Abstract
We have investigated use of a conserved non-canonical GA 5′ splice site present in vertebrate fibroblast growth factor receptor (FGFR) genes. Despite previous studies suggesting that GA at the beginning of an intron is incompatible with splicing, we observe efficient utilization of this splice site for human FGFR1 gene constructs. We show that use of the GA splice site is dependent on both a conventional splice site six nucleotides upstream and sequence elements within the downstream intron. Furthermore, our results are consistent with competition between the tandem 5′ splice sites being mediated by U6 snRNP, rather than U1 snRNP. Thus the GA 5′ splice site represents an extension of the adjacent conventional 5′ splice site, the first natural example of such a composite 5′ splice site.
Keywords: alternative splicing/fibroblast growth factor receptor/intron sequence element/non-consensus 5′ splice site
Introduction
Eukaryotic pre-mRNAs contain intervening sequences (introns) that must be removed (spliced out) to generate a mature messenger RNA. This process requires three partially conserved sequences on the substrate: the 5′ and 3′ splice sites, which span the exon–intron and intron– exon junctions, and the branchpoint sequence within the intron itself. During the splicing reaction these elements are recognized by a multicomponent complex, the spliceosome (reviewed by Sharp, 1994; Staley and Guthrie, 1998), which is responsible for catalysing the two steps required to remove the intron and join the flanking exons. In step 1, the 2′ hydroxyl of an adenosine in the branchpoint attacks the phosphodiester bond corresponding to the exon–intron junction. This generates a free 5′ exon and an intron–3′ exon lariat held by a 2′–5′ phosphodiester bond. Attack of the 3′ hydroxyl of the upstream exon on the phosphodiester bond at the intron– exon junction (step 2) results in exon joining and release of the intron lariat.
The central components of the major spliceosome are the small nuclear ribonucleoprotein particles (snRNPs), which make RNA–RNA and protein–RNA contacts with the pre-mRNA. In addition, a large number of non-snRNP proteins are also required during the splicing reaction, facilitating splice site recognition, spliceosome assembly and recycling of the snRNP components following removal of the intron (Staley and Guthrie, 1998; Hastings and Krainer, 2001). Initially, all introns were believed to begin and end with invariant dinucleotides: GT and AG, respectively. The discovery that a small number of introns contain AT and AC dinucleotides led to the characterization of a variant spliceosome, at the core of which are a distinct set of snRNPs, structurally and functionally similar to those in the ‘classical’ spliceosome (Tarn and Steitz, 1997). Surprisingly, each spliceosome is capable of recognizing both GT–AG and AT–AC introns, with the sequences of the splice sites and branchpoint dictating which complex recognizes which intron (Sharp and Burge, 1997).
Correct gene expression is not only reliant on the accurate selection of the phosphodiester bonds to be cleaved (reviewed by Reed 1996, 2000), but is often also dependent upon the inclusion of a specific subset of exons. Such alternative splicing is a common mechanism for the generation of multiple related but structurally or functionally distinct products from a single gene (reviewed by Lopez, 1998; Black, 2000). Alternative splicing is tightly regulated by trans-acting factors that bind to specific elements on the pre-mRNA, ensuring that the desired protein isoform is expressed at the correct developmental stage or in the correct location. Examples of such splicing enhancers and silencers have been found, often in combination, within both the introns and the flanking exons (for reviews on splicing control elements see Blencowe, 2000; Smith and Valcárcel, 2000). One important family of trans-acting factors, the SR proteins, function in both constitutive and regulated splicing (Graveley, 2000).
Fibroblast growth factor receptors (FGFRs) are paralogous members of the transmembrane receptor tyrosine kinase family, signalling by which can induce a diverse range of biological processes including mesoderm induction, mitogenesis, angiogenesis, chemotaxis and neuronal survival (Mason, 1994; Marshall, 1995). Although only four vertebrate FGFRs have been identified, 22 distinct fibroblast growth factors have been characterized (Ornitz and Itoh, 2001). Ligand specificity of the FGFRs is controlled by alternative splicing of the extracellular Ig-like domains (reviewed by Johnson and Williams, 1993). In addition, for FGFR1–FGFR3 (but not FGFR4), two competing 5′ splice sites are present at the 3′ end of exon 10 (Hou et al., 1991). These splice sites direct inclusion or exclusion of 6 nucleotides (nt) (GTAACA) encoding valine and threonine (VT) (Figure 1A). This VT motif is required for interaction of the receptor with the FRS2 signalling adaptor protein (Burgar et al., 2002). Furthermore, transient transfection experiments have shown that only the VT+ isoform of human FGFR2 is competent to initiate signalling via the mitogen-activated protein kinase (MAPK) signalling pathway (Twigg et al., 1998). Thus a relatively minor change in the sequence encoded by the mRNA has a dramatic effect on the function of the resulting protein.
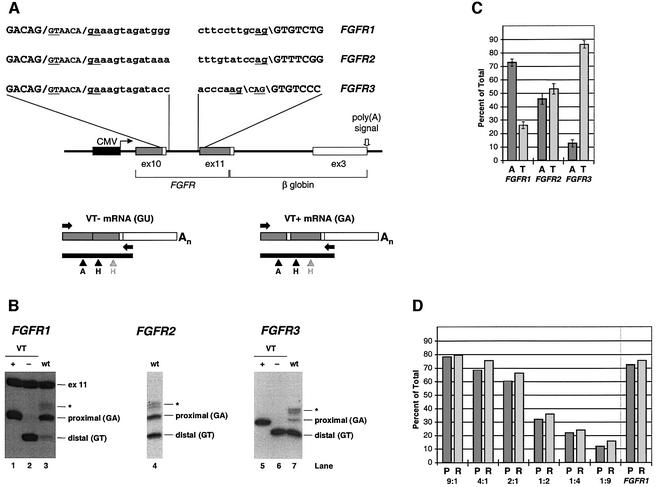
Fig. 1. Alternative splicing of the FGFR juxtamembrane region. (A) A cartoon of the alternative splicing event, showing the two alternative codons (for valine and threonine; GTA ACA) and the two species of mRNA generated. (B) Alignment of FGFR1, FGFR2 and FGFR3 sequences from a variety of species that, as far as is known, contain the tandem 5′ splice sites as part of a conserved 18 nt sequence. Splice junctions are denoted by /, perfectly conserved sequences are highlighted in black and purine or pyrimidine conservation is boxed. Sources: DDBJ/EMBL/GenBank accession No.; † identified by genome BLAST search; * ditto, but confirmed cloning and sequencing; # this work; ‡ Gillespie et al. (1995).
