Abstract
The majority of DNA damage-induced mutagenesis in the yeast Saccharomyces cerevisiae arises as a result of translesion replication. This process is critically dependent on the deoxycytidyl transferase Rev1p, which forms a complex with the subunits of DNA polymerase ζ, Rev3p and Rev7p. To examine the role of Rev1 in vertebrate mutagenesis and the DNA damage response, we disrupted the gene in DT40 cells. Rev1-deficient DT40 grow slowly and are sensitive to a wide range of DNA-damaging agents. Homologous recombination repair is likely to be intact as basal and damage induced sister chromatid exchange and immunoglobulin gene conversion are unaffected. How ever, the mutant cells show a markedly reduced level of non-templated immunoglobulin gene mutation, indicating a defect in translesion bypass. Furthermore, ultraviolet exposure results in marked chromosome breakage, suggesting that replication gaps created in the absence of Rev1 cannot be efficiently repaired by recombination. Thus, Rev1-dependent translesion bypass and mutagenesis is likely to be a trade-off for the ability to complete replication of a damaged template and thereby maintain genome integrity.
Keywords: chromosome instability/immunoglobulin diversification/mutagenesis/Rev1/translesion synthesis
Introduction
DNA damage that stalls replication poses a major threat to genomic stability and cellular viability. Several strategies have evolved to allow a cell to tolerate or repair such damage, thereby allowing replication to be completed. These strategies can be broadly divided into those based on recombination and those based on lesion bypass. In yeast, the genes involved in lesion bypass fall into the RAD6 post-replication repair epistasis group (reviewed in Broomfield et al., 2001). The relationships within this group of genes are complex and incompletely understood. On genetic grounds, there appear to be three subpathways (Liefshitz et al., 1998; Xiao et al., 2000; Torres-Ramos et al., 2002), or possibly four (Cejka et al., 2001). These pathways can be further divided into those which result in largely error-free bypass and those which are error prone. Each has a requirement for RAD6 and RAD18. Rad6p is an E2 ubiquitin conjugating enzyme and in post-replication repair functions with Rad18p, which is a DNA-binding protein and E3 ubiquitin ligase.
Error-free damage avoidance can be accomplished in two ways: use of an alternative undamaged template or translesion synthesis on the damaged template with restoration of the original coding sequence. These approaches are reflected in two of the genetic subpathways in the RAD6 group. The first error-free pathway is thought to involve template switching. Here the nascent DNA strand that has encountered a block switches transiently to the newly synthesized daughter strand of the undamaged template returning once the lesion is bypassed (Higgins et al., 1976). It has been proposed that this template switch may be promoted by the recently demonstrated sequential mono-ubiquitylation of proliferating cell nuclear antigen (PCNA; the DNA polymerase sliding clamp) by Rad6p/Rad18p followed by the formation of Lys63-linked polyubiquitin chains. These variant ubiquitin chains, which are not linked to protein degradation, are extended from the initial single ubiquitin moiety by Rad5p (which genetically defines this pathway) acting in concert with two other members of the RAD6 group, the E2 ubiquitin ligase Ubc13p and the non-canonical E2-like protein, MMS2p (Hoege et al., 2002).
A second error-free pathway is defined by RAD30 (DNA polymerase η). This polymerase is able to bypass accurately some of the major products of UV-induced DNA damage, such as cis–syn TT dimers and CC or TC 6–4 photoproducts (Johnson et al., 1999; Yu et al., 2001).
In addition to these largely accurate bypass pathways, there also exists an error-prone system defined by the REV1, REV3 and REV7 genes. Rev3p and Rev7p form DNA polymerase ζ (Rev3p containing the catalytic domain), which associates with an accessory protein, Rev1p. This pathway also involves direct bypass of the lesion, but with a trade-off: the ability to synthesize across, or extend from, damaged DNA that would otherwise stall the replicative polymerases is paid for by the frequent introduction of mutations. This occurs both because the lesions are non- or mis-instructional, and because the polymerases themselves are intrinsically more error prone (reviewed in Goodman, 2002).
REV1 was identified in yeast through a screen to detect mutants that are hypomutable in response to ultraviolet (UV) light (Lemontt, 1971). rev1 mutants are also sensitive to UV light and functional Rev1p is required for the vast majority of spontaneous and induced base pair substitution mutation found in yeast, as well as playing a role in more complex frameshift events (Lawrence and Christensen, 1976, 1978; Lawrence et al., 1984; Harfe and Jinks-Robertson, 2000). REV1 is a member of the Y-type polymerase family, which also includes UmuC and DinB in Escherichia coli, RAD30 in Saccharomyces cerevisiae and polymerases η, ι and κ in vertebrates (Ohmori et al., 2001). Although it shares five highly conserved motifs with these other polymerases, it possesses only limited bona fide polymerase activity in vitro (Haracska et al., 2002). Yeast Rev1p has been shown to possess deoxycytidyl transferase activity and to be capable of inserting a C opposite an abasic site in vitro, a mismatch that is relatively efficiently extended by polymerase ζ (Nelson et al., 1996). More recently the human and mouse proteins have been shown to display similar biochemical behaviour (Lin et al., 1999; Masuda et al., 2001, 2002).
However, REV1 is essential for mutagenic bypass of a wide variety of lesions, a function which does not appear to be dependent on its deoxycytidyl transferase activity: the yeast rev1-1 mutant, which retains a substantial amount of its transferase activity in vitro, is deficient for induced mutagenesis (Nelson et al., 2000). Furthermore, strains that carry a complete disruption of the deoxycytidyl transferase activity of Rev1 display no defect in mutagenicity induced by abasic sites (Haracska et al., 2001). Interestingly, the rev1-1 mutation is within the enzyme’s N-terminal BRCT motif, a domain that has been implicated in structural interactions between DNA repair proteins (Callebaut and Mornon, 1997).
The importance of translesion bypass in higher eukaryotes has been difficult to assess. Disruption of Rev3 results in lethality during early to mid-gestation in mice due to a failure of cellular proliferation (Bemark et al., 2000; Esposito et al., 2000; Kajiwara et al., 2001; O-Wang et al., 2002; Van Sloun et al., 2002). Furthermore, there is evidence that Rev3 has a role in recombination as well as translesion bypass (Holbeck and Strathern, 1997). Indeed the Drosophila mutant of Rev3 (mus205) appears not to be involved in induced mutagenesis (Eeken et al., 2001), and yet is required for tolerance of DNA damage by alkylating agents and UV light. Conversely, experiments in human cell lines using antisense against Rev1 demonstrated a decrease in UV light-induced mutation at the HPRT locus, but showed no effect on survival in response to UV light (Gibbs et al., 2000).
