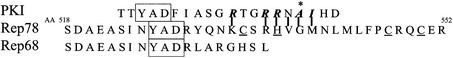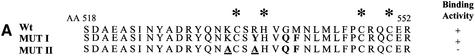Abstract
Interference between viruses occurs when infection by one virus results in the inhibition of replication of another virus. Adeno-associated virus (AAV2) is a human parvovirus with the unique characteristics of a dependence upon a helper virus for a productive infection and the ability to interfere with the replication of the helper virus. Previously, we demonstrated that AAV2 Rep78 and Rep52 interact and inhibit cAMP-dependent protein kinase A (PKA) and its novel homolog PrKX. We hypothesized that modulation of PKA activity by AAV2 may be responsible for inhibition of helper virus replication. In this study we demonstrate that adenovirus replication is sensitive to PKA activity and that AAV2 Rep78/Rep52 proteins contain an inhibitory domain similar to that of the heat-stable PKA inhibitor. This domain, while not directly necessary for AAV2 replication and packaging, is necessary to preserve AAV2 replication fitness during an Ad co-infection. Furthermore, a mutant AAV2 virus lacking this region fails to inhibit adenovirus replication. Thus, inhibition of PKA activity by AAV2 constitutes a novel form of viral interference.
Keywords: AAV/adenovirus/PKA/viral interference
Introduction
Viral interference is a frequent consequence following infection of an animal cell with more than one related or unrelated virus. Characterization of this interaction during influenza infection led to the discovery of the interferons and their role in viral interference (Isaacs and Lindenmann, 1957). Classically, viral interference can be either direct or interferon mediated (indirect). Direct interference occurs when a viral protein or a viral-induced protein disrupts the replication of another virus, whereas indirect interference takes place when a virus induces secretion of interferon and other cytokines that protect cells from subsequent infection. While the molecular mechanisms of interferon-mediated interference are well known (Sen, 2001), mechanisms of direct interference have not been as extensively characterized. Examples of direct interference include the inhibition of vaccinia (a pox virus) and vesicular stomatitis virus (VSV) by poliovirus and vaccinia by Frog virus 3 (an iridovirus) (Aubertin et al., 1970; Garry, 1988; Aldabe et al., 1995). Recently, non-interferon-mediated interference was described in patients infected with HIV and the non-lytic virus hepatitis G (Xiang et al., 2001).
A hallmark of the adeno-associated virus serotype 2 (AAV2) life cycle is its interaction and inhibition of replication of other viruses. AAV2 is a dependent virus and requires the presence of a helper virus for productive replication. Helper viruses include adenovirus (Ad) (Atchison et al., 1965), cytomegalovirus (McPherson et al., 1985), herpesvirus (Buller et al., 1981; Bauer and Monreal, 1986), Epstein–Barr virus, varicella virus (Georg-Fries et al., 1984), pseudorabies virus (Parks et al., 1967), vaccinia virus (Schlehofer et al., 1986) and papillomavirus (Walz et al., 1997; Ogston et al., 2000). However, during a co-infection, AAV can inhibit both Ad and simian virus 40 (SV40) propagation (Casto et al., 1967; Carter et al., 1979), while expression of AAV Rep proteins is sufficient to inhibit the DNA replication of HSV (Bantel-Schaal and zur Hausen, 1988), BPV (Hermonat, 1992), HPV (Hermonat, 1994) and HIV (Antoni et al., 1991; Rittner et al., 1992). In addition to inhibiting helper virus replication, AAV co-infection can also suppress transformation by a number of viruses and viral oncoproteins (for a review see Schlehofer, 1994). In the absence of a helper virus, AAV2 is able to establish a latent infection by integrating in a locus specific manner on the Q arm of human chromosome 19 (Kotin et al., 1990). The four related viral non-structural Rep proteins (Rep78, 68, 52 and 40) are essential for both aspects of the AAV2 life cycle.
Previously we described a stable interaction between the unspliced Rep proteins, Rep78 and Rep52, and the cAMP-dependent protein kinase A (PKA) and its novel homolog PrKX (Chiorini et al., 1998; Di Pasquale and Stacey, 1998). This interaction resulted in the inhibition of PKA kinase activity. No interaction was detected with the spliced Rep proteins, Rep68 and Rep40. A biological effect of Rep78 inhibition of PKA and PrKX activity is the decreased expression of cAMP responsive genes in Rep78-transfected cells (Chiorini et al., 1998). No inhibition is observed in Rep68 transfected cells (Chiorini et al., 1998). The interaction between Rep78/52 and PKA was found to have a physiologically relevant dissociation constant of 320 nM (Chiorini et al., 1998) and additional studies using steady state kinetic analysis and mutagenesis of Rep78 demonstrated it could act as a competitive inhibitor with respect to a peptide kinase substrate (Schmidt et al., 2002).
In addition to its role in cellular signal transduction, the PKA pathway also has a role in viral infection. cAMP-responsive elements (CRE) regulate the expression of numerous viral genes (for a review see Gilchrist et al., 1996). Recently, Ad has been shown to require cAMP-dependent protein kinase activation to facilitate its transport to the nucleus (Suomalainen et al., 2001). In addition, the E1a, E3 and E4 promoters of Ad contain CREs and are PKA responsive (Leza and Hearing, 1989).
Thus, like their cellular homologs, viral genes also may be affected by Rep78 inhibition of PKA and PrKX activity. We hypothesized that during an AAV2/Ad co-infection, Rep78 inhibition of cellular PKA and PrKX activities could result in both the down-regulation of specific Ad genes and an inhibition of Ad replication.
In this study, we demonstrate that AAV2 Rep78/52 proteins contain an inhibitory domain similar to that of the heat-stable PKA inhibitor PKI (Walsh et al., 1971). This domain, while not directly necessary for AAV replication and packaging, is necessary for AAV2 Rep78/52 inhibition of PKA and PrKX. Furthermore, mutated AAV2 genomes lacking this region fail to inhibit Ad replication, suggesting that modulation of PKA activity is a mechanism by which AAV2 interferes with helper virus replication. Finally, we show that AAV2’s ability to inhibit Ad replication via modulation of PKA activity is necessary to preserve its replication fitness during an Ad co-infection.
Results
PKA activity changes during AAV2 and Ad infection
Transfection of an AAV2 Rep78 expression plasmid is able to suppress CREB-dependent transcription activation by both PKA and its homolog PrKX (Di Pasquale and Stacey, 1998). In order to establish whether Rep78 modulation of kinase activity occurs during viral infection, PKA kinase activity was measured by fluorescent kemptide kinase assay (Di Pasquale and Stacey, 1998) in infected HeLa cells.
