Abstract
Recently, we reported the isolation and characterization of an anti-laminin antibody that modulates the extracellular matrix-dependent morphogenesis of endothelial cells. Here we use this antibody to precisely map the binding site responsible for mediating this biologically important interaction. By using a phage display-assisted mapping strategy to preserve protein structure, we demonstrate for the first time that the coiled-coil region of laminin contains a cell binding site. The adhesion motif is formed by residues contributed by both α and γ chains, and is located in the middle part of the rod-like portion in a highly flexible area, which corresponds to a protease-susceptible site. Based on this information, a peptide mimotope was used to characterize the cognate receptor. Although we can not rule out the implication of other receptors, our results demonstrate that the laminin helical rod active site interacts with α2β1 integrin on the surface of endothelial cells. These findings provide new insight into the complex mechanisms regulating capillary morphogenesis.
Keywords: coiled-coil/extracellular matrix/integrin/laminin
Introduction
Angiogenesis, the growth of new blood vessels from pre-existing ones, is critical for normal growth, development and repair. However, aberrant angiogenesis is an important contributing factor in several pathological disorders (Carmeliet and Jain, 2000). The control of angiogenesis and the molecular regulation of each step of the process are areas of intense investigation, since the ability to inhibit capillary growth has the potential of arresting progression of solid tumors. A recent strategy has been to identify endogenous angiogenesis inhibitors and use these molecules to control angiogenesis. Endostatin, which is a 20 kDa C-terminal cleavage product of collagen XVIII, is a potent antagonist of angiogenesis that exerts its inhibitory effects on angiogenic blood vessels by blocking α5 and αv integrin-dependent endothelial cell (EC) functions (Rehn et al., 2001). Modulation of integrin function and signaling is emerging as a common functional theme among various angiogenesis inhibitors. However, with the exception of the αv integrins, relatively little is known about the mechanisms by which cell adhesion molecules function in angiogenesis (Bischoff, 1997).
We have previously reported the isolation and characterization of an anti-laminin antibody (L36) that modulates the ECM-dependent morphogenesis of ECs (Sanz et al., 2001, 2002a). The ECM provides structural support and regulates cellular morphogenesis by acting both as a repository for growth factors and as a ligand for cellular receptors. However, a complete mechanistic explanation for this fundamental phenomenon of morphogenesis is lacking, since our knowledge about the contribution of adhesion systems to such a complex process is very limited.
Basement membranes (BM) are highly specialized ECMs, usually found underlying epithelial and endothelial cells (Timpl, 1996). BM deposition is known to be crucial in the functional organization and maturation of newly formed vessels (Stupack and Cheresh, 2002). Typical components of BM include laminins (LN), a few characteristic collagens, proteoglycans and nidogen/entactin, with LN playing a central role in the interaction between cells and extracellular matrix (Belkin and Stepp, 2000). The known receptors and receptor-like molecules for LN are various β1 and β4 integrins, dystroglycan, leukocyte antigen-related protein (LAR), heparan sulfates and other cell surface molecules. By virtue of these receptor interactions, intracellular signaling events are triggered that regulate cell proliferation and determine cell and tissue development, differentiation and function (Belkin and Stepp, 2000).
L36 antibody treatment at the time of EC plating on ECM preparations (Matrigel) strongly inhibits assembly into tubular structures, with cells remaining dispersed and resembling the morphology of cells plated on plastic (Sanz et al., 2002a). These morphological changes are similar to those observed when EC cultures plated on matrigel substrates are treated with the collagen XVIII NC1/endostatin domain (Kuo et al., 2001). In fact, it has recently been reported that these properties of endostatin are primarily mediated by laminin (Javaherian et al., 2002). Furthermore, L36 inhibits angiogenesis in vivo in the chick embryo chorioallantoic membrane assay and prevents the establishment and growth of subcutaneous tumors in mice (Sanz et al., 2002a). In the current work we have used this antibody to precisely map the binding site responsible for mediating this biologically relevant interaction. Based on this information, peptide mimotopes were used to characterize the cognate receptor, thereby gaining general insight into the processes leading to angiogenesis.
Our results demonstrate that the adhesion motif is located in the middle part of the triple coiled-coil domain and is formed by residues contributed by both α and γ chains. This finding implies that the heterotrimeric portion of the LN molecule plays a critical functional role, beyond the structural one. Furthermore, our data reveal the implication of integrin α2β1 in EC adhesion to the LN helical rod peptide mimotope.
Results and discussion
Topographic localization of the L36 epitope within the laminin molecule
The structure of LN-1, as seen by electron microscopy after rotary shadowing, revealed an unusual extended, four-armed cruciform shape, with three short arms and a long arm (Figure 1A). While N-terminal regions of the three chains form each short arm, more C-terminal portions of these three chains associate in a triple coiled-coil α-helix, forming the rod-like part of the long arm. The long arm appears as a rather flexible rod with a large terminal globular domain composed by the C-terminal region of the α subunit (Engvall and Wewer, 1996; Tunggal et al., 2000).
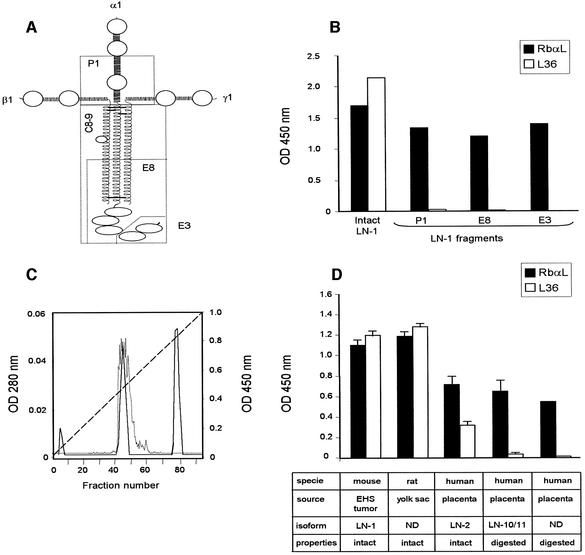
Fig. 1. (A) Schematic model of laminin-1 (LN-1) with localizations of fragments P1, E8, E3 and C8-9. Horizontal lines connecting the chains represent disulfide bridges. (B) ELISA to assay binding of L36 antibody (10 µg/ml) and a rabbit anti-LN (RbαL) antiserum (1 µg/ml) to intact LN-1 or to purified fragments (P1, E8 and E3). (C) Separation of cathepsin G-derived LN fragments (solid line) and ELISA to assay binding of L36 to cathepsin G-derived fractions (dotted line). Broken line represent the NaCl gradient. (D) ELISA to assay binding of L36 and RbαL to purified laminins directly attached to plastic.