To study the role of Rev1 and of translesion bypass in vertebrates we have inactivated the gene in chicken DT40 cells. We find that the cells are viable but exhibit increased spontaneous apoptosis and are sensitive to a wide variety of DNA damaging agents with UV irradiation, leading to an increase in chromosomal aberrations. Furthermore, we find that non-templated mutation in the constitutively diversifying immunoglobulin light chain locus is dependent on Rev1, while templated changes (gene conversions) are unaffected. Together these data suggest that, as in yeast, Rev1 is central to the DNA damage response in DT40 and contributes to the maintenance of chromosomal integrity by allowing replication to be completed despite a damaged template, even though this may be at the expense of an increase in mutagenesis. The data also suggests that Rev1 is likely to play a role in the generation of sequence diversity in the immunoglobulin locus of vertebrates.
Results
Disruption of Rev1 in DT40
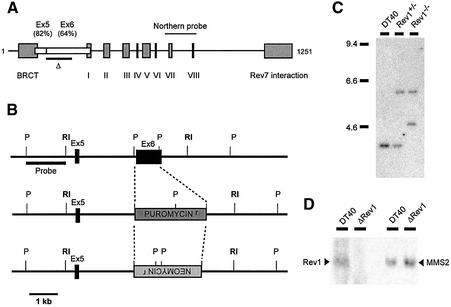
We wished to generate a targeting construct for chicken Rev1 that disrupted the gene in its N-terminal region. Since, at the time, no expressed-sequence tags (ESTs) in the database extended this far 5′, we opted to isolate a genomic clone from a chicken phage λ genomic DNA library. Filter hybridization of the library at low stringency with the first 1541 bp of mouse Rev1 cDNA identified four clones. A 4.5 kb EcoRI fragment, which contained regions homologous to human Rev1 exons 5 (82% amino acid identity) and 6 (64% amino acid identity) was used to generate the targeting construct. The 5′ intron/exon boundary and a large part of exon 6 (equivalent to 195 amino acids) were disrupted by replacement of a 900 bp PstI fragment with a drug resistance cassette (Figure 1A–C; Supplementary figure 1 available at The EMBO Journal Online). This disrupts the protein upstream of the motifs conserved in translesion polymerases and the putative Rev7 interacting domain but downstream of the BRCT domain (Figure 1A). Northern blotting for Rev1 using a cDNA probe spanning polymerase motifs VII and VIII (Figure 1A) confirmed loss of the Rev1 transcript (Figure 1D).
Fig. 1. Generation of ΔRev1-DT40. (A) Schematic of the human Rev1 cDNA. The grey boxes represent sequence features: BRCT, homology to BRCA-1 C-terminus. Boxes I–VIII also show significant homology between species and contain motifs common to translesion polymerases. ‘Rev7 interaction’ denotes the region mapped in human Rev1 to interact with Rev7 (Murakumo et al., 2001). The white boxes show the positions of exons 5 and 6 with the amino acid identity between chicken and human indicated (see also Supplementary figure 1). The black bar shows the region deleted in the ΔRev1-DT40 in this study. (B) Restriction map of the Rev1 genomic locus in DT40 containing exons homologous to human exons 5 and 6. The targeting construct is derived from the 4.5 kb EcoRI fragment defined by the bold RI sites. P, PstI; R, EcoRI. (C) Southern blot of genomic DNA digested with PstI and probed with the PstI–EcoRI fragment indicated in (B). (D) Northern blot for Rev1. The left-hand two lanes are probed with a chicken Rev1 cDNA fragment spanning polymerase motifs VII and VIII. The right hand two lanes shows the same blot hybridized with chicken MMS2 cDNA.
ΔRev1-DT40 expands slowly and exhibit increased spontaneous apoptosis
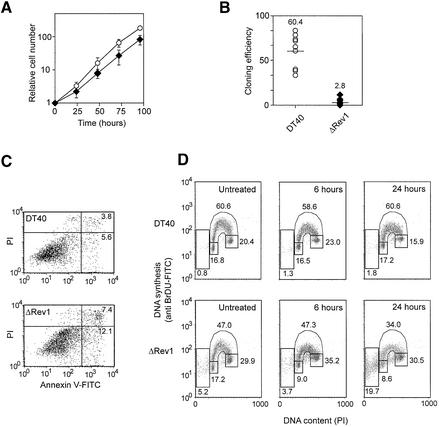
ΔRev1-DT40 expands with noticeably slower kinetics than wild-type DT40. The doubling time was estimated by exponential curve fit to be 15–16 h compared with 12 h for wild-type cells (Figure 2A) at 37°C. At 42°C, the doubling times were reduced to ∼10 h for wild type and 13–14 h for the mutants (data not shown), suggesting that the ΔRev1-DT40 are not significantly temperature sensitive. They are, however, sensitive to freezing, exhibiting noticeably prolonged cycle times in the first few days after thawing. Two further independently targeted clones exhibited similar growth characteristics, but Rev1 heterozygotes grew at the same rate as wild-type cells (data not shown). Cloning efficiency, as assessed by survival of cells seeded in plates at 1 cell/well, was also markedly reduced compared with wild-type cells, with a median of 2.8% compared with >60%, suggesting that ΔRev1-DT40 cells have a much reduced chance of successfully completing a cell cycle than their wild-type counterparts (Figure 2B). This correlates with an increased level of apoptosis in an exponentially expanding culture of ΔRev1-DT40, demonstrated by Annexin V staining (Figure 2C). Examination of cell cycle profiles in asynchronous populations confirmed this relative increase in the apoptotic (sub-G1) population in ΔRev1-DT40 (Figure 2D). In the mutant there is also a significant reduction in S phase cells accompanied by an increase in the G2/M population. Although the reduction in S phase cells is accompanied by an increase in non-replicating cells with S phase DNA content, it is not possible to distinguish in this experiment whether these are cells arrested mid-S phase or are cells that are apoptosing from G2. Following treatment with cisplatin, a DNA interstrand crosslinking agent to which ΔRev1 is sensitive (see below; Figure 3), all these features become accentuated: an increase in the G2/M population is seen at 6 h followed by, at 24 h after treatment, a further decrease in the number of cells in S phase associated with continued G2/M arrest and massive apoptosis (Figure 2D). These observations are consistent with enforcement of a G2 DNA-damage checkpoint known to be present in DT40 (Sudo et al., 2001), suggesting that loss of Rev1 results in a failure to remove or bypass DNA damage during S phase resulting in the transmission of an incompletely replicated genome to G2. It is, however, not possible from these data to distinguish the relative contribution of Rev1 and translesion synthesis to rescuing stalled forks in S phase from a role in gap filling in G2 or whether cells are initiating apoptosis both in G2/M and S phase. These issues are the subject of further investigation.
