Abstract
In most organisms, tRNA aminoacylation is ensured by 20 aminoacyl-tRNA synthetases (aaRSs). In eubacteria, however, synthetases can be duplicated as in Thermus thermophilus, which contains two distinct AspRSs. While AspRS-1 is specific, AspRS-2 is non-discriminating and aspartylates tRNAAsp and tRNAAsn. The structure at 2.3 Å resolution of AspRS-2, the first of a non-discriminating synthetase, was solved. It differs from that of AspRS-1 but has resemblance to that of discriminating and archaeal AspRS from Pyrococcus kodakaraensis. The protein presents non-conventional features in its OB-fold anticodon-binding domain, namely the absence of a helix inserted between two β-strands of this fold and a peculiar L1 loop differing from the large loops known to interact with tRNAAsp identity determinant C36 in conventional AspRSs. In AspRS-2, this loop is small and structurally homologous to that in AsnRSs, including conservation of a proline. In discriminating Pyrococcus AspRS, the L1 loop, although small, lacks this proline and is not superimposable with that of AspRS-2 or AsnRS. Its particular status is demonstrated by a loop-exchange experiment that renders the Pyrococcus AspRS non-discriminating.
Keywords: archaea/asparaginyl-tRNA synthetase/aspartyl-tRNA synthetase/eubacteria/X-ray crystallography
Introduction
Aminoacyl-tRNA synthetases (aaRSs) constitute a family of enzymes of structural and functional diversity (Martinis et al., 1999; Ibba et al., 2003). Their pivotal role is in protein synthesis where they ensure translation of the genetic code into proteins. Correct translation relies on specific charging by each of the 20 synthetases of the cognate isoaccepting tRNAs with the homologous amino acid (First, 1998; Ibba and Söll, 2000). However, exceptions to the rule of unity of tRNA aminoacylation systems, namely one synthetase for each of the 20 amino acids, appeared with the discovery in some organisms of duplicated or missing synthetases (Becker and Kern, 1998; Ibba and Söll, 2001). This is the case in the thermophilic eubacterium Thermus thermophilus, which contains two genetically distinct aspartyl-tRNA synthetases (AspRSs) (Becker et al., 1997). The distinction is also functional, since AspRS-1 charges solely tRNAAsp in strong contrast to AspRS-2, which aspartylates both tRNAAsp and tRNAAsn with similar catalytic efficiencies, despite the presence in T.thermophilus of a fully functional asparaginyl-tRNA synthetase (AsnRS). Aspartate mischarged on tRNAAsn is then converted into asparagine by a tRNA-dependent amidotransferase. Altogether, this establishes coexistence in T.thermophilus of two distinct pathways of both tRNA aspartylation and tRNA asparaginylation. While AspRS-1 shares functional and structural features with other eubacterial AspRSs, the sequence of AspRS-2 strikingly resembles those of the non-discriminating AspRSs present in archaea lacking AsnRS (Curnow et al., 1996; Becker et al., 2000; Tumbula et al., 2000; Tumbula-Hansen et al., 2002). Thus, the two distinct systems, one archaeal-like and the other one eubacterial, are probably related to ancestral and modern aminoacylation pathways.
This study addresses the question of the structural basis accounting for the relaxed specificity of the archaeal-type AspRS-2 from T.thermophilus. To this aim, the crystal structure of this protein belonging to class IIb synthetases was solved. It is the first one of a non-discriminating synthetase. Structural analysis reveals features that distinguish AspRS-2 from the crystal structure of AspRS-1 (Delarue et al., 1994; Ng et al., 2002) and from that of other AspRSs (Giegé and Rees, 2003). Despite its eubacterial origin, non-discriminating AspRS-2 presents architectural features found in eukaryotic (Ruff et al., 1991; Sauter et al., 2000) and archaeal (Schmitt et al., 1998) AspRSs. The most prominent difference from conventional AspRSs lies in the conformation of the anticodon-recognizing domain, and more precisely in that of two loops joining β-strands within its OB-fold, characteristic of class IIb synthetases (see Figure 6B). One shares strong conformational resemblance with a homologous loop present in AsnRSs. Altogether, the new structural data combined with functional experiments account for the relaxed tRNA recognition of AspRS-2 and shed new light on the structure–function relationship of archaeal-type and archaeal AspRSs.
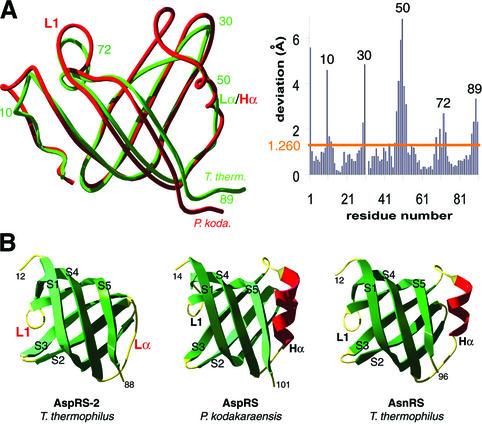
Fig. 6. Comparison of anticodon-binding domains in T.thermophilus AspRS-2, P.kodakaraensis AspRS and T.thermophilus AsnRS. (A) Superimposition of the two domains in Thermus (green) and Pyrococcus (red) AspRSs (left) and r.m.s. deviations (right). (B) Ribbon representation of the anticodon-binding domain in AspRS-2 (left), Pyrococcus AspRS (middle) and Thermus AsnRS (right). Notice in Pyrococcus AspRS and in Thermus AsnRS a standard OB-fold formed by a five-stranded β-barrel with an α-helix (Hα displayed in red) between strands S3 and S4. In AspRS-2, α-helix Hα is replaced by the Lα loop. Loops and strands are coloured in yellow and green, respectively; Lα and L1 loops that are specific to AspRS-2 are emphasized by red labels (notice that L1 corresponds to L45 in the conventional OB-fold nomenclature; Murzin, 1993).
Results and discussion
Structure determination
Because of significant sequence similarities, the structure of native AspRS-2 was solved initially by molecular replacement using AspRS from the archeon Pyrococcus kodakaraensis (Schmitt et al., 1998) as the starting model. Since the quality of the electron density did not allow an easy trace of the protein fold in a few regions of the electron density map, the multiwavelength anomalous dispersion (MAD) method was used on a selenomethionine-substituted protein to ensure an unbiased structure.
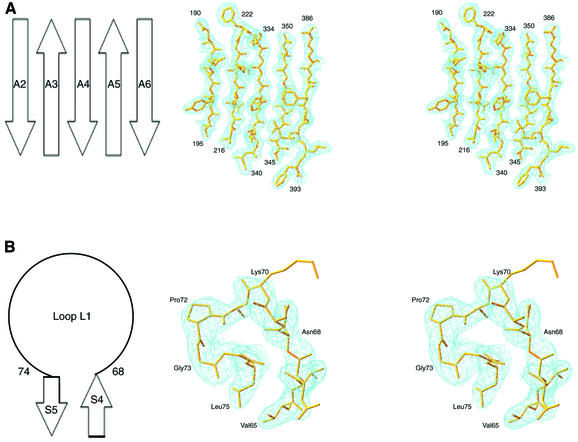
The final model of AspRS-2 was refined to an Rfactor of 22.7% and an Rfree of 26.2% between 30 and 2.3 Å resolution and has tightly restrained geometry (r.m.s. bond and angle deviations of 0.007 Å and 1.227°, respectively; Table I). As examples, Figure 1 shows the quality of the electron density map in the active site domain and anticodon-binding region of the enzyme. Over 91% of the residues are within the most favoured regions in a Ramachandran plot, as defined by PROCHECK (Laskowski et al., 1993). Only well-ordered Glu191 of each subunit has unfavourable main-chain torsion angles, as found in P.kodakaraensis AspRS for the equivalent acidic residue Asp204.