To study the relative position of the L36 epitope within the LN molecule, we tested the reactivity of this antibody against purified fragments. As shown in Figure 1B, the antibody recognized affinity-purified intact LN-1, but did not show any reactivity with the LN-derived fragments P1, E8 and E3. Plastic immobilized fragments were recognized by a rabbit anti-LN-1 antiserum, and the absorbance profiles were similar to those observed with intact LN-1 (Figure 1B). Fragment P1 is comprised of the inner part of the three disulfide-bonded short arms, and fragment E8 together with fragment E3 comprises the complete terminal half of the long arm (Figure 1A). These results indicate that the epitope recognized by L36 is located either on the upper long-arm region or on the outer segment of one of the three short arm structures of the molecule.
To further investigate the topographic localization of the L36 epitope, mouse LN-1 was treated with cathepsin G, and purification of native fragments was achieved by ion-exchange chromatography (Figure 1C). L36 reactivity was confined to fragment C8-9, eluted from the Mono Q column as a peak at 0.25 M salt (Bruch et al., 1989). Fragment C8-9 contains the entire triple coiled-coil region and the major part of the terminal globular domain of the long arm (Figure 1A). L36 showed no reactivity with other cathepsin G-derived fragments (Figure 1C). By contrast, both peak fractions were recognized by the polyclonal anti-LN-1 antiserum (data not shown). Taken together, our results indicate that the epitope recognized by L36 is located on the upper long-arm region (domains II and I).
Epitope mapping and sequence analysis
To precisely map the epitope recognized by L36, plastic immobilized antibody was used to screen a phage display peptide library fused to the N-terminal region of M13 coat protein III; each peptide consisted of seven random amino acids flanked by two cysteines. Bound phages were either eluted by adding an excess of native LN-1 in solution (competitive elution), or by decreasing the pH to 2.2 (non-specific elution). After three rounds of biopanning, 12 randomly selected phage clones from each group were sequenced. The amino acid sequences could be arranged into two groups on the basis of their similarity. All phages eluted with LN-1 encoded an identical displayed peptide sequence: LPKHARS. The second group was characterized by the consensus sequence IXWNXXD, where X represents an unrestricted residue (see Supplementary table I, available at The EMBO Journal Online). Eight clones displayed the sequence IRWNYND. Selected phage clones displaying the sequence LPKHARS or IRWNYND were assayed for binding to immobilized L36 by ELISA. Both recognized L36 specifically, with phages displaying the sequence IRWNYND giving higher absorbance signals, indicating that these phages bound with higher affinity, in agreement with the fact that these were obtained by the harsh non-specific elution (data not shown). Phage binding to a control recombinant antibody (CGS-1) was insignificant (data not shown).
Two important features are readily apparent from the selected sequences. First, the central residues (KHARS) of the sequence LPKHARS exactly match amino acids 524–528 of the human LN α3 chain, located in domain I of the long arm (see Supplementary table II). Domains II and I span 600 residues, and are proposed to fold into an elongated helical structure arranged in a heterotrimeric parallel coiled-coil, making up the rod-like portion of the long arm of the molecule. Given that the sequence of both domains is poorly conserved among the LN subunits (20–40%) (Engvall and Wewer, 1996), it is noteworthy that the selected sequence is located in the only highly conserved region among α subunits, apart from the N- and C-terminal Cys residues implicated in the disulfide bridges that stabilize the trimeric coiled-coil structure (see Supplementary table II). The central residues of the sequence (His and Ala) are conserved among subunits α1–5 and among α subunits from different species. The consensus sequence IXWNXXD corresponds closely (with the exception of the Trp residue) to amino acids 1366–1372 of the human LN γ1 chain and amino acids 1364–1370 of the mouse γ1 chain. Interestingly, the sequence is also located in domain I and is highly conserved in the γ subunits. Both sequences are located in two distinct chains, but closely linked spatially, as indicated by the quaternary structure of LN (see below). These results suggest that the epitope recognized by L36 involves amino acids from both the α and γ chains, and that the two families of selected peptides (see Supplementary table I) are mimotopes of two different parts of a structurally related epitope (Luzzago et al., 1993).
Molecular modeling
The sequences of LN domains II and I contain ∼80 heptad repeats of the form (abcdefg)n, typical of coiled-coil structures. The mean block of consecutive heptads is ∼50 residues long, with residues at positions a, d usually apolar and buried, and residues at positions e, g frequently charged (Beck et al., 1993). This sequence pattern is imperfect, or is interrupted, at several locations along the sequence, which suggests that the structure might not be a long continuous coiled-coil rod, but a succession of coiled-coil blocks separated by short, irregular or flexible regions. In the β chains only, domains II and I are separated by an additional domain (B domain) of ∼35 residues. Domains II and I guide the intramolecular assembly and determine the subunit composition of the particular LN isoform. Beck and coworkers proposed that the specificity of the coiled-coil interaction in LN lies in the interhelix ionic interactions, and found them to be optimized in a parallel trimeric coiled-coil structure with an anticlockwise order of the α, β and γ chains when viewed from the N-termini (Beck et al., 1993).
We have used these findings to analyze the potential to adopt coiled-coil structures by the sequences of LN chains containing the peptide sequences identified in this work (see Supplementary table I) and to construct a model of this LN region. Alignment of the sequences of the α3, β1 and γ1 chains of human LN-6 assumes that the coiled-coil region starts after the pair of Cys residues at the beginning of domain II. Deletion of the additional domain in chain β1 (33 residues) produces an alignment (see Supplementary figure 1) in which the regions around the sequences identified in selected phages have continuous stretches of heptad repeats. However, this particular alignment does not match the heptad repeats in the three chains. A first match is obtained by shifting chain β1 by one amino acid to the N-terminus, and chain γ1 by two residues to the C-terminal end, with respect to chain α3 (β–1, γ+2). An alternative alignment matching the heptads can be achieved with the shifts β–8, γ–5. In both alignments, the regions around the sequences identified in selected phages have a high probability of adopting a coiled-coil structure (see Supplementary figure 1). The second alignment pairs the sequence KHA in chain α3 with a region in β1 that has no clear heptad pattern and has high coiled-coil potential.