Sequencing of this region has shown that the downstream (proximal) 5′ splice site differs from the 5′ splice site consensus, having an A at position +2 (/GAAAGT) (Gillespie et al., 1995). Unlike GC and AT 5′ splice sites, present in 0.5–1% (Thanaraj and Clark, 2001) and 0.05% (Burset et al., 2000) of introns, respectively, GA (and GG) 5′ splice sites have previously been shown to be non-functional. To date, 43 pathological GT→GA mutations have been reported in the Human Gene Mutation Database (Krawczak and Cooper, 1997), while in vitro splicing experiments with model substrates have shown that GA 5′ splice sites block splicing after step 1 (Aebi et al., 1986, 1987). The sequence of the FGFR GA splice site has now been confirmed in a number of species (see below), negating the possibility that it is a trivial sequencing error. In addition, where the full sequence of the intron is known, it can be shown that the additional 6 nt present in the VT+ mRNA are not generated by a separate micro-exon.
Beyond identification of the VT+ and VT– isoforms, little quantitative analysis has been reported. Develop mental regulation of the Xenopus laevis FGFR1 gene has been observed during embryogenesis, with production of the VT+ form predominating for many of the early stages of blastula development (Gillespie et al., 1995; Paterno et al., 2000). Similarly, analysis of cDNA clones for the rat FGFR1 gene suggests that expression of the VT+ form predominates in several tissues (heart, lung, liver, intestine and kidney), with more equal expression of the two forms seen in the brain (Yazaki et al., 1993). These results suggest that not only is the FGFR GA 5′ splice site active, but it may be the dominant of the two 5′ splice sites in some situations.
In the work reported here, we have undertaken experiments to address the question of why the FGFR genes are able to utilize a supposedly ‘dead-end’ 5′ splice site. Our results extend previous studies on yeast and mammalian splicing that suggested that U6 snRNP could dictate the location of a 5′ splice junction at mutationally weakened splice sites (Kandels-Lewis and Séraphin, 1993) or when the U6 snRNA itself was modified (Hwang and Cohen, 1996). Specifically, we demonstrate that the endogenous U6 snRNP can dictate the location of a 5′ splice junction in the context of naturally occurring competing 5′ splice sites.
Results
To confirm the generality of the FGFR GA splice site, we performed GenBank searches for FGFR sequences from a variety of species. Strikingly, for almost all of these genes the splice sites are present in a well conserved sequence GNCAG/GTA/CACA/GAAAGTA (Figure 1B) (Twigg et al., 1998), with the majority of sequences apparently conforming to GACAG/GTAACA/GAAAGTA). In addition, for a given FGFR gene, the homology of the splice sites extends further into the intron; for the mammalian genes, for example, the conservation extends to positions +28, +25 and +24 (for FGFR1, FGFR2 and FGFR3, respectively). Analysis of GenBank cDNA entries for the mammalian genes shows FGFR1 cDNAs to be primarily GA spliced (VT+) and FGFR3 mRNA to be all GT spliced, but the two FGFR2 isoforms are slightly more even (Table I).
Table I. Breakdown of FGFR cDNA isoforms reported in GenBanka.
| Gene | Isoform | Mouse | Rat | Human | Total |
|---|---|---|---|---|---|
| FGFR1 | VT+ | 7/7 | 1/2 | 16/16 | 24/25 |
| VT– | 0/7 | 1/2 | 0/16 | 1/25 | |
| FGFR2 | VT+ | 3/3 | 1/2 | 6/8 | 10/13 |
| VT– | 0/3 | 1/2 | 2/8 | 3/13 | |
| FGFR3 | VT+ | 0/4 | 0/1 | 0/2 | 0/7 |
| VT– | 4/4 | 1/1 | 2/2 | 7/7 |
aData abstracted from LocusLink (Pruitt and Maglott, 2001).
Creation of model FGFR GA-splicing minigenes
To investigate use of the FGFR GA splice site, we constructed minigenes based on human FGFR1, FGFR2 and FGFR3 (Figure 2A). While the complete sequences from FGFR1 and FGFR3 have been used (with intron sizes of ∼1.1 kb and ∼300 bp, respectively), the size of the FGFR2 intron (11 189 bp) has necessitated the construction of a shortened minigene with only 150 nt from each end of the intron. In addition, FGFR3 differs from the other two FGFR genes in that the presence of two 3′ splice sites (AAG\CAG\) allows production of Q+ (CAG+) and Q– forms (Twigg et al., 1998). Therefore, to simplify the analysis of the spliced products generated by the FGFR3 minigene, the minor upstream 3′ splice site has been inactivated by mutation (AAG→TTC). This mutation does not affect the relative use of the two 5′ splice sites (data not shown). Expression of these constructs in transiently transfected 293T cells is detected by reverse transcription polymerase chain reaction (RT–PCR). The relative use of the two 5′ splice sites is determined by cutting the cDNA on both sides of the splice junction, end-labelling with 32P and resolving the products on native polyacrylamide gels. The quantitative nature of this assay has been confirmed by amplification of varying ratios of control FGFR cDNAs and comparison with the results of RNase protection assays (a typical experiment is shown in Figure 2D).
Fig. 2. Alternative splicing of human FGFR1, FGFR2 and FGFR3 minigenes in 293T cells. (A) Schematic of the basic construct used to transiently transfect 293T cells showing the sequences around the 5′ and 3′ splice sites. Splice site dinucleotides are underlined, 5′ and 3′ splice junctions are denoted by / and \, respectively, and the cytomegalovirus promoter is denoted by CMV. The FGFR3 sequence shows the unmutated upstream 3′ splice site (see text for details). Schematics of the VT+ and VT– cDNAs are also shown; the arrowheads denote the locations of the ApaLI (A) and HinfI (H) restriction sites (the grey arrowhead denotes a second HinfI site in FGFR1 exon 11), and horizontal arrows indicate the locations of the RT–PCR primers. (B) Digestion of the RT–PCR fragments for FGFR1, FGFR2 and FGFR3. Digestions of control VT+ and VT– cDNA are shown for FGFR1 (lanes 1 and 2) and FGFR3 (lanes 5 and 6). The FGFR1 fragment labelled ex 11 represents the HinfI fragment from exon 11, and the asterisk indicates the heteroduplex of the two cDNAs that form during the PCR (data not shown). The gels have been cropped to exclude the flanking fragments generated by the double digest. (C) Quantitation of the relative frequencies of GT (denoted T) and GA (denoted A) splicing for the minigenes (average and standard deviations of four independent experiments). (D) Comparison of quantitation by RT–PCR (P) and by RNase protection (R) of a typical transfection of the FGFR1 minigene and varying VT+:VT– ratios of cDNA expression constructs.
Figure 2B shows analysis of cDNAs from the three transiently expressed minigenes; quantitation is shown in Figure 2C. Use of the GA 5′ splice site (relative to the upstream site) differs significantly between the constructs: just over 70% for FGFR1 (lane 3), ∼45% for FGFR2 (lane 4) and only ∼12% for FGFR3 (lane 7). Thus, despite the sequence conservation, the level of GA splicing differs for the three genes in our model system. Based on these results, we have chosen to study the FGFR1 and FGFR3 minigenes as these show the greatest difference in GA splice site use.