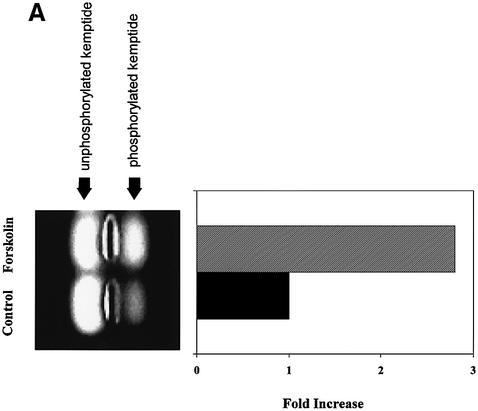
As shown in Figure 1A, this assay is quite sensitive to activation of PKA. Extracts from HeLa cells treated with the PKA activator forskolin had an almost 3-fold increase in PKA activity compared with control cell lysates (Figure 1A). Furthermore, addition of 10 µM of PKI, a psuedosubstrate inhibitor of PKA, blocked >90% of the detectable kinase activity (data not shown).
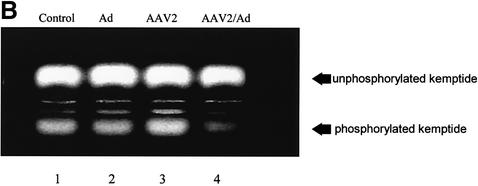
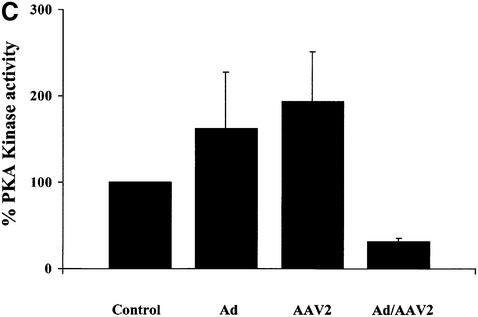
Fig. 1. Fluorescent kemptide kinase assay of PKA activity during viral infection. (A) To demonstrate that the kemptide kinase assay is sensitive to variation in cellular PKA activity, whole-cell HeLa extracts of forskolin treated and untreated cells were incubated for 10 min in a kinase buffer together with the fluorescent-labeled PKA substrate kemptide (LRRASLG; Promega). Phosphorylated kemptide was separated from unphosphorylated substrate by agarose gel electrophoresis and visualized on a UV transilluminator. The photograph of a typical agarose gel used for the kemptide assay is on the left and the quantification of that assay is on the right. Kinase activity is represented as relative to that of untreated cells. (B and C) Whole HeLa cell extracts of uninfected, Ad5, AAV2 or AAV2/Ad5 (m.o.i. 50/50) [lanes 1, 2, 3 and 4, respectively, in (B)] were incubated, for 10 min, in a kinase buffer together with the PKA substrate kemptide. This experiment was repeated three times, the gel photograph shows one of the three experiments. (C) The quantification of the three experiments, means ± SD. Kinase activity is represented as relative to that of uninfected cells.
HeLa cells were infected with either Ad5 or AAV2, or co-infected with both viruses (m.o.i. 50/50) and kinase activity measured 48 h post-infection. Ad5 or AAV2 infection alone had a small stimulatory affect on kinase activity compared with control uninfected cell extract (Figure 1B, lanes 1, 2 and 3, and C, columns 1, 2 and 3, respectively). However, kinase activity was decreased 4-fold in extracts from cells infected with both Ad5 and AAV2 compared with control cell extracts (lane 4 versus 1, and column 4 versus 1, respectively). The result that PKA activity is inhibited during a co-infection is consistent with the expression pattern of AAV2 Rep proteins. Previous research has demonstrated that in the absence of Ad helper functions, AAV2 Rep proteins are poorly expressed (Kyöstiö et al., 1995; Pereira and Muzyczka, 1997). Conversely AAV2 Rep protein expression dramatically increases during Ad/AAV co-infection (Kyöstiö et al., 1995; Pereira and Muzyczka, 1997).
Ad replication is sensitive to modulators of PKA activity
The biological effects of Rep78/52-mediated inhibition of PKA on the AAV2 and Ad5 life cycles are unknown. We hypothesized that during an AAV2/Ad co-infection, Rep78 inhibition of cellular PKA activity could be a mechanism by which AAV2 interferes with the replication of its helper virus. In order to test whether Ad replication is sensitive to modulation of PKA activity, Ad infections were carried out in the presence of drugs that are known to activate or inhibit PKA activity and their effects on Ad5 replication were measured by plaque assay.
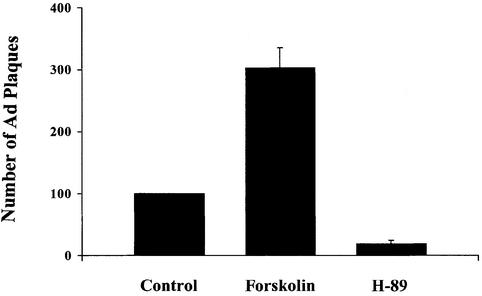
HeLa cells were infected with Ad (50 m.o.i.) in presence or absence of 20 µM of the PKA specific inhibitor H-89 or 20 µM of the PKA activator forskolin. Forty-eight post-infection cells were lyzed and infectious Ad viral particles quantified by plaque assay in 293 cells (Figure 2). Addition of the PKA inhibitor H-89 inhibited plaque formation by 85% compared with virus produced in control, untreated cells. However, the number of plaques observed in forskolin treated cells increased by 3-fold. Thus, Ad5 replication appears to be sensitive to these pharmacological modulators of PKA activity. Addition of these modulators of PKA activity were not toxic at the tested concentrations (data not shown).
Fig. 2. Effect of modulators of PKA activity on formation of Ad5 infective particles. HeLa cells were infected with Ad (m.o.i. 50) and treated with or without forskolin (20 µM) or H-89 (20 µM). Forty-eight hours post-infection cells were lyzed and the number of infectious Ad particles counted in a plaque assay on 293 cells. The results are presented as relative to the number of plaques obtained from untreated Ad infected cells. Mean values ± SD of four experiments are shown.
Fine mapping of a PKA/PrKX interacting and inhibiting Rep78/68 domain
Our previous research determined that Rep78, but not Rep68, interacts with and inhibits both PKA and PrKX activities (Chiorini et al., 1998; Di Pasquale and Stacey, 1998). The difference in modulator activity was localized to the unique C-terminus of Rep78 (Di Pasquale and Stacey, 1998). Subsequent experiments confirmed this region and more precisely localized the PKA binding domain to amino acids (aa) 526–621 of Rep78 (Schmidt et al., 2002).