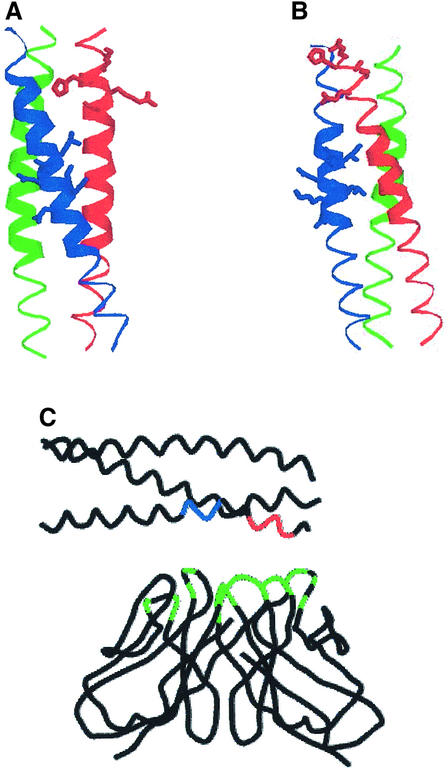
We have built model structures of this region, using as template the crystal structure of a designed heterotrimeric parallel coiled-coil (Nautiyal and Alber, 1999; Protein Data Bank entry 1bb1). The model corresponding to the second alignment (not shown) contains a large number of steric clashes involving the two proline residues at the N-terminal end of chain β1. These clashes would strongly destabilize the coiled-coil structure, so that the local structure might be somewhat different, but we have no basis for modeling it on any other type of structure. The model corresponding to the third alignment (Figure 2A) complies with the criteria for a reasonable structure (see Materials and methods). In particular, the distribution of residue types over the inside and the outside of the molecule and the average structural packing environment scores are within normal ranges and do not show large steric clashes. This model has a region (indicated by thick ribbons) where a knobs-into-holes packing of side chains typical of coiled-coils exists. In this structure, the two peptide regions appear in a continuous patch on the rod, a disposition of the residues that would facilitate the binding of L36 to both sequences, as shown in Figure 2C. The His residue in the sequence KHARS corresponds to one of the d positions in the corresponding heptad and would be buried. While it is not unusual to find polar residues buried in coiled-coils (Burkhard et al., 2001), the conservation of this residue in LN α chains (see Supplementary table II) suggests a functional role, implying that the histidine side chain is either more exposed than observed in the model structure, or the structure is distorted upon binding so that the histidine becomes accessible to its receptor. To test wheter this region of the coil is less stable than the rest, we examined the structure after a short (7 psec) molecular dynamics trajectory and energy minimization. Both ends of the coil were distorted, but to a larger extent at the N-terminus, and the His residue side chain was exposed to the solvent. As discussed above, if the correct alignment is the second one, the region where the His is located is not in a coiled-coil conformation and may well be solvent-exposed. We do not have enough information to select one of the two models as the best representation of the actual structure of this LN region, but it seems that at least part of it is highly likely to be in a coiled-coil structure. We have followed a similar strategy to analyze the sequences and build structural models for other LNs considered in this work (see Supplementary figure 2). The structure of mouse LN-1 (Figure 2B) places the His residue in position g, exposed to the solvent, and also has a region with the typical side-chain packing of coiled coils. In human LN-2 the His is also exposed (see Supplementary figure 2), but the packing of the side chains is not as regular as the one in mouse LN. No models have been built for human LN-10 or LN-11, since the sequence of the α5 chain does not have a clearly defined pattern and it is uncertain how to pair the three chains; but the probability for adopting a coiled coil is still higher than those of known coiled-coil sequences.
Fig. 2. Model structures of the region of the long arm containing the peptide sequences found to be involved in binding to L36. (A) Model structure of human LN-3 using the third alignment (see Supplementary data). The Cα traces of chains α3, β1 and γ1 are shown as red, green and blue ribbons, respectively. The side chains of residues KHARS in chain α3 and ILNNLKD in chain γ1 are represented with sticks. The thick ribbon indicates the region of the structure where the packing of side chains in the core is recognized as the one typical for coiled-coils by the program SOCKET (Walshaw and Woolfson, 2001). (B) An analogous model for mouse LN-1 second alignment (see Supple mentary data). (C) A possible arrangement of the model structure shown in (A) on the surface of the L36 antibody fragment. Only the Cα trace is shown, with red and blue colors indicating the regions of sequences KHARS and ILNNLKD, respectively. The antibody structure used for this figure is the Fv fragment of the D1.3 monoclonal murine antibody against hen egg-white lysozyme. The residues colored in green are those that make contacts with the antigen in the crystal structure (PDB code 1vfa). Plots (A) and (B) were prepared with RASMOL (Sayle and Milner-White, 1995) and plot (C) with SYBYL (Tripos Inc., St Louis, MO).
In summary, the region where the binding site for L36 is located has a large potential to adopt a coiled-coil structure, with a possible departure from a regular coiled-coil structure or increased flexibility at residues KHARS. This model structure provides an explanation for the identification of two peptide sequences in the α and γ chains interacting simultaneously with the variable regions of the antibody. Furthermore, the analysis of several LN chain sequences suggests that these general conclusions can be extended to other isoforms of LN.
L36 binding profile
In the light of this model structure, we could anticipate: (i) a conformation dependency on L36 binding to LN; and (ii) a broad reactivity profile. To verify this hypothesis, we tested the reactivity of L36 to all the purified LN preparations commercially available. Figure 1D shows that L36 recognized all the intact LNs tested, in spite of their different origins: LN-1 (α1β1γ1) extracted from mouse EHS tumors; LN from rat L2 yolk-sac tumors; and LN-2 (α2β1γ1) from human placenta. L36 gave no significant signals on two LNs purified from pepsinized human placenta: LN-10/LN-11 (α5β1γ1/α5β2γ1) and a non-characterized LN preparation (Figure 1D). Pepsin digestion of LN gave rise to two major fragments, designated P1 (Figure 1A) and P2 (outer segment of the short arms), but degraded ∼70% of the molecule (including the heterotrimeric rod-like structure) into small peptides (Rohde et al., 1980; Ott et al., 1982). Thus, the L36 binding profile (conformation-dependency and LN panreactivity) is consistent with the model structures we have constructed. The physiological significance of our data is further supported by the fact that L36 recognizes vascular-associated BMs in various human and murine tissues (Sanz et al., 2001, 2002a).