Table I. Crystallographic statistics.
| Data collection and MAD phasing |
|
|
|
|
|---|---|---|---|---|
| Native | Se edge | Se peak | Se remote | |
| Space group | P212121 | |||
| Unit cell constants (Å) | a = 57.4, b = 122.6, c = 167.1 | |||
| Wavelength (Å) | 0.9330 | 0.9798 | 0.9796 | 0.9150 |
| Resolution (Å) | 30–2.3 | 30–2.3 | 30–2.3 | 30–2.3 |
| Completeness (%)a | 99.8 (99.9) | 98.5 (99.5) | 99.8 (99.9) | 96.5 (97.2) |
| Rsym (%)a,b | 4.5 (22.1) | 5.3 (28.1) | 6.1 (28.2) | 5.3 (24.1) |
| Multiplicitya | 4.2 (4.4) | 3.4 (3.4) | 4.8 (4.8) | 3.1 (3.1) |
|
I/σ(I)a |
11.5 (3.1) |
9.9 (2.2) |
8.8 (2.5) |
10.1 (2.9) |
| Refinement |
|
|
|
|
| Resolution (Å) | 30–2.3 | |||
| Reflections work set | 45 048 | |||
| Reflections test set | 3443 | |||
| Rfactor (%)c | 22.7 | |||
| Rfree (%)d | 26.2 | |||
| R.m.s.d. bonds (Å) | 0.007 | |||
| R.m.s.d. angles (°) | 1.2 | |||
| Mean B values (Å2) | 38.1 | |||
| No. of protein atoms | 5769 | |||
| No. of solvent molecules | 160 |
aNumber in parentheses corresponds to the last resolution shell 2.36–2.30 Å.
bRsym = Σ|I – <I>|ΣI
cRfactor = Σ||Fobs| – |Fcalc||/Σ|Fobs|
dFor Rfree calculation, 7% of data were selected.
Fig. 1. Stereoviews of the MAD electron density map (contoured at 1.0 σ) of part of the catalytic site (A) and anticodon-binding (B) domains of T.thermophilus AspRS-2. In (A), the β-strands A2, A3, A4, A5, and A6 are labelled as in the P.kodakaraensis AspRS structure (Schmitt et al., 1998). In (B), displaying an amino acid stretch (residues 65–75) comprising loop L1, notice the functionally important Pro72 and the lack of density for the side chain of Lys70 (see text).
The T.thermophilus AspRS structure visualizes residues 1–93, 111–150, 179–199 and 213–414 of subunit A, residues 1–97, 111–150, 180–199 and 213–414 of subunit B and 160 water molecules solvating the dimer. As already suspected in the model obtained by molecular replacement, a few amino acid stretches covering 14% of the AspRS-2 sequence are not seen in the final MAD model because of non-existent density. They correspond to the last nine C-terminal amino acids of the synthetase (residues 415–422) and to three loops. One non-defined loop is located in the hinge region connecting the anticodon-binding and active site domains (residues 98–110) and the two others are in the active site domain. They are a region comprising the so-called mobile flipping loop (as defined in Schmitt et al., 1998) (residues 151–178) and the loop of class II consensus motif 2 (residues 200–212). It is likely that these mobile loops will acquire fixed conformations when liganded with substrates. Further, some side chains of surface residues are not well defined (Lys9, Lys51, Lys70, Tyr111, Lys280, Glu305, Glu330, Glu361, Glu371); Figure 1B shows the example of Lys70 in the anticodon-binding domain of the synthetase.
Mobile regions were observed in other class IIb synthetases. In the apo form of T.thermophilus AsnRS, the homologues of these loops only became defined when AsnRS was complexed with a non-hydrolysable analogue of asparaginyl-adenylate (Berthet-Colominas et al., 1998). In Escherichia coli LysRS (LysU) complexed with lysine, seven residues in the hinge region could not be built because of lack of density (Onesti et al., 1995; Desogus et al., 2000). Since this region is in contact with tRNAAsp in the yeast (Ruff et al., 1991; Cavarelli et al., 1993) and T.thermophilus (Briand et al., 2000) complexes, it is suggested that stabilization of its non-built homologue in AspRS-2 will be brought about by tRNA binding. The movement of the hinge domain in yeast AspRS, which allows binding of the tRNA D-stem, supports this view (Sauter et al., 2000).
Overall description of the structure of non-discriminating AspRS-2
AspRS-2 from T.thermophilus is a homodimer with each subunit comprising 422 amino acids (Becker et al., 2000). The protein has a modular architecture. Its N-terminal β-sheet-rich anticodon-binding domain resembles an OB-fold as defined by Murzin (1993) and is linked by a short interconnection to a C-terminal active-site domain, comprising the three class II consensus motives, which is built around a six-stranded antiparallel β-sheet surrounded by α-helices (Figure 2A). This architecture is characteristic of class IIb synthetases.
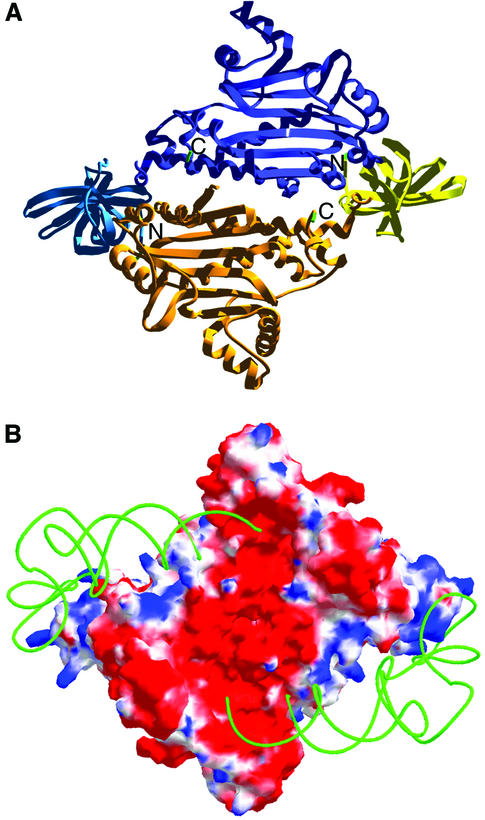
Fig. 2. Structure of T.thermophilus AspRS-2. (A) Ribbon representation of the dimeric synthetase. Subunit A is drawn in yellow (N-terminal domain) and in orange (catalytic domain), and subunit B is in blue (N-terminal domain) and purple (catalytic domain). The N- and C-terminal ends of each subunit are labelled. (B) Electrostatic potential mapped on the molecular surface of dimeric AspRS-2, as computed with Swiss-Pdb Viewer (Guex and Peitsch, 1997). Blue, white and red regions correspond to positive, neutral and negative electrostatic potentials, respectively. The putative location of a backbone model of tRNAAsp, as in the complex with T.thermophilus AspRS-1 (Briand et al., 2000), covering ‘blue’ regions of positive potential, is indicated. The orientation of the synthetase is as in (A).
AspRS-2 has a dimeric interface surface of 5829 Å2, the largest interface known so far in an AspRS (to be compared with 4600 Å2, the smallest interface as found in yeast AspRS; see Sauter et al., 2001). The structure is well defined in the anticodon-binding domain but contains a few disordered regions in the rest of the molecule. When analysing these regions, faint differences appear between the two subunits. The most prominent one concerns residues 94–97 in the hinge domain, which could be traced in one subunit and are not seen in the other one. Indeed, the connection between active site and anticodon-binding domains is only partly seen in the electron density map of AspRS-2. Of the 24 amino acids making the connection, only seven upstream of the active-site domain of both subunits and four downstream of the anticodon-binding domain in subunit B are seen. Altogether, this is indicative of the pseudo-homodimeric nature of AspRS-2, reflected by the conformational heterogeneity of the monomers. The differences can be quantitated by superimposition of the main-chain atoms of subunits A and B of the dimer and correspond to an r.m.s. deviation of 0.424 Å.