Several Rep-associated activities have been mapped to the C-terminal region of Rep78/52. This region contains a zinc finger domain and is able to transactivate reporter plasmids in a yeast two-hybrid system (Hörer et al., 1995; Di Pasquale and Stacey, 1998). The C-terminus of Rep 78/52 also is implicated in Rb-mediated cell cycle arrest (Saudan et al., 2000). Computer pattern searches identified a potential PKI like motif in the C-terminal region of Rep78 (Figure 3). This region spans aa 523–542 of Rep78 and overlaps one of the four Rep78 zinc finger domains [CxxC (H) underlined].
Fig. 3. AAV2 Rep homology to the PKI oligopeptide. A computer pattern search (PCGENE) identified a unique region in the Rep78/52 C-terminal region with homology to the PKA interaction domain of PKI. The homologous region spans aa 523–542 of Rep78/52 and partially overlaps one of the Rep zinc finger domains [CxxC (H) underlined]. Only the N-terminal half of the PKI-like motif is present on Rep68, which does not interact with PKA/PrKX. Amino acids in bold have been identified as important for stable interaction between PKA and PKI based on analysis of co-crystals (Knighton et al., 1991).
X-ray crystallographic study of the PKA–PKI complex has confirmed a number of critical amino acid interactions for the high affinity, pseudo-substrate interaction between PKA and PKI (Figure 3) (Knighton et al., 1991). While some of the contributing amino acids are found in both Rep68 and Rep 78, the pseudosubstrate phosphorylation domain is unique to Rep78 and absent in Rep68.
Amino acid mutations in the Rep78/52 PKI-like motif relieve Rep78/52 from PKA binding and kinase inhibition
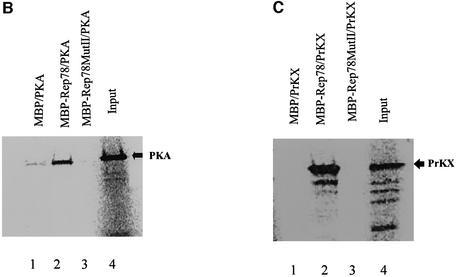
To confirm the role of this region in Rep78-mediated inhibition of PKA, two amino acid substitutions mutants were generated. In MutI, Arg536 (–3p), Gly539 (p) and Met540 (+1p) in the surrounding the pseudosubstrate phosphorylation site were substituted with Tyr, Gln and Phe, respectively (Figure 4A). In MutII, Lys533 (–6p) and Tyr536 (–3p) of MutI were also substituted with Ala (Figure 4A). In contrast to previous mutations in this region, the Cys and His residues participating in the zinc finger domain formation were not changed to preserve the zinc finger motif (Schmidt et al., 2002). The ability of the mutant Rep proteins to interact with PKA or PrKX was first tested using co-precipitation experiments (Figure 4B and C). Maltose-binding protein (MBP)–Rep78 wild type (wt) or the mutant proteins were bound to amylose beads via the MBP portion of the fusion protein and incubated with 35S to radiolabel in vitro translated PKA or PrKX, respectively. After extensive washing, the bound protein was denatured and separated by PAGE and the labeled precipitated protein detected using a phosphoimager. The amino acid substitutions in MutI had only a minor effect on the ability of Rep78 to interact with PKA compared with wt Rep78, and was not further characterized (data not shown). However, the amino acid substitutions made in MutII resulted in a loss of binding activity for both PKA and PrKX compared with that seen in the wt Rep78 (Figure 4B and C, lanes 3 versus lanes 2).
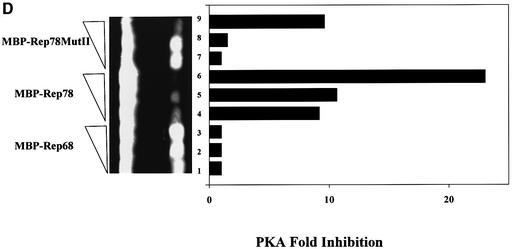
Fig. 4. Interaction of Rep78/52 mutants with PKA or PrKX and their effect on kinase activity. (A) Two mutants were generated in the Rep78 open reading frame contained in the pMal-Rep78 plasmid and are referred to as MutI and MutII. In MutI, Arg536, Gly539 and Met540 were substituted with Tyr, Gln and Phe, respectively. MutII contains additional substitutions of Lys533 with Ala and Tyr536 with Ala. Cys and His residues participating in the zinc finger domain formation were not changed. (B and C) MBP fusion proteins expressed in bacteria containing Rep68, Rep78 or Rep78 MutII were immobilized on amylose beads and then incubated with 35S-labeled PKA or PrKX proteins synthesized in vitro. After washing, the protein complexes were separated in 4–20% gradient PAGE and the labeled proteins visualized by autoradiography. Lane 4 contains 0.1× the amount of PKA or PrKX used in the co-precipitations shown in lanes 1–3. Arrows indicate the bands corresponding to the 42 kDa PKA and 46 kDa PrKX proteins. (D) One unit of purified PKA was added to a 10 min kemptide assay, along with 0.5, 1 or 1.5 µg of MBP–Rep68 (lanes 1–3), MBP–Rep78 (lanes 4–6) or MBP–Rep78 MutII (lanes 7–9) eluted fusion proteins. Phosphorylated kemptide was separated by agarose gel electrophoresis and visualized by UV transillumination. The photograph of the agarose gel used for the kemptide assay is on the left, and the quantification of the assay is on the right. Kinase inhibition is represented as relative to that of kinase activity in the presence of MBP–Rep68.
To demonstrate that the loss of binding to PKA also resulted in a loss of inhibitory activity, the effect of MutII on PKA kinase activity was tested in the kemptide kinase assay (Figure 4D). One unit of purified PKA was added to a kemptide assay, along with 0.5, 1.0 or 1.5 µg of MBP–Rep68 (lanes 1–3), MBP–Rep78 (lanes 4–6) or MBP–Rep78 MutII (lanes 7–9) maltose-eluted Rep fusion proteins. Consistent with the co-precipitation results in Figure 4B, MutII had significantly reduced inhibitory activity compared with wt Rep78 (lanes 7–9 versus lanes 4–6, respectively). As previously reported, Rep68 has no inhibitory activity (lanes 1–3) (Chiorini et al., 1998; Di Pasquale and Stacey, 1998). Taken together, these experiments show that the amino acid substitutions of MutII in the Rep78/52 PKI-like motif disrupt the interaction of Rep78 with PKA/PrKX and substantially alter the ability of Rep78 to inhibit kinase activity.
Mutation of the PKA/PrKX binding domain does not affect AAV2 production using Ad helper plasmid transfection
While amino acid substitutions of MutII disrupt the interaction of Rep78 with PKA/PrKX, it is not clear whether these changes would alter other biochemical activities associated with Rep78. To test possible effects of these mutations on Rep78 activities required for AAV2 rescue, replication and packaging, the amino acid substitutions in MutII were introduced into the AAV2 genome and the yield of virus measured by a quantitative PCR (Q-PCR) assay of CsCl purified virus. Virus yield from the pAV2 MutII vector was very similar to that obtained with pAV2 wt plasmid (1.3 × 1012 ± 1.3 × 1011 versus 2 × 1012 ± 1 × 1011, respectively). These data demonstrate that the amino acid changes in MutII do not affect the ability of Rep78/52 to direct rescue, replication, and packaging of AAV2.