Helical conformation-dependency of cell binding activity is a characteristic feature of many LN isoforms (Aumailley and Smyth, 1998). The current interpretation is that cell binding sites are located on adjacent globular domains, which, for correct folding, require the presence of the adjacent helical rod (Sung et al., 1993). To date, structure–function relationships have come largely from studies with proteolytic fragments and synthetic overlapping peptides covering the entire sequence of individual LN subunits (Nomizu et al., 1998). However, it is evident that with these reductionist approaches, functionally active sites located either in protease-sensitive regions or in highly structured areas will not be identified. The central portion of the long arm fulfils both criteria, and this could explain why it was assumed to have an exclusively structural role. However, it has recently been reported that the long arm of LN contains a site for binding to the N-terminal domain of agrin, a protein involved in synapse formation at the neuromuscular junction (Kammerer et al., 1999). Agrin binds to the γ1 chain of LN-1, and the coiled structure seems to be required for high-affinity binding. This interaction is probably of ionic nature, since the agrin domain is highly charged and the exposed residues in coiled-coils are usually polar or charged. Therefore, domains II and I of LN have a functional role beyond the structural one.
Biological activity of a synthetic peptide mimotope
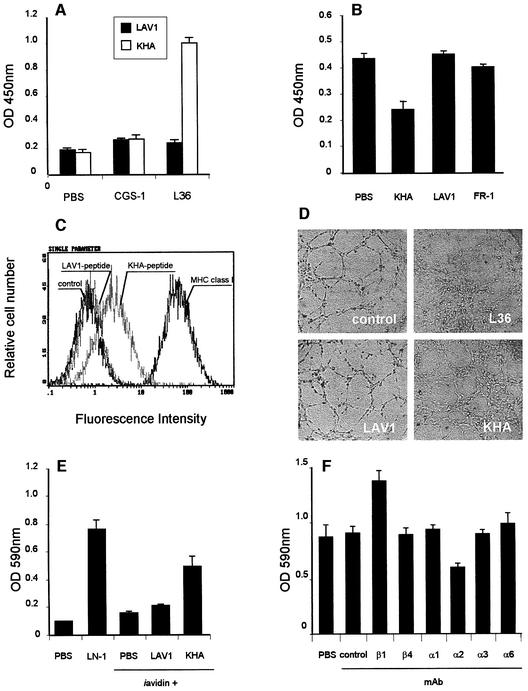
To determine the activity of the LN region that we have modeled, a cyclic peptide, whose sequence corresponds to the insert of phages eluted with soluble LN-1 (KHA peptide) and includes a short spacer segment (GGGS) present in the phage, was chemically synthesized (CLPKHARSCGGGSK) and biotinylated at the C-terminus (Luzzago et al., 1993). As seen in Figure 3A, KHA retained its reactivity independently of the phage context and bound specifically to plastic-immobilized L36. There was no significant cross-reactivity between an irrelevant biotinylated peptide (CSRARKQAASIKVAVSADR) acting as control (LAV1) and L36, nor between KHA or LAV1 and CGS-1 (Figure 3A). In competition ELISA, L36 and a molar excess of KHA, LAV1 or FR-1 (a cyclic peptide based on the cell-attachment sequences of fibronectin: CRGDSPASSC), were incubated for 1 h and then added to the wells of a plate precoated with LN-1. The binding of L36 to LN-1 was inhibited by KHA, but not by the control, biotinylated (LAV1) or cyclic (FR-1) peptide (Figure 3B). The results indicate that, in spite of representing only a portion of the L36 epitope (incomplete epitope), the amount of binding energy contributed by the fraction of contact points involving amino acids present in the KHA peptide sequence is sufficient for the formation of a detectable complex. The peptide binds to the antigen-binding site of L36, and is by itself capable of displacing the antibody from the LN.
Fig. 3. (A) Binding of biotinylated LAV1 or KHA peptide (10 µg/ml) to plastic-immobilized L36 or CGS-1 antibodies. One representative experiment out of three independent ones is shown. (B) Competition ELISA of LAV1, KHA or FR-1 peptide (100 µg/ml) with L36 (0.2 µg/ml) binding to plastic-immobilized LN-1. One representative experiment out of three independent ones is shown. (C) FACS profiles of HMEC-1 cells labeled with biotin-conjugated anti-MHC class I mAb (2 µg/ml), biotin-conjugated KHA peptide or biotin-conjugated LAV-1 peptide (10 µg/ml). Fluorescence intensity (abscissa) is plotted against relative cell number (ordinate). (D) Light microscopic photos of HMEC-1 cells plated for 16 h on a Matrigel monolayer in the presence of medium (control), L36 (62.5 µg/ml), LAV1 or KHA (62.5 µg/ml). (E) Cell attachment to different substrates. HMEC-1 cells were allowed to adhere for 30 min at 37°C to 96-well covalynk wells coated with LN-1, avidin or avidin plus biotinylated peptide (LAV1 or KHA). Each value represents the mean of four separate determinations ± SD. (F) Effect of anti-β and anti-α integrin mAb (10 µg/ml) on the attachment of HMEC-1 cells to KHA. Assays were performed on untreated controls, or in the presence of a control anti-MHC class I mAb (W6/32), or anti-integrin mAb, and after 30 min the attached cells were assessed by crystal violet staining. Each value represents the mean of three separate determinations ± SD.
To determine the effect of KHA on cell atachment, we tested its ability to interfere with EC adhesion to plastic-immobilized LN. Cell attachment to LN-1 is a complex process in which different integrins and several other non-integrin cell surface molecules cooperate (Mercurio, 1995). Therefore, L36 like anti-integrin antibodies only partially blocked cell adhesion to LN-1 (Sanz et al., 2001). Cultures of HMEC-1 endothelial cells treated with KHA gave levels of inhibition similar to those observed with L36 (data not shown). In contrast, no significant inhibition of adhesion to LN-1 was observed with LAV1 or FR-1 (data not shown). The interaction of KHA with the membrane receptor molecule was further studied by immunofluorescence and flow cytometry. Figure 3C shows one representative experiment performed on HMEC-1 cells incubated with either KHA or LAV1. Specific staining was evident with KHA, but not with LAV1. We also explored the behavior of KHA in an in vitro Matrigel morphogenesis assay. In the absence of KHA, ECs spontaneously aggregate and assemble into multicellular capillary-like structures (CLS). When KHA was added, EC assembly into CLS was strongly inhibited. The cells still attached to Matrigel but they assumed a scattering morphology (Figure 3D). Results are similar to those obtained with L36-treated cultures. Control peptide (LAV1) had no effect on endothelial cell CLS formation (Figure 3D). We conclude that the KHA peptide reproduces the activity of the LN region that is mimicking and binds to a putative membrane receptor molecule/s with enough affinity to disrupt that critical interaction/s that take place during the BM-dependent capillar morphogenesis.
