Abstract
Ornithine decarboxylase (ODC) is regulated by its metabolic products through a feedback loop that employs a second protein, antizyme 1 (AZ1). AZ1 accelerates the degradation of ODC by the proteasome. We used purified components to study the structural elements required for proteasomal recognition of this ubiquitin-independent substrate. Our results demonstrate that AZ1 acts on ODC to enhance the association of ODC with the proteasome, not the rate of its processing. Substrate-linked or free polyubiquitin chains compete for AZ1-stimulated degradation of ODC. ODC–AZ1 is therefore recognized by the same element(s) in the proteasome that mediate recognition of polyubiquitin chains. The 37 C-terminal amino acids of ODC harbor an AZ1-modulated recognition determinant. Within the ODC C terminus, three subsites are functionally distinguishable. The five terminal amino acids (ARINV, residues 457–461) collaborate with residue C441 to constitute one recognition element, and AZ1 collaborates with additional constituents of the ODC C terminus to generate a second recognition element.
Keywords: antizyme/ornithine decarboxylase/proteasome/protein degradation/ubiquitin
Introduction
Protein degradation performs an important role in cellular regulation. Degradation not only disposes of defective proteins, but also imposes control over the levels of intrinsically and conditionally labile proteins involved in cell cycle control, transcription, apoptosis and other processes that demand precise temporal adjustment (Ciechanover et al., 2000). Cells have elaborated complex mechanisms to destroy appropriate substrates and spare other proteins. Three elements are commonly found (Baumeister et al., 1998): (i) the sites of catalytic proteolysis are segregated within the interior of a hollow proteolytic machine; (ii) authentic substrates are equipped with tags that license degradation; and (iii) the proteolytic machine distinguishes marked proteins and admits them to its interior, where they are processed to peptides.
The proteasome, the major neutral protease of eukaryotes, exemplifies these principles. Its two structural components are a barrel-shaped catalytic chamber (20S proteasome or proteolytic core particle) capped at one or both ends by a regulatory complex (19S regulatory particle or PA700; Voges et al., 1999). Substrates are most commonly marked by covalent attachment to multiple copies of ubiquitin (Pickart, 2001), a highly conserved protein of 76 amino acids. Enzyme-catalyzed reactions link the C-terminal glycine of ubiquitin through isopeptide bonds to the free amino group of lysines within target proteins; further additions to internal lysine residues of ubiquitin extend the modification to form polyubiquitin chains, which are required for the targeting of many proteasome substrates (Chau et al., 1989). The 20S proteasome alone is incapable of degrading folded proteins, but can associate with the 19S particle to form a 26S proteasome fully active for protein degradation. The 19S particle distinguishes proteins with ubiquitin chains and additionally can unfold substrates (Liu et al., 2002), strip ubiquitin chains for recycling (Verma et al., 2002; Yao and Cohen, 2002) and open an axial portal into the 20S chamber (Kohler et al., 2001). A subcomplex of the 19S particle containing 8–10 proteins (the lid) can be removed from the 26S proteasome in vitro (Glickman et al., 1998a); the remaining subcomplex of the 19S regulator (the base) is juxtaposed to the end of the 20S particle. The base contains six ATPases (Rpt1 through Rpt6) and two other proteins (Rpn1 and Rpn2). Protein degradation by the 26S proteasome requires ATP. The composition and position of the base is consistent with a requirement for ATP-dependent processing of substrates for insertion into the core particle.
Polyubiquitin chains may be of considerable length, but four ubiquitins (Ub4) linked through lysine 48 appear to constitute the minimal structure that is effective in directing degradation (Thrower et al., 2000). Such a chain confers on polyubiquitylated substrates the capacity to be recognized at sub-micromolar concentrations. Polyubiquitin chains can direct proteins to the proteasome or, if detached from substrates, can act as competitive inhibitors of the degradation of polyubiquitylated substrates (Amerik et al., 1997; Thrower et al., 2000). Crosslinking experiments have shown Rpt5/S6′, one of the ATPase proteins of the base, to be a direct binding site for the chain (Lam et al., 2002).
A small number of proteins have been identified that are substrates of the proteasome but that do not require ubiquitin for their degradation (Verma and Deshaies, 2000). The best characterized of these is the protein pair ornithine decarboxylase (ODC) and antizyme 1 (AZ1) (Coffino, 2001). In vivo and in vitro experiments have shown that AZ1-stimulated proteasomal degradation of ODC does not involve ubiquitin (Glass and Gerner, 1987; Bercovich et al., 1989; Murakami et al., 1992a). Together with the proteasome, ODC and AZ1 mediate an unusual form of metabolic feedback regulation. ODC is the initial enzyme in the biosynthesis of polyamines, small abundant polycations that are essential for life. AZ1 levels rise in response to excess cellular polyamines (Murakami and Hayashi, 1985). AZ1 binds to ODC, displacing the weakly associated ODC homodimer to form the enzymatically inactive ODC–AZ1 heterodimer (Murakami and Hayashi, 1985; Mitchell and Chen, 1990; Li and Coffino, 1992). ODC has a short half-life in the absence of AZ1, but turns over still faster when associated with AZ1 (Murakami and Hayashi, 1985). By inactivating ODC and catalyzing its destruction, polyamine-induced AZ1 reduces polyamine biosynthesis, thus averting the toxicity of polyamine excess. Forced expression of AZ1 in cells mimics the effect of polyamine excess on ODC degradation (Murakami et al., 1992b), implying that AZ1 is the sole intermediary through which the polyamines act to enhance ODC turnover.
The proteasome- and ATP-dependent stimulatory effect of AZ1 on proteolysis of ODC can by reconstituted in vitro using purified components (Murakami et al., 1992a, 1999; Elias et al., 1995). Here we use such a system to investigate the following questions: what specific step in ODC turnover is stimulated by AZ1? What element within the proteasome recognizes ODC–AZ1, and is it the same or different from that which recognizes ubiquitylated proteins? What structural elements of ODC are recognized by the proteasome, and how do these compare in the presence or absence of AZ1? What are the steps of substrate proteolysis and which are rate-limiting? Our results demonstrate that the C terminus of ODC harbors an AZ1-modulated recognition determinant that is recognized by the same element(s) in the proteasome that mediate recognition of polyubiquitin chains.
Results
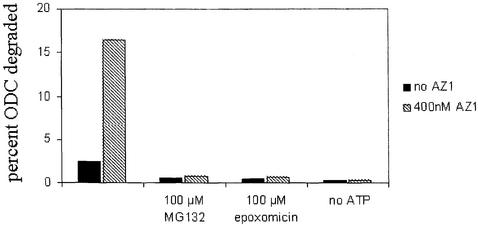
ODC is a favorable substrate for studies of proteasomal specificity because it has a well-defined structure, undergoes rapid proteasomal degradation in vivo and is extensively degraded by purified proteasomes in association with AZ1. We sought to determine the signals within ODC that are recognized by the proteasome both in the presence of AZ1 and in its absence. Because ODC degradation is much slower when AZ1 is absent, we required assay conditions sufficiently sensitive to measure both AZ1-stimulated and AZ1-independent turnover. Recombinant ODC was metabolically labeled in Escherichia coli to high specific activity and purified by use of an N-terminal affinity tag (Materials and methods). AZ1 was similarly prepared, but in unlabeled form. After incubation with highly purified 26S rat proteasomes (see Supplementary figure 1, available at The EMBO Journal Online), degradation was assessed by measuring production of acid-soluble radiolabel. ODC degradation was observed in the absence of AZ1 and was ∼8-fold more extensive in its presence (Figure 1). Production of acid-soluble counts was linear with time (data not shown). Both AZ1-independent and AZ1-stimulated degradation were strongly diminished by the proteasome-specific inhibitors MG132 and epoxomicin and dependent on the addition of ATP (Figure 1). The characteristics of the degradation system we employed therefore reflected salient general properties of proteasomal degradation and the specific substrate ODC.