Fig. 2. Growth and cell cycle characteristics of ΔRev1-DT40. (A) Growth characteristics of DT40 (open circles) and its ΔRev1 derivative (filled symbols). (B) Cloning efficiency. Data were derived from eight plates for DT40 and 13 for ΔRev1 from two separate sorting sessions. (C) Annexin V staining. The upper right quadrant represents dead apoptotic cells [Annexin V positive, propidium iodide (PI) positive]. The lower right quadrant incipiently apoptotic cells (Annexin V positive, PI negative). (D) 2D cell cycle analysis in respsonse to cisplatin. Representative plots of fixed cells pulsed with BrDU counterstained with PI. The cell population is gated to exclude clumps of two or more cells and the proportion of cells in sub-G1, G1, S and G2/M is indicated by the appropriate gate. The three panels from left to right are untreated and 6 and 24 h after a 1 h pulse of 10 µM cisplatin.
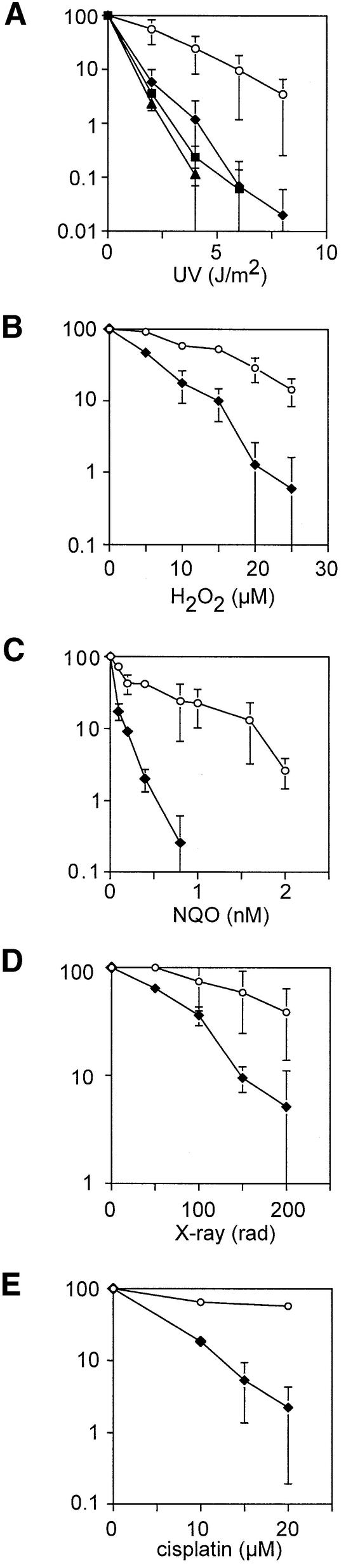
Fig. 3. Sensitivity of ΔRev1-DT40 to DNA-damaging agents. (A) UV light (254 nm); (B) hydrogen peroxide; (C) NQO; (D) X-rays; and (E) cisplatin. Curves are derived from at least three separate experiments. Wild-type DT40 open circles, ΔRev1-DT40 filled diamonds. For UV curves data are shown for two additional, independently targeted ΔRev1-DT40 (filled squares and triangles). Error bars are 1 SD.
ΔRev1-DT40 is sensitive to a wide range of DNA-damaging agents
We asked whether ΔRev1-DT40 cells are more sensitive to DNA damage than their wild-type counterparts. A previous study had not demonstrated increased sensitivity of a human fibroblast line expressing high levels of Rev1 antisense to UV light (Gibbs et al., 2000). In contrast, three independently derived clones of ΔRev1-DT40 are just over 3-fold more sensitive to 254 nm UV irradiation than wild type (D10 1.8 versus 5.8 J/m2) (Figure 3A). A characteristic of the RAD6 post-replication repair pathway mutants in yeast is sensitivity to a broad range of DNA-damaging agents that probably share a common ability to stall DNA replication (reviewed in Friedberg et al., 1995). In this regard, ΔRev1-DT40 cells mirror both the yeast RAD6 epistasis group and DT40 cells deficient in another key member of the RAD6 pathway, RAD18 (Yamashita et al., 2002).
Hydrogen peroxide increases the load of base damage in a cell, including creation of thymine glycol, 8-oxoguanine and abasic sites through the production of oxygen free radicals. Indeed, abasic sites that pose a potential block to replication if unrepaired are probably one of the most commonly occurring lesions in DNA, each cell being estimated to generate ∼10 000 per day through spontaneous depurination alone (Lindahl, 1979). ΔRev1-DT40 are nearly 3-fold more sensitive to this agent than wild-type cells (D10 12 versus 33 µM), suggesting that Rev1 is required for tolerating oxidative DNA damage (Figure 3B). Exposure to 4-nitroquinoline-1-oxide (NQO) results in the generation of bulky quinoline-purine monoadducts that appear to be processed in a similar fashion to 254 nm UV light-induced pyrimidine dimers (reviewed in Friedberg, 1995). However, NQO is also likely to result in the generation of reactive oxygen intermediates (Ramotar et al., 1998) and hence induce lesions similar to those seen with hydrogen peroxide. Interestingly, the ΔRev1-DT40 cells are more sensitive to NQO (∼6-fold: D10 0.2 versus 1.4 nM) than either 254 nm UV light or hydrogen peroxide alone. A sensitivity to X-rays of 3.4-fold is also evident (Figure 3C) (D10 160 rad for the mutant versus 530 rad for wild type; Figure 3D). It is possible that this reflects sensitivity to X-ray induced base damage rather than a response to double-strand breaks. Of the mutagens tested, ΔRev1-DT40 appeared to be the most sensitive (6.8-fold) to the DNA interstrand crosslinking agent cisplatin (D10 12 versus 82 µM), indicating a significant role in the repair of these lesions (Figure 3E). This observation is considered further in the Discussion.