Integrin antibodies modulate cell adhesion to KHA peptide
To investigate whether KHA also promotes adhesion, cell attachment experiments were performed using peptide-coated plastic plates. KHA showed strong EC attachment activity, similar to that of LN-1, whereas no significant effect was observed with LAV1 (Figure 3E). Cell adhesion to ECM and BM is mostly mediated by transmembrane αβ heterodimers of the integrin superfamily, which is subdivided into eight families on the basis of the β subunit. Since integrins that are known to interact with LN belong to the β1 and β4 families (Belkin and Stepp, 2000) we first examined the functional effects of anti-β1 and anti-β4 integrin mAbs on cell attachment mediated by KHA peptide. As shown in Figure 3F, the anti-β1 Lia 1/2 mAb promoted KHA-mediated HMEC-1 cell attachment, whereas no significant effect was exerted by the anti-β4 ASC-3 mAb. It has been previously reported that Lia 1/2 mAb triggers cell aggregation of different leukocyte cell lines (Campanero et al., 1992); however, we did not find induction of homotypic cell aggregation after treatment of HMEC-1 cells with this mAb (data not shown). The effect of anti-β1 mAb Lia 1/2 was not only aggregation independent, but also substratum specific, since no significant induction of cell attachment was observed to plates coated with LAV1 peptide or BSA (data not shown). Possible mechanisms accounting for the β1-mediated enhancement of EC-KHA peptide interaction include: (i) the binding of anti-β1 mAb may trigger intracellular signals; and (ii) a conformational change of the αβ heterodimer as a result of the binding of anti-β1 mAb resulting in a higher-affinity form of the integrin that enables the interaction with the ligand.
Next we examined the functional effects of various blocking anti-α integrin antibodies. HMEC-1 cell attachment to KHA was reduced by anti-α2, but not by anti-α1, -α3 or -α6 mAb (Figure 3F). Anti-α2 P1E6 mAb inhibited HMEC-1 cell attachment on KHA-coated wells to 70% of the control levels. These findings suggest that α2β1 integrin contributes to cell attachment to KHA, although the participation of other receptors can not be ruled out since the inhibition by anti-α2 integrin mAb is partial. Similar results were obtained when using HT1080 cells (data not shown).
Expression of α2β1 integrin on the cell surface mediates the recognition of the KHA peptide
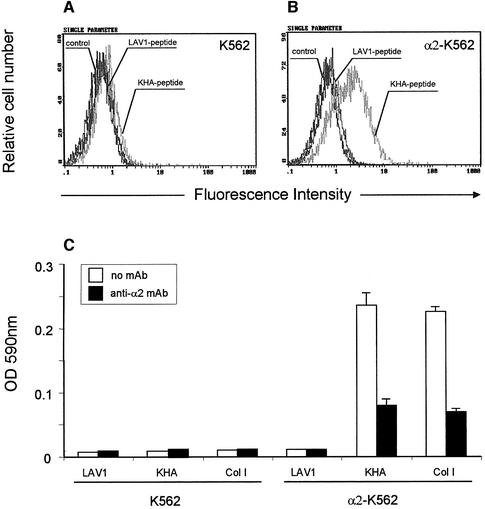
To determine the contribution of α2β1 integrin to cell binding to KHA, we assayed for adhesion with K562 cells. These cells only express the integrin α5β1 (Elices et al., 1990) so therefore could not bind to KHA (Figure 4C). Accordingly, specific cell-surface staining with KHA was not evident (Figure 4A). We then studied the attachment of K562 cells transfected with a cDNA encoding α2 chain to KHA. K562 cells expressing α2β1 integrin bound to KHA and to type I collagen (Figure 4C). Specific cell surface staining was evident, with no demonstrable binding using the LAV1 peptide (Figure 4B). Anti-α2 integrin antibody was able to decrease cell adhesion on both KHA and on collagen type I (Figure 4C). No specific binding of KHA was observed to α4 chain transfected K562 cells (data not shown). These experiments showed that the expression of α2β1 integrin is necessary and sufficient for the recognition of the KHA peptide.
Fig. 4. FACS profiles of K562 cells (A) and α2-K562 cells (B) labeled with LAV1 peptide or KHA peptide (10 µg/ml). Fluorescence intensity (abscissa) is plotted against relative cell number (ordinate). (C) K562 and α2-K562 cell adhesion on control LAV1 peptide, KHA peptide and type I collagen (Col I) in the absence or presence of the anti-α2 integrin P1E6 mAb (10 µg/ml). One representative experiment out of three is shown.
Binding of recombinant I-domain fragments to the KHA peptide
To further investigate the implication of α2β1 integrin in cell binding to this region and to determine the nature of the interaction, a recombinant human integrin α2 I-domain was generated as a GST fusion protein. The α2 subunit is a member of a subset of α integrin chains that contain an ∼200 residue long insertion located near the N-terminus, often referred to as the I (or inserted) domains. Crystal structures of several different α I-domains have been solved, showing great similarities among them and suggesting that they bind their cognate ligands in similar fashions (Tulla et al., 2001). The complex of the α2 I-domain to a segment of the triple helix of collagen is an example of receptor binding to an elongated helical structure with predominantly structural roles (Emsley et al., 2000). The integrin–collagen complex contains a metal ion coordinated by residues of three loops of the I-domain and a glutamate residue of collagen. A similar binding could involve one of the 14 glutamate side chains of the model structures we have constructed, some of them spatially close to the sequence KHARS (Figure 2).
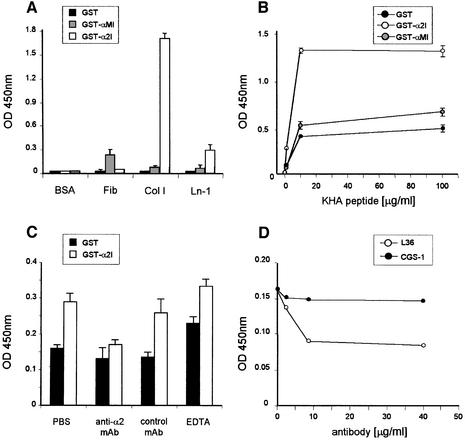
As a control I-domain, a GST human integrin αM I-domain fusion protein was generated. As reported previously (Dickeson et al., 1997), recombinant GST– I-domain fusion proteins bound ligands specifically in a cation-dependent manner. Integrin αM I-domain bound to fibrinogen, while α2 I-domain bound to both collagen I and LN (Figure 5A). However, in this case, collagen I was a better ligand than LN, as already reported (Calderwood et al., 1997). No binding to fibronectin (or to a range of other proteins not present in BMs) was observed (data not shown). We first examinated the binding of KHA to the I-domain fusion proteins. Figure 5B shows that the GST–α2I protein supports dose-dependent, saturable binding of KHA, while GST–αMI and wild-type GST exhibit very little binding. There was no significant cross-reactivity between the LAV1 peptide and the recombinant I-domain fragments (data not shown). KHA binding to GST–α2I was reduced to the levels of the GST control by the anti-α2 blocking mAb P1E6, but not by an irrelevant control mAb (Figure 5C). These data indicate that KHA binding to recombinant α2I is specific.