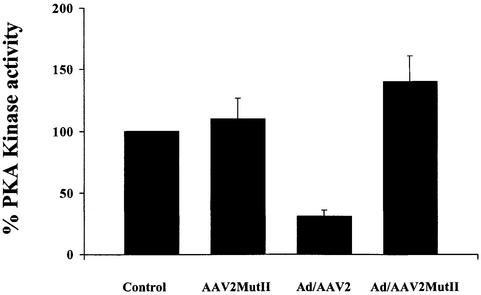
Furthermore, we also tested the ability of MutII to inhibit PKA activity during AAV2/Ad5 co-infection. HeLa cells were infected with AAV2MutII alone or with Ad, or AAV2wt and Ad (m.o.i. 50/50) and kinase activity measured using the kemptide assay (Figure 5). As shown in Figure 1C, kinase activity is inhibited during an AAV2wt/Ad co-infection. However, kinase activity is not inhibited during an AAV2MutII/Ad co-infection, but slightly elevated over control level (Figure 5). These results are consistent with our in vitro observations with MBP–Rep78 MutII and further define the PKI-like Rep78/52 motif as a PKA/PrKX regulatory domain.
Fig. 5. Effects of AAV2-MutII infection on PKA kinase activity. Forty-eight hour HeLa cell extracts of uninfected, or cells infected either with AAV2MutII, Ad/AAV2 or Ad/AAV2MutII (m.o.i. 50/50), were incubated, for 10 min, in a kinase buffer together with the fluorescence- labeled PKA substrate kemptide (LRRASLG). Phosphorylated kemptide was separated from unphosphorylated substrate by agarose gel electrophoresis and visualized on a UV transilluminator (Figure 1). Data shown are the result of three independent experiments, means ± SD. Kinase activity is represented as relative to that of uninfected cells control.
Amino acid mutations in the Rep78/52 PKI-like motif block AAV2 interference with Ad replication
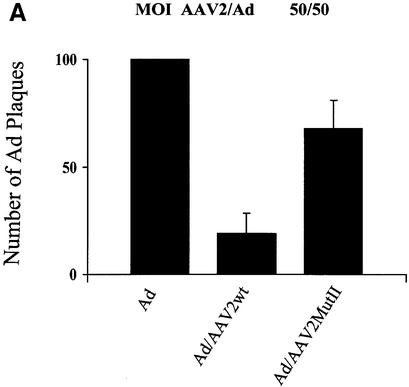
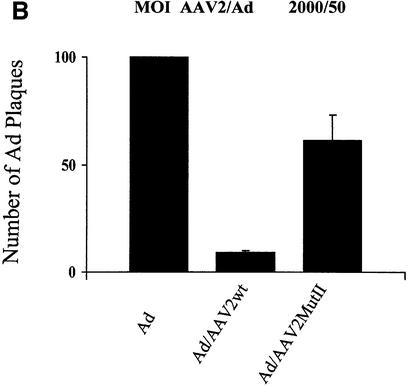
One characteristic of AAV2’s life cycle is the inhibition of helper virus replication. The finding that AAV2 Rep78/52 interferes with PKA/PrKX kinase activity and the observation that Ad replication is sensitive to cellular PKA activity together suggest that AAV2 may interfere with Ad replication by modulating cellular PKA activity. Therefore, we hypothesized that a mutation of the PKA/PrKX inhibitory domain in Rep78/52 should relieve AAV2’s inhibition of Ad replication. To compare the effect of AAV2wt versus AAV2MutII on Ad replication, HeLa cells were infected with Ad, AAV2wt/Ad or AAV2MutII/Ad at an m.o.i. of either 50/50 or 2000/50 AAV2/Ad5 per cell, respectively. Forty-eight hours post-infection, cells were lyzed and the Ad titered by a plaque assay in 293 cells (Figure 6). Co-infection of AAV2wt/Ad inhibited Ad plaque formation at both ratios (Figure 6). The degree of inhibition depended upon the ratio of wtAAV2 to Ad present. Ad plaques were inhibited by 90% at an AAV2/Ad ratio of 2000/50 and by 81% with a ratio of 50/50 (Figure 6). However, during a co-infection with AAVMutII, Ad plaque formation was only slightly inhibited. Ad5 replication was inhibited by 39% at an AAV2MutII/Ad ratio of 2000/50 and by 31% at a ratio of 50/50 (Figure 6). These results suggest that mutation of the PKI-like motif in Rep78/52 can blunt AAV2’s inhibitory effect on production of Ad5. Furthermore, the data confer a biological role for the PKI-like motif in AAV2’s life cycle, probably as a mechanism to interfere with helper virus replication.
Fig. 6. Effects of AAV2wt or AAV2MutII on formation of infectious Ad particles. (A) HeLa cells were infected with Ad5, Ad5/AAV2wt or Ad5/AAV2MutII with 50 particles/cell for each virus. Forty-eight hours post-infection CVL was prepared for the infected cells and the number of infectious Ad particles measured by plaque assay on 293 cells. Results are presented as relative to the number of plaques obtained from cells infected with Ad alone. Data shown are the means ± SD of four experiments. (B) The same as in (A) except that Ad5 and AAV2 particles per cell were 50 and 2000, respectively.