ΔRev1-DT40 shows a normal induction of sister chromatid exchange following DNA damage
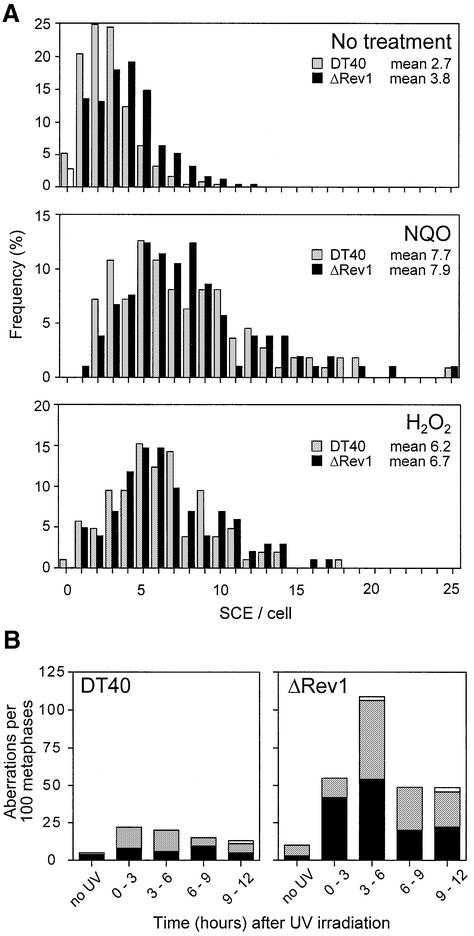
The predominant mode of recombination repair in mammalian cells is likely to be gene conversion (Johnson and Jasin, 2000). Instances of post-replicative gene conversion that resolve with crossing over are thought to be the basis for cytologically visible sister chromatid exchanges (SCE). We asked whether Rev1 is necessary for SCE or whether loss of Rev1 leads to an increase in use of SCE, as is seen in ΔRAD18-DT40 (Yamashita et al., 2002). We examined the level of SCE in metaphase arrested cells with and without treatment with the DNA-damaging agents NQO and hydrogen peroxide. ΔRev1-DT40 cells have a small but not significant increase in basal level of sister chromatid exchange (an average of 3.8 versus 2.7 per metaphase for wild-type cells) (Figure 4A). Following treatment with NQO, this level rises to an average of 7.9 per cell. Interestingly, this is not more than is seen in wild-type cells (average of 7.7 per cell), and is in contrast to DT40 cells deficient in RAD18 (Yamashita et al., 2002) where there is increased basal and NQO induced level of SCE relative to wild-type cells. A similar result was seen with hydrogen peroxide. Following treatment, wild-type cells showed an average of 6.2 SCE per cell compared with 6.7 for ΔRev1-DT40. These data suggest that Rev1 is not required for SCE, and loss of Rev1 does not lead to an increased use of recombination mediated repair.
Fig. 4. (A) Induction of SCE following DNA damage. The histograms represent the percentage of metaphases examined (y-axis) containing a given number of SCE (x-axis) with wild type in grey and ΔRev1-DT40 in black. From top to bottom the three panels show SCE in metaphases from untreated cells, NQO-treated and hydrogen peroxide-treated cells, respectively. The mean number of SCE per metaphase is indicated in the top right of each panel. Each database contains at least 100 metaphases. (B) Chromosomal aberrations following UV irradiation. For each time point aberrations are shown as stacked histogram bars with chromatid breaks in black, chromosome (isochromatid) breaks in grey and exchanges (radial structures) in white. More than 50 metaphases were scored blind for each time point.
UV irradiation leads to marked chromosome breakage in the absence of Rev1
That SCE is not increased in the mutant cells relative to wild type suggests the possibility that lesions are not diverted into recombination pathways in the absence of Rev1. We asked whether this might leave UV-induced lesions unrepaired such that chromosomal breaks were generated at increased frequency in ΔRev1-DT40. This appears to be the case (Figure 4B). ΔRev1-DT40 cells exhibit a moderately elevated resting level of chromosomal aberrations compared with wild type. However, following UV irradiation the mutant cells show a marked increase in the number of chromosome and chromatid type breaks compared with wild type, with a peak between 3 and 6 h after irradiation. This time course supports the idea that Rev1 is needed predominantly for tolerance of UV damage in late S phase or early G2, where it may be involved in rescuing stalled forks and/or gap filling following replication. It is also consistent with a model in which other pathways cannot completely compensate for loss of Rev1 in DT40.
Immunoglobulin gene conversion in ΔRev1-DT40 is normal but non-templated mutation is markedly reduced
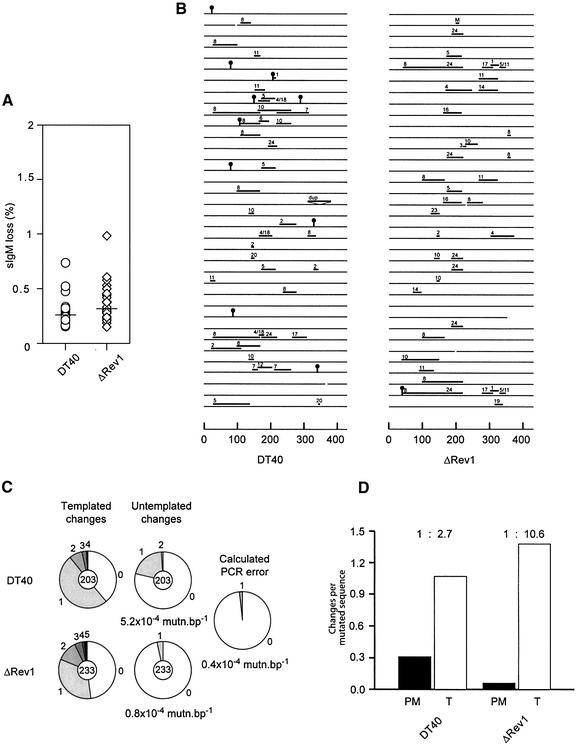
DT40 diversifies its immunoglobulin loci constitutively predominantly by gene conversion but also by non-templated mutation (Buerstedde et al., 1990; Kim et al., 1990). The immunoglobulin gene diversification in DT40 results in the generation of surface immunoglobulin loss variants, and this can be used as an assay for the extent of gene conversion and mutation (Sale et al., 2001). Multiple immunoglobulin positive clones of wild type and ΔRev1-DT40 were expanded for 28 days, following which the percentage of immunoglobulin negative cells in each subclone was assayed by flow cytometry. Wild-type cells generated a median loss-variant population of 0.27%, with ΔRev1-DT40 behaving similarly, generating 0.31% (Figure 5A), suggesting that there is no major difference in the overall diversification of the two lines. As non-templated mutation accounts for a relatively small proportion of immunoglobulin sequence changes, loss of this component would not be expected to have a major impact on the generation of loss-variants. We therefore performed sequence analysis of the rearranged light chain from selected immunoglobulin loss-variants from five independent clones of DT40 and three independently targeted clones of ΔRev1-DT40. These populations had been expanded for 1–3 months. We designated sequence changes as templated (i.e. gene conversion tracts and single base changes with donors in the database of light chain pseudogenes) and non-templated (i.e. base changes with no donor) (Sale et al., 2001). Following this analysis, the striking feature was markedly diminished non-templated mutation in the Rev1 mutant clones (Figure 5B). Examination of a larger sequence database confirmed a >6-fold diminution in the number of non-templated mutations in ΔRev1-DT40 (Figure 5C), each of the three clones behaving in a similar fashion. Indeed, the number of point mutations in light chain sequences from the mutant cells is close to the calculated PCR error. In order to be certain that clonality in the sequence databases could play no role in skewing the results and to correct for any bias that might be introduced through length of time in culture of different clones, we examined the ratio of templated to non-templated mutations in only the mutated clones of each database. In this analysis each sequence change was counted only once from an individual clone (Figure 5D). In wild-type cells there is one non-templated mutation for every 2.7 templated changes in mutated sequences. In ΔRev1-DT40, this ratio rises to 1 to 10.6, confirming that for a given level of gene conversion these cells do indeed incorporate significantly fewer point mutations.