Fig. 1. ODC degradation by the 26S proteasome in vitro. The extent of degradation was determined after 30 min in reaction mixtures with 50 nM proteasomes, 50 nM ODC and in the presence or absence of 400 nM AZ1. The effects of the proteasome inhibitors MG132 and epoxomicin and of ATP depletion were tested.
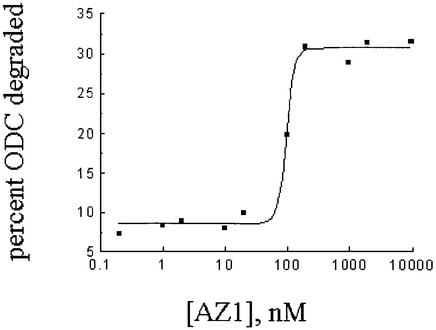
To determine how the rate of ODC degradation depends on AZ1 concentration, we used various stoichiometric ratios of AZ1 to ODC, encompassing a range from no AZ1 to a 200-fold excess with respect to both ODC and proteasome (Figure 2). Maximal stimulation of ODC degradation by AZ1 was observed at a concentration of AZ1 slightly in excess of ODC. These two proteins interact strongly: using the same buffer conditions, association of recombinant ODC and AZ1 has an equilibrium constant (Kd) of 1.0–2.0 × 10–9 M (results not shown). The data depicted in Figure 2 are therefore consistent with previous investigations, which concluded that AZ1 and ODC form a 1:1 molecular complex and that this complex is a more efficient substrate of the proteasome than is the ODC homodimer. The stimulatory effect of AZ1 remained little changed even at the highest AZ1 concentrations tested. The failure of AZ1 to change the extent of ODC degradation when present in great stoichiometric excess with respect to both ODC and the proteasome has two implications: (i) AZ1 does not stimulate the proteasome other than by interacting with ODC, else AZ1 should have further augmented ODC degradation when present in great excess. (ii) A second model can also be rejected based on these data. In this model, the proteasome has two recognition elements, one for AZ1 and the second for ODC, each of which recognizes its ligand with identical affinity regardless of whether AZ1 and ODC are free or associated with each other. If that were the case, excess free AZ1 should have occupied the first of these sites, diminishing ODC degradation to the level observed when AZ1 is absent or present in an amount not saturating for ODC. This too was not observed. Therefore, free AZ1 has no significant functional proteasome association and instead confers increased substrate affinity by forming a new recognition element in association with ODC.
Fig. 2. Dependence of ODC degradation on AZ1 concentration. The extent of degradation was determined after 50 min in reaction mixtures with 50 nM proteasomes, 50 nM ODC and the indicated concentrations of AZ1.
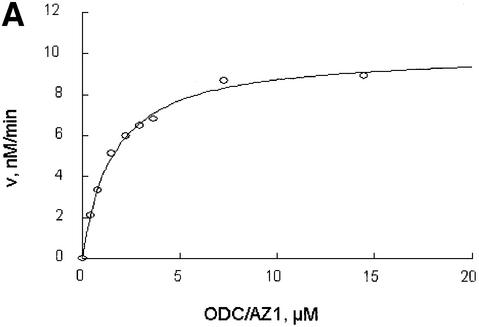
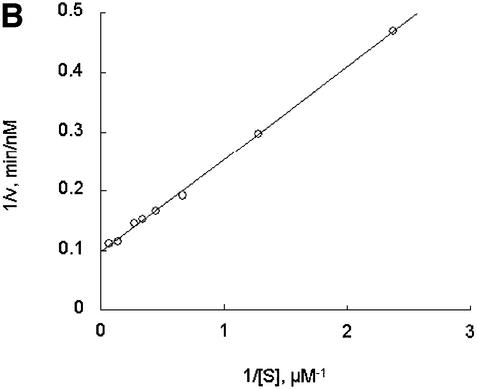
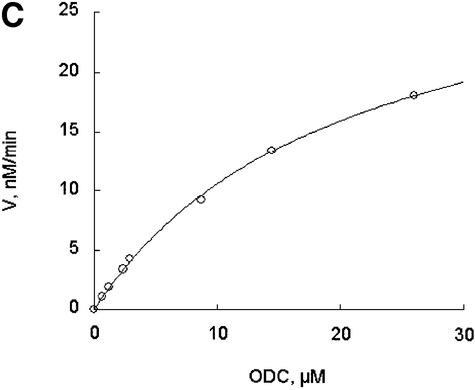
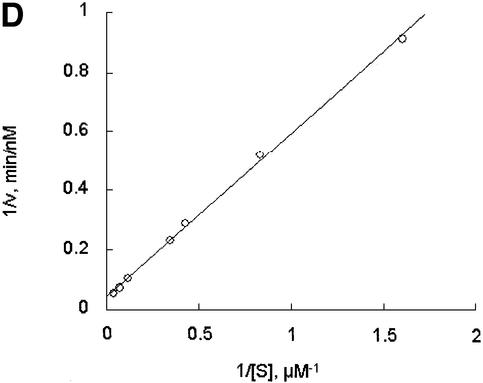
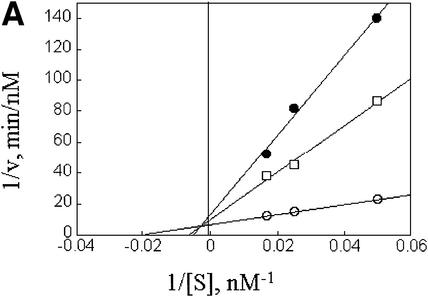
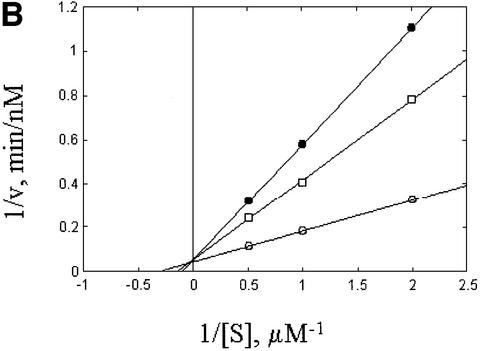
Further exploring the molecular characteristics of ODC–AZ1 interactions with the proteasome required first determining the kinetic characteristics of proteolysis in the presence or absence of AZ1. In the simplest and most readily analyzed case, one could treat the proteasome as an enzyme, ODC as a substrate, and AZ1 as a ligand that perturbs either the substrate association or the catalytic velocity of the enzyme. This approach proved to be feasible. The initial rate of degradation of ODC was determined using various concentrations of the ODC substrate and a stoichiometric excess of AZ1 (Figure 3A and B) or no AZ1 (Figure 3C and D). Saturation was observed in both cases and double reciprocal plots produced straight lines (R2 = 0.99), consistent with simple Michaelis–Menten kinetics. The value of kcat was little affected by AZ1 (0.22 and 0.20 min–1, respectively, without or with AZ1). In contrast, AZ1 decreased the value of Km, from 13 to 1.6 µM, consistent with the 8-fold stimulation of ODC degradation by AZ1 observed in Figure 1. AZ1 therefore improves the association of ODC with the proteasome, not the rate of its processing.