Mutation on Rep78 PKI-like domain reduces AAV2 propagation fitness
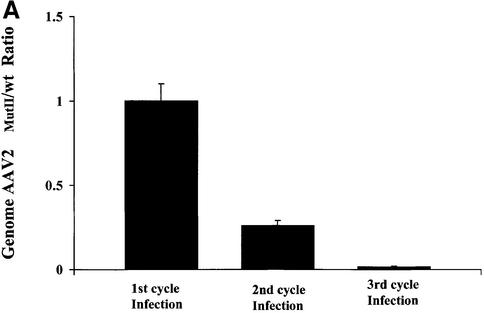
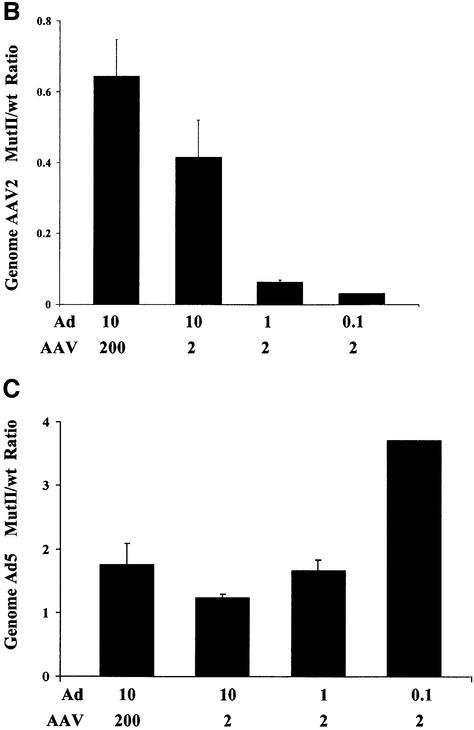
Our experiments have addressed the biological function of the PKI-like inhibitory motif in the C-terminus of Rep78/52 and defined a role for this kinase inhibitory activity in the AAV2-mediated inhibition of Ad production. Furthermore, mutation of this region has no apparent effect on AAV2 MutII replication and encapsidation in a recombinant system utilizing a minimal Ad helper plasmid. However, since AAV2’s life cycle is interrelated with that of its helper virus, it was not clear whether in a co-infection with Ad, failure to inhibit Ad would ultimately affect AAV2 MutII propagation. Therefore, we monitored the titer of both wt and AAV2MutII during Ad co-infection through three rounds of serial passages. HeLa cells were co-infected with either AAV2wt/Ad5 or AAV2MutII/Ad5 at an AAV/Ad m.o.i. of 50/50 per cell. Virus was harvested from cells 48 h post-infection, quantified by Q-PCR, and a dilution of the lysate used to initiate another round of co-infection. While a similar level of DNA replication was observed for both AAV2wt and AAV2MutII after the first round of replication, after the second round of replication with diluted virus from the first round, AAV2MutII replication was 4-fold less compared with AAV2wt (Figure 7A). Moreover, after the third round of passage, AAV2MutII replication had decreased almost 100-fold compared with AAV2wt. Therefore, under conditions of high a m.o.i. (Figure 7A, 1st cycle), mutation of the PKI-like motif has no effect on AAV2 replication. However, under conditions of limiting virus, mutation of this region inhibits AAV2’s ability to replicate efficiently (Figure 7A, 2nd and 3rd cycles). In order to investigate further the conditions where AAV2 MutII propagation is impaired with respect to the wt, we monitored AAV2 and Ad5 DNA replication in cells co-infected at various m.o.i. combinations. HeLa cells were infected with either AAV2wt or MutII at an m.o.i. of 200 or 2 together with Ad5 at an m.o.i. of 10, 1 or 0.1, respectively. Seventy-two hours post-infection AAV2 or Ad5 virus DNA was quantified by Q-PCR. The replication of either virus was then compared in cells infected with MutII or wt AAV2 (MutII/wt) (Figure 7B and C, respectively). AAV2 MutII replication in cells infected at m.o.i. 200/10 (AAV2/Ad) yielded results very similar to those of AAVwt (Figure 7B, column 1). However, when either AAV or Ad m.o.i. was decreased, AAV2MutII DNA replication decreased 30-fold compared with AAV2wt (Figure 7B, column 4). While the amount of AAV2MutII DNA replication decreased compared with AAV2wt with limiting virus as shown in Figure 7B, the replication of Ad DNA in the AAV2MutII samples increased 3.5-fold compared with AAV2wt samples (Figure 7C, column 4). Taken together, these data strongly suggest that mutation of the PKI-like motif in Rep78/52 severely affect the inter-relationship of AAV2 with Ad5. While under some circumstances AAV2MutII replication is similar to that of AAV2wt, under conditions of limiting virus AAV2MutII is not able to effectively compete with Ad, and thus would eventually be lost from a co-culture. Therefore, we conclude that retention of this activity is necessary to maintain the fitness of AAV2 during Ad co-infection.
Fig. 7. Replication properties of AAV2MutII during repeated infection cycles and at different m.o.i. combinations. (A) HeLa cells were co- infected with AAV2wt/Ad or AAV2MutII/Ad with 50 particles/cell for each virus. Forty-eight hours post-infection wt and mutant AAV2 genomes were quantified by Q-PCR (1st cycle infection). A 1:100 fold dilution of the crude cell lysate from the first cycle of infection was used to initiate the second cycle of HeLa cell infections. Forty-eight hours post-infection wt and mutant AAV2 genomes were again quantified (2nd cycle infection). This process was repeated again (3rd cycle infection). Data are presented as the ratio of AAV2MutII/AAV2wt genomes, and are the means ± SD of four experiments, each conducted in duplicate. (B and C) HeLa cells were infected with either AAV2wt or MutII at an m.o.i. of 200 or 2, together with Ad at an m.o.i. of 10, 1 or 0.1. Seventy-two hours post-infection AAV2 (B) or Ad (C) viral DNA was quantified by Q-PCR. The data are presented as a ratio (AAV2MutII/AAV2wt) of the AAV2 or Ad viral DNA detected in the corresponding co-infection. Means ± SD of two independent experiments, each conducted in duplicate, are shown.
Discussion
The results presented in this paper describe a novel mechanism by which one virus, in order to efficiently propagate, interferes with the life cycle of another virus during co-infection. Here we describe a virus–host interaction that creates a cellular environment which is inhibitory to the replication of other viruses and results in direct interference.
A number of viruses require stimuli from the cellular environment for regulation of their gene expression and productive replication. Examples of viruses that use the PKA/CREB pathways include HTLVI, HSV, Ad, Epstein–Barr virus and HIV (Lee et al., 1987; Lillie et al., 1989; Leib et al., 1991; Nokta and Pollard, 1992; Flamand and Menezes, 1996; Kwok et al., 1996; Cole et al., 1998). The cAMP response pathway components are frequently used to control switches from early to late stages in the virus life cycle. Ad E1a protein can transactivate genes containing CREs via an interaction with CBP/p300 (Mishra and Rose, 1990). In addition, the E1a, E3 and E4 promoters of Ad contain CREs (Leza and Hearing, 1989; Meyer et al., 1993). In HSV, ICP4 is a substrate for PKA and its phosphorylation promotes HSV lytic replication (Xia et al., 1996).
In addition to requiring stimuli from the host cell, viruses are also able to regulate cellular pathways by direct interaction between viral proteins and cellular factors. Bovine papillomavirus type 1 E5 oncoprotein binds to the platelet-derived growth factor receptor, mimics its dimerization and activates its tyrosine kinase activity (Mattoon et al., 2001). In contrast, SV40 large T antigen, Ad E1B protein and the X protein of hepatitis B virus all form a complex with p53 and inactivate it. Several examples of PKA pathway regulation have also been described. Poliovirus 3C protease inhibits the PKA pathway by cleaving and inactivating CREB (Weidman et al., 2001). HCV contains a sequence that resembles the heat-stable PKA inhibitor PKI. While the role of PKA inhibition in HCVs life cycle is not clear, this sequence is able to block the translocation of the catalytic subunit into the nucleus (Borowski et al., 1999).
