scholar]
- Grimm M., Brunen-Nieweler,C., Junker,V., Heckmann,K. and Beier,H. (1998) The hypotrichous ciliate Euplotes octocarinatus has only one type of tRNACys with GCA anticodon encoded on a single macronuclear DNA molecule. Nucleic Acids Res., 26, 4557–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu N., Kuchino,Y., Susumu,N. and Beier,H. (1986) Dramatic events in ciliate evolution: alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs Gln. EMBO J., 5, 1307–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper D.S. and Jahn,C.L. (1989) Differential use of termination codons in ciliated protozoa. Proc. Natl Acad. Sci. USA, 86, 3252–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y. and Doolittle,W.F. (2001) Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res., 29, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y., Blouin,C., Doolittle,W.F. and Roger,A.J. (2002) Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: the evolution of stop codon specificity. Nucleic Acids Res., 30, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Ebihara,K., Uno,M. and Nakamura,Y. (1996) Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA–protein mimicry hypothesis. Proc. Natl Acad. Sci. USA, 93, 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Ebihara,K. and Nakamura,Y. (1998) The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA, 4, 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Uno,M. and Nakamura,Y. (2000) A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Ito K., Frolova,L., Seit-Nebi,A., Karamyshev,A., Kisselev,L. and Nakamura,Y. (2002) Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl Acad. Sci. USA, 99, 8494–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamyshev A.L., Ito,K. and Nakamura,Y. (1999) Polypeptide release factor eRF1 from Tetrahymena thermophila: cDNA cloning, purification and complex formation with yeast eRF3. FEBS Lett., 457, 483–488. [DOI] [PubMed] [Google Scholar]
- Kervestin S., Frolova,L., Kisselev,L. and Jean-Jean,O. (2001) Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO rep., 2, 680–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S., Garnier,O.A., Karamyshev,A., Ito,K., Nakamura,Y., Meyer,E. and Jean-Jean,O. (2002) Isolation and expression of two genes encoding eukaryotic release factor 1 from Paramecium tetraurelia. J. Eukaryot. Microbiol., 49, 374–382. [DOI] [PubMed] [Google Scholar]
- Kisselev L.L. and Buckingham,R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- Klaholz B.P., Pape,T., Zavialov,A.V., Myasnikov,A.G., Orlova,E.V., Vestergaard,B., Ehrenberg,M. and van Heel,M. (2003) Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature, 421, 90–94. [DOI] [PubMed] [Google Scholar]
- Knight R.D. and Landweber,L.F. (2000) The early evolution of the genetic code. Cell, 101, 569–572. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu,N., Tashiro,F. and Nishimura,S. (1985) Tetrahymena thermophila glutamine tRNA and its gene that corresponds to UAA termination codon. Proc. Natl Acad. Sci. USA, 82, 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X., Philippe,M. and Jean-Jean,O. (1997) Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol. Cell. Biol., 17, 3164–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang A., Brunen-Nieweler,C., Muramatsu,T., Kuchino,Y., Beier,H. and Heckmann,K. (2001) The ciliate Euplotes octocarinatus expresses two polypeptide release factors of the type eRF1. Gene, 262, 161–168. [DOI] [PubMed] [Google Scholar]
- Lozupone C.A., Knight,R.D. and Landweber,L.F. (2001) The molecular basis of nuclear genetic code change in ciliates. Curr. Biol., 11, 65–74. [DOI] [PubMed] [Google Scholar]
- Merkulova T.I., Frolova,L.Y., Lazar,M., Camonis,J. and Kisselev,L.L. (1999) C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett., 443, 41–47. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Heckmann,K., Kitanaka,C. and Kuchino,Y. (2001) Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett., 488, 105–109. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. and Ito,K. (2002) A tripeptide discriminator for stop codon recognition. FEBS Lett., 514, 30–33. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito,K. and Ehrenberg,M. (2000) Mimicry grasps reality in translation termination. Cell, 101, 349–352. [DOI] [PubMed] [Google Scholar]
- Pape T., Wintermeyer,W. and Rodnina,M.V. (1998) Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E.coli ribosome. EMBO J., 17, 7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preer J.R. Jr, Preer,L.B., Rudman,B.M. and Barnett,A.J. (1985) Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature, 314, 188–190. [DOI] [PubMed] [Google Scholar]
- Rawat U.B., Zavialov,A.V., Sengupta,J., Valle,M., Grassucci,R.A., Linde,J., Vestergaard,B., Ehrenberg,M. and Frank,J. (2003) A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature, 421, 87–90. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Seit-Nebi A., Frolova,L. and Kisselev,L. (2002) Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO rep., 3, 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Mugnier,P., Das,A.K., Webb,H.M., Evans,D.R., Tuite,M.F., Hemmings,B.A. and Barford,D. (2000) The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Stansfield I. et al. (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J., 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno M., Ito,K. and Nakamura,Y. (2002) Polypeptide release at sense and noncognate stop codons by localized charge-exchange alterations in translational release factors. Proc. Natl Acad. Sci. USA, 99, 1819–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard B., Van,L.B., Andersen,G.R., Nyborg,J., Buckingham,R.H. and Kjeldgaard,M. (2001) Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell, 8, 1375–1382. [DOI] [PubMed] [Google Scholar]
- Yoshimura K., Ito,K. and Nakamura,Y. (1999) Amber (UAG) suppressors affected in UGA/UAA-specific polypeptide release factor 2 of bacteria: genetic prediction of initial binding to ribosome preceding stop codon recognition. Genes Cells, 4, 253–266. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova,L., Le Goff,X., Le Guellec,R., Inge-Vechtomov,S.G., Kisselev,L. and Philippe,M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]