Fig. 3. AZ1 increases the apparent affinity of ODC but not the rate of catalysis. Incubations were carried out for 30 min and contained 50 nM 26S proteasomes (A and B) or 100 nM proteasomes (C and D), 50 nM [35S]ODC and various amounts of total ODC (labeled plus unlabeled), as indicated. (A and B) Reactions with 12 µM AZ1. (C and D) Reactions with no AZ1 present. (A and C) Velocity of degradation versus substrate concentration. The curves are a least-squares fit of the Michaelis–Menten equation assuming a Km of 1.6 µM (A) or of 13 µM (C). (B and D) Double reciprocal (Lineweaver–Burk) plots of data in (A and C), respectively.
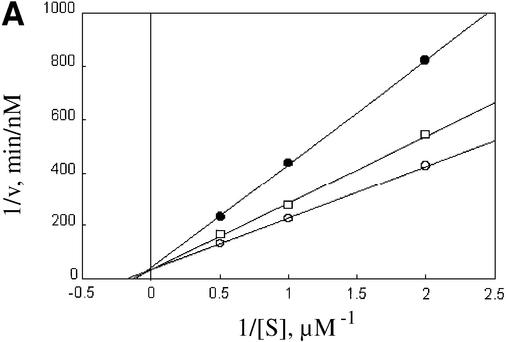
A ubiquitin chain provides the means for recognizing most substrates of the proteasome (Chau et al., 1989; Thrower et al., 2000). Although no sequence similarity is apparent between ubiquitin and ODC or AZ1, this does not exclude the participation of a common proteasomal recognition element for these apparently disparate signals, especially since polyubiquitin chains are not recognized based on primary sequence motifs. One way to investigate this question is to carry out experiments in which both types of substrate are presented simultaneously to the proteasome. The observation of competitive cross inhibition would constitute evidence for competitive occupancy of a single functionally important site. In these experiments we used the well characterized oligo-ubiquitylated substrate Ub5DHFR (Thrower et al., 2000). This consists of dihydrofolate reductase carrying an N-terminal ubiquitin, with the addition of a Ub4 chain linked through internal lys48 isopeptide bonds. Degradation of ODC:AZ1 was inhibited by Ub5DHFR in a competitive manner (Figure 4A). The inverse experiment, in which ODC:AZ1 was used to perturb Ub5DHFR degradation, revealed a similar competitive effect (Figure 4B).
Fig. 4. Competition between ODC–AZ1 and Ub5DHFR. (A) Com petitive inhibition of ODC–AZ1 by Ub5DHFR. Incubations contained 50 nM rat 26S proteasomes, 12 µM AZ1 with 0.5, 1 or 2 µM [35S]ODC and 0 (open circles), 100 nM (squares) or 500 nM (filled circles) Ub5DHFR. (B) Competitive inhibition of Ub5DHFR by ODC–AZ1. Incubations contained 2.5 nM rat 26S proteasomes, with 20, 40 or 60 nM [32P]Ub5DHFR, 12 µM AZ1, and 0 (open circles), 2.5 µM (squares) or 5 µM (filled circles) ODC–AZ1.
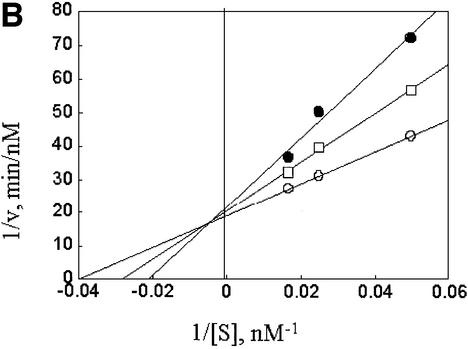
The observation of competitive cross inhibition implies that the two substrates tested compete at some step required for proteolysis, but that step need not be the initial substrate interaction step. The two substrates could, for example, compete for a downstream process required for unfolding or insertion into the 20S complex. This question was investigated in further competition experiments using an unanchored Ub4 chain as a mock substrate. Such ubiquitin chains can compete with polyubiquitylated substrates only in the initial binding step, as they are unable to undergo downstream reactions (Thrower et al., 2000). In confirmation of previously reported results, the Ub4 chain functioned as a competitive inhibitor of Ub5DHFR proteolysis (Figure 5A). It also competitively inhibited the degradation of ODC:AZ1 (Figure 5B). Importantly, the Ub4 chain had similar inhibitory constants with both substrates: 0.82 µM for Ub5DHFR and 0.67 µM for ODC:AZ1, indicating that competition involves the binding of the unanchored chain to a common, chain-recognizing site. Therefore, both ODC–AZ1 and Ub5DHFR interact with a common binding site of the proteasome, one which recognizes the ubiquitin chain.
Fig. 5. Competition by Ub4. (A) Competitive inhibition of Ub5DHFR by Ub4. Incubations contained 2.5 nM rat 26S proteasomes, with 20, 40 or 60 nM [32P]Ub5DHFR, and 0 (open circles), 0.8 µM (squares) or 2 µM (filled circles) Ub4. (B) Competitive inhibition of ODC–AZ1 by Ub4. Incubations contained 50 nM rat 26S proteasomes, with 12 µM AZ1 and 0.5, 1 or 2 µM [35S]ODC, and no (open circles), 0.8 µM (squares) or 2 µM (filled circles) Ub4.
Within ODC, what signal is recognized by the proteasome and by what means does AZ1 enhance that recognition? The C-terminal region of ODC is critical for its turnover (Ghoda et al., 1989). In vivo and in vitro studies have examined the effect of truncating or mutating this region. Two structural features within this domain have been shown to influence turnover, the last five C-terminal amino acids (Ghoda et al., 1992) of the 461 amino acid protein and Cys441 (Miyazaki et al., 1993). AZ1-stimulated degradation of ODC has been previously studied in a purified in vitro system (Elias et al., 1995; Murakami et al., 1992a, 1999), but AZ1-independent turnover has not. As we could now measure both types of degradation (Figure 1), it became possible to ask whether they impose similar or different structural requirements on the C terminus. To answer this question, we compared the extent of degradation of wild-type ODC to that of ODC with various mutations of the C terminus (Figure 6). Reactions were performed in the presence or absence of AZ1.
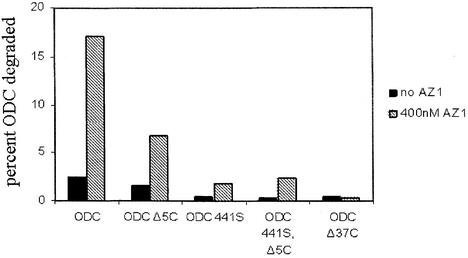
Fig. 6. Effect of C-terminal truncations and mutation on ODC degradation. The extent of degradation was determined after 30 min in incubations containing 50 nM proteasome, with or without 400 nM AZ1, as indicated.