Because of the unique life cycle of AAV, it has interactions with both the host cell and the helper virus. Our results suggest that AAV2 regulates PKA activity as a mechanism of interfering with helper virus propagation and promoting its own replication. Ad replication is sensitive to cellular PKA activity. Alteration of PKA activity via chemical modulators can either stimulate or inhibit Ad replication. While no alteration of PKA activity is detected during Ad or AAV2 infection alone, a co-infection results in a dramatic decrease in PKA activity. Our previous research demonstrated that AAV2 Rep78/52 could inhibit PKA activity, but very little Rep is expressed during AAV2 infection alone. Co-infection with Ad transactivates Rep expression, which then inhibits PKA activity, thus inhibiting Ad replication. Unlike Ad, AAV2 replication is not dependent upon PKA activity. Mutant viruses that lack this region are able to replicate and be packaged with similar efficiency to AAV2wt when Ad early gene functions are provided on a plasmid. This suggests the mutation has not affected any of the Rep activities necessary for replication. However, during a co-infection with Ad, the mutant AAV2 is no longer able to inhibit Ad replication and as a result is out competed by Ad and eventually lost from the co-infection. Thus, a selective pressure exists for retaining this function in the AAV2 virus.
Mammalian cells have developed defensive strategies against viral infection. These include secretion of interferon, induction of apoptosis and inhibition of cellular translation (Sen, 2001). In response, viruses have developed schemes to interfere with and evade the host cell defense mechanisms. Virus proteins can inhibit interferon synthesis or inactivate interferon-induced proteins. Hence the outcome of a viral infection depends on the complex interactions of viral and cellular factors (Sen, 2001). AAV2-mediated inhibition of the PKA pathway as a means of inhibiting helper virus replication without inducing a classic antiviral response is a novel antiviral strategy and underlines the complex nature of the relationship between AAV2, its helper virus and the host cell.
Materials and methods
Cell culture
HeLa cells were cultivated in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS), 100 mg/l penicillin and 100 U/ml of streptomycin (Gibco). Infection, depending on the experiment, was performed in T70 flasks (kinase assay) or 24-well plates when nearly confluent. 293 cell lines used for Ad plaque assays were plated in 12-well plates and cultivated in EMEM supplemented with 10% FCS, 100 µg/ml penicillin and 100 U/ml of streptomycin.
Ad type 5 plaque assay
293 cells were infected with serial dilutions of infected HeLa cell lysates. After infection, cells were over-layered with agar-medium mix. Plaques were counted 6–10 days later.
PKA specific kinase assay of infected HeLa cells
Forty-eight hours post-infection cells were washed with phosphate-buffered saline (PBS), and then scraped and recovered in PBS. Cytoplasmatic and nuclear proteins were released by sonication and then equal amounts of lysate were assayed for PKA-specific kinase activity. The same amount of whole sonicate protein contained in 5 µl was incubated in PepTag PKA kinase buffer (20 mM Tris–HCl pH 7.4, 10 mM MgCl2, 1 mM ATP) with 5 µl of PepTag A1 Peptide (fluorescent kemptide, 0.4 µg/µl in water; Promega) and 1 µl of Peptide Protection Solution (protease inhibitor mix) in 25 µl final volume for 10 min at room temperature (Promega). The reaction was terminated by boiling for 5 min, and then 1 µl of 80% glycerol was added. Phosphorylated and unphosphorylated substrate were separated in agarose gel electrophoresis (0.8% agarose in 50 mM Tris–HCl pH 8.0 buffer). The gels were photographed after placing on a ultraviolet transilluminator. The extent of phosphorylation was determined by quantifying the peptide bands after scanning images of the fluorescent gels (Stratagene). For experiments with forskolin, cells were treated with the drug for 48 h prior to cell lysis. For PKI inhibition, the inhibitor was added to cell lysate prior to the addition of substrate.
Virus production
AAV2wt and AAV2MutII were produced by a co-transfection method described by Kaludov et al. (2001). Briefly, nearly confluent 293 cells were transfected by calcium phosphate with two plasmids; an Ad helper plasmid (pAd12) and a plasmid containing a full-length AAV2 genome (pAV2) or a mutated AAV2 genome (pAV2-MutII); see description below. Forty-eight hours post-transfection cells were harvested by scraping in TD buffer (140 mM NaCl, 5 mM KCl, 0.7 mM K2HPO4, 25 mM Tris–HCl pH 7.4) and concentrated by centrifugation. Helper virus free AAV2 was concentrated using isopycnic CsCl gradient centrifugation.
Site-directed mutagenesis
Amino acid substitutions were introduced into MBP–Rep78 fusion protein using the QuikChange Kit (Stratagene) and the plasmid pMal-Rep78 as backbone (Di Pasquale and Stacey, 1998). The forward primer 5′-CCAAAACAAATGTTCTTATCACGCGCAGTTCAATCTGATGC TGTTTCCC and reverse primer 5′-GGGAAACAGCATCAGATTG AACTGCGCGTGATAAGAACATTTGTTTTGG were used to introduce the amino acid modification present in pMal-Rep78MutI. A second set of primers was used to introduce additional mutations in the pMal-Rep78MutI plasmid. Forward primer 5′-GACAGGTACCAAAACG CATGTTCTGCTCACGCGCAGTTC and the reverse primer 5′-GAA CTGCGCGTGAGCAGAACATGCGTTTTGGTACCTGTC were used to obtain pMal-Rep78MutII.
The pAV2MutII vector was generated by subcloning of a NcoI–EcoRI fragment (1369 bp, nucleotides 625–1985 AAV2 genome) from the pMal-Rep78MutII into NcoI–EcoRI-digested pAV2 vector (Laughlin, 1983).
Expression and purification of MBP–Rep78, MBP–Rep78MutII and MBP–Rep68 fusion proteins in Escherichia coli
The expression and purification of MBP–Rep78 or MBP–Rep68 fusion proteins were carried out as described in the Expression and Purification Manual supplied by the manufacturers of the pMalc2 vector (New England BioLabs). When required, fusion proteins were eluted from the amylose beads by washing with 10 mM maltose/PBS buffer.
Co-precipitation binding assay
MBP fusion proteins bound to beads (Rep78, Rep78MutII and Rep68, containing 1 µg of each fusion protein) were incubated at 4°C for 15 min with 20 µl of rabbit reticulocyte lysate (RRL) containing 35S-labeled PrKX or PKAc expressed by coupled in vitro transcription/translation from the vector pcDNA-PrKX (Di Pasquale and Stacey, 1998) or pcDNA-PKA (gift from S.Ghosh), respectively. The reaction was made to a final volume of 250 µl with PBS containing 1 mg/ml of bovine serum albumin (BSA) and 0.1% NP-40. Incubation under agitation was followed by five washings in PBS–BSA–NP-40; in each wash the supernatant was completely removed by suction using a pipette with microcapillary tip. Radiolabeled protein bound to beads was analyzed by SDS–PAGE and visualized by phosphoimager analysis.