Fig. 5. Diversification of the immunoglobulin light chain variable region in ΔRev1-DT40. (A) Fluctuation analysis of IgM-loss variants in wild-type DT40 and ΔRev1. The median Ig loss population for DT40 in these experiments was 0.27% and for ΔRev1-DT40 0.31%. (B) Graphic representation of consecutive sequences from sorted Ig negative cells from DT40 and ΔRev1-DT40. Each line represents an individual sequence. Black ‘lollipops’ are non-templated mutations and bars gene conversion tracts. The pseudogene donor for each tract is indicated above it (M, multiple possible donors; dup, duplication). Deletions are indicated by gaps in the sequence line. (C) Analysis of the whole database of DT40 and Rev1-DT40 immunoglobulin light chain sequence variation in selected surface Ig loss-variants. The pie charts represent the proportion of sequences with the indicated number of changes. The total number of sequences analysed is indicated in the centre of the pies. PCR error was calculated for 30 cycles of PCR assuming an error rate for PfuTurbo of 1.3 × 10–6/bp/cycle. (D) Ratio of non-templated to templated changes in DT40 IgM-loss variants. The bars show the number of changes per mutated sequence: black bars non-templated (PM); white bars templated (T). The figures above the bars represent the ratio of non-templated point mutations to templated changes (gene conversions).
Discussion
Rev1 is not essential for viability in DT40 but is required for tolerating wide range of DNA insults
Rev1 is not essential for viability in DT40, but the low plating efficiency of ΔRev1-DT40 suggests that the cells have a markedly reduced chance of completing a cell cycle than wild type. Deletion of Rev1 renders the cells susceptible to a wide variety of DNA-damaging agents with a pattern sensitivity similar to that seen in ΔRAD18-DT40 (Yamashita et al., 2002) as well as yeast mutants in the RAD6 group (reviewed in Friedberg et al., 1995), consistent with a defective response to replication blocking lesions. Indeed, assuming that it does not have intrinsically higher repair rates, DT40 might be particularly sensitive to such damage since a larger proportion of cells are in S phase (usually 40–60% in an asynchronous population) than in many vertebrate cell lines. This would be expected to increase the chance that a lesion will meet a replication fork. Failure to complete replication and to resolve the resulting gaps will lead to many cells undergoing apoptosis or acquiring chromosomal aberrations. Indeed, the response of Rev1-deficient DT40 to UV irradiation differs from that of Rev1 antisense expressing human fibroblasts (Gibbs et al., 2000). While this may be partly explained by an increased reliance of DT40 on S phase DNA-damage response pathways, it may also reflect only partial inhibition of Rev1 activity in the antisense expressing cells. Further studies will be needed to address this paradox.
Of particular note is the marked sensitivity of ΔRev1-DT40 to the interstrand crosslinking agent cisplatin. DNA interstrand crosslinks pose a considerable problem to cells as both strands of DNA are damaged simultaneously and, unrepaired, will present an absolute block to the completion of replication. Although the mechanism of crosslink repair in vertebrates remains incompletely understood, clues have come from mutants which are hypersensitive to DNA crosslinking agents. In vertebrates, extreme sensitivity to DNA crosslinks is seen in cells lacking both the ERCC1/XPF exinuclease (Hoy et al., 1985) and the homologous recombination factors XRCC2 and XRCC3 (Jones et al., 1987; Fuller and Painter, 1988). Cells from patients with Fanconi’s anaemia are also highly DNA crosslink sensitive (reviewed in Joenje and Patel, 2001), although it remains unclear whether this reflects a direct role of the Fanconi proteins in crosslink repair. While a requirement for translesion synthesis in the tolerance of crosslinks has been shown for yeast deficient in Rev3 (Henriques and Moustacchi, 1980), it has only been inferred in vertebrate cells through the demonstration of a mode of crosslink repair that results in mutations at the site of the crosslink (Sage et al., 1993; Wang et al., 2001). Evidence from yeast suggests that Rev3 is important for tolerating crosslinks in non-dividing cells (McHugh et al., 2000), and the mutagenic repair pathway demonstrated by Wang et al. (2001) is recombination independent, but nucleotide excision repair (NER) dependent. This led these authors to suggest a simple model: the crosslink is uncoupled by the NER machinery, the resulting gap filled is in by translesion synthesis and a further round of NER results in complete excision of the lesion. However, they also suggest that this is unlikely to be a major mechanism for crosslink repair in mammalian cells, since most mammalian NER mutants do not display a severe sensitivity to this form of damage. While this is likely to be the case, the results presented here suggest that, in vertebrates, Rev1 and translesion synthesis are critical for efficient tolerance or repair of interstrand crosslinks, and thus are likely to also be involved in other pathways.
Non-templated point mutations in the immunoglobulin loci of DT40 are generated by translesion synthesis
A particularly striking example of point mutagenesis in vertebrates is seen in the diversifying immunoglobulin loci of humans and mice. This somatic hypermutation is dependent on activation-induced deaminase (AID) (Muramatsu et al., 2000; Revy et al., 2000). AID is a cytidine deaminase that appears to work directly on the immunoglobulin V gene DNA. This results in the generation of uracil which is then acted upon by uracil DNA glycosylase to generate abasic sites (Di Noia and Neuberger, 2002; Petersen-Mahrt et al., 2002; Rada et al., 2002). Immunoglobulin diversification in the avian cell line DT40 is also dependent on AID (Arakawa et al., 2002; Harris et al., 2002) and therefore on the local generation of abasic sites. However, instead of exclusive point mutation, DT40 also employs gene conversion, in which stretches of upstream immunoglobulin pseudogenes are copied into the rearranged and expressed heavy and light chain variable regions by a process that is dependent on the homologous recombination factor RAD54 (Bezzubova et al., 1997). Thus immunoglobulin diversification in DT40 results from a combination of templated diversification and non-templated point mutations (Buerstedde et al., 1990; Kim et al., 1990). Recently, we showed that ablation of the recombination promoting factors XRCC2 or XRCC3, which are paralogues of the strand exchange protein RAD51, markedly shifted the balance of diversification towards non-templated point mutation (Sale et al., 2001). One interpretation of this result is that replication fork-stalling abasic sites that would have been repaired by recombination are instead processed by an alternative mutagenic pathway, such as translesion bypass. The present finding that Rev1 is required for the non-templated mutation observed in the immunoglobulin loci of DT40 can be most easily explained if vertebrate Rev1 is, like its yeast counterpart, central to efficient translesion bypass, and if this mechanism is largely responsible for the somatic mutation at the immunoglobulin locus in DT40. Clearly, the double mutant of Rev1 and XRCC2 or XRCC3 will be informative in testing this hypothesis, although it will not be surprising if the combination of gene disruptions is lethal.
In the absence of Rev1, non-templated point mutations are still found in the light chain variable regions and, although close to the level ascribable to PCR, it remains possible that not all somatic mutation in wild-type DT40 is dependent on Rev1. Either these mutations result from residual Rev1 independent translesion bypass or from the ultimate repair of the abasic site.