Calculation of the electrostatic potential reveals two symmetrical positive zones at the molecular surface of the AspRS-2 dimer, which span from the anticodon-binding domain to the catalytic centre of the enzyme. They are similar to the footprints of tRNAAsp on T.thermophilus AspRS-1, as deduced from the crystal structure of the complex (Briand et al., 2000), and thus most likely correspond to the binding areas of tRNA (Figure 2B).
Structural and functional comparison of AspRS-2 with other AspRSs of known crystal structure
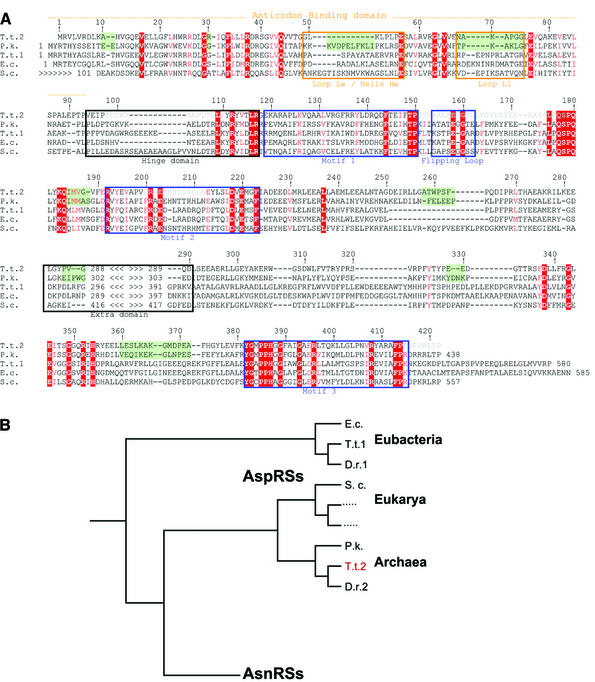
The structural view. A sequence alignment based on the superimposition of the Cα traces in AspRS-2 and in the four other known crystallographic structures of AspRSs shows 49 strictly conserved residues (Figure 3A). Overall, the comparison reveals a greater relatedness of AspRS-2 with AspRS of P.kodakaraensis than with other AspRSs (e.g. in the anticodon-binding and hinge domains). This relatedness is clearly seen in the phylogenetic tree of AspRSs (Figure 3B).
Fig. 3. Structure-based sequence alignment of AspRSs of known crystal structure and simplified phylogenetic tree of AspRSs. (A) Subdomains in AspRS-2 showing structural deviations with P.kodakaraensis AspRS are emphasized on a green background (Figure 5B). Strictly conserved residues in the five crystal structures are on a red background (only Leu237 is not highly conserved); semi-conserved residues are in red; (–) missing residues. Residues in AspRS-2 not built in the crystal structure because of lack of density are in light grey. Strategic regions (see text) are boxed. Numbering corresponds to the AspRS-2 sequence (Becker et al., 2000). (B) The phylogenetic tree is adapted from Becker et al. (2000) and Woese et al. (2000). Notice that AsnRSs arise from the archaeal genre of AspRSs. Organisms are abbreviated as follows: D.r. (D.radiodurans), E.c. (E.coli), P.k. (P.kodakaraensis), S.c. (Saccharomyces cerevisiae) and T.t. (T.thermophilus); D.r.1 or 2 and T.t.1 or 2 refer to the eubacterial or archaeal forms of AspRS. Sequences are retrieved from DDBJ/EMBL/GenBank.
Among the 36 conserved residues in the active site domain, eight were shown to ensure specific aspartate binding in the AspRSs from yeast (Cavarelli et al., 1994), E.coli (Eiler et al., 1999), T.thermophilus (Poterzman et al., 1994) and P.kodakaraensis (Schmitt et al., 1998). They are Glu158, Gln180, Lys183, Arg201, Asp218, Glu220, Glu345 and Arg352 (with the numbering of AspRS-2 sequence). For instance in P.kodakaraensis AspRS, Lys183 and Arg352 hydrogen bond with the aspartate carboxylate group, which is further stabilized by Arg201, Asp218 and Glu220. Given the conservation of these important functional amino acids, it is likely that recognition of aspartate by AspRS-2 occurs as in the four other AspRSs. Notice, however, in AspRS-2 the flexibility of Glu158 and of class IIb invariant Arg201, which are not seen, but are clearly visible in the apo form of other AspRSs (Eiler et al., 1999; Sauter et al., 2000; Ng et al., 2002).
In the N-terminal domain, three conserved residues (Phe33, Gln44 and Glu76) participate in recognition of anticodon bases G34 and U35 by discriminating yeast (Cavarelli et al., 1993) and E.coli (Briand et al., 2000; Moulinier et al., 2001) AspRSs. Here also, it is likely that AspRS-2 recognizes these two bases, as do discriminating AspRSs. Likewise, the aromatic ring of Phe33 would stack the ring of U35 while side chains of Gln44 and Glu76 would interact with tRNAAsp via hydrogen bonds to U35 and G34, respectively.
Several residues in AspRS-2 are only conserved in AspRSs originating from one or two of the three phylogenetic kingdoms of life. For instance, Ser348 is specific to archaeal and eukaryotic AspRSs and was shown to participate in stabilization of the transition state of the tRNA aspartylation reaction in the yeast and Pyrococcus enzymes (Cavarelli et al., 1994; Schmitt et al., 1998). This residue is absent in most eubacterial AspRSs. On the other hand, some conserved residues in most AspRSs are absent in AspRS-2. This is the case for Tyr214, which position in class II synthetases is generally occupied by a phenylalanine that stacks to the adenine ring of ATP (Cavarelli et al., 1994; Eiler et al., 1999). The absence of a phenylalanine at this position was also reported for hamster class IIa HisRS (Delarue and Moras, 1993) and P.kodakaraensis AspRS (Schmitt et al., 1998). Unlike in AspRS-2, this residue is almost always a hydrophobic amino acid (alanine, isoleucine or valine) in all known archaeal AspRS sequences (Becker et al., 2000).
In contrast to other eubacterial AspRSs, AspRS-2 contains neither a C-terminal extension nor a large insertion domain between motifs 2 and 3 in its active site module (which would correspond to residues 296–391 in T.thermophilus AspRS-1). By these features it resembles archaeal and eukaryotic AspRSs. However, and in contrast to eukaryotic class IIb synthetases, AspRS-2 is missing the N-terminal extension upstream of the anticodon-binding domain. In yeast, this extension helps to bind the anticodon stem of tRNAAsp on the AspRS core (Frugier et al., 2000). In summary, non-discriminating eubacterial AspRS-2 reveals hybrid eukaryal and archaeal characteristics that are found in both its catalytic and its anticodon-binding domains.
The functional view. Table II illustrates that AspRS-2 from T.thermophilus is non-discriminating and aspartylates tRNAAsp and tRNAAsn with comparable catalytic efficiency. This peculiar functionality is likely to rely on the relatedness of aspartate and asparagine identity sets that share the same discriminator base (G73) and similar GUC/U anticodons differing only by base 36 (C in tRNAAsp and U in tRNAAsn) (Giegé et al., 1998). As a likely consequence, the non-discriminating or discriminating nature of AspRSs should be linked to alternative recognition patterns of identity base 36.