PKA kinase inhibition assay
The kinase assay kit PepTag (Promega) was used to determine the amount of purified PKA (Promega) required to phosphorylate 50% of the kinase substrate in a given time (units). To this standard kinase reaction were added increasing concentrations (0.5, 1, 1.5 µg) of maltose-eluted MBP–Rep68, MBP–Rep78 or MBP–Rep78MutII, respectively, and the reaction mixture then incubated for 10 min in kinase buffer. Phosphorylated and unphosphorylated substrate was separated in 0.8% agarose by electrophoresis. The amount of phosphorylation was determined by quantifying the fluorescent intensity of the peptide bands after scanning images of the fluorescent gels.
Quantification of AAV2 and Ad DNA by quantitative PCR
Infected cells or viral stocks were lyzed using PCR Lyse-N-Go (Pierce). One microliter of lysate was added to a PCR containing 1× SYBR Green master mix (ABI), 0.25 pmol/µl of forward and reverse primers and amplification detected on using an ABI 7700 sequence detector (ABI). Specific primers for AAV2 and Ad were designed by using the primer express program (ABI): AAV2 forward primer 5′-ACCAACATC GCGGAGGC, reverse primer 5′-CCCTCCTCCCACCAGATCA; Ad type 5: forward primer 5′-AACCGAAGGCTGCATTCACT, reverse primer 5′-ACCGCACAGGGTCTTAATAGAG. Following a 96°C, 10 min denaturing step, cycling conditions were 96°C for 15 s, and 60°C for 1 min for 40 cycles. The viral DNA in each sample was quantified by comparing the fluorescence profiles with a set of DNA standards.
Acknowledgments
Acknowledgements
We thank Beverly Handelman for excellent technical assistance and Roland Owens, Robert Kotin, and Bruce Baum for helpful discussion. We also thank Sankar Ghosh for providing critical reagents.
References
- Aldabe R., Feduchi,E., Novoa,I. and Carrasco,L. (1995) Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: effects on vaccinia virus. Biochem. Biophys. Res. Commun., 215, 928–936. [DOI] [PubMed] [Google Scholar]
- Antoni B.A., Rabson,A.B., Miller,I.L., Trempe,J.P., Chejanovsky,N. and Carter,B.J. (1991) Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J. Virol., 65, 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison R.W., Casto,B.C. and Hammon,W.M. (1965) Adeno-associated defective virus particles. Science, 149, 754–755. [DOI] [PubMed] [Google Scholar]
- Aubertin A.M., Guir,J. and Kirn,A. (1970) The inhibition of vaccinia virus DNA synthesis in KB cells infected with frog virus 3. J. Gen. Virol., 8, 105–111. [DOI] [PubMed] [Google Scholar]
- Bantel-Schaal U. and zur Hausen,H. (1988) Adeno-associated viruses inhibit SV40 DNA amplification and replication of herpes simplex virus in SV40-transformed hamster cells. Virology, 164, 64–74. [DOI] [PubMed] [Google Scholar]
- Bauer H.J. and Monreal,G. (1986) Herpesviruses provide helper functions for avian adeno-associated parvovirus. J. Gen. Virol., 67, 181–185. [DOI] [PubMed] [Google Scholar]
- Borowski P., Kuhl,R., Laufs,R., Schulze zur Wiesch,J. and Heiland,M. (1999) Identification and characterization of a histone binding site of the non-structural protein 3 of hepatitis C virus. J. Clin. Virol., 13, 61–69. [DOI] [PubMed] [Google Scholar]
- Buller R.M., Janik,J.E., Sebring,E.D. and Rose,J.A. (1981) Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol., 40, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B.J., Laughlin,C.A., de la Maza,L.M. and Myers,M. (1979) Adeno-associated virus autointerference. Virology, 92, 449–462. [DOI] [PubMed] [Google Scholar]
- Casto B.C., Atchison,R.W. and Hammon,W.M. (1967) Studies on the relationship between adeno-associated virus type I (AAV-1) and adenoviruses. I. Replication of AAV-1 in certain cell cultures and its effect on helper adenovirus. Virology, 32, 52–59. [DOI] [PubMed] [Google Scholar]
- Chiorini J.A., Zimmermann,B., Yang,L., Smith,R.H., Ahearn,A., Herberg,F. and Kotin,R.M. (1998) Inhibition of PrKX, a novel protein kinase and the cAMP-dependent protein kinase, PKA, by the regulatory proteins of Adeno-associated virus type 2. Mol. Cell. Biol., 18, 5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S.W., Korin,Y.D., Fahey,J.L. and Zack,J.A. (1998) Norepinephrine accelerates HIV replication via protein kinase A-dependent effects on cytokine production. J. Immunol., 161, 610–616. [PubMed] [Google Scholar]
- Di Pasquale G. and Stacey,S.N. (1998) Adeno-associated virus Rep78 protein interacts with protein kinase A and its homolog PRKX and inhibits CREB-dependent transcriptional activation. J. Virol., 72, 7916–7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamand L. and Menezes,J. (1996) Cyclic AMP-responsive element-dependent activation of Epstein–Barr virus zebra promoter by human herpesvirus 6. J. Virol., 70, 1784–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry R.F. (1988) Poliovirus protease 2A is required for interference with vesicular stomatitis virus-specified protein synthesis. Arch. Virol., 103, 133–137. [DOI] [PubMed] [Google Scholar]
- Georg-Fries B., Biederlack,S., Wolf,J. and zur Hausen,H. (1984) Analysis of proteins, helper dependence and seroepidemiology of a new human parvovirus. Virology, 134, 64–71. [DOI] [PubMed] [Google Scholar]
- Gilchrist C.A., Orten,D.J. and Hinrichs,S.H. (1996) Evidence for the role of cyclic AMP-responsive elements in human virus replication and disease. J. Biomed. Sci., 3, 293–306. [DOI] [PubMed] [Google Scholar]
- Hermonat P.L. (1992) Inhibition of bovine papillomavirus plasmid DNA replication by adeno-associated virus. Virology, 189, 329–333. [DOI] [PubMed] [Google Scholar]
- Hermonat P.L. (1994) Adeno-associated virus inhibits human papillomavirus type 16: a viral interaction implicated in cervical cancer. Cancer Res., 54, 2278–2281. [PubMed] [Google Scholar]
- Hörer M., Weger,S., Butz,K., Hoppe-Seyler,F., Geisen,C. and Kleinschmidt,J.A. (1995) Mutational analysis of adeno-associated virus rep protein-mediated inhibition of heterologous and homologous promoters. J. Virol., 69, 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A. and Lindenmann,J. (1957) Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci., 147, 258–267. [PubMed] [Google Scholar]