For wild-type ODC, the extent of degradation was 8-fold greater with AZ1. Truncating the 37 C-terminal amino acids (ODCΔ37C) reduced degradation to the same baseline level, regardless of whether AZ1 was present. Truncating the last five amino acids (ODCΔ5C) had a weaker effect than removing all 37 amino acids, and attenuated degradation in either the absence or presence of AZ1 to a similar 2- or 3-fold extent. This implies that these last five amino acids may constitute one subsite, which will be termed S1, of a composite proteasomal recognition element. An isosteric Cys441Ser mutation (ODC441S) completely prevented AZ1-independent degradation, reducing it as much as the full Δ37C truncation. These data imply that Cys441 may constitute a second component, termed S2, of a composite proteasome recognition element. Because mutating Cys441 has as strong an effect as mutating both Cys441 and removing the last five amino acids (ODC 441S, Δ5C), the last five amino acids must depend on the function of Cys441 (a relationship that can be represented as S1→S2). The Cys441Ser mutation also had a strong effect on AZ1-stimulated degradation, reducing it ∼8-fold, but failed to reduce it to baseline level: AZ1 still stimulated degradation and did so even in the context of the Cys441Ser, Δ5C double mutation. Therefore, a distinct element, S3, is created by the interaction of AZ1 and some part of the 37 amino acid C-terminal region, and neither Cys441 nor the last five amino acids are an essential part of the postulated recognition element S3. In summary, for wild-type ODC, the terminal 37 amino acids contain subsites S1, S2 and S3. If AZ1 is not present, S2 is active, its activity augmented by the influence of S1. When AZ1 is present, it provides an 8-fold improvement in Km by adding the effect of S3 to that of S1→S2.
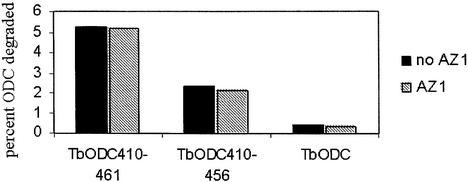
The adequacy of this model, proposing a tripartite collection of subsites, was further assessed by appending the ODC C terminus to other proteins and testing their properties as substrates or competitors. Although AZ1 acts on the C terminus of ODC to create (or augment) subsite S3, AZ1 requires a binding domain outside the C terminus contained within amino acids 117–142 of mammalian ODC to bind tightly to ODC (Li and Coffino, 1992). Therefore, proteins constructed as fusions to the ODC C terminus but which lack the requisite region for AZ1 association are expected to exhibit subsites S1 and S2, but not S3. ODC from the African parasite Trypanosoma brucei is highly homologous to vertebrate ODC, but is stable both in vitro and in vivo. It lacks the capacity to bind to AZ1 (Li and Coffino, 1992) and contains no homolog of the C terminus found in vertebrate ODC (Ghoda et al., 1989, 1990). We constructed a chimera in which T.brucei ODC is extended at its natural C terminus by amino acids 410–461 of mouse ODC and evaluated its degradation (Figure 7). The degradation of this chimeric molecule (TbODC410–461) was not augmented by AZ1, because a high affinity binding site for AZ1 is not present, a result consistent with the operation of S1→S2 without S3. Truncation of the last five amino acids (creating TbODC410–456) stabilized the protein about two-fold, which may be represented as converting S1→S2 to S2. Trypanosoma brucei ODC without addition of the ODC C terminus, and therefore with none of the putative subsite binding elements, was stable.
Fig. 7. Degradation of T.brucei ODC with mouse ODC C-terminal extensions. The extent of degradation was determined after 30 min in incubations containing 100 nM proteasome, with or without 400 nM AZ1, as indicated.
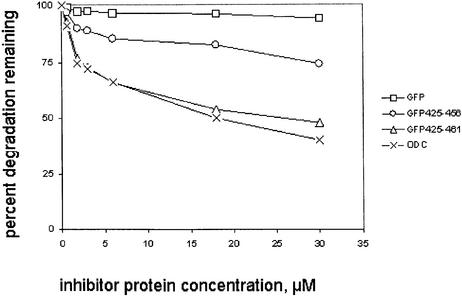
The degradation properties of a protein are dependent not only on its initial proteasome association but also on subsequent events. The rate-limiting event for properly folded natural substrates is likely unfolding rather than initial binding (Braun et al., 1999; Thrower et al., 2000; Lee et al., 2001; Liu et al., 2002). We fused amino acids 425–461 of mouse ODC to GFP, a tightly folded β barrel protein (Tsien, 1998). This fusion has been found to destabilize GFP in vivo in animal cells (Li et al., 1998) and in yeast (Hoyt et al., 2003). However, no destabilizing effect of the ODC C terminus on GFP was observed using the same in vitro system employed in the experiments described above (data not shown). This may reflect the absence from the in vitro system of chaperone proteins extrinsic to the proteasome that are available in vivo to participate in unfolding. Despite the failure of the ODC C terminus to enhance in vitro degradation of GFP, its interaction with the proteasome could be assessed by examining its inhibitory effect on the degradation of ODC (Figure 8). GFP with no ODC-derived extension had essentially no inhibitory capacity in this competition assay, but GFP425–461 diminished the degradation of wild-type ODC in a dose-dependent manner. Tellingly, the competitive potency of GFP425–461 was equal to that of full-length ODC containing native amino acids 1–461. The similar potency of full-length ODC and GFP425–461 implies that the terminal 37 amino acid region of ODC, containing putative binding element S1→S2, fully embodies the proteasome interaction properties displayed by ODC in the absence of AZ1. Absent AZ1, which is required to generate subsite S3, the 37 amino acid ODC C terminus can act on fully equal terms with the complete ODC molecule. The similar affinity of GFP425–461 and ODC for the proteasome also implies that the failure to degrade the former is due to a difference in kcat, not in Km, and further implies that the Km is a good approximation to the dissociation constant of the ODC C terminus for the proteasome. The potency of GFP425–456, which lacks the last five amino acids, was intermediate between that of GFP and GFP425–461. The lesser potency of GFP425– 456 in this inhibition assay compared with GFP425–461 is consistent with the presence of S2 in the former and the presence of S1→S2 in the latter.
Fig. 8. Inhibition of ODC degradation by GFP with mouse ODC C-terminal extensions. Data are normalized to ODC degradation observed in the absence of competing inhibitor protein and is expressed as percent residual degradation. Incubations were for 30 min and contained 100 nM proteasome, 50 nM ODC and various concentrations of inhibitors, as indicated. The extent of ODC degradation in the absence of inhibitors was 5.4%.