Table II. Aspartylation of T.thermophilus tRNAAsp and tRNAAsn by native or engineered AspRSs.
| AspRSs | tRNAAsp charging |
tRNAAsn charging |
||||
|---|---|---|---|---|---|---|
| kcat s–1 (×10–2) | Km µM | kcat/Km (relative) | kcat s–1 (×10–2) | Km µM | kcat/Km (relative) | |
| T.t.2-nativea | 4.2 | 0.14 | 1 | 0.56 | 0.20 | 0.09 |
| P.k.-native | 1.17 | 0.2 | 0.17 | no detectable charging | ||
| P.k.-mutantb | 0.83 | 1.0 | 0.03 | 0.08 | 0.90 | 0.003 |
Aminoacylation conditions are as described in Materials and methods. Km and kcat values were determined at 37°C.
aAt 70°C, the kcat/Km for tRNAAsp is only 2-fold higher than for tRNAAsn (Becker and Kern, 1998).
bThe Pyrococcus mutant has the L1 loop exchanged with that from Thermus AspRS-2.
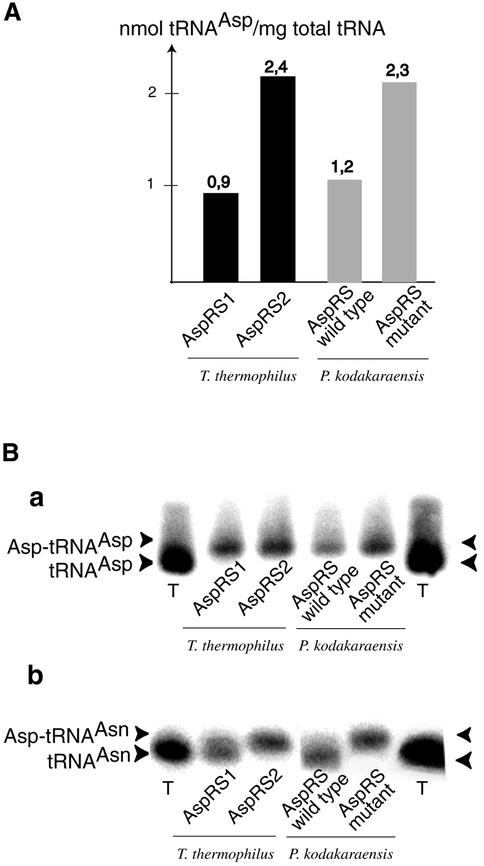
As to the enzyme level, it is known that AspRSs recognize aspartate identity determinants and in particular anticodon determinant C36 (Giegé and Rees, 2003). In archaeal P.kodakaraensis AspRS, identification of a peculiar loop homologous to those recognizing the third anticodon base in other AspRSs led to the suggestion that this synthetase should be able to accommodate either a GUC or a GUU anticodon (Schmitt et al., 1998). Further, the conformational resemblance of non-discriminating AspRS-2 with archaeal AspRSs and the lack of AsnRS in some archaea, suggests that archaeal AspRSs, including AspRS from P.kodakaraensis, would be non-discriminating. Aminoacylation assays and biochemical characterization of the charged tRNAs, however, indicate that the Pyrococcus enzyme is discriminating and solely charges tRNAAsp (Figure 4A; Table II). This conclusion was also reached by others as the result of genome analysis and functional assays (Tumbula-Hansen et al., 2002). In addition, these authors showed the existence of both discriminating and non-discriminating forms of AspRS among the archaea, despite the high sequence identity of archaeal AspRSs. Following these considerations, the non-discriminating nature of AspRS-2 should be searched among the structural similarities and differences it has with Pyrococcus AspRS (and with other archaeal AspRSs), with the expectation that the features important for dual tRNA recognition are located in the anticodon-binding domain.
Fig. 4. Specificity of tRNA charging of T.thermophilus AspRSs and archaeal P.kodakaraensis AspRS and effect of the L1 loop on tRNA discrimination. (A) Aspartylation levels (plateau values) of unfractionated T.thermophilus tRNA with AspRS-1 or AspRS-2 and with native or mutated AspRS from Pyrococcus. (B) Identification by hybridization assays of tRNAAsp (a) and tRNAAsn (b) aspartylated by the various AspRSs. The AspRS mutant from P.kodakaraensis has the L1 loop exchanged by that from Thermus AspRS-2; C, control with uncharged tRNA.
Similarities and differences between Thermus AspRS-2 and Pyrococcus AspRS
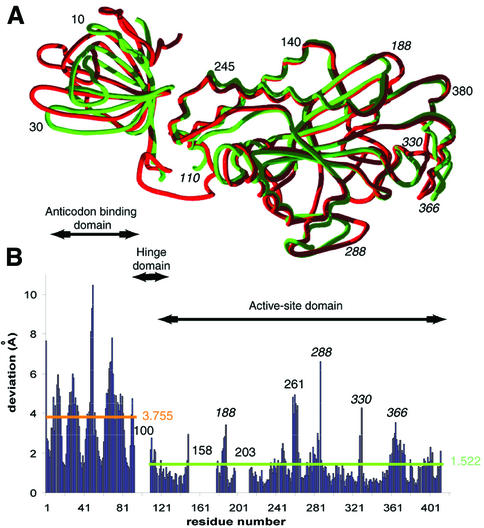
Overall comparison. Comparison of AspRS-2 with AspRS from P.kodakaraensis reveals 41% sequence identity. This high level of identity suggests an overall conformational similarity of the two synthetases and has justified initial use of molecular replacement for solving the structure of AspRS-2. However, comparison of the two structures yields an r.m.s. deviation of superimposed Cα positions of 2.196 Å. This high value is indicative of conformational differences between the two proteins (Figure 5A), a conclusion in apparent contradiction with the high sequence identity. The contradiction is explained when comparing individual domains of the modular AspRS monomers. For the catalytic domain the r.m.s. deviation is reduced to 1.522 Å (Figure 5B) while for the anticodon-binding domains it becomes 1.260 Å (Figure 6A). The higher r.m.s. deviation for the entire monomer originates from a rigid-body movement of the anticodon-binding domain. Altogether, most residues in individual domains superimpose well, with deviations in the ∼1 Å range, but several local conformational changes, with structural deviations that can reach 6 Å, are identified at the protein surfaces (Figures 5 and 6A).
Fig. 5. Comparison of the 3D models of T.thermophilus AspRS-2 and of P.kodakaraensis AspRS (ATP form, Schmitt et al., 1998). (A) Superimposition of subunit A of AspRS-2 (green) and subunit A from P.kodakaraensis AspRS (red). Surface residues of the AspRS-2 active-site domain that show largest structural deviations with P.kodakaraensis AspRS are in italics. (B) Values of r.m.s. deviations calculated by least squares minimized superimposition of the active-site domains. Notice that the N-terminal domain of AspRS-2 deviates from that of P.kodakaraensis AspRS by a rigid body rotation of 4°.
Active-site domain. Structural differences between AspRS-2 and Pyrococcus AspRS are found in five regions centred on residues 188, 261, 288, 330 and 366 (Figure 5B). Four differences are due to insertions or deletions in sequence patches around residues 188, 261, 288 and 330 and lead to distortions of between 3 and 8 Å (Figures 3 and 5B). That occurring around Gly188 in AspRS-2 (Ala200 in Pyrococcus) near the N-terminus of β-sheet A2 of the catalytic domain, comprises deletion of the homologue to Ser201 in Pyrococcus AspRS. This deletion may affect the overall flexibility of the region comprising the flipping-loop. The three other differences concern regions at the AspRS surfaces that are not involved in catalysis, namely around Pro261 (Leu273 in Pyrococcus), Gly288 (Gly302 in Pyrococcus) and Glu330 (Lys346 in Pyrococcus). Sequence inspection shows that these insertions or deletions are characteristics of AspRS-2. Their putative role remains to be deciphered. Two other differences concern regions not seen in AspRS-2 because of high flexibility in the absence of bound ligands, but visible under different conformations in several crystal forms of Pyrococcus AspRS corresponding to specific functional states of the protein (Schmitt et al., 1998). One, with conserved Glu158, comprises the flipping-loop that anchors the aspartate substrate in the catalytic pocket. The other, centred on Glu203 and class II conserved Arg201, is the loop joining strands A2 and A3 in motif 2. It is involved in aminoacyl-adenylate formation and tRNA acceptor stem recognition. Interestingly, when comparing the structure of free or substrate-bound AspRSs from E.coli or yeast, the main conformational changes in the active-site domain concern precisely the flipping-loop and the loop of motif 2 (Rees et al., 2000; Sauter et al., 2000).