- Kaludov N., Brown,K.E., Walters,R.W., Zabner,J. and Chiorini,J.A. (2001) Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol., 75, 6884–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton D.R., Zheng,J., ten Eyck,L.F., Ashford,V.A., Xuong,N.-H., Taylor,S.S. and Sowadski,J.M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science, 253, 407–413. [DOI] [PubMed] [Google Scholar]
- Kotin R.M. et al. (1990) Site-specific integration by adeno-associated virus. Proc. Natl Acad. Sci. USA, 87, 2211–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok R.P., Laurance,M.E., Lundblad,J.R., Goldman,P.S., Shih,H., Connor,L.M., Marriott,S.J. and Goodman,R.H. (1996) Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature, 380, 642–646. [DOI] [PubMed] [Google Scholar]
- Kyöstiö S.R.M., Wonderling,R.S. and Owens,R.A. (1995) Negative regulation of the adeno-associated virus (AAV) p5 promoter involves both the p5 rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J. Virol., 69, 6787–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin C.A., Tratschin,J.D., Coon,H. and Carter,B.J. (1983) Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene, 23, 65–73. [DOI] [PubMed] [Google Scholar]
- Lee K.A., Hai,T.Y., SivaRaman,L., Thimmappaya,B., Hurst,H.C., Jones,N.C. and Green,M.R. (1987) A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc. Natl Acad. Sci. USA, 84, 8355–8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib D.A., Nadeau,K.C., Rundle,S.A. and Schaffer,P.A. (1991) The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proc. Natl Acad. Sci. USA, 88, 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leza M.A. and Hearing,P. (1989) Independent cyclic AMP and E1A induction of adenovirus early region 4 expression. J. Virol., 63, 3057–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie J.W., Hai,T., Coukos,W.J., Lee,K.A., Martin,K.J. and Green,M.R. (1989) Transcriptional activation of adenoviral early genes. Curr. Top. Microbiol. Immunol., 144, 191–195. [DOI] [PubMed] [Google Scholar]
- Mattoon D., Gupta,K., Doyon,J., Loll,P.J. and DiMaio,D. (2001) Identification of the transmembrane dimer interface of the bovine papillomavirus E5 protein. Oncogene, 20, 3824–3834. [DOI] [PubMed] [Google Scholar]
- McPherson R.A., Rosenthal,L.J. and Rose,J.A. (1985) Human cytomegalovirus completely helps adeno-associated virus replication. Virology, 147, 217–222. [DOI] [PubMed] [Google Scholar]
- Meyer T.E., Waeber,G., Lin,J., Beckmann,W. and Habener,J.F. (1993) The promoter of the gene encoding 3′,5′-cyclic adenosine monophosphate (cAMP) response elements binding protein contains cAMP response elements: evidence for positive autoregulation of gene transcription. Endocrinology, 132, 770–780. [DOI] [PubMed] [Google Scholar]
- Mishra L. and Rose,J.A. (1990) Adeno-associated virus DNA replication is induced by genes that are essential for HSV-1 DNA synthesis. Virology, 179, 632–639. [DOI] [PubMed] [Google Scholar]
- Nokta M.A. and Pollard,R.B. (1992) Human immunodeficiency virus replication: modulation by cellular levels of cAMP. AIDS Res. Hum. Retroviruses, 8, 1255–1261. [DOI] [PubMed] [Google Scholar]
- Ogston P., Raj,K. and Beard,P. (2000) Productive replication of adeno-associated virus can occur in human papillomavirus type 16 (HPV-16) episome-containing keratinocytes and is augmented by the HPV-16 E2 protein. J. Virol., 74, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks W.P., Melnick,J.L., Rongey,R. and Mayor,H.D. (1967) Physical assay and growth cycle studies of a defective adeno-satellite virus. J. Virol., 1, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira D.J. and Muzyczka,N. (1997) The adeno-associated virus type 2 p40 promoter requires a proximal Sp1 interaction and a p19 CArG-like element to facilitate Rep transactivation. J. Virol., 71, 4300–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittner K., Heilbronn,R., Kleinschmidt,J.A. and Sczakiel,G. (1992) Adeno-associated virus type 2-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) replication: involvement of p78rep/p68rep and the HIV-1 long terminal repeat. J. Gen. Virol., 73, 2977–2981. [DOI] [PubMed] [Google Scholar]
- Saudan P., Vlach,J. and Beard,P. (2000) Inhibition of S-phase progression by adeno-associated virus Rep78 protein is mediated by hypophosphorylated pRb. EMBO J., 19, 4351–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlehofer J.R. (1994) The tumor suppressive properties of adeno-associated viruses. Mutat. Res., 305, 303–313. [DOI] [PubMed] [Google Scholar]
- Schlehofer J.R., Ehrbar,M. and zur Hausen,H. (1986) Vaccinia virus, herpes simplex virus and carcinogens induce DNA amplification in a human cell line and support replication of a helpervirus dependent parvovirus. Virology, 152, 110–117. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Chiorini,J.A., Afione,S. and Kotin,R. (2002) Adeno-associated virus type 2 Rep78 inhibition of PKA and PRKX: fine mapping and analysis of mechanism. J. Virol., 76, 1033–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.C. (2001) Viruses and interferons. Annu. Rev. Microbiol., 55, 255–281. [DOI] [PubMed] [Google Scholar]
- Suomalainen M., Nakano,M.Y., Boucke,K., Keller,S. and Greber,U.F. (2001) Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J., 20, 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D.A., Ashby,C.D., Gonzalez,C., Calkins,D., Fischer,E.H. and Kerbs,E.G. (1971) Purification and characterization of a protein inhibitor of adenosine 3′,5′-monophosphate-dependent protein kinases. J. Biol. Chem., 246, 1977–1987. [PubMed] [Google Scholar]
- Walz C., Deprez,A., Dupressoir,T., Durst,M., Rabreau,M. and Schlehofer,J.R. (1997) Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J. Gen. Virol., 78, 1441–1452. [DOI] [PubMed] [Google Scholar]
- Weidman M.K., Yalamanchili,P., Ng,B., Tsai,W. and Dasgupta,A. (2001) Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology, 291, 260–271. [DOI] [PubMed] [Google Scholar]
- Xia K., Knipe,D.M. and DeLuca,N.A. (1996) Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J. Virol., 70, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Wunschmann,S., Diekema,D.J., Klinzman,D., Patrick,K.D., George,S.L. and Stapleton,J.T. (2001) Effect of coinfection with GB virus C on survival among patients with HIV infection. N. Engl. J. Med., 345, 707–714. [DOI] [PubMed] [Google Scholar]