Fig. 5. (A) Binding of GST, GST–α2 I and GST–αM I (10 µg/ml) to different substrates in a solid phase assay (LN-1, laminin 1; Col I, collagen I). (B) Binding of KHA peptide to plastic-immobilized GST, GST–α2 I or GST–αM I. (C) Inhibition of the binding of KHA (5 µg/ml) to immobilized GST–α2 I by the anti-α2 integrin P1E6 mAb (10 µg/ml) or EDTA (10 mM). (D) Competition ELISA of L36 or CGS-1 antibody with α2I (10 µg/ml) binding to plastic-immobilized LN-1. Graphs show the results of a representative experiment performed in triplicate; the experiments were repeated at least three times.
We next examined the divalent cation requirement for KHA–α2I interaction. Figure 5C shows that the addition of EDTA (up to 10 mM) does not inhibit the interaction. The KHA peptide contains no acidic residues, indicating that its mode of binding to integrin must be different from the binding of collagen. α2 I-domain binding to LN-1 occurs via both cation-dependent and cation-independent mechanisms (Dickeson et al., 1998), indicating that other interactions besides those involving the MIDAS loops are involved in the recognition of both molecules. A binding site distinct from the MIDAS would be consistent with the KHA peptide binding to the I-domain in the presence of EDTA (Figure 5C; data not shown).
It has been reported that a cyclic peptide with the sequence CTRKKHDNAC, derived from a snake venom metalloprotease, binds to the recombinant I-domain of human α2β1 integrin and prevents the binding of collagen types I and IV and LN-1 (Ivaska et al., 1999). Mutational and modeling analyses have indicated that the essential residues for this binding are RKKH, and that the recognition site is mediated by acidic residues on the surface of the I-domain in the vicinity of the MIDAS (Pentikainen et al., 1999). Although the sequence similarity between the KHA peptide mimotope and the snake venom RKKH peptide suggests that their binding mode is similar, this may not be the case, since RKKH binding needs magnesium, while KHA does not. Although the binding mode may be different, it is interesting that RKKH interferes with the binding of collagen by interacting with a site that is close to, but different from, the collagen binding site.
To further analyze whether α2I could recognize the same site in the LN molecule as L36, a competitive ELISA was designed. LN-coated wells were preincubated with L36 at different concentrations prior to α2I addition. The binding of α2I to LN-1 was inhibited in a concentration-dependent fashion by L36, but not by a control antibody (Figure 5D). The competition of KHA with LN binding to both integrin and L36, and the direct binding of KHA to both the integrin I-domain and L36, strongly suggests that the binding sites are very similar. However, whether the integrin binding site exactly matches the antibody binding site is not clear, especially since the sequence on the γ1 chain is not necessary for a measurable binding of the integrin domain. The inhibitory effect of L36 on EC morphogenesis would also be consistent with overlapping binding sites, the antibody blocking only part of the integrin binding site or impeding integrin binding by steric hindrance.
In summary, by using an indirect phage display-assisted mapping strategy to preserve protein structure, we have demonstrated for the first time that the long arm of LN contains a cell binding site. According to our data, the α2β1 integrin is implicated in this binding. The adhesion motif is formed by residues contributed by both α and γ chains, and is located in the middle part of the helical rod. Indeed, electron microscopy studies indicate that a protruding kink is located in this area, a structural feature that facilitates the putative cell-interactive properties of the region (Bruch et al., 1989). Furthermore, this flexible region corresponds to a susceptible site cleaved by several proteases, including porcine pancreatic elastase, which is responsible for the creation of the E8 fragment. The level of sequence homology of this region, together with the binding data reported here, suggest that the identified cell binding motif could be functional in other LN isoforms expressed in different localizations. We propose that this assembly-dependent motif would act as a ‘sentinel’ domain, whose integrity is checked by specific cell-surface receptors. This interaction would provide a feedback mechanism, acting as a biosensor to fit the differentiation state of the cell to the physical nature of the surrounding extracellular matrix.
Materials and methods
Epitope mapping of L36 scFv antibody
A phage peptide library (Ph.D.-C7C, New England Biolabs, Hitchin, UK) that consists of filamentous phage-displaying randomized 7-mer peptides was panned against L36. Purified L36 (5 µg/well) was coated to Maxisorp ELISA plate wells (Nunc, Roskilde, Denmark), and binding phages were selected from the library according to the manufacturer’s protocol. Single clones displaying relevant sequences were tested for binding to L36 and a control antibody by ELISA (Sanz et al., 2001).
Molecular modeling
The program Multicoil (Wolf et al., 1997) was used to look for the heptad repeats in the sequences of LN chains and to examine their potential to adopt parallel coiled-coil structures. The model structures of the LN sequences were constructed using the procedures implemented in the program WHATIF (Vriend, 1990). The quality of the models was evaluated with the WHATCHECK option of WHATIF. Molecular dynamics simulations were performed with the program SYBYL (Tripos Inc., St Louis, MO). Briefly, after adding hydrogens to the model, the structure was refined by 200 steps of Powell energy minimization, subjected to 7 psec of molecular dynamics in vacuo at 300 K, and energy minimized by another 200 Powell steps. The unrefined model was manually docked to the structure of the Fv fragment of D1.3 murine antibody bound to hen egg-white lysozyme after deletion of the lysozyme chain (Bhat et al., 1994), PDB code 1vfb. Only gross disposition of both molecules attending to the possible simultaneous recognition of the sequences in chains α3 and γ1 was intended, ignoring details about energy or precise shape complementarity.