A last conformational change corresponds to a global movement of a helix and part of a loop and occurs in AspRS-2 in the region comprising Lys366 (Lys382 in Pyrococcus). In subunit A of AspRS-2, this region is involved in a packing contact that does not occur in subunit B. Therefore, even if partly resulting from packing effects, it is likely that the conformation in the 360–370 region is an intrinsic characteristic of AspRS-2.
Articulation between active site and anticodon-binding domains. Flexibility of the hinge domain, leading to different relative orientations of the two functional protein modules, is a characteristic of AspRSs and was shown to be associated with tRNA binding. In yeast AspRS the initial recognition of the tRNA anticodon loop necessitates a 6° rotation of the anticodon-binding domain relative to the active-site domain, so as to prevent a steric clash between the tRNA and the hinge domain of the synthetase (Sauter et al., 2000). Similar pathways were proposed for T.thermophilus AspRS-1 (Briand et al., 2000) and E.coli AspRS (Rees et al., 2000). In AspRS-2, the hinge domain also shows a great potential of flexibility, since part of its sequence is not seen in the crystal structure. When its catalytic domain is superimposed on that of P.kodakaraensis AspRS, the flexibility of the hinge is evaluated by a rigid-body rotation of 4° needed to superimpose the N-terminal domains.
Anticodon-binding domain. The OB-fold of the anticodon-binding domain of AspRS-2 shares strong resemblance with its homologue in P.kodakaraensis AspRS and in all other known 3D AspRS structures. It differs however from a standard OB-fold [formed by a five-stranded β-barrel with a α-helix between strands S3 and S4, as defined by Murzin (1993)] by the absence of the α-helix joining strands S3 and S4 (Figure 6B). This absence is correlated with a much shorter amino acid sequence joining these two strands. The length of the connection is eight residues in AspRS-2 instead of 18 in AspRS from P.kodakaraensis (Schmitt et al., 1998). This difference precisely corresponds to the deletion of helix Hα formed by 10 amino acids. Following these considerations, the connection in the pseudo OB-fold in AspRS-2 is named loop Lα. A further difference between the two AspRSs lies in the orientation and sequence of the so-called L1 loop that joins strands S4 and S5 (Figure 6B). Notice that the L1 loop is located opposite to Hα or Lα in the OB-fold and that both subdomains are directly linked via strand S4. In AspRS-2, the fact that Lα is short as compared with the Hα helix in P.kodakaraensis AspRS, introduces a structural constraint that may mediate via the S4 strand the different orientation of the L1 loop in AspRS-2 as compared with that in the Pyrococcus enzyme (Figure 6).
Non-discrimination by AspRS-2 lies in its anticodon-binding domain
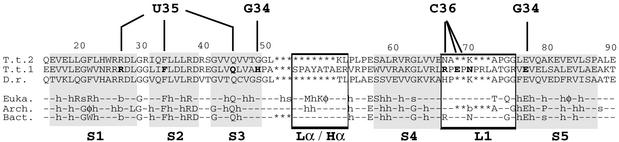
Replacement of the Hα helix by a loop in non-discriminating AspRS-2. First, as discussed above, the Hα helix located between strands S3 and S4 in canonical OB-folds is absent in AspRS-2 and is replaced by loop Lα (Figure 6B). This missing α-helix, present in the crystal structure of other AspRSs, is seen in those of LysRS from E.coli (Onesti et al., 1995, 2000) and of AsnRS from T.thermophilus (Berthet-Colominas et al., 1998) (Figure 6B), two other class IIb synthetases. Sequence alignment predicts the presence of an Hα helix in almost all known AspRSs, although without significant amino acid conservation (Figure 7). The exception arises in the bacterium Deinococcus radiodurans (White et al., 1999), phylogenetically related to T.thermophilus, which also encompasses two AspRS genes, the second closely related to that of AspRS-2 (Figure 3B). The corresponding AspRS lacking the Hα helix is non-discriminating, and like AspRS-2, produces mischarged aspartyl-tRNAAsn involved in an alternative pathway of tRNA asparaginylation, including amidotransferase activity (Curnow et al., 1998; Min et al., 2002). Notice that absence of the Hα helix in an AspRS does not necessarily imply dual specificity of the synthetase, since sequence comparison of archaeal non-discriminative AspRSs suggests the presence of such a helix in some of these enzymes.
Fig. 7. Peculiarities in the anticodon-binding domain of AspRS-2 from T.thermophilus as revealed by sequence comparison with other AspRSs. (T.t.2, T.t.1) AspRSs from T.thermophilus, (D.r.) D.radiodurans and (Euka., Arch., Bact.) consensus sequences from eukarya, archaea and eubacteria. (–) Non-conserved residues; semi-conserved residues with φ, h, a, b, s representing, respectively, aromatic, hydrophobic, acid, basic and small (Gly, Ala, Ser) side chains; (*) missing amino acids. Residues belonging to β-strands (S1, S2, S3, S4 and S5) of the OB-fold are on a grey background. Lα/Hα and L1 regions are boxed. Amino acids in AspRS-1 that contact the three anticodon identity determinants of tRNAAsp (G34, U35 and C36) are in bold. Notice the differences in length and sequence in L1.
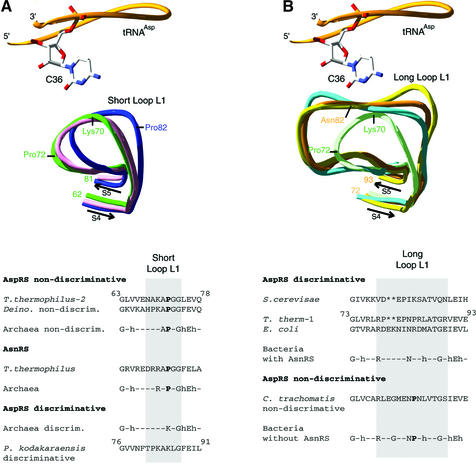
Peculiarities of L1 loops in AspRSs. Previous structural investigations have shown the crucial role of tRNA recognition of the L1 loop that joins strands S4 and S5 in the anticodon-binding OB-fold of AspRSs. Sequence comparisons indicate variable sizes of this loop and analysis of crystal structures show their well-defined conformations (Figures 1B and 8). When structures of the complex with tRNAAsp are available, interaction of the L1 loop with aspartate identity determinant C36 is always clearly seen (Cavarelli et al., 1993; Eiler et al., 1999; Briand et al., 2000; Moulinier et al., 2001). So, in discriminating T.thermophilus AspRS-1, the L1 loop has an extended size (Figure 8B) and three of its amino acids contact three different atoms from C36 (Nδ of Asn82 with O2, Nη of Arg78 with N3 and O of Glu80 with N4 of the cytosine ring; Briand et al., 2000). Two of these amino acids (Arg78 and Asn82) are conserved within discriminating bacterial AspRSs, suggesting that this recognition pattern is essentially conserved. From the functional viewpoint, C36 is a universal aspartate identity determinant (Pütz et al., 1991; Becker et al., 1996; Giegé et al., 1998) and, as shown with yeast AspRS, mutating amino acids contacting this determinant strongly affects tRNAAsp binding (Eriani and Gangloff, 1999).