The role of Rev1 in mammalian immunoglobulin somatic hypermutation remains an open question, although it would not be unreasonable to assume that it, and translesion bypass, will be involved to some extent. Indeed, evidence from antisense experiments to Rev3 in hypermutating B cell lines and in mice implies that it is (Diaz et al., 2001; Zan et al., 2001). A feature of the mutations in XRCC2/3-deficient DT40 is the very striking preponderance of mutations to G and C, consistent with C being inserted opposite an abasic site generated in consequence of programmed cytidine deamination by AID. This initially suggested to us that Rev1 might be involved, since this type of mutation could be accomplished by a deoxycytidyl transferase. However, the fact that in DT40 Rev1 is needed for the whole spectrum of point mutation, and not just mutation to G/C, suggests that, as in yeast, this protein plays a more fundamental role than just deoxycytidyl transfer.
Recombination-based repair does not appear to be able to substitute for loss of Rev1
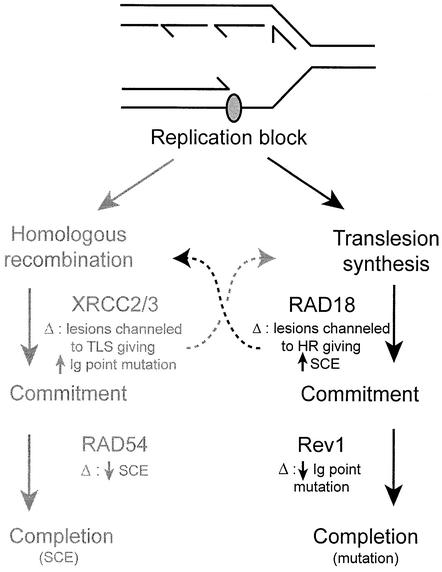
A notable feature of the response of ΔRev1-DT40 to DNA damage is a normal induction of SCE. Clearly this implies that Rev1 is not required for SCE. More interestingly, SCE is not elevated in the ΔRev1 background relative to wild type in undamaged or damaged cells. This is in contrast to DT40 deficient in RAD18, an E3 ubiquitin ligase that, in yeast, is epistatic to Rev1 (reviewed in Broomfield et al., 2001) and which is thought to play a key role in the regulation of both the error-prone and error-free branches of the RAD6 pathway through ubiquitylation of key target proteins including PCNA (Hoege et al., 2002). It has been proposed that the increased level of SCE in ΔRAD18-DT40 is due to increased use of recombination (Yamashita et al., 2002). A simple model would suggest that this is due to lesions that might normally have been processed by RAD18-dependent translesion bypass being ‘channelled’ into recombination pathways. Extending this idea, an explanation for SCE not being increased in Rev1-deficient cells relative to wild type is that the requirement for Rev1 in translesion bypass comes, unlike RAD18, after the commitment to avoidance of a lesion by this pathway and that an intermediate is generated that is not a substrate for ‘rescue’ by homologous recombination (Figure 6). We propose that such lesions go unresolved, leaving incompletely replicated daughter strands. This would promote apoptosis or result in an increase in DNA-damage-induced chromosome aberrations similar to those seen in recombination defective cells. The alternative explanation that Rev1 has a direct role in recombination is not really supported by the observation of intact SCE and immunoglobulin gene conversion. Thus, loss of a component of the translesion bypass machinery in a vertebrate cell may reduce damage induced point mutation at the expense of increasing the risk of chromosomal instability.
Fig. 6. Simplified model illustrating lesion channelling between homologous recombination and translesion synthesis. TLS, translesion synthesis; HR, homologous recombination. The effect of deleting (Δ) each gene is given under the gene name. Damage stalling a replication fork can be dealt with by either homologous recombination or post- replication repair. Ablation of genes involved in the ‘early’ stages of either of these pathways (e.g. XRCC2/3 or RAD18) results in lesions being channelled into the other pathway with a consequent increase in the detectable endpoints of those pathways (SCE for homologous recombination, Ig point mutation for translesion synthesis). However, loss of genes in the ‘later’ stages of either pathway (e.g. RAD54 and Rev1) is proposed to result in an intermediate that cannot be subsequently processed by the other pathway.
An inverse situation may be envisaged with respect to the perversion of homologous recombination that follows loss of RAD54 or the RAD51 paralogues XRCC2 or XRCC3. At the immunoglobulin loci of DT40, translesion bypass appears to be unable to compensate for loss of RAD54, where gene conversion is reduced 5-fold, leaving point mutation relatively preserved, but not elevated (Bezzubova et al., 1997; Sale et al., 2001). In contrast, the appearance of large numbers of point mutations in ΔXRCC2- or ΔXRCC3-DT40 (Sale et al., 2001) may reflect failure of recombination at a stage when translesion bypass can still take over (Figure 6).
This model is open to further testing and we believe that the diversification of the DT40 immunoglobulin locus provides a unique system in which to study the vertebrate RAD6 pathway and its interaction with recombination-mediated repair.
Materials and methods
Assembly of the Rev1 targeting construct and analysis of clones
To isolate a phage λ genomic DNA clone containing chicken Rev1, the first 1541 bp of the mouse cDNA probe were used to probe a White Leghorn genomic library (Stratagene) by hybridization at 45°C with washing in 2× SSC/0.1% SDS at 65°C. Four clones were identified and mapped. An ∼8 kb SalI–XhoI fragment was cloned into pBluescript from which a 4.5 kb EcoRI fragment was isolated that contained exons homologous to human exons 5 and 6 (Figure 1A and B; Supplementary figure 1). Exon 6 was largely contained within a 900 bp PstI fragment. The targeting construct was made using the EcoRI fragment cloned into pBluescript containing a modified polylinker. The PstI fragment was replaced with a BamHI linker and selection cassettes for puromycin and neomycin cloned in as BamHI fragments. Linearization of the construct was with MluI, which had been engineered into the modified polylinker. Targeted integration was detected by Southern blotting of PstI-digested DNA. Northern blotting was performed with 25 µg total RNA. The chicken Rev1 cDNA probe spanned a region homologous to amino acids 690–829 of human Rev1 (Figure 1D) and the chicken MMS2 probe comprised the entire open reading frame.
Cell culture and transfection
DT40 cells were obtained from Dr John Young at the Institute of Animal Health, Compton, Berkshire UK. DT40 cells and derivatives were propagated at 37°C in RPMI 1640 supplemented with 7% fetal calf serum, 3% chicken serum, 50 µM 2-mercaptoethanol and penicillin/streptomycin. Transfection was carried out by electroporation at 550 V and 25 µF in 0.4 cm cuvettes.
Estimation of cloning efficiency
Cells were seeded at 1 cell/well into 96-well plates using a MoFlo cell sorter (Cytomation) and gating on forward and side scatter to select for viability. Clones arising in the central 60 wells were counted.