Contribution of exon and intron sequences to the frequency of GA splicing
As a first step to understanding the mechanism of GA splicing, we have sought to locate the elements responsible for controlling the relative use of the tandem 5′ splice sites. Hybrids were made between the FGFR1 and FGFR3 minigenes, assembling the intron and associated exons in all possible combinations. Analysis of four of these minigenes (the names of which reflect the origin of the upstream exon, the intron and the downstream exon) is shown in Figure 3A. Replacing one or both of the FGFR1 exons with the corresponding exons from FGFR3 does not affect the frequency of GA splicing [constructs 3-1-1 (lane 3), 1-1-3 (lane 4) and 3-1-3 (lane 5)]. Similarly, little effect is seen when the FGFR3 exons are replaced with their FGFR1 counterparts [construct 1-3-3 (lane 6), and constructs 3-3-1 and 1-3-1 (data not shown)]. Thus the major element(s) responsible for controlling the relative use of the GA splice site must be located within the introns.
Fig. 3. Alternative splicing of hybrid constructs. (A) Alternative splicing of selected hybrid constructs containing sequences from FGFR1 (grey) and FGFR3 (black). The names of these reflect the origin of the upstream exon, intron and downstream exon (thus, FGFR1 is 1-1-1). To allow alignment of the various products (those containing the upstream exon from FGFR3 are 3 nt smaller than those with the corresponding exon from FGFR1), the lanes of a single gel have been separated (lanes 3–6 only). The frequency of GA splicing (and standard deviation for four replicates) is given below each lane. (B) Alternative splicing of the FGFR1 exon replacement hybrids. The numbers in parentheses denote the sizes of the GT- and GA-spliced products. Restriction sites used: H, HinfI; Ap, ApaLI; Ac, AccI. Again, the lanes shown represent a single gel.
Although the FGFR1 and FGFR3 flanking exons appear to be neutral in terms of determining the level of GA splicing, they share considerable sequence homology. Thus it is possible that elements controlling GA splicing are present. To investigate this, we replaced the FGFR1 exons with exons 1 and 2 from human β globin: minigenes β-1-1, 1-1-β and β-1-β (Figure 3B). All three constructs give at least as much GA product as the wild-type FGFR1 construct. Thus GA splicing of the FGFR genes is controlled primarily by sequences located within the introns.
Identification of intronic regulatory elements
Having established the importance of the intron in controlling the level of GA splicing, we undertook to define the locations of active sequences present within the FGFR1 intron. Deletion of the central ∼800 bp of the intron, reducing its length to ∼310 bp, had little effect on the frequency of GA splicing (data not shown). Thus the intron elements must be present within the first 160 and/or the final 150 nt of the intron. To define the locations of these putative 5′ and 3′ intronic splicing enhancers (ISEs) more precisely, we created hybrids between the introns of FGFR1 and human β globin (Figure 4A). For the first of these hybrids (construct 1-13β-1), all but the first 13 nt of FGFR1 intron (to the end of the conserved 18 nt sequence) was replaced by the globin intron (from position 14). This construct gave only a trace of GA-spliced mRNA (∼3%) (Figure 4A, lane 1). Extending the FGFR1 sequence present by 8 nt (construct 1-21β-1, lane 2) has little or no effect on the level of GA splicing. In contrast, the presence of 24 nt (construct 1-24β-1, lane 3) and 28 nt (construct 1-28β-1, lane 4) of FGFR1 allows sequentially more GA splicing (14 and 33%, respectively). However, additional increases in the extent of the FGFR1 5′ intron sequence (to 54, 105 or 160 nt) have no significant effect on the extent of GA splicing (lanes 5–7). Thus, a splicing enhancer must be present in the first 28 nt of the FGFR1 intron.
Fig. 4. Definition of the FGFR1 intron splicing enhancers. (A) The schematic shows construction of the hybrid intron. The FGFR1 intron was replaced with the first intron of β globin (from position +14), and increasing lengths of sequence from the 5′ end of the FGFR1 intron were included between the 5′ exon and the globin intron (lanes 1–7). Similarly, 150, 100, 50 and 25 nt of sequence from the 3′ end of the FGFR1 intron were used to replace the 3 nt of the globin 3′ splice site in the presence of varying amounts of FGFR1 5′ intron sequence (lanes 8–13). Constructs are named for the amount of FGFR1 intron sequence present upstream and downstream of the remnants of globin intron (e.g. 1-54β100-1 has 54 nt of 5′ sequence and 100 nt of 3′ sequence). (B) Sequence of the FGFR1 splice site region and 5′ ISE, showing the changes introduced. (C) Splicing of the 5′ ISE mutants in the absence (lanes 1–5) and presence (lanes 6–10) of the 3′ ISE.
The partial effect of the FGFR1 5′ sequence implies the presence of an additional ISE in the last 150 nt of the intron. To identify the location of this 3′ ISE, we replaced the final 3 nt of the β globin intron (TAG\) with sequences from the 3′ end of the FGFR1 intron. Initially, we replaced the globin 3′ splice site with the final 150 nt of the FGFR1 intron in combination with the 13 nt of the 5′ sequence, generating construct 1-13β150-1. The presence of this additional FGFR1 intron sequence had little or no effect on the level of GA splicing (Figure 4A, lane 8). However, in combination with 54 nt of 5′ sequence (construct 1-54β150–1, lane 9), the 150 nt of 3′ intron sequence promoted very efficient use of the GA splice site (over 60%). Restricting the 3′ intron sequence to 100 nt (construct 1-54β100-1, lane 10) or 50 nt (construct 1-54β50-1, lane 11) did not reduce the level of GA splicing. However, the extent of GA splicing was slightly reduced when 25 nt of 3′ intron sequence was included (construct 1-54β25-1, lane 12). Thus, the 3′ ISE resides within the final 50 nt of the intron. Strikingly, combination of the minimal 5′ and 3′ sequences (construct 1-28β50-1, lane 13) results in a level of GA splicing identical with that of the wild-type FGFR gene.
Dissection of the 5′ ISE
To gain further insight into how the 5′ ISE functions, we introduced base changes at various positions (detailed in Figure 4B) and tested their effects in the absence and presence of the 3′ ISE (Figure 4C). The most striking feature of the FGFR1 5′ ISE is the G5 tract beginning at position +17 and present in all FGFR1 introns so far sequenced (Figure 1B). Mutation of this sequence to C5 or A5 decreased the level of GA splicing for both 1-28β-1 (lanes 2 and 3) and 1-28β100-1 (lanes 7 and 8) to 10% or less. Thus this conserved sequence is vital for the 5′ ISE to function. However, this sequence alone is not active as an enhancer; the FGFR1 sequence present in construct 1-21β-1 ends at the end of the G5 tract and does not activate the GA splice site (Figure 4A, lane 2). It is also important to note that the 5′ element does not act as a classical splicing enhancer; separating it from the tandem splice site region by even 6 nt abolishes all activity (data not shown).