Fig. 8. L1 loop regions in AspRS and AsnRS anticodon-binding domains. (A and B, top) Comparison of L1 loops resulting from least square minimized superimpositions of the entire anticodon-binding domains. (A) Short L1 loops in T.thermophilus AspRS-2 (green), P.kodakaraensis AspRS (blue) and T.thermophilus AsnRS (pink). (B) Long L1 loops in T.thermophilus AspRS-1 (orange), yeast (yellow) and E.coli (light blue) AspRSs (for purposes of comparison, the short L1 loop region of AspRS-2 is displayed in light green). Notice in both panels the trace of the tRNAAsp backbone (orange) with identity element C36 (Becker et al., 1996) in contact (or proximity) with L1 loops as seen in the crystal structure of the complex with AspRS-1 (Briand et al., 2000). Notice also the exact superimposition of either the S4 or the S5 antiparallel β-strands that anchor the L1 loops. (A and B, bottom) Structure-based sequence comparison of the regions encompassing short (A) or long (B) L1 loops in AspRSs and AsnRSs. (–) Non- conserved residues, (*) missing residues and (h) residues with hydrophobic side chains. Residues belonging to the loop are on a grey background; conserved Pro72 in short L1 loops of non-discriminating AspRSs and of AsnRSs is emphasized in bold; notice also the presence of a conserved proline (in bold) in long L1 loops of non-discriminating AspRSs.
Non-discriminating AspRS-2, like the non-discriminating AspRS from eubacterial Deinococcus radiodurans, possesses an L1 loop of small size. This feature is reminiscent of what was first discovered in the structure of P.kodakaraensis AspRS (Schmitt et al., 1998) and later in the sequence of other archaeal AspRSs (Figure 7). The small size of the loop as such, however, cannot account for the non-discriminating nature of AspRS-2, since the Pyrococcus enzyme is discriminating. In agreement with this fact are the different conformations of the L1 loops in both AspRSs that do not superimpose (Figure 8A). Strikingly, however, the L1 loop from AspRS-2 superimposes perfectly with that of class IIb-related T.thermophilus AsnRS (Berthet-Colominas et al., 1998). In contrast, strong sequence and conformation deviations occur between the L1 loop of AspRS-2 and those of the discriminating AspRSs from S.cerevisiae, E.coli and T.thermophilus such that all three are very similar and superimpose (Figure 8B). Notice that in class IIb LysRS, size and conformation of the L1 loop differ (Cusack et al., 1996).
Taking into account both sequence and conformation features, it appears that L1 loops are short (seven residues long) in archaeal non-discriminating AspRSs. This holds true in T.thermophilus and D.radiodurans non-discriminating AspRSs as well as in T.thermophilus and archaeal AsnRSs. In contrast, most discriminating AspRSs present a long L1 loop (12–14 residues long). However, the correlation between short L1 loops and the non-discriminating nature of AspRSs, or between long L1 loops and the discriminating AspRSs is not absolute (Figure 8). Indeed, several discriminating AspRSs with short L1 loops have been identified in archaeal organisms, and likewise, non-discriminating AspRSs were characterized in eubacteria lacking AsnRS (Tumbula-Hansen et al., 2002), in particular in the eubacterium Chlamidia trachomatis (Raczniak et al., 2001). A closer view of the sequences indicates that non-discrimination and tRNAAsn recognition are accompanied by conservation of a proline residue either in short L1 loops (homologue to Pro72 in AspRS-2) or long L1 loops, while discrimination relies on the absence of such a residue in short loops (Figure 8). In particular, in P.kodakaraensis and other discriminating archaeal AspRSs, Pro72 is replaced by Lys72. From the structural point of view, the presence or absence of a proline at position 72 imposes different stiffnesses to the L1 loops. Interestingly, in P.kodakaraensis AspRS, the differences may even be enhanced by the presence of a proline residue at position 68 on the opposite side of the loop (Figure 8A). Notice that these considerations find some support when comparing the B-factors of the L1 loop in the two structures (40.4 Å2 in Thermus AspRS-2 versus 30.1 Å2 in Pyrococcus AspRS).
Functional implications. The same conformation of L1 loops in non-discriminating AspRS-2 and in AsnRS, and the fact that anticodon residues C36 in tRNAAsp and U36 in tRNAAsn are aspartate (Becker et al., 1996) and asparagine (Shimizu et al., 1992; Li et al., 1993) identity determinants, suggests a direct functional role of the L1 loop in AspRS-2. This view gains support on considering that identity determinant U36 in tRNAAsn contains two chemical groups (O2 and N3) present in C36, a base recognized by conventional AspRSs (Cavarelli et al., 1993; Eiler et al., 1999; Briand et al., 2000; Moulinier et al., 2001). Thus, recognition of U36 within the tRNAAsn anticodon could account for the efficient mischarging of tRNAAsn by AspRS-2. The dual recognition by an AspRS of both tRNAAsp and tRNAAsn, however, would not be possible if its L1 loop would deviate in conformation from that present in AsnRS, like the one present in the AspRS from P.kodakaraensis, which is discriminating (Table II). This statement implies, if non-discrimination lies in the structure of the L1 loop, that the Pyrococcus enzyme can be rendered non-discriminating upon replacement of its L1 loop by that of Thermus AspRS-2. To verify this implication, a mutant AspRS from P.kodakaraensis was engineered with an L1 loop swap and its aspartylation ability tested on crude tRNA. Direct characterization of the charged tRNAs by hybridization experiments demonstrated that the mutant enzyme, as anticipated, aspartylates both tRNAAsp and tRNAAsn (Figure 4B). Kinetic measurements show that tRNA charging occurs with catalytic efficiencies almost as high as for the reactions catalysed by T.thermophilus AspRS-2 (Table II). The slightly reduced activity for tRNAAsn charging suggests that the discrimination process is not solely mediated by the L1 loop and may be tuned via indirect effects by additional AspRS regions such as the Hα/Lα domain (Figure 6). These results with AspRSs can be related to the specificity change of T.thermophilus GluRS obtained after a single amino acid mutation in the anticodon-binding domain of the synthetase (Sekine et al., 2001). Replacing the amino acid recognizing the anticodon base C36 in tRNAGlu by the homologous residue in GlnRS allows the mutant GluRS to charge a tRNAGlu mutant with C36 replaced by its tRNAGln homologue G36. In both aspartate and glutamate systems, aminoacylation by the mutant synthetase is Km-dependent and exhibits kcats comparable to those of the wild-type reactions.
Altogether, this work presents further evidence showing the significance of anticodon in tRNA identity, and consequently of the functional importance of architectural features in the anticodon-binding domains of synthetases. The knowledge originating from the Thermus AspRS-2 structure, and its comparison with other class IIb synthetases, now provides the background necessary for rational design of AspRSs with altered tRNA anticodon recognition, i.e with altered specificity, and for a deeper understanding of the structure–function relationships between aspartate and asparagine tRNA aminoacylation systems.
Materials and methods
Protein purification and crystallization
AspRS-2 from T.thermophilus was overexpressed in E.coli BL21 transformed by the pET-3b vector (Becker et al., 2000) and purified as previously described (Charron et al., 2001). Crystals suitable for X-ray analysis were obtained by vapour phase diffusion and grew in a mother liquor containing 10% (v/v) PEG 8000 as the crystallizing agent, 100 mM CHES buffer at pH 9.5 and 200 mM NaCl. They belong to space group P212121 with unit cell dimensions a = 57.4, b = 122.6 and c = 167.1 Å with one dimer per asymmetric unit (Charron et al., 2001). Crystals were flash-frozen in liquid ethane in the mother liquor with addition of 20% glycerol as a cryoprotectant. Selenomethionyl- (SeMet) AspRS-2 was overexpressed in E.coli B834(DE46) (Met-auxotroph; Coli Genetic Stock Centre, Yale, CT) and purified as for the native protein.