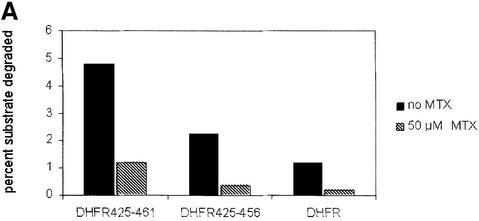
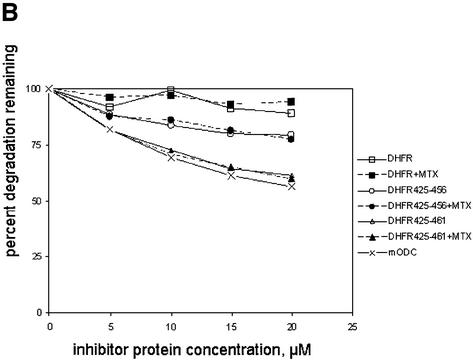
The hypothesis that unfolding limits the degradation of proteins with an ODC C-terminal extension can be tested directly using a substrate and a ligand that upon binding can impair unfolding. Methotrexate (MTX), a DHFR ligand, has been shown previously to stabilize Ub5DHFR (Thrower et al., 2000). We tested the unfolding requirement in the degradation of ODC-targeted substrates using fusions of the ODC C terminus to DHFR. We examined the effect of MTX on DHFR425–461, DHFR425–456, which lacks the last five amino acids of the ODC C terminus, and on DHFR (Figure 9A). If methotrexate was absent, the extent of degradation of the three proteins was DHFR425–461 > DHFR425–456 > DHFR. Metho trexate markedly impaired the turnover of each protein. However, in a competition inhibition assay, methotrexate did not change the inhibitory capacity of DHFR425–461 or DHFR425–456 (Figure 9B). Consistent with the inhibitory properties observed with fusions of the ODC C terminus to GFP, the inhibitory potency of the proteins examined in this experiment were ODC = DHFR425– 461 = DHFR425–461+MTX > DHFR425–456 = DHFR425–456+MTX > DHFR = DHFR +MTX. This demonstrates directly that for DHFR with ODC C-terminal extensions, methotrexate alters degradation by acting on unfolding, not by changing affinity for the proteasome.
Fig. 9. DHFR with ODC C-terminal extensions: degradation and competitive inhibition. (A) Degradation of DHFR with ODC C-terminal extensions. The extent of degradation was determined after 30 min in incubations containing 100 nM proteasome, with or without 50 µM methotrexate, as indicated. (B) Inhibition of ODC degradation by DHFR with mouse ODC C-terminal extensions. Incubations were performed in the absence of methotrexate (open symbols, ×) or in the presence of 50 µM methotrexate (filled symbols). Conditions of incubation and presentation of data are as in Figure 8. The extent of ODC degradation in the absence of inhibitors was 6.3%.
Discussion
The proteasome recognition elements of ODC lie within its 37 C-terminal amino acids. The functional importance of this region and of AZ1 expression have been documented previously by in vivo experiments (Ghoda et al., 1989; Murakami et al., 1992b, 1994; Miyazaki et al., 1993; Li et al., 1998; Gandre and Kahana, 2002) and by in vitro experiments using cell extracts (Ghoda et al., 1992; Li and Coffino, 1992; Murakami et al., 1993; Mamroud-Kidron et al., 1994), and have been reviewed recently (Coffino, 2001). In the present studies, we have used defined components to determine the kinetic characteristics of ODC degradation. We have shown here that deletion and mutagenesis of this region distinguishes three subsites, S1–S3. S1 consists of the last five amino acids. S2 consists of a single residue, cysteine 441. S1 acts by augmenting the effect of S2, because the Cys441Ser mutation of S2 alone has as strong an effect as mutating both S2 and deleting S1. S3 is produced by the joint action of AZ1 and the 37 amino acid C terminus. AZ1 per se is not a recognition element for the proteasome, because, as noted, excess free AZ1 does not stimulate or inhibit ODC:AZ1 degradation (Figure 2), or degradation of a chimeric form of ODC, which lacks the ability to bind AZ1 with high affinity (Figure 7). Neither S1 nor S2 is required to generate S3, because ODC with dual mutations in S1→S2 still responds to AZ1. By comparing the relative proteasome affinity of molecules with S1→S2 + S3, S1→S2, S2 alone, S3 alone and S2 + S3 we can evaluate the changes in affinity associated with disabling individual subsites. Disabling either S1→S2 or S3 produces in each case about an 8-fold effect on affinity compared with S1→S2 + S3 (Figure 6, compare the effect on degradation of ODC–AZ1 of withholding AZ1 versus mutating Cys441 to Ser441). Therefore S1→S2 and S3 make approximately equal contributions to association. Disabling S1 causes an ∼2- to 3-fold effect on affinity compared with S1→S2 (Figures 6–9). The effect on affinity of disabling S1 is approximately similar in effect (2- to 3-fold), regardless of whether or not S3 is functional. This observation implies that S1→S2 and S3 do not interact functionally.
The last 40 amino acids of the ODC C terminus do not contribute significant electron density in the crystal structure, (Kern et al., 1999; Almrud et al., 2000) providing evidence of conformational disorder. Antibody obtains greater access to the ODC C terminus in the presence of AZ1, antibody directed at the C terminus inhibits degradation and topologic constraint of the C terminus also impairs degradation (Li and Coffino, 1993). These data together imply that the C terminus is solvent accessible and that AZ1 can alter its conformation. However, AZ1 does not enhance degradation simply by dissociating the native ODC:ODC homodimer to form an ODC:AZ1 heterodimer. Mutating amino acids 131 and 145 in AZ1 abolishes its capacity to enhance ODC degradation, but does not alter its ability to form a ODC:AZ1 heterodimer (Chen et al., 2002). It will be of interest to determine the role of these two residues, and whether they are contact sites for the ODC C terminus or for the proteasome. The biochemical function of Cys441 is also not known. Cysteine thiols are chemically reactive, can participate in such reactions as formation of disulfide bonds or thioesters (Law and Dodds, 1997) and can be oxidized and nitrosylated (Stamler and Hausladen, 1998). The Cys441Ser substitution utilized in the experiments reported here is isosteric but completely abolishes subsite S1→S2 activity. It is possible that chemical modification of Cys441 is intrinsic to the action of subsite S2. Whatever the mechanistic function of S1→S2 in degradation, it has the capacity to interact productively with phylogenetically distant proteasomes. Its essential features are recognized by the yeast proteasome both in vivo and in vitro (Hoyt et al., 2003), despite the absence from yeast ODC of any C-terminal extension corresponding to that found in mouse ODC.
Kinetic analysis of ubiquitylated substrates of the proteasome has yielded Km values in the sub-micromolar range: 25–35 nM for Ub5DHFR (this study; Thrower et al., 2000). By this measure, polyubiquitylated substrates appear to be preferred substrates compared with ODC– AZ1. The kinetic properties of several model polyubiquitylated substrates have demonstrated that unfolding is rate limiting for their degradation. For these proteins, measures of Km are therefore good approximations of Kd, the dissociation constant. ODC has a well ordered structure, (Kern et al., 1999) suggesting that for this substrate as well, unfolding is rate limiting for proteasomal degradation. Consistent with this model, the Km of ODC–AZ1 is within a factor of two of its Ki as determined by inhibition of Ub5DHFR degradation (Figures 3 and 4). Also supportive of this model are the properties of distinct proteins with common ODC C termini: the presence of the 37 amino acid ODC C terminus confers on ODC, GFP and DHFR similar affinities for the proteasome, but ODC, GFP425–461 and DHFR425–461 have markedly different rates of degradation, differences presumably due to different rates of unfolding. Methotrexate, a DHFR ligand that impairs its unfolding by heat (Gaume et al., 1998) or by the 19S complex (Liu et al., 2002), reduced the degradation rate of DHFR425–461 and DHFR425–456, but did not change their affinity, directly sustaining this conclusion. Similarly, folic acid, a substrate of DHFR, modulates the rate of degradation of Ub5DHFR without affecting the interaction of this substrate with proteasomes (Thrower et al., 2000).