Cell cycle analysis
Cells were pulsed with 10 µM bromodeoxyuridine (BrDU; Sigma) for 10 min, washed twice with phosphate-buffered saline (PBS) and fixed overnight in 70% ethanol at –20°C. The cells were then washed and resuspended in 2 N HCl/0.5% Triton X-100 for 30 min at room temperature. Following neutralization in 0.1 M Na tetraborate pH 8.5 for 1 min, the cells were washed with PBS/0.5% Tween 20/1% BSA and the stained with anti-BrDU-FITC (PharMingen) for 30 min, then washed and resuspended in PBS with propidium iodide 5 µg/ml. Analysis was performed on a FACSCalibur cytometer (Beckton Dickinson) with gating on FL2-A and FL2-W to exclude cell clumps. For examination of the cell cycle response to cisplatin, cells were treated for 1 h with 10 µM cisplatin followed by harvest 6 and 24 h later.
Mutagen sensitivity assays
Mutagen sensitivity assays were carried out in methylcellulose medium. The curves were plotted relative to the surviving fraction of cells in the untreated population. D10 values were calculated following curve fitting with Cricket Graph III (Computer Associates). UV light at 254 nm was delivered using a Stratalinker (Stratagene). X-rays were delivered using a Torrex 150 generator. Hydrogen peroxide was freshly diluted from an ∼30% solution (BDH). Cisplatin and NQO were obtained from Sigma.
Analysis of SCE
For measurement of SCE, cells were treated with 10 µM BrDU for 21 h with colcemid (0.1 µg/ml) added 2 h before harvest. For measurement of damage-induced SCE, NQO (0.2 ng/ml) or hydrogen peroxide (20 µM) was added 8 h before harvest. Hypotonic swelling was performed in 75 mM KCl before fixation in ice-cold methanol:acetic acid (3:1) for 30 min. Cells were dropped onto glass slides and stained with Hoechst 33258 (10 µg/ml in phosphate buffer pH 6.8) for 20 min. The slides were rinsed in MacIlvaine solution [164 mM Na2HPO4, 16 mM citric acid (pH 7)] before being exposed to ‘black light’ (λ 365 nm) for 1 h followed by incubation in 2× SSC at 62°C for 1 h. The slides were finally stained for 1 min in Leishman’s stain.
Analysis of UV-induced chromosomal aberrations
Aliquots of 107 cells were washed, resuspended in 0.5 ml PBS and irradiated in 6-well plates with 5 J/m2 254 nm UV light delivered by a Stratalinker. The cells were recovered to medium and incubated in the 3 h prior to fixing with 1 µg/ml colcemid. Cells were fixed in methanol:acetic acid (3:1), dropped onto cold wet glass slides and stained with Leishman’s stain.
Ig gene conversion assay and sequencing
The Ig loss-variant assay was performed as described previously (Sale et al., 2001). Surface Ig was stained with goat anti-chicken IgM (Bethyl Laboratories). Cell cloning and Ig loss-variant cell sorting was performed using a MoFlo cell sorter (Cytomation). For sequence analysis, 30 cycles of PCR amplification with Pfu Turbo (Stratagene) was performed from with the primers CVLF5 (5′-CAGGAGCTCGGCTCTGTCCCATTGCTGCGCGG) and CVLR3 (5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG). The resulting products were cloned into either pBluescript or M13 and sequenced with the T3 and –20 primers respectively. The extension products were analysed on an ABI 377 sequencer.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Anna-Laura Ross, K.J.Patel and Cristina Rada for discussions and critical reading of the manuscript, Richard Grenfell for help with cell sorting, Wojciech Niedzwiedz for assistance with cell cycle analysis, Jean-Marie Buerstedde for selection cassettes, and Shunichi Takeda for sharing unpublished lines and results.
References
- Arakawa H., Hauschild,J. and Buerstedde,J.M. (2002) Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science, 295, 1301–1306. [DOI] [PubMed] [Google Scholar]
- Bemark M., Khamlichi,A.A., Davies,S.L. and Neuberger,M.S. (2000) Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol., 10, 1213–1216. [DOI] [PubMed] [Google Scholar]
- Bezzubova O., Silbergleit,A., Yamaguchi-Iwai,Y., Takeda,S. and Buerstedde,J.M. (1997) Reduced X-ray resistance and homologous recombination frequencies in a RAD54–/– mutant of the chicken DT40 cell line. Cell, 89, 185–193. [DOI] [PubMed] [Google Scholar]
- Broomfield S., Hryciw,T. and Xiao,W. (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res., 486, 167–184. [DOI] [PubMed] [Google Scholar]
- Buerstedde J.M., Reynaud,C.A., Humphries,E.H., Olson,W., Ewert,D.L. and Weill,J.C. (1990) Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J., 9, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I. and Mornon,J.P. (1997) From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett., 400, 25–30. [DOI] [PubMed] [Google Scholar]
- Cejka P., Vondrejs,V. and Storchová,Z. (2001) Dissection of the functions of the Saccharomyces cerevisiae RAD6 postreplicative repair group in mutagenesis and UV sensitivity. Genetics, 159, 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M., Verkoczy,L.K., Flajnik,M.F. and Klinman,N.R. (2001) Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase ζ. J. Immunol., 167, 327–335. [DOI] [PubMed] [Google Scholar]
- Di Noia J. and Neuberger,M.S. (2002) Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature, 419, 43–48. [DOI] [PubMed] [Google Scholar]
- Eeken J.C., Romeijn,R.J., de Jong,A.W., Pastink,A. and Lohman,P.H. (2001) Isolation and genetic characterisation of the Drosophila homologue of (SCE)REV3, encoding the catalytic subunit of DNA polymerase zeta. Mutat. Res., 485, 237–253. [DOI] [PubMed] [Google Scholar]
- Esposito G., Godindagger,I., Klein,U., Yaspo,M.L., Cumano,A. and Rajewsky,K. (2000) Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr. Biol., 10, 1221–1224. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Fuller L.F. and Painter,R.B. (1988) A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res., 193, 109–121. [DOI] [PubMed] [Google Scholar]
- Gibbs P.E., Wang,X.D., Li,Z., McManus,T.P., McGregor,W.G., Lawrence,C.W. and Maher,V.M. (2000) The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl Acad. Sci. USA, 97, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M.F. (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem., 71, 17–50. [DOI] [PubMed] [Google Scholar]
- Haracska L., Unk,I., Johnson,R.E., Johansson,E., Burgers,P.M., Prakash,S. and Prakash,L. (2001) Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev., 15, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Prakash,S. and Prakash,L. (2002) Yeast Rev1 protein is a G template-specific DNA polymerase. J. Biol. Chem., 277, 15546–15551. [DOI] [PubMed] [Google Scholar]
- Harfe B.D. and Jinks-Robertson,S. (2000) DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell, 6, 1491–1499. [DOI] [PubMed] [Google Scholar]
- Harris R.S., Sale,J.E., Petersen-Mahrt,S.K. and Neuberger,M.S. (2002) AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol., 12, 435–438. [DOI] [PubMed] [Google Scholar]