AspRS from P.kodakaraensis was purified from overproducing E.coli strains JM103 transformed with pUC18 recombined with native or mutated AspRS genes. After sonication, cell debris and ribosomes were removed by centrifugation at 105 000 g and most proteins floculated by incubating the extract for 30 min at 70°C. The dialysed supernatant was adsorbed on a DEAE-cellulose column that was resolved with a linear gradient of K phosphate buffer from 20 mM pH 7.5 to 400 mM pH 6.5. Native AspRS was further purified on a phosphocellulose column eluted with a linear gradient from 0 to 0.5 M KCl in 20 mM K phosphate buffer pH 6.8. Mutated AspRS, not retained on phosphocellulose, was fully purified by a second chromatography on DEAE-cellulose. Purity of AspRSs, as estimated by gel electrophoresis, was >95%. Protein concentrations were determined using E280nm values of 0.96 and 1.09 (mg/ml)–1.cm–1, respectively, for P.kodakaraensis and T.thermophilus AspRSs.
Data collection and structure determination
A native data set at 2.3 Å resolution was collected at 100 K on beamline ID14-4 at the European Synchrotron Radiation Facility (ESRF, Grenoble), with incident radiation at a wavelength of 0.933 Å and a crystal-to-detector distance of 180 mm. Diffraction spots were recorded on an ADSC-Q4 CCD detector with a 1.0° oscillation and a 2 s exposure per CCD image over a range of 120°. Data were indexed and scaled using MOSFLM and SCALA (CCP4, 1994). Indexed intensities were converted to structure factors using TRUNCATE (CCP4, 1994) without any σ cutoff.
Initial attempts to solve the structure of AspRS-2 were performed by molecular replacement with the program AMoRe (Navaza, 1994). The dimeric P.kodakaraensis AspRS (Schmitt et al., 1998), which shows 41% identity with T.thermophilus AspRS-2, served as the structural model. It gave one major peak with a correlation of 19.8% (Charron et al., 2001). From this solution, a map was calculated at 2.3 Å resolution. After rigid-body refinement using CNS (Brünger et al., 1998), the crystallographic R-factor was 52.4%. The refinement was carried out using CNS (Brünger et al., 1998). A total of 7% of the data were selected for Rfree calculations. Manual corrections of the model were performed using O (Jones et al., 1991). Nevertheless, the quality of the electron density did not allow an easy trace of the protein structure in several regions of the protein.
The MAD data from a SeMet AspRS-2 crystal were collected at absorption edge, absorption peak and remote up to 2.3 Å resolution at the ID-29 beamline at ESRF. Data sets were indexed and scaled using MOSFLM and SCALA (CCP4, 1994). Eleven of 14 possible selenium sites were found and refined at 2.3 Å resolution using SOLVE (Terwilliger and Berendzen, 1999), which produced a mean figure of merit of 0.34 and an overall score of 58. After density modification with RESOLVE, the mean figure of merit was 0.58 and all of the traceable residues of the AspRS-2 molecule were easily modelled into the experimental map. Coordinates of the AspRS-2 structure have been submitted to the PDB Bank (accesssion number 1N9W).
Biochemical and protein engineering methods
Aminoacylation assays were conducted in reaction mixtures containing 100 mM Na-HEPES buffer pH 7.2, 2 mM ATP, 10 mM MgCl2, 30 mM KCl, 6 µM [3H]Asp (3000 c.p.m./pmol for Km measurements of tRNA) or 20 µM [14C]Asp (280 c.p.m./pmol, for kcat measurements), T.thermophilus tRNAAsp or tRNAAsn (accepting capacities 30 nmol/mg), purified from overexpressing E.coli strains, in concentration ranges varying from 0.02 to 2 µM for Km determinations, or at a concentration of 10 Km for kcat determinations, and appropriate amounts of AspRSs for initial velocity measurements. Reactions were conducted at 37°C and Asp-tRNA formed was determined as described (Becker and Kern, 1998).
Identity of charged tRNAs was verified according to Varshney et al. (1991), using 5′-32P-labelled oligonucleotides complementary to nucleotides 6–26 of tRNAAsp and nucleotides 7–25 of tRNAAsn from T.thermophilus. Unfractionated and deacylated T.thermophilus tRNA (3.5 µg) was charged by the various AspRSs (Thermus AspRS-1 or AspRS-2 and native or mutated Pyrococcus AspRS) present in such amounts as to reach plateaux in <10 min. Reaction products were fractionated by acidic PAGE with urea. After transfer on a Hybond-XL membrane (Amersham), tRNAs were identified by hybridization with the labelled probes and revealed on an Image Plate.
Mutagenesis of Pyrococcus AspRS was performed by PCR using the pUC18AspKOD1 vector (Schmitt et al., 1998), two 50 nucleotide-long synthetic oligonucleotides, each complementary to one strand and containing the appropriate mutations, and the QuikChange Site-Directed Mutagenesis Kit (Stratagene). DNA sequencing controlled the presence of the mutations.
Acknowledgments
Acknowledgements
We are grateful to ESRF for access to the ID14 and ID29 beamlines and we thank its staff for assistance with data collection. We thank E.Schmitt (Ecole Polytechnique, Palaiseau) for the gift of plasmid pUC18AspKOD1 and for access to the coordinates of the various crystal forms of Pyrococcus AspRS and D.Söll (Yale University) for access to manuscripts before publication. This work was supported by Centre National de la Recherche Scientifique and Université Louis Pasteur, Strasbourg, and by grants from the Association de la Recherche contre le Cancer, Centre National d’Etudes Spatiales and the European Community (BIO-CT98-0086). C.C. was the recipient of EC and CNES fellowships, and H.R. and M.B. of fellowships from Ministère de l‘Education Nationale, de la Recherche et de la Technologie.