In vivo, ODC may in part evade this kinetic limitation by undergoing degradation before the protein completes folding. Pulse labeling of animal cells induced to express AZ1 demonstrated loss of about half of the ODC produced during a 4 min label by the end of that period (van Daalen Wetters et al., 1989). This very rapid post-translational degradation suggests that ODC may be degraded before it fully folds, as has been observed for model polyubiquitin-targeted proteasome substrates (Turner and Varshavsky, 2000). Pulse–chase experiments in yeast also support this conclusion (Toth and Coffino, 1999). The degradation rate of [35S]methionine-labeled yeast ODC was faster after a 3 min pulse label time than after a longer 2.5 h label time. In cells treated with polyamines to augment degradation, the half-life of newly synthesized short-labeled ODC was 10 min but the half-life of long-labeled steady-state ODC was 60 min. This shows that yeast ODC is rapidly degraded immediately after synthesis, and is still susceptible to turnover if it escapes this fate, but at a slower rate. It is plausible but not proven that maturation to a more stable form of yeast ODC is accompanied by folding to a native conformation. If a significant fraction of ODC is degraded before folding, its inefficient capture by the proteasome (high Km compared with ubiquitylated proteins) may be compensated by more rapid downstream processing (fast kcat compared with proteins that fold before ubiquitylation).
The present studies demonstrate that ODC and ubiquitylated proteins share a binding site in the proteasome. Crosslinking of oligo-ubiquitin to proteasomes has demonstrated that Rpt5/S6′, an ATPase of the proteasome base, is a component of this recognition site (Lam et al., 2002). The competition experiments imply that this ATPase is likely to contribute to recognition of both ubiquitylated and non-ubiquitylated substrates of the proteasome. Further work will be needed to understand this surprising congruence at the molecular level. Several lines of evidence suggest that polyubiquitin chains are recognized in part through the presentation of specific recognition determinants, including a series of hydrophobic patches, on the chain surface (reviewed in Thrower et al., 2000). Whether the ODC–AZ1 complex displays analogous determinants, and whether they are unique to the ODC–AZ1 complex or displayed by ODC alone, remains to be determined.
The path that substrates of the proteasome trace subsequent to their initial association is unknown, but for the fact that they must eventually enter the interior catalytic chamber of the 20S complex. Does the proteasome employ uniform mechanisms to process ubiquitylated proteins and ODC, and, by extension, other proteasome substrates that do not depend on ubiquitin for their recognition? Do both types of substrates trace a similar path through the proteasome subsequent to initial association? Both kinds of substrate must share common general requirements, including unfolding and insertion, but key molecular events may differ. For example, the Ubn modification may provide a molecular handle in conformational manipulation subsequent to the initial binding event. If so, a substitute handle must be present (or alternative mechanisms utilized) to deal with substrates that lack ubiquitin modification.
One characteristic that distinguishes the two classes is the mechanism of tag clearance: ubiquitin chains are cleared by enzymatic cleavage of peptide bonds, but the tag is intrinsic for ODC (or merely requires sufficient unfolding to liberate AZ1, which does not enter the proteasome along with ODC). Two enzymes that cleave ubiquitin are known to be associated with the yeast proteasome. Rpn11 is an intrinsic subunit protein of the lid (Verma et al., 2002; Yao and Cohen, 2002), while UBP6 binds through its ubiquitin-like (Ubl) domain to the 19S complex, perhaps through association with the Rpn1 protein in the base (Leggett et al., 2002). A lidless proteasome can degrade denatured proteins, but not folded ubiquitylated proteins (Glickman et al., 1998a). No specific biochemical function has as yet been attributed to the lid, aside from the deubiquitylating activity of Rpn11 (which is essential for yeast viability); the lid may therefore prove redundant for the degradation of non-ubiquitylated substrates. This notion is testable. If correct, further investigations of proteasome-substrate interactions may be facilitated by the use of a structurally simplified proteasome and the substrate ODC.
Materials and methods
Reagents
Ubiquitin aldehyde (Ubal) was purchased from Boston Biochem (Cambridge, MA), methionine assay medium from Difco, protease inhibitor cocktail tablets from Roche. Epoxomicin was from Affiniti Research Products, UK. Other reagents were purchased from Sigma.
Purification of rat liver 26S proteasomes
26S proteasomes were purified from the livers of Sprague Dawley rats. Livers were stored frozen prior to use. All steps were performed at 0–4°C, using a modification of a published procedure (Reidlinger et al., 1997). Briefly, 150 g of rat livers were homogenized in 500 ml buffer A [20 mM Tris–HCl pH 7.5, containing 2 mM ATP, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 50 mM NaCl and 20%(v/v) glycerol]. After removing cell debris by centrifugation (RCFmax = 10 410 g, 80 min) and filtration through cheesecloth, the sample was further cleared by centrifugation (30 000 r.p.m. in a Beckman Type 35 rotor, RCFmax = 105 000 g, 1 h). The supernatant was filtered through Whatman filter paper, and the pH was adjusted to 7.5. The 26S proteasomes were sedimented by centrifugation (35 000 r.p.m. in a Beckman Type 35 rotor, RCFmax = 143 000 g, 18 h) and the pellet was resuspended in 25 ml buffer A without glycerol. The pH was again adjusted to 7.5, if required. The sample was then layered onto a glycerol gradient (15–40% in buffer A, 32 ml/tube × 6) and centrifuged for 16 h (25 000 r.p.m. in a Beckman SW27 rotor, RCFmax = 110 000 g). Fractions containing peptidase activity were pooled and 10 mg of protein per run loaded onto a Mono Q HR 5/5 column (Amersham Pharmacia) equilibrated with 20 mM Tris–HCl pH 7.5, 1 mM DTT, 50 mM NaCl and 10% (v/v) glycerol. Proteasomes were eluted using a linear salt gradient (50–800 mM NaCl, 40 column volumes) in the equilibration buffer. Active fractions were pooled and 26S proteasomes collected by centrifugation (31 000 r.p.m. in a Beckman type 35 rotor, RCFmax = 112 000 g, 12 h). The purified 26S proteasomes (24 mg) were resuspended in 1.5 ml of the above equilibration buffer, and stored at –70°C. The 26S proteasomes were analyzed by SDS–PAGE fractionation and showed a characteristic pattern of protein bands which closely resembled that previously reported for highly purified preparations. Purity and 26S mobility was further confirmed by nondenaturing gel and substrate overlays (Glickman et al., 1998b).