- Henriques J.A. and Moustacchi,E. (1980) Isolation and characterisation of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics, 95, 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N.P., Kato,K. and Strauss,B. (1976) A model for replication repair in mammalian cells. J. Mol. Biol., 101, 417–425. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander,B., Moldovan,G.L., Pyrowolakis,G. and Jentsch,S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature, 419, 135–141. [DOI] [PubMed] [Google Scholar]
- Holbeck S.L. and Strathern,J.N. (1997) A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics, 147, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy C.A., Thompson,L.H., Mooney,C.L. and Salazar,E.P. (1985) Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res., 45, 1737–1743. [PubMed] [Google Scholar]
- Joenje H. and Patel,K.J. (2001) The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet., 2, 446–457. [DOI] [PubMed] [Google Scholar]
- Johnson R.D. and Jasin,M. (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J., 19, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Jones N.J., Cox,R. and Thacker,J. (1987) Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res., 183, 279–286. [DOI] [PubMed] [Google Scholar]
- Kajiwara K. et al. (2001) Sez4 gene encoding an elongation subunit of DNA polymerase ζ is required for normal embryogenesis. Genes Cells, 6, 99–106. [DOI] [PubMed] [Google Scholar]
- Kim S., Humphries,E.H., Tjoelker,L., Carlson,L. and Thompson,C.B. (1990) Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol. Cell. Biol., 10, 3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W. (2002) Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair, 1, 425–435. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W. and Christensen,R. (1976) UV mutagenesis in radiation-sensitive strains of yeast. Genetics, 82, 207–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W. and Christensen,R.B. (1978) Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. I. rev1 mutant strains. J. Mol. Biol., 122, 1–21. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W., O’Brien,T. and Bond,J. (1984) UV-induced reversion of his4 frameshift mutations in rad6, rev1 and rev3 mutants of yeast. Mol. Gen. Genet., 195, 487–490. [DOI] [PubMed] [Google Scholar]
- Lemontt J.F. (1971) Mutants of yeast defective in mutation induced by ultraviolet light. Genetics, 68, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefshitz B., Steinlauf,R., Friedl,A., Eckardt-Schupp,F. and Kupiec,M. (1998) Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res., 407, 135–145. [DOI] [PubMed] [Google Scholar]
- Lin W., Xin,H., Zhang,Y., Wu,X., Yuan,F. and Wang,Z. (1999) The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res., 27, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. (1979) DNA glycosylases, endonucleases for apurinic/apyrimidinic sites and base excision repair. Prog. Nucleic Acid Res. Mol. Biol., 22, 135–192. [DOI] [PubMed] [Google Scholar]
- Masuda Y., Takahashi,M., Tsunekuni,N., Minami,T., Sumii,M., Miyagawa,K. and Kamiya,K. (2001) Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J. Biol. Chem., 276, 15051–15058. [DOI] [PubMed] [Google Scholar]
- Masuda Y., Takahashi,M., Fukuda,S., Sumii,M. and Kamiya,K. (2002) Mechanisms of dCMP transferase reactions catalyzed by mouse Rev1 protein. J. Biol. Chem., 277, 3040–3046. [DOI] [PubMed] [Google Scholar]
- McHugh P.J., Sones,W.R. and Hartley,J.A. (2000) Repair of intermediate structuress produced at DNA interstrand crosslinks in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakumo Y., Ogura,Y., Ishii,H., Numata,S., Ichihara,M., Croce,C.M., Fishel,R. and Takahishi,M. (2001) Interactions in the error-prone postreplication repair proteins hRev1, hRev3 and hRev7. J. Biol. Chem., 276, 35644–35651. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita,K., Fagarasan,S., Yamada,S., Shinkai,Y. and Honjo,T. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell, 102, 553–563. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Gibbs,P.E., Nowicka,A.M., Hinkle,D.C. and Lawrence,C.W. (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol., 37, 549–554. [DOI] [PubMed] [Google Scholar]
- Ohmori H. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- O-Wang J., Kajiwara,K., Kawamura,K., Kimura,M., Miyagishima,H., Koseki,H. and Tagawa,M. (2002) An essential role for REV3 in mammalian cell survival: absence of REV3 induces p53-independent embryonic death. Biochem. Biophys. Res. Commun., 293, 1132–1137. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris,R.S. and Neuberger,M.S. (2002) AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature, 418, 99–103. [DOI] [PubMed] [Google Scholar]
- Rada C., Williams,G.T., Nilsen,H., Barnes,D.E., Lindahl,T. and Neuberger,M.S. (2002) Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol., 12, 1–12. [DOI] [PubMed] [Google Scholar]
- Ramotar D., Belanger,E., Brodeur,I., Masson,J.-Y. and Drobetsky,E.A. (1998) A yeast homologue of the human phosphotyrosyl phosphatase activator PTPA is implicated in protection against oxidative DNA damage induced by the model carcinogen 4-nitroquinoline-1-oxide. J. Biol. Chem., 273, 21489–21496. [DOI] [PubMed] [Google Scholar]
- Revy P. et al. (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell, 102, 565–575. [DOI] [PubMed] [Google Scholar]
- Sage E., Drobetsky,E.A. and Moustacchi,E. (1993) 8-methoxypsoralen induced mutations are highly targeted at crosslinkable sites of photoaddition on the non-transcribed strand of a mammalian chromosomal gene. EMBO J., 12, 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J.E., Calandrini,D.M., Takata,M., Takeda,S. and Neuberger,M.S. (2001) Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature, 412, 921–926. [DOI] [PubMed] [Google Scholar]
- Sudo T., Ota,Y., Kotani,S., Nakao,M., Takami,Y., Takeda,S. and Saya,H. (2001) Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. EMBO J., 20, 6499–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos C.A., Prakash,S. and Prakash,L. (2002) Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sloun P.P., Varlet,I., Sonneveld,E., Boei,J.J., Romeijn,R.J., Eeken,J.C. and De Wind,N. (2002) Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol. Cell. Biol., 22, 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Peterson,C.A., Zheng,H., Nairn,R.S., Legerski,R.J. and Li,L. (2001) Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol. Cell. Biol., 21, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Chow,B.L., Broomfield,S. and Hanna,M. (2000) The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics, 155, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Okada,T., Matsusaka,T., Sonoda,E., Zhao,G.Y., Araki,K., Tateishi,S., Yamaizumi,M. and Takeda,S. (2002) RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J., 21, 5558–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.-L., Johnson,R.E., Prakash,S. and Prakash,L. (2001) Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol., 21, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H., Komori,A., Li,Z., Cerutti,A., Schaffer,A., Flajnik,M.F., Diaz,M. and Casali,P. (2001) The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity, 14, 643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]