Specific ELISA
The ability of purified soluble antibody (L36 or CGS-1) to bind LNs or LN-1-derived fragments was studied by ELISA as described (Sanz et al., 2001). To study the capacity of the synthetic peptides (KHA or LAV1) to bind to L36 or CGS-1, Maxisorp 96-well plates were coated ON (1 µg/well) with purified antibody. Then 100 µl of a peptide solution in PBS–1% BSA–0.05% Tween-20 was added for 1 h at RT and developed with HRP-conjugated streptavidin. For competition ELISA, a 0.2 µg/ml antibody solution of L36 was incubated with increasing concentrations of KHA, LAV1 or FR-1 peptide. After 1 h at 37°C, 100 µl per well of each mixture was added to the wells of a plate precoated with LN-1 and incubated for 1 h at RT.
Recombinant integrin I-domains generation and binding assays
Cloning and expression of recombinant integrin α2 I- and αΜ I-domains was performed as described (Dickeson et al., 1997; Griggs et al., 1998). For binding assays, the wells of a Maxisorp plate were coated ON at 4°C with 100 µl of 20 µg/ml LN-1, collagen I or fibrinogen. Wells were blocked with 4% BSA in TBS for 1 h, and then 20 µg/ml of GST fusion proteins (α2 I or αM I) or wild-type GST was added in 1% BSA, 0.05% Tween-20 TBS for 1.5 h at RT. Wells were washed and anti-GST antibody was added followed by incubation with a HRP-conjugated anti-goat/sheep mAb. Binding of soluble ligands to plastic immobilized α I-domains was measured using assays adapted from Calderwood et al. (1997). In competition experiments a 5 µg/ml KHA peptide solution was incubated with a 10 µg/ml antibody solution for 1 h at RT before transfer to the wells. The GST–α2 I fusion protein was cleaved with Factor Xa (Novagen, Madison, WI) that was removed with Xarrest agarose (Novagen), and the cleavage mixture was passed a second time down the glutathione–Sepharose column. The rα2I remained in the flow-through, as assessed by SDS–PAGE, and was dialyzed against PBS.
L36 blocking assay
The wells of a Maxisorp plate were coated ON at 4°C with 100 µl of 20 µg/ml LN-1. After blocking, different concentrations of L36, ranging from 40 to 1.6 µg/ml were added. The CGS-1 antibody was used as control. After washing, cleaved and purified α2 I (20 µg/ml) was added for 1.5 h and detected with 100 µl of TEA 1/41 mAb (anti-human α2 integrin), followed by incubation with a HRP-conjugated goat anti-mouse IgG (Fc specific).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank E.Ades, M.E.Hemler, D.Neri, A.Rodriguez-Tebar, F.Sánchez-Madrid, J.Teixido and R.Timpl for providing reagents. We greatly appreciate the critical reading of this manuscript by A.G.Arroyo and R.Vile. Special thanks to S.Jones for his editorial assistance. This study was supported by grants from the Fondo de Investigación Sanitaria (grant PI021144), from the Ministerio de Ciencia y Tecnología (grant BIO2001-0385) and from the 5th framework of the European Community (grant EC QLK3-CT-1999-00386) to L.A-V., and by grants from the Danish Research Council and the Carlsberg Foundation to P.K. L.S. and L.G-B. were supported by the European Community. B.B. is a recipient of a Comunidad Autónoma de Madrid training grant (01/0369/2000).
References
- Arroyo A.G., Sanchez-Mateos,P., Campanero,M.R., Martin-Padura,I., Dejana,E. and Sanchez-Madrid,F. (1992) Regulation of the VLA integrin-ligand interactions through the β1 subunit. J. Cell Biol., 117, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumailley M. and Smyth,N. (1998) The role of laminins in basement membrane function. J. Anat., 193, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K., Dixon,T.W., Engel,J. and Parry,D.A. (1993) Ionic interactions in the coiled-coil domain of laminin determine the specificity of chain assembly. J. Mol. Biol., 231, 311–323. [DOI] [PubMed] [Google Scholar]
- Belkin A.M. and Stepp,M.A. (2000) Integrins as receptors for laminins. Microsc. Res. Tech., 51, 280–301. [DOI] [PubMed] [Google Scholar]
- Bhat T.N. et al. (1994) Bound water molecules and conformational stabilization help mediate an antigen-antibody association. Proc. Natl Acad. Sci. USA, 91, 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff J. (1997) Cell adhesion and angiogenesis. J. Clin. Invest., 100, S37–S39. [PubMed] [Google Scholar]
- Bruch M., Landwehr,R. and Engel,J. (1989) Dissection of laminin by cathepsin G into its long-arm and short-arm structures and localization of regions involved in calcium dependent stabilization and self-association. Eur. J. Biochem., 185, 271–279. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Stetefeld,J. and Strelkov,S.V. (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol., 11, 82–88. [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Tuckwell,D.S., Eble,J., Kuhn,K. and Humphries,M.J. (1997) The integrin α1 A-domain is a ligand binding site for collagens and laminin. J. Biol. Chem., 272, 12311–12317. [DOI] [PubMed] [Google Scholar]
- Campanero M.R. et al. (1992) Functional role of α2/β1 and α4/β1 integrins in leukocyte intercellular adhesion induced through the common β1 subunit. Eur. J. Immunol., 22, 3111–3119. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. and Jain,R.K. (2000) Angiogenesis in cancer and other diseases. Nature, 407, 249–257. [DOI] [PubMed] [Google Scholar]
- Dickeson S.K., Walsh,J.J. and Santoro,S.A. (1997) Contributions of the I and EF hand domains to the divalent cation-dependent collagen binding activity of the α2β1 integrin. J. Biol. Chem., 272, 7661–7668. [DOI] [PubMed] [Google Scholar]
- Dickeson S.K., Walsh,J.J. and Santoro,S.A. (1998) Binding of the α2 integrin I domain to extracellular matrix ligands: structural and mechanistic differences between collagen and laminin binding. Cell Adhes. Commun., 5, 273–281. [DOI] [PubMed] [Google Scholar]
- Elices M.J., Osborn,L., Takada,Y., Crouse,C., Luhowskyj,S., Hemler,M.E. and Lobb,R.R. (1990) VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell, 60, 577–584. [DOI] [PubMed] [Google Scholar]