Experiments with FGFR3-based hybrids (analogous to those in Figure 4A) revealed the presence of a single ISE in the FGFR3 intron (data not shown). This enhancer, also located within the first 28 nt of the intron, is weaker than its FGFR1 counterpart. Introducing eight base changes to the FGFR1 5′ enhancer in 1-28β-1 to recreate the FGFR3 enhancer (mutation R3; Figure 4C, lane 5) reduces the level of GA splicing to 18%, only slightly greater than that of the FGFR3 minigene. Strikingly, however, we see no increase in the level of GA splicing when the R3 mutation is combined with the FGFR1 3′ ISE (lane 10), suggesting that the FGFR1 3′ ISE is tailored to interact with the specific sequences present in the FGFR1 5′ enhancer.
The presence of enhancers close to the tandem 5′ splice sites of both FGFR1 and FGFR3 suggests that such elements could be a general feature of the FGFR genes. To investigate this, we introduced seven base changes to recreate the putative FGFR2 5′ ISE. Mutation R2 results in ∼25% GA splicing in the absence of the FGFR1 3′ ISE (lane 4), approximately half that seen for the FGFR2 minigene (Figure 2B). As with the FGFR3 ISE, the presence of the FGFR1 3′ ISE did not significantly enhance the level of GA splicing seen for the R2 mutation (32%, lane 9).
Mutation of the conserved splice site region
To understand more about the sequences required for GA splicing, we introduced point mutations at various positions through the conserved 18 nt splice site region. Transversion of each of the first four positions (–5 to –2) had very little effect on the relative use of the two 5′ splice sites (Figure 5A, lanes 2–5). Indeed, mutation –4A→T resembles the X.laevis FGFR2 gene which also differs from the 18 nt consensus at this position (5′-GCCAG-3′) (Figure 1B). Surprisingly, however, mutation of the fifth conserved nucleotide (–1) activates a cryptic GT 5′ splice site at the end of the conserved sequence (+11), with use of this site accounting for ∼50% of the mRNA produced (Figure 5A, lane 6).
Fig. 5. Mutagenesis of the FGFR1 5′ splice site region. (A) Effects of transversion of the first five bases of the conserved sequence on use of the two splice sites. Positions of the base changes are shown (left) and numbered relative to the first base of the intron. The band corresponding to use of the cryptic 5′ splice site at +11 (lane 6) co-migrates with the heteroduplex. (B) RT–PCR detection of expression of FGFR1-based constructs by transiently transfected 293T cells. Lane 1 shows a 100 bp ladder. (C) Alternative splicing of the constructs shown in (B) (lanes 1–5), and the spacing constructs (lanes 6–8). Note that % GA denotes the frequency of use of the proximal splice site (GA, GU, GC or GG, depending on lane).
Single-point mutations were introduced to change the second base of each 5′ splice site (positions +2 and +8). Mutation +2A changes the upstream splice site from GT to GA, and abolishes the accumulation of correctly spliced mRNA (Figure 5B, lane 4). The fact that this mutation appears to block both splice sites suggests that use of the downstream GA splice site is dependent upon recognition of the GT splice site upstream, although effects later in gene expression cannot be excluded. All of the +8 mutations (converting the GA splice site to GT, GC and GG) allowed accumulation of mRNA (lanes 5–7) but with altered splicing profiles. Both mutations +8T and +8C (Figure 5C, lanes 2 and 3) dramatically increase use of the upstream splice site (from ∼26% with the wild-type minigene to >70% for the mutants). Previous studies have shown that GG 5′ splice sites (like GA) are blocked after step 1 (Aebi et al., 1987). The +8G mutation (lane 4) results in approximately equal use of the two 5′ splice sites, demonstrating that the particular context of the tandem 5′ splice sites in the FGFR1 gene permits splicing of both ‘dead-end’ 5′ splice sites. Finally, combination of mutations +2A and +8T (Figure 5C, lane 5) results in splicing from only the downstream (now GT) splice site, perhaps suggesting that the GA splice site must lie downstream of the GT splice site to be recognized.
Figure 5C also shows the effect of increasing the spacing between the two splice sites. No GA splicing is seen when the spacing between the GT and GA splice sites is increased by 3 nt (construct SP+3, lane 6). This cannot result from either nonsense-mediated decay or changes to the splice site sequence, as the increased spacing was achieved by duplicating positions +4 to +6, preserving both the reading frame and the sequence of the GA splice site. An identical effect is seen when the spacing increase is introduced into the +8G mutation (lane 8). However, introducing the 3 nt spacer in the context of the +8T mutation results in a modest increase in splicing from the downstream splice site (compare lane 7 with lane 2).
Use of two closely spaced 5′ splice sites is strongly affected by steric hindrance: U1 snRNP bound at one site will prevent factors binding to the other (Cunningham et al., 1991). Thus a 3 nt increase in the spacing of the two FGFR 5′ splice sites would be expected to increase use of the minor 5′ splice site only slightly. Such a modest effect is only seen for the +8T mutant, however. The observation that use of the GA and GG sites is prevented by the spacing increase suggests that splice site selection differs for these ‘dead-end’ sites. Indeed, competition for U1 binding may not be a factor in the selection of the tandem FGFR 5′ splice sites, i.e. U1 snRNP may bind only to the upstream GT splice site.
The role of U6 in FGFR 5′ splice site selection
Prior to the first catalytic step of splicing, the 5′ splice site is sequentially recognized by the U1 and U6 snRNPs (Staley and Guthrie, 1998). Thus, the 5′ splice site consensus sequence is capable of base pairing with both U1 snRNA, and, in a more limited manner, U6 snRNA. For the FGFR 5′ splice sites, the upstream GT splice site has the greatest potential to base pair with U1 snRNA, while the potential for base pairing to U6 snRNA is greater at the downstream GA splice site (Figure 6A). Two results shown in Figure 5 suggest that competition for U1 snRNP binding is not involved in dictating the relative use of these tandem 5′ splice sites. First, improving the potential of the GA splice site to base pair with the U1 snRNA (mt+8A→T) actually reduces use of this splice site. Secondly, increasing the spacing of the splice sites abolishes use of the GA 5′ splice site. If the GA splice site does not interact with the U1 snRNP, it may be that the binding of U6 snRNP controls the alternative use of the two FGFR 5′ splice sites. To investigate this, we introduced mutations to the U6 binding sites in each splice site (Figure 6B). Increasing the base pairing potential of the GT splice site by changing positions +5 (C→G) and +6 (A→T), either alone (lanes 2 and 3) or in combination (lane 4), abolishes GA splicing. The effect of these base changes on the strength of the GA splice site is likely to be minimal, as CA and GT both represent poor terminal exon sequences. Reducing the match of the GA site to the splice site consensus by changing positions +11 (G→C) and +12 (T→A), alone (lanes 5 and 6) or in combination (lane 7), also abolishes GA splicing. Thus efficient use of the GA splice site depends upon both its own match to the splice site consensus and the presence of a suboptimal GT splice site upstream.