Protein expression and purification
All DNA manipulations used standard molecular methods. Constructions that utilized PCR steps were verified by sequencing; constructions that relied on restriction–ligation utilized fully sequenced constituents. The ODC gene was from mouse (Mus musculus) and AZ1 from rat (Rattus norvegicus). For constructs with ODC C-terminal extensions, dihydrofolate reductase (DHFR) was from humans (Homo sapiens) and variant Aequorea green fluorescent protein (GFPuv) from the plasmid pBAD-GFPuv (Clontech). Expression plasmids for ODC and its derivatives were generated from vector pQE30 (Qiagen) as described (Chen et al., 2002). Point mutations were introduced following the QuikChange™ protocol (Stratagene). Recombinant proteins (with a His6 tag at the N-terminus) were expressed in M15[pREP4] by induction with 1 mM isopropyl-β-d-galactopyranoside (IPTG) for 5 h and purified by TALON metal affinity resin (Clontech) following the protocol provided by the manufacturer. The version of His10-UbDHFR used in the present work was modified relative to that used previously (Thrower et al., 2000) as follows: (i) an Ala4 linker was inserted between V76 of Ub and M1 of (mouse) DHFR and (ii) a protein kinase A site (RASVQ) was placed after the C-terminal residue of DHFR. Synthesis of (Ub)4 and Ub5DHFR were as described previously (Thrower et al., 2000). Concentrations of unlabelled proteins were determined by Bradford assay (Bio-Rad) using bovine serum albumin (BSA) as standard and their molarity calculated using as molecular weight 2.0 × 106 for 26S proteasomes, 5.2 × 104 (the monomer molecular weight) for ODC, 3.2 × 104 for Ub4 and 6.4 × 104 for Ub5DHFR.
Radiolabeling of proteins
Ub5DHFR was enzymatically labeled with [γ-32P]ATP using protein kinase A (Sigma). Unincorporated label was removed by means of a spin column. Specific activity was ∼1 × 105 c.p.m./pmol. Other proteins used as substrates for the proteasome were metabolically 35S-labeled in E.coli strain M15[pREP4] at 37°C (Thrower et al., 2000). Cells (80 ml) were grown in LB medium to OD600 = 0.5, washed once with M9 medium, and resuspended in 40 ml of M9 medium containing 0.4% glucose and 0.063% methionine assay medium. After 30 min, IPTG was added. One hour after induction with IPTG, [35S]methionine (800 µCi) was added for 5 min, followed by unlabeled methionine (1 mM) for a further 10 min. Clarified lysate was applied to a 0.5 ml Talon column and the labeled proteins were purified by standard procedures, except 0.2 mg/ml BSA was included as a carrier during elution. Specific activity for 35S-labeled proteins was about 5 × 104 c.p.m./pmol. After buffer exchange to 50 mM Tris–HCl pH 7.5, 10 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM ATP and 10% glycerol, the concentration of labeled proteins was estimated by SDS–PAGE and Coomassie Blue staining, using unlabeled proteins as a standard.
Proteasome degradation assays
Assays of the degradation of [32P]Ub5DHFR were performed in a volume of 20 µl at 37°C and contained: 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM ATP, 10 mM KCl, 10% glycerol, a ATP regenerating system (2 mM DTT, 10 mM creatine phosphate, 1.6 mg/ml creatine kinase), 2 mg/ml BSA and 2.5 nM rat 26S proteasomes. The degradation of [35S]mODC and variants was performed similarly, except 50 nM proteasomes were used. Ubal (1 µM) was included in the reactions whenever Ub5DHFR or Ub4 were present to prevent chain disassembly. Concentrations of substrate or competitor proteins are given in the legends to figures. Reactions were preincubated for 10 min, initiated by addition of proteasomes and quenched by adding 140 µl of 20% (w/v) trichloroacetic acid. MG132 and epoxomicin when present were preincubated with all the other components of the reaction for 30 min at 37°C before substrates were added. For [32P]Ub5DHFR, the reaction time was 20 min (released counts accumulated linearly during the first 30 min), and for [35S]mODC and variants the reaction time was 30 min (released counts accumulated linearly during the first 60 min). After microcentifugation for 30 min at 14 000 g, 150 µl of the supernatant was removed for scintillation counting to determine released counts. Total counts were obtained using water in place of trichloroacetic acid. Background of released counts without proteasomes was usually about 0.5% of total counts. Percentage degradation of labeled proteins was determined by the formula: percent of degradation = (released c.p.m. – background c.p.m.)/total c.p.m. The initial velocity of degradation was determined by: v = [(percent of degradation) × (labeled + unlabeled substrate)]/incubation period. Kinetic parameters were determined using the curve fitting utilities of Kaleidagraph (Synergy Software).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Susumu Tomita from David Bredt’s laboratory for providing rat livers, H.Chen for making the plasmids to express mODC441S, J.Wang for assistance in preparation of rat proteasomes and S.McDonough for preparation of recombinant AZ1. The work was supported by USA National Institutes of Health grants R01 GM-45335 (to P.C.) and DK-46984 (to C.M.P.).
References
- Almrud J.J., Oliveira,M.A., Kern,A.D., Grishin,N.V., Phillips,M.A. and Hackert,M.L. (2000) Crystal structure of human ornithine decarboxylase at 2.1 Å resolution: structural insights to antizyme binding. J. Mol. Biol., 295, 7–16. [DOI] [PubMed] [Google Scholar]
- Amerik A., Swaminathan,S., Krantz,B.A., Wilkinson,K.D. and Hochstrasser,M. (1997) In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J., 16, 4826–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The protea some: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Bercovich Z., Rosenberg-Hasson,Y., Ciechanover,A. and Kahana,C. (1989) Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J. Biol. Chem., 264, 15949–15952. [PubMed] [Google Scholar]
- Braun B.C., Glickman,M., Kraft,R., Dahlmann,B., Kloetzel,P.M., Finley,D. and Schmidt,M. (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol., 1, 221–226. [DOI] [PubMed] [Google Scholar]
- Chau V., Tobias,J.W., Bachmair,A., Marriott,D., Ecker,D.J., Gonda,D.K. and Varshavsky,A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science, 243, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Chen H., MacDonald,A. and Coffino,P. (2002) Structural elements of antizymes 1 and 2 required for proteasomal degradation of ornithine decarboxylase. J. Biol. Chem., 277, 45957–45961. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Orian,A. and Schwartz,A.L. (2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays, 22, 442–451. [DOI] [PubMed] [Google Scholar]
- Coffino P. (2001) Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol., 2, 188–194. [DOI] [PubMed] [Google Scholar]
- Elias S., Bercovich,B., Kahana,C., Coffino,P., Fischer,M., Hilt,W., Wolf,D.H. and Ciechanover,A. (1995) Degradation of ornithine decarboxylase by the mammalian and yeast 26S proteasome complexes requires all the components of the protease. Eur. J. Biochem., 229, 276–283. [PubMed] [Google Scholar]
- Gandre S. and Kahana,C. (2002) Degradation of ornithine decarboxylase in Saccharomyces cerevisiae is ubiquitin independent. Biochem. Biophys. Res. Commun., 293, 139–144. [DOI] [PubMed] [Google Scholar]