- Emsley J., Knight,C.G., Farndale,R.W., Barnes,M.J. and Liddington,R.C. (2000) Structural basis of collagen recognition by integrin α2β1. Cell, 101, 47–56. [DOI] [PubMed] [Google Scholar]
- Engvall E. and Wewer,U.M. (1996) Domains of laminin. J. Cell. Biochem., 61, 493–501. [DOI] [PubMed] [Google Scholar]
- Griggs D.W., Schmidt,C.M. and Carron,C.P. (1998) Characteristics of cation binding to the I domains of LFA-1 and MAC-1. The LFA-1 I domain contains a Ca2+-binding site. J. Biol. Chem., 273, 22113–22119. [DOI] [PubMed] [Google Scholar]
- Ivaska J., Kapyla,J., Pentikainen,O., Hoffren,A.M., Hermonen,J., Huttunen,P., Johnson,M.S. and Heino,J. (1999) A peptide inhibiting the collagen binding function of integrin α2I domain. J. Biol. Chem., 274, 3513–3521. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Park,S.Y., Pickl,W.F., LaMontagne,K.R., Sjin,R.T., Gillies,S. and Lo,K.M. (2002) Laminin modulates morphogenic properties of the collagen XVIII endostatin domain. J. Biol. Chem., 277, 45211–45218. [DOI] [PubMed] [Google Scholar]
- Kammerer R.A., Schulthess,T., Landwehr,R., Schumacher,B., Lustig,A., Yurchenco,P.D., Ruegg,M.A., Engel,J. and Denzer,A.J. (1999) Interaction of agrin with laminin requires a coiled-coil conformation of the agrin-binding site within the laminin γ1 chain. EMBO J., 18, 6762–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.J. et al. (2001) Oligomerization-dependent regulation of motility and morphogenesis by the collagen XVIII NC1/endostatin domain. J. Cell Biol., 152, 1233–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzago A., Felici,F., Tramontano,A., Pessi,A. and Cortese,R. (1993) Mimicking of discontinuous epitopes by phage-displayed peptides I. Epitope mapping of human H ferritin using a phage library of constrained peptides. Gene, 128, 51–57. [DOI] [PubMed] [Google Scholar]
- Mercurio A.M. (1995) Laminin receptors: achiving specificity through cooperation. Trends Cell Biol., 5, 419–423. [DOI] [PubMed] [Google Scholar]
- Nautiyal S. and Alber,T. (1999) Crystal structure of a designed, thermostable, heterotrimeric coiled coil. Protein Sci., 8, 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomizu M., Kuratomi,Y., Malinda,K.M., Song,S.Y., Miyoshi,K., Powell,S.K., Hoffman,M.P., Kleinman,H.K. and Yamada,Y. (1998) Cell binding sequences in mouse laminin α1 chain. J. Biol. Chem., 273, 32491–32499. [DOI] [PubMed] [Google Scholar]
- Ott U., Odermatt,E., Engel,J., Furthmayr,H. and Timpl,R. (1982) Protease resistance and conformation of laminin. Eur. J. Biochem., 123, 63–72. [DOI] [PubMed] [Google Scholar]
- Pentikainen O., Hoffren,A.M., Ivaska,J., Kapyla,J., Nyronen,T., Heino,J. and Johnson,M.S. (1999) ‘RKKH’ peptides from the snake venom metalloproteinase of Bothrops jararaca bind near the metal ion-dependent adhesion site of the human integrin α2 I-domain. J. Biol. Chem., 274, 31493–31505. [DOI] [PubMed] [Google Scholar]
- Rehn M., Veikkola,T., Kukk-Valdre,E., Nakamura,H., Ilmonen,M., Lombardo,C., Pihlajaniemi,T., Alitalo,K. and Vuori,K. (2001) Interaction of endostatin with integrins implicated in angiogenesis. Proc. Natl Acad. Sci. USA, 98, 1024–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H., Bachinger,H.P. and Timpl,R. (1980) Characterization of pepsin fragments of laminin in a tumor basement membrane. Evidence for the existence of related proteins. Hoppe Seylers Z. Physiol. Chem., 361, 1651–1660. [DOI] [PubMed] [Google Scholar]
- Sanz L., Kristensen,P., Russell,S.J., Ramírez García,J.R. and Álvarez-Vallina,L. (2001) Generation and characterization of recombinant human antibodies specific for native laminin epitopes. Potential application in cancer therapy. Cancer Immunol. Immunother., 50, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Kristensen,P., Blanco,B., Facteau,S., Russell,S.J., Winter,G. and Álvarez-Vallina,L. (2002a) Single-chain antibody-based gene therapy: inhibition of tumor growth by in situ production of phage-derived human antibody fragments blocking functionally active sites of cell-associated matrices. Gene Therapy, 9, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Sanz L., Pascual,M., Muñoz,A., González,M.A., Salvador,C.H. and Álvarez-Vallina,L. (2002b) Development of a computer-assisted high-throughput screening platform for anti-angiogenic testing Microvascular Res., 63, 335–339. [DOI] [PubMed] [Google Scholar]
- Sayle R.A. and Milner-White,E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- Stupack D.G. and Cheresh,D.A. (2002) ECM remodeling regulates angiogenesis: endothelial integrins look for new ligands. Sci. STKE, 2002, PE7. [DOI] [PubMed] [Google Scholar]
- Sung U., O’Rear,J.J. and Yurchenko,P.D. (1993) Cell and heparin binding in the distal long arm of laminin: identification of active and cryptic sites with recombinant and hybrid glycoprotein. J. Cell Biol., 123, 1255–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixido J., Parker,C.M., Kassner,P.D. and Hemler,M.E. (1992) Functional and structural analysis of VLA-4 integrin α4 subunit cleavage. J. Biol. Chem., 267, 1786–1794. [PubMed] [Google Scholar]
- Timpl R. (1996) Macromolecular organization of basement membranes. Curr. Opin. Cell Biol., 8, 618–621. [DOI] [PubMed] [Google Scholar]
- Tulla M., Pentikainen,O.T., Viitasalo,T., Kapyla,J., Impola,U., Nykvist,P., Nissinen,L., Johnson,M.S. and Heino,J. (2001) Selective binding of collagen subtypes by integrin α1I, α2I and α10I domains. J. Biol. Chem., 276, 48206–48212. [DOI] [PubMed] [Google Scholar]
- Tunggal P., Smyth,N., Paulsson,M. and Ott,M.-T. (2000) Laminins: structure and genetic regulation. Micros. Res. Tech., 51, 214–227. [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- Walshaw J. and Woolfson,D.N. (2001) Socket: a program for identifying and analysing coiled-coil motifs whithin protein structures. J. Mol. Biol., 307, 1427–1450. [DOI] [PubMed] [Google Scholar]
- Wolf E., Kim,P.S. and Berger,B. (1997) MultiCoil: a program for predicting two- and three-stranded coiled-coils. Protein Sci., 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]