Fig. 6. The role of U6 in GA splicing. (A) Predicted base pairing of U1 (top) and U6 (bottom) snRNAs with the GT and GA splice sites. The asterisk denotes an interaction that has been identified biochemically but not genetically (Sontheimer and Steitz, 1993). (B) Mutation of key positions in each 5′ splice site. (C) Potential base pairing of the GT and GA splice sites with the modified U6 snRNAs employed. (D) Alternative splicing of the FGFR1 wt construct in the presence of the wild-type (lane 1), GT (lane 2) and GA (lane 3) U6 expression constructs. (E) Alternative splicing of the FGFR3 wt construct in the presence of the three U6 expression constructs. (F) Alternative splicing of various 1-13β-1 (lanes 1 and 2) and 1-13β150-1 (lanes 3 and 4) in the presence of the wild-type (lanes 1 and 3) and GA (lanes 2 and 4) U6 expression constructs.
The mutations we have introduced here also affect the potential of the splice sites to interact with the binding of U1 snRNP. However, specifically mutating positions +5 and +6 to AG (so that the GA splice site is now 5′-AAG/GAAAGTA-3′) actually increases use of the GT splice site (to ∼30%) (Figure 6B, lane 8), again suggesting that U1 snRNA does not base pair with the GA splice site. To obtain direct proof that U6 is the factor responsible for controlling alternative splicing, we have cotransfected our minigenes with plasmids expressing so-called ‘shift’ snRNAs. These shift snRNAs are identical with their endogenous counterparts, except for changes to the regions that base pair to the pre-mRNA. Previous reports have shown that it is possible to rescue use of mutationally weakened 5′ splice sites by cotransfection of plasmids expressing U1 snRNAs with modified recognition sequences (Zhuang and Weiner, 1986; Cohen et al., 1994). Furthermore, in the presence of a given shift U1, competition between different potential (/GT) splice sites can be controlled by the presence of shift U6 snRNAs (Hwang and Cohen, 1996).
Initially, we employed two shift U1 snRNA expression constructs, U1 GT and U1 GA, which contain mutations in the region that base pairs with the 5′ splice site (positions 3–11, wild type: 5′-ACUUACCUG-3′) to allow 9 bp at either the GT or GA splice sites. Neither construct is able to alter the alternative splicing of either the FGFR1 or FGFR3 minigenes, despite expressing snRNA to similar levels (data not shown). Therefore we created two shift U6 snRNAs (again designated GT and GA) with changes in positions 41–46 that increase their potential to base pair with the GT and GA splice sites (Figure 6C). We cotransfected expression constructs for these snRNAs with the wild-type FGFR1 minigene (Figure 6D), using a wild-type U6 snRNA expression construct as a control. No effect is seen with the wild-type (lane 1) and U6 GA (lane 3) constructs, while the U6 GT construct (lane 2) reduces the amount of GA-spliced mRNA. RNase protection assays confirm the accumulation of similar levels of snRNA from each of these constructs (data not shown). With the FGFR3 minigene (Figure 6E) both U6 constructs are active: U6 GT (lane 2) induces a slight decrease in the amount of GA spliced mRNA, while U6 GA (lane 3) doubles the amount of GA splicing. No effect of the U6 GA construct is seen with constructs 1-13β-1 and 1-13β150-1 (Figure 6F). Modest increases in the level of GA splicing were seen for constructs 1-24β-1 and 1-28β-1 in the presence of U6 GA (data not shown), suggesting that the ability of the U6 GA snRNA to influence splicing depends upon the presence of the 5′ ISE.
Discussion
We have investigated use of a novel GA 5′ splice site present in three of the four human FGFR genes, as well as the FGFR genes of a diverse range of vertebrate species. The frequency of GA splicing differs for the minigenes derived from the three human FGFR genes, and these reflect well the frequency with which the two mRNA species have been reported for the three mammalian genes (Table I). These results suggest that the GT:GA splicing ratios of the three human FGFR genes may be static in many tissues, perhaps fine tuning the functioning of each receptor for its given role. However, developmental regulation of the X.laevis FGFR1 gene (Paterno et al., 2000) and tissue-specific regulation of the rat FGFR1 gene (Yazaki et al., 1993) suggest that GT/GA splicing of the human FGFR genes may be similarly dynamic.
Importantly, our results rule out the possibility that RNA editing prior to splicing modifies the GA splice site to GT (or GC), analogous to the creation of a 3′ splice site by A→I editing of the rat ADAR2 transcript (Rueter et al., 1999). If this were the case, the frequency of splice site use would depend at least partly on the frequency with which the unlikely A→T (or C) transversion occurred. Such a scenario is ruled out by the result of the +8A→T mutation (Figure 5C). This change would simply obviate the requirement for the editing event and should not result in the dramatic decrease we observe in utilization of the downstream (now GT) 5′ splice site.
A model for use of the FGFR GA splice sites
Mutation of a GT 5′ splice site at positions +1 and +2 can block splicing after step 1 for single intron substrates (Aebi et al., 1986). In the context of multiple introns, however, these mutations give different results: a +1G→A mutation causes skipping of the associated exon, while a +2T→A mutation (generating a GA splice site) leads to accumulation of lariat-exon intermediate both in vitro and in vivo (Aebi et al., 1987). This suggests that +2 mutations are less detrimental than +1 mutations for the initial recognition of the splice site by U1 snRNP. In addition, for the mammalian spliceosome, U6 A45 (5′-ACAGAG-3′) can be cross-linked to the second intron nucleotide following the first step of splicing (Sontheimer and Steitz, 1993), and this interaction may form part of the putative proof-reading of the 5′ splice site prior to the completion of the splicing reaction (Burgess et al., 1990; Fabrizio and Abelson, 1990; Burgess and Guthrie, 1993; Luukkonen and Séraphin, 1998).