References
- Becker H.D. and Kern,D. (1998) Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl Acad. Sci. USA, 95, 12832–12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H.D., Reinbolt,J., Kreutzer,R., Giegé,R. and Kern,D. (1997) Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry, 36, 8785–8797. [DOI] [PubMed] [Google Scholar]
- Becker H.D., Roy,H., Moulinier,L., Mazauric,M.-H., Keith,G. and Kern,D. (2000) Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase. Biochemistry, 39, 3216–3230. [DOI] [PubMed] [Google Scholar]
- Becker H.D., Giegé,R. and Kern,D. (1996) Identity of prokaryotic and eukaryotic tRNAAsp for aminoacylation by aspartyl-tRNA synthetase from Thermus thermophilus. Biochemistry, 35, 7447–7458. [DOI] [PubMed] [Google Scholar]
- Berthet-Colominas C., Seignovert,L., Härtlein,M., Grotli,M., Cusack,S. and Leberman,R. (1998) The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J., 17, 2947–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand C., Poterszman,A., Eiler,S., Webster,G., Thierry,J.-C. and Moras,D. (2000) An intermediate step in the recognition of tRNAAsp by aspartyl-tRNA synthetase. J. Mol. Biol., 299, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees,B., Ruff,M., Thierry,J.-C. and Moras,D. (1993) Yeast tRNAAsp recognition by its cognate class-II aminoacyl-transfer RNA synthetase. Nature, 362, 181–184. [DOI] [PubMed] [Google Scholar]
- Cavarelli J. et al. (1994) The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J., 13, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron C., Roy,R., Lorber,B., Kern,D. and Giegé,R. (2001) Crystallization and preliminary X-ray diffraction data of the second and archaebacterial-type aspartyl-tRNA synthetase from Thermus thermophilus. Acta Crystallogr. D, 57, 1177–1179. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Curnow A.W., Tumbula,D.L., Pelaschier,J.T., Min,B. and Söll,D. (1998) Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl Acad. Sci. USA, 95, 12838–12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnow A.W., Ibba,M. and Söll,D. (1996) tRNA-dependent asparagine formation. Nature, 382, 589–590. [DOI] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (1996) The crystal structures of T.thermophilus lysyl-tRNA synthetase complexed with E.coli tRNALys and a T.thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J., 15, 6321–6324. [PMC free article] [PubMed] [Google Scholar]
- Delarue M. and Moras,D. (1993) The aminoacyl-tRNA synthetase family: modules at work. BioEssays, 15, 675–687. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poterszman,A., Nikonov,S., Garber,M., Moras,D. and Thierry,J.-C. (1994) Crystal structure of a prokaryotic aspartyl-tRNA synthetase. EMBO J., 13, 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desogus G., Todone,F., Brick,P. and Onesti,S. (2000) Active site of lysyl-tRNA synthetase: Structural studies of the adenylation reaction. Biochemistry, 39, 8418–8425. [DOI] [PubMed] [Google Scholar]
- Eiler S., Dock-Bregeon,A., Moulinier,L., Thierry,J.-C. and Moras,D. (1999) Synthesis of aspartyl-tRNAAsp in Escherichia coli—a snapshot of the second step. EMBO J., 18, 6532–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G. and Gangloff,J. (1999) Yeast aspartyl-tRNA synthetase residues interacting with tRNAAsp identity bases connectively contribute to tRNAAsp binding in the ground and transition-state complex and discriminate against non-cognate tRNAs. J. Mol. Biol., 291, 761–773. [DOI] [PubMed] [Google Scholar]
- First E.A. (1998) Catalysis of tRNA aminoacylation by class I and class II aminoacyl-tRNA synthetases. In Sinnott,M. (ed.), Comprehensive Biological Catalysis. Vol. 1. Academic Press, London, UK, pp. 573–607.
- Frugier M., Moulinier,L. and Giegé,R. (2000) A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J., 19, 2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Sissler,M. and Florentz,C. (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res., 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R. and Rees,B. (2003) Aspartyl-tRNA synthetases. In Ibba,M., Francklyn,C. and Cusack,S. (eds), The Aminoacyl-tRNA Synthetases. Landes Bioscience, Georgetown, TX, in press.
- Guex N. and Peitsch,M.C. (1997) Swiss-model and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Ibba M and Söll,D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., 69, 617–650. [DOI] [PubMed] [Google Scholar]
- Ibba M and Söll,D. (2001) The renaissance of aminoacyl-tRNA synthesis. EMBO rep., 2, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M., Francklyn,C. and Cusack,S. (2003) The Aminoacyl-tRNA Synthetases. Landes Bioscience, Georgetown, TX, in press.
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Li S., Pelka,H. and Schulman,L.H. (1993) The anticodon and discriminator base are important for aminoacylation of Escherichia coli tRNAAsn. J. Biol. Chem., 268, 18335–18339. [PubMed] [Google Scholar]
- Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999) Aminoacyl-tRNA synthetases: a new image for a classical family. EMBO J., 18, 4591–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B., Pelaschier,J.T., Graham,D.E., Tumbula-Hansen,D. and Söll,D. (2002) Transfer RNA-dependent amino acid biosynthesis: An essential route to asparagine formation. Proc. Natl Acad. Sci. USA, 99, 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulinier L., Eiler,S., Eriani,G., Gangloff,J., Thierry,J.-C., Gabriel,K., McClain,W.H. and Moras,D. (2001) The structure of an AspRS–tRNAAsp complex reveals a tRNA-dependent control mechanism. EMBO J., 20, 5290–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G. (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Ng J.D., Sauter,C., Lorber,B., Kirkland,N., Arnez,J. and Giegé,R. (2002) Space-grown crystals are more useful for structure determination: A case for Thermus thermophilus aspartyl-tRNA synthetase-1. Acta Crystallogr. D, 58, 645–652. [DOI] [PubMed] [Google Scholar]
- Onesti S., Miller,A.D. and Brick,P. (1995) The crystal structure of the lysyl-tRNA synthetase (LysU) from Escherichia coli. Structure, 3, 163–176. [DOI] [PubMed] [Google Scholar]
- Onesti S., Desogus,G., Brevet,A., Chen,J., Plateau,P., Blanquet,S. and Brick,P. (2000) Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding. Biochemistry, 39, 12853–12861. [DOI] [PubMed] [Google Scholar]
- Poterzman A., Delarue,M., Thierry,J.-C. and Moras,D. (1994) Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase. J. Mol. Biol., 244, 158–167. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi,J.D., Florentz,C. and Giegé,R. (1991) Identity elements for specific aminoacylation of yeast tRNAAsp by cognate aspartyl-tRNA synthetase. Science, 252, 1696–1699. [DOI] [PubMed] [Google Scholar]
- Raczniak G, Becker,H.D., Min,B. and Söll,D. (2001) A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J. Biol. Chem., 276, 45862–45867. [DOI] [PubMed] [Google Scholar]
- Rees B., Webster,G., Delarue,M., Boeglin,M. and Moras,D. (2000) Aspartyl tRNA-synthetase from Escherichia coli: flexibility and adaptability to the substrates. J. Mol. Biol., 299, 1157–1164. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy,S., Boeglin,M., Poterszman,A., Mitschler,A., Podjarny,A., Rees,B., Thierry,J.-C. and Moras,D. (1991) Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science, 252, 1682–1689. [DOI] [PubMed] [Google Scholar]
- Sauter C., Lorber,B., Cavarelli,J., Moras,D. and Giegé,R. (2000) The free yeast aspartyl-tRNA synthetase differs from the tRNAAsp-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain. J. Mol. Biol., 299, 1333–1324. [DOI] [PubMed] [Google Scholar]
- Sauter C., Lorber,B., Dietrich-Théobald,A. and Giegé,R. (2001) Crystallogenesis in tRNA aminoacylation systems: how packing accounts for crystallization drawbacks with yeast aspartyl-tRNA synthetase. J. Cryst. Growth, 232, 399–408. [Google Scholar]
- Sekine S.-i., Nureki,O., Shimada,A., Vassylyev,D.G. and Yokoyama,S. (2001) Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat. Struct. Biol., 8, 203–206. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Asahara,H., Tamura,K., Hasegawa,T. and Himeno,H. (1992) The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNA by their aminoacyl-tRNA synthetases. J. Mol. Evol., 35, 436–443. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Moulinier,L., Fujiwara,S., Imanaka,T., Thierry,J.-C. and Moras,D. (1998) Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD1: archaeon specificity and catalytic mechanism of adenylate formation. EMBO J., 17, 5227–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbula D.L., Becker,H.D., Chang,W.Z. and Söll,D. (2000) Domain-specific recruitment of amide amino acids for protein synthesis. Nature, 407, 106–110. [DOI] [PubMed] [Google Scholar]
- Tumbula-Hansen D., Feng,L., Toogood,H., Stetter,K.O. and Söll,D. (2002) Evolutionary divergence of the archaeal aspartyl-tRNA synthetases into discriminating and non-discriminating forms. J. Biol. Chem., 277, 37184–37190. [DOI] [PubMed] [Google Scholar]
- Varshney U., Lee,C.P. and RajBhandary,U.L. (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of E. coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem., 266, 24712–24718. [PubMed] [Google Scholar]
- White O. et al. (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science, 286, 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R., Olsen,G.J., Ibba,M. and Söll,D. (2000) Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev., 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]