- Gaume B., Klaus,C., Ungermann,C., Guiard,B., Neupert,W. and Brunner,M. (1998) Unfolding of preproteins upon import into mitochondria. EMBO J., 17, 6497–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoda L., van Daalen Wetters,T., Macrae,M., Ascherman,D. and Coffino,P. (1989) Prevention of rapid intracellular degradation of ODC by a C-terminal truncation. Science, 243, 1493–1495. [DOI] [PubMed] [Google Scholar]
- Ghoda L., Phillips,M.A., Bass,K.E., Wang,C.C. and Coffino,P. (1990) Trypanosome ornithine decarboxylase is stable because it lacks sequences found in the C terminus of the mouse enzyme which target the latter for intracellular degradation. J. Biol. Chem., 265, 11823–11826. [PubMed] [Google Scholar]
- Ghoda L., Sidney,D., Macrae,M. and Coffino,P. (1992) Structural elements of ornithine decarboxylase required for intracellular degradation and polyamine-dependent regulation. Mol. Cell. Biol., 12, 2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass J.R. and Gerner,E.W. (1987) Spermidine mediates degradation of ornithine decarboxylase by a non-lysosomal, ubiquitin-independent mechanism. J. Cell Physiol., 130, 133–141. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubin,D.M., Coux,O., Wefes,I., Pfeifer,G., Cjeka,Z., Baumeister,W., Fried,V.A. and Finley,D. (1998a) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubin,D.M., Fried,V.A. and Finley,D. (1998b) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol., 18, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Zhang,M. and Coffino,P. (2003) Ubiquitin-independent mechanisms of mouse ornithine decarboxylase degradation are conserved between mammalian and fungal cells. J. Biol. Chem., 278, 12135–12143. [DOI] [PubMed] [Google Scholar]
- Kern A.D., Oliveira,M.A., Coffino,P. and Hackert,M.L. (1999) Structure of mammalian ornithine decarboxylase at 1.6 Å resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure, 7, 567–581. [DOI] [PubMed] [Google Scholar]
- Kohler A., Cascio,P., Leggett,D.S., Woo,K.M., Goldberg,A.L. and Finley,D. (2001) The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell, 7, 1143–1152. [DOI] [PubMed] [Google Scholar]
- Lam Y.A., Lawson,T.G., Velayutham,M., Zweier,J.L. and Pickart,C.M. (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature, 416, 763–767. [DOI] [PubMed] [Google Scholar]
- Law S.K. and Dodds,A.W. (1997) The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci., 6, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Schwartz,M.P., Prakash,S., Iwakura,M. and Matouschek,A. (2001) ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell, 7, 627–637. [DOI] [PubMed] [Google Scholar]
- Leggett D.S., Hanna,J., Borodovsky,A., Crosas,B., Schmidt,M., Baker,R.T., Walz,T., Ploegh,H. and Finley,D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell, 10, 495–507. [DOI] [PubMed] [Google Scholar]
- Li X. and Coffino,P. (1992) Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol. Cell. Biol., 12, 3556–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. and Coffino,P. (1993) Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol. Cell. Biol., 13, 2377–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhao,X., Fang,Y., Jiang,X., Duong,T., Fan,C., Huang,C.C. and Kain,S.R. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem., 273, 34970–34975. [DOI] [PubMed] [Google Scholar]
- Liu C.W., Millen,L., Roman,T.B., Xiong,H., Gilbert,H.F., Noiva,R., DeMartino,G.N. and Thomas,P.J. (2002) Conformational remodeling of proteasomal substrates by PA700, the 19S regulatory complex of the 26S proteasome. J. Biol. Chem., 277, 26815–26820. [DOI] [PubMed] [Google Scholar]
- Mamroud-Kidron E., Omer-Itsicovich,M., Bercovich,Z., Tobias,K.E., Rom,E. and Kahana,C. (1994) A unified pathway for the degradation of ornithine decarboxylase in reticulocyte lysate requires interaction with the polyamine-induced protein, ornithine decarboxylase antizyme. Eur. J. Biochem., 226, 547–554. [DOI] [PubMed] [Google Scholar]
- Mitchell J.L.A. and Chen,H.J. (1990) Conformational changes in ornithine decarboxylase enable recognition by antizyme. Biochim. Biophys. Acta, 1037, 115–121. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Matsufuji,S., Murakami,Y. and Hayashi,S. (1993) Single amino-acid replacement is responsible for the stabilization of ornithine decarboxylase in HMOA cells. Eur. J. Biochem., 214, 837–844. [DOI] [PubMed] [Google Scholar]
- Murakami Y. and Hayashi,S. (1985) Role of antizyme in degradation of ornithine decarboxylase in HTC cells. Biochem. J., 226, 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Kameji,T., Hayashi,S., Igarashi,K., Tamura,T., Tanaka,K. and Ichihara,A. (1992a) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature, 360, 597–599. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Miyazaki,Y. and Hayashi,S. (1992b) Destabilization of ornithine decarboxylase by transfected antizyme gene expression in hepatoma tissue culture cells. J. Biol. Chem., 267, 13138–13141. [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Tanaka,K., Ichihara,A. and Hayashi,S. (1993) Involvement of the proteasome and antizyme in ornithine decarboxylase degradation by a reticulocyte lysate. Biochem. J. 295, 305–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Miyazaki,Y. and Hayashi,S. (1994) Forced expression of antizyme abolishes ornithine decarboxylase activity, suppresses cellular levels of polyamines and inhibits cell growth. Biochem. J., 304, 183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Hayashi,S.I., Tanahashi,N. and Tanaka,K. (1999) ATP-Dependent inactivation and sequestration of ornithine decarboxylase by the 26S proteasome are prerequisites for degradation. Mol. Cell. Biol., 19, 7216–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Reidlinger J., Pike,A.M., Savory,P.J., Murray,R.Z. and Rivett,A.J. (1997) Catalytic properties of 26S and 20S proteasomes and radiolabeling of MB1, LMP7, and C7 subunits associated with trypsin-like and chymotrypsin-like activities. J. Biol. Chem., 272, 24899–24905. [DOI] [PubMed] [Google Scholar]
- Stamler J.S. and Hausladen,A. (1998) Oxidative modifications in nitrosative stress. Nat. Struct. Biol., 5, 247–249. [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Pickart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth C. and Coffino,P. (1999) Regulated degradation of yeast ornithine decarboxylase. J. Biol. Chem., 274, 25921–25926. [DOI] [PubMed] [Google Scholar]
- Tsien R.Y. (1998) The green fluorescent protein. Annu. Rev. Biochem., 67, 509–44. [DOI] [PubMed] [Google Scholar]
- Turner G.C. and Varshavsky,A. (2000) Detecting and measuring cotranslational protein degradation in vivo. Science, 289, 2117–2120. [DOI] [PubMed] [Google Scholar]
- van Daalen Wetters T., Macrae,M., Brabant,M., Sittler,A. and Coffino,P. (1989) Polyamine-mediated regulation of mouse ornithine decarboxylase is posttranslational. Mol. Cell. Biol., 9, 5484–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R. and Deshaies,R.J. (2000) A proteasome howdunit: the case of the missing signal. Cell, 101, 341–344. [DOI] [PubMed] [Google Scholar]
- Verma R., Aravind,L., Oania,R., McDonald,W.H., Yates,J.R.,III, Koonin,E.V. and Deshaies,R.J. (2002) Role of Rpn11 metallo protease in deubiquitination and degradation by the 26S proteasome. Science, 298, 611–615. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Yao T. and Cohen,R.E. (2002) A cryptic protease couples deubiquitin ation and degradation by the proteasome. Nature, 419, 403–407. [DOI] [PubMed] [Google Scholar]