The reduction we observe in the use of the downstream 5′ splice site following the +8A→T mutation argues against a role for U1 in recognizing the GA splice site, as this change actually increases the potential for base pairing with the U1 snRNA. Similarly, increasing the spacing of the unmutated splice sites should partially relieve steric hindrance resulting from U1 snRNP binding to both sites, rather than abolish all use of the downstream splice site. Therefore we propose that the FGFR GA splice site constitutes an extension of the conventional GT splice site immediately upstream. For a conventional 5′ splice site, the role of U1 is simply to mark the approximate location of the exon–intron boundary, while the actual splice junction is defined by the base pairing of both U5 (Newman and Norman, 1991, 1992) and U6 (Kandels-Lewis and Séraphin, 1993; Lesser and Guthrie, 1993) with the pre-mRNA. As mentioned, the FGFR GA 5′ splice site represents a better U6 binding site than the GT splice site (Figure 6B). Thus the upstream GT splice site contains the only U1 binding site at the end of the exon, and is presumably used for exon definition (Berget, 1995). When U6 enters the spliceosome, it can associate with either the poor sequences associated with the GT splice site close to the site of U1 interaction or the more favourable sequences downstream at the GA splice site.
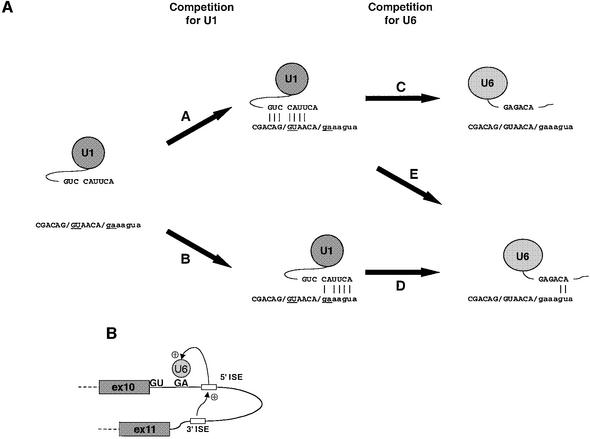
Figure 7A shows a comparison of this model [competition for the binding of U6 snRNP (pathway C versus pathway E)] with the conventional view of 5′ splice site selection [competition for the binding of U1 snRNP (pathway A versus pathway B)]. Evidence in support of this model is provided by the experiments presented in Figure 6, where a modified U6 snRNA could increase use of the GA splice site. Importantly, this effect is not an intrinsic ability of the U6 GA snRNA, as the 5′ ISE was required; furthermore, we presume that it is the enhancers that allow our extensively modified shift U6 snRNAs (Figure 6C) to function. Our model is consistent with previous work demonstrating that the location of a 5′ splice junction can be controlled by U6 snRNP. For mammalian splice sites, such effects have only been seen with mutationally weakened splice sites in the presence of specifically modified shift U1 and U6 snRNAs (Hwang and Cohen, 1996). In yeast, aberrant cleavage 3–4 nt upstream of the RP51A intron 5′ splice site, observed when the splice site was mutationally weakened, is enhanced when the cryptic splice site is mutated to GA or GG (Séraphin and Rosbash, 1990). Strikingly, this cryptic splicing results from U6 misaligning with the pre-mRNA, as improving the base pairing of the U6 snRNA with the genuine splice site restores its use (Kandels-Lewis and Séraphin, 1993). However, our results represent the first description of alternative splice site choice being mediated by the binding of the endogenous U6 snRNP at naturally occurring splice sites.
Fig. 7. A model for use of the human FGFR GA splice site. (A) Comparison of models of splice site selection. For conventional 5′ splice sites, competition is dependent upon the location of U1 snRNP binding (i.e. pathway A competes with pathway B), with subsequent replacement of U1 by U6 (pathways C or D). For the FGFR splice sites, only the upstream GT splice site represents a U1 binding site. Thus, initial definition of the splice site region occurs only via pathway A; competition between the splice sites represents the competition for U6 binding (i.e. pathway C competes with pathway E). (B) A model illustrating the functional interdependence of the two FGFR1 ISEs.
Sequences required for GA splicing of FGFR1
Use of the FGFR GA splice sites is controlled by sequences present within the introns. For FGFR1, we have mapped the 5′ element to the 15 nt immediately downstream of the GA splice site. Although we call this element the 5′ intron splicing enhancer, we note that it does not function as a conventional enhancer. The observation that this element cannot be separated from the GA splice site means that it is more accurate to consider it an integral part of the GA splice site. The core of the 5′ ISE is the G5 tract, present in all of the FGFR1 introns sequenced to date (Figure 1B), although this sequence is not sufficient to act as an enhancer on its own. We have also identified a single ISE in the FGFR3 intron in a corresponding position to the FGFR1 element. Changing the FGFR1 5′ enhancer in 1-28β-1 to that present in FGFR3 results in a level of GA splicing comparable to that seen for the FGFR3 minigene (Figure 4C). Of the eight changes in the R3 mutation, most of the reduction in GA splicing can be attributed to the lack of the G5 tract (data not shown). The generality of these 5′ enhancers is suggested by the conservation of the 5′ intron sequences of a given FGFR gene between different species
We have yet to investigate the FGFR1 3′ ISE in such detail, but it is tempting to speculate that this element reflects some feature of the 3′ splice site region. Indeed, preliminary studies indicate that the FGFR1 branchpoint (5′-GTGAC-3′) lies 19 nt upstream of the 3′ splice site (data not shown) and is preceded by a polypyrimidine sequence conserved in both rat and mouse. Two aspects of the FGFR1 3′ ISE are of interest. First, it is not functionally equivalent to the 5′ ISE, being incapable of increasing use of the GA splice site when present alone (Figure 4A). This suggests that the 3′ enhancer acts via the 5′ enhancer (Figure 7B). Secondly, the 3′ ISE synergizes with the various 5′ ISE derivatives to different extents (Figure 4C). Thus, while activity of the FGFR1 5′ ISE is doubled by the 3′ ISE, activities of the FGFR3 ISE and the putative FGFR2 5′ ISE were not affected. The results of the C5 and A5 mutations suggest that the G5 tract is critical for the synergy between the two elements.
If our model of FGFR 5′ splice site selection is correct, the function of the ISEs may be to allow the realignment of the U6 snRNP on the pre-mRNA, perhaps by stabilizing binding at the GA splice site. Indeed, such stabilization of U6 may be enough to overcome the step 2 block normally associated with GA splice sites, and would explain why shift U1 snRNAs were unable to influence use of the GA splice site. Our working hypothesis for FGFR1 is that the 5′ ISE represents the site of interaction of one or more specific trans-acting factors, the binding of which is stabilized by the 3′ ISE either directly or via additional factors. The presence of the purine-rich element in the FGFR1 5′ ISE could indicate a role for the SR proteins in activating the GA splice site. Indeed, splicing in vitro in the absence of U1 can be observed for conventional splice sites in the presence of increased concentrations of SR proteins (Tarn and Steitz, 1994; Crispino and Sharp, 1995). However, we do not observe any consistent effects when various SR protein expression plasmids are cotransfected with our constructs (S.Brackenridge and G.R.Screaton, unpublished results).
Other GA splice sites
Our results demonstrate efficient splicing from a supposedly ‘dead-end’ 5′ splice site. Therefore, it is important to consider just how widespread such splice sites are. The original report of a GA 5′ splice site was from the Drosophila Antennapaedia gene where, as for the FGFR genes, a conventional splice site lies upstream of a GA 5′ splice site: TTC/GTAAGTGTCAAC/GAAAGTGATC (Hooper et al., 1992). Crucially, the alternatively spliced mRNA was isolated from a strain different to that supplying the genomic DNA, raising the possibility of a strain-specific DNA polymorphism (Jackson, 1991). In addition, the separation of the two splice sites is significantly greater than for the FGFR genes. More recently, intron 13 of the human heparanase gene was reported to contain a GA 5′ splice site and a GG 3′ splice site (Dong et al., 2000), although an earlier report did not include this intron (Vlodavsky et al., 1999). The only GA splice site to have been investigated in any detail is a cryptic GA 5′ splice site in the intron of the Schizosaccharomyces pombe cdc2 gene that is used at low frequency (<10%) when the natural 5′ splice site is weakened by mutation (Romfo et al., 2000). Although this splice site superficially resembles the FGFR GA splice site, it is specifically recognized by U1 snRNP (Alvarez and Wise, 2001), perhaps reflecting differences between the yeast and mammalian spliceosomes.
Our results suggest that use of the FGFR GA 5′ splice site depends upon both the specific sequence of the tandem 5′ splice sites and the presence of intronic enhancers. With the exception of the Antennapaedia gene GA splice site, none of the other GA 5′ splice sites resemble those present in the FGFR genes. However, it may be that other examples of such unusual 5′ splice sites will be identified when the human genome is fully annotated. Furthermore, it is tempting to speculate that competition between very closely spaced conventional 5′ splice sites may also be controlled by U6 snRNP.
Materials and methods
Plasmid constructions
Transfection constructs were based on pCG (Tanaka and Herr, 1990), with fragments inserted using XbaI and BamHI. The snRNA expression plasmids were pG3U1 (Ashe et al., 1997), and pGEM1U6, which contains the human U6 gene cloned by Kunkel and coworkers (Kunkel et al., 1986). All mutations and hybrid fragments were generated by PCR with Pfu and confirmed by sequencing.
Transient transfection and RNA extraction
293T cells were cultured in six-well plates in RPMI-1640 media with 10% fetal calf serum, 50 units/ml penicillin, 50 µg/ml streptomycin and 2 mM l-glutamine. Between 30 and 70% of subconfluent cells in Dulbecco’s modified Eagle’s medium (DMEM), supplemented as before, were transfected with calcium phosphate precipitates of 1 µg of plasmid. For the cotransfection experiments shown in Figure 6, 4 µg of the appropriate snRNA expression construct was also included. The DMEM was replaced with RPMI-1640 after 6–8 h, and after a further 18–36 h RNA was extracted as described previously (Brackenridge et al., 1997).
Reverse transcription PCR
Ten microlitre RT reactions contained AMV-RT buffer, 500 µM dNTPs, 10 pmol primer, 10 units of RNasin (Promega), 4 units of AMV-RT (Cambio) and 4 µl of cytoplasmic RNA, and were incubated at 42°C for 1 h and then at 90°C for 10 min. The reverse primer was βex3R (5′-GTGATGGGCCAGCACACAGACCAG-3′), which anneals to the rabbit β globin exon 3 in pCG, except for the RT–PCRs shown in Figure 3B where the reverse primer was βex2R (5′-CACCTTAGGGTTGCCCATAACAGC-3′), which anneals to human β globin exon 2. PCRs (1 × Taq buffer, 1.5 mM MgCl2, 100 µM dNTPs, 10 pmol of each primer, 1 unit of Taq polymerase, 1 µl of RT reaction) were performed on a Hybaid Touchdown PCR machine using 28 cycles of 1 min at 94°C, 1 min at 65°C (or 60°C; Figure 3B) and 2 min at 72°C. A final 10 min at 72°C was included at the end of the programme. Reverse primers were as for the RT, while the forward primers were hR1F (5′-CCCTGGAAGAGAGGCCGGCAG-3′, FGFR1), hR2F (5′-CGCCTGGAAGAGAAAAGGAGA-3′, FGFR2), hR3F (5′-CCGAGGAGGAGCTGGTGGAG-3′, FGFR3) and βex1F (5′-GCACCTGACTCCTGAGGAGAAG-3′, human β globin).
Digestion and labelling of RT–PCR products
Aliquots of the RT–PCRs were checked by electrophoresis on 2% agarose gels; the remainder of the product was recovered by ethanol precipitation and centrifugation (13 000 r.p.m., 20 min, 4°C). Pellets were air dried and dissolved in 20 µl of distilled water. Five microlitres of DNA was digested (1 h, 37°C) in a final volume of 15 µl with 10 units each of ApaLI and HinfI (FGFR1, FGFR2 and FGFR3; 1-1-β), HinfI (β-1-1) or AccI and HinfI (β-1-β). Reactions were adjusted to 20 µl with the addition of 0.1 units of sequencing-grade Klenow polymerase (Roche) and 0.037 MBq of [α-32P]dATP (111 TBq/mmol; Amersham Pharmacia) and were incubated for 20 min at room temperature, with all four dNTPs added to 125 µM after the first 10 min. Five microlitres of loading dye (10 mM EDTA, 1% w/v sodium dodecyl sulfate, 10 mg/ml bromophenol blue and xylene cyanol) was added and 5 µl aliquots were loaded onto 5.8% native acrylamide gels (18 × 40 cm). Gels were run in 1× TBE at 30 W for 75 min, dried onto 3MM chromatography paper (Whatman) and exposed at –70°C to Kodak BioMax MR-1 film or at room temperature to Fuji BAS-2000 phosphoimager screens for quantitation.
Acknowledgments
Acknowledgements
We thank S.Murphy, A.Furger and N.J.Proudfoot for kind gifts of plasmids pGEM1U6 and pGEM3U1, members of the Screaton Group for helpful discussions and J.F.Caceres for critical reading of the manuscript. This work was supported by BBSRC project grant G11717 to G.R.S. and A.O.M.W.
References
- Aebi M., Hornig,H., Padgett,R.A., Reiser,J. and Weissman,C. (1986) Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell, 47, 555–565. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig,H. and Weissmann,C. (1987) 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice site region, not by the conserved 5′ GU. Cell, 50, 237–246. [DOI] [PubMed] [Google Scholar]
- Alvarez C.J. and Wise,J.A. (2001) Activation of a cryptic 5′ splice site by U1 snRNA. RNA, 7, 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., Pearson,L.H. and Proudfoot,N.J. (1997) The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J., 16, 5752–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S.M. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem., 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- Black D.L. (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell, 103, 367–370. [DOI] [PubMed] [Google Scholar]
- Blencowe B.J. (2000) Exonic splicing enhancers: mechanisms of action, diversity and role in human genetic disorders. Trends Biochem. Sci., 25, 106–110. [DOI] [PubMed] [Google Scholar]
- Brackenridge S., Ashe,H.L., Giacca,M. and Proudfoot,N.J. (1997) Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res., 25, 2326–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgar H.R., Burns,H.D., Elsden,J.L., Lalioti,M.D. and Heath,J.K. (2002) Association of the signalling adapter FRS2 with fibroblast growth factor receptor 1 (Fgfr1) is mediated by alternative splicing of the juxtamembrane domain. J. Biol. Chem., 277, 4018–4023. [DOI] [PubMed] [Google Scholar]
- Burgess S.M. and Guthrie,C. (1993) A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell, 73, 1377–1391. [DOI] [PubMed] [Google Scholar]
- Burgess S., Couto,J.R. and Guthrie,C. (1990) A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell, 60, 705–717. [DOI] [PubMed] [Google Scholar]
- Burset M., Seledtsov,I.A. and Solovyev,V.V. (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res., 28, 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.B., Snow,J.E., Spencer,S.D. and Levinson,A.D. (1994) Suppression of mammalian 5′ splice site defects by U1 small nuclear RNA from a distance. Proc. Natl Acad. Sci. USA, 91, 10470–10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino J.D. and Sharp,P.A. (1995) A U6 snRNA:pre-mRNA interaction can be rate-limiting for U1-independent splicing. Genes Dev., 9, 2314–2323. [DOI] [PubMed] [Google Scholar]
- Cunningham S.A., Else,A.J., Potter,B.V. and Eperon,I.C. (1991) Influences of separation and adjacent sequences on the use of alternative 5′ splice sites. J. Mol. Biol., 217, 265–281. [DOI] [PubMed] [Google Scholar]
- Dong J., Kukula,A.K., Toyoshima,M. and Hakalima,M. (2000) Genomic organisation and chromosomal localisation of the newly identified human heparanase gene. Gene, 253, 171–178. [DOI] [PubMed] [Google Scholar]
- Fabrizio P. and Abelson,J. (1990) Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science, 250, 404–409. [DOI] [PubMed] [Google Scholar]
- Gillespie L.L., Chen,G. and Paterno,G.D. (1995) Cloning of a fibroblast growth factor receptor 1 splice variant from Xenopus embryos that lacks a protein kinase C site important for the regulation of receptor activity. J. Biol. Chem., 270, 22758–22763. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M.L. and Krainer,A.R. (2001) Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol., 13, 302–309. [DOI] [PubMed] [Google Scholar]
- Hooper J.E., Perez-Alonso,M., Bermingham,J.R., Prout,M., Rocklein,B.A., Wagenbach,M., Edstrom,J.-E., De Fritos,R. and Scott,M.P. (1992) Comparative studies of Drosophila Antennapedia genes. Genetics, 132, 453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Kan,M., McKeehan,K., McBride,G., Adams,P. and McKeehan,W.L. (1991) Fibroblast growth factor receptors from liver vary in their structural domains. Science, 251, 665–668. [DOI] [PubMed] [Google Scholar]
- Hwang D.-Y. and Cohen,J.B. (1996) U1 snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes Dev., 10, 338–350. [DOI] [PubMed] [Google Scholar]
- Jackson I.J. (1991) A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res., 19, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.E. and Williams,L.T. (1993) Structural and functional diversity in the FGF receptor multigene family. In Vande Woude,G.F. and Klein,G. (eds), Advances in Cancer Research, Vol. 60. Academic Press, San Diego, CA, pp. 1–41. [DOI] [PubMed]
- Kandels-Lewis S. and Séraphin,B. (1993) Role of U6 in 5′ splice site selection. Science, 262, 2035–2039. [DOI] [PubMed] [Google Scholar]
- Krawczak M. and Cooper,D.N. (1997) The human gene mutation database. Trends Genet., 13, 121–122. [DOI] [PubMed] [Google Scholar]
- Kunkel G.R., Maser,R.L., Calvet,J.P. and Pederson,T. (1986) U6 small nuclear RNA is transcribed by RNA polymerase III. Proc. Natl Acad. Sci. USA, 83, 8575–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser C.F. and Guthrie,C. (1993) Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science, 262, 1982–1988. [DOI] [PubMed] [Google Scholar]
- Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- Luukkonen B.G.M. and Séraphin,B. (1998) Genetic interaction between U6 snRNA and the first intron nucleotide in Saccharomyces cerevisiae. RNA, 4, 167–180. [PMC free article] [PubMed] [Google Scholar]
- Marshall C.J. (1995) Specificity of receptor kinase signalling: transient versus sustained extracellular signal-regulated kinase activation. Cell, 80, 179–185. [DOI] [PubMed] [Google Scholar]
- Mason I.J. (1994) The ins and outs of fibroblast growth factors. Cell, 78, 547–552. [DOI] [PubMed] [Google Scholar]
- Newman A. and Norman,C. (1991) Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell, 65, 115–123. [DOI] [PubMed] [Google Scholar]
- Newman A.J. and Norman,C. (1992) U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell, 68, 743–754. [DOI] [PubMed] [Google Scholar]
- Ornitz D.M. and Itoh,N. (2001) Fibroblast growth factors. Genome Biol., 2, 3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterno G.D., Ryan,P.J., Kao,K.R. and Gillespie,L.L. (2000) The VT+ and VT– isoforms of the fibroblast growth factor receptor type 1 are differentially expressed in the presumptive mesoderm of Xenopus embryos and differ in their ability to mediate mesoderm formation. J. Biol. Chem., 275, 9581–9586. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. (1996) Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev., 6, 215–220. [DOI] [PubMed] [Google Scholar]
- Reed R. (2000) Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol., 12, 340–345. [DOI] [PubMed] [Google Scholar]
- Romfo C.M., Alvarez,C.J., van Heeckeren,W.J., Webb,C.J. and Wise,J.A. (2000) Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe. Mol. Cell. Biol., 20, 7955–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter S.M., Dawson,R.T. and Emeson,R.B. (1999) Regulation of alternative splicing by RNA editing. Nature, 399, 75–79. [DOI] [PubMed] [Google Scholar]
- Séraphin B. and Rosbash,M. (1990) Exon mutations uncouple 5′ splice site selection from U1 snRNA pairing. Cell, 63, 619–629. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (1994) Split genes and RNA splicing. Cell, 77, 805–815. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. and Burge,C.B. (1997) Classification of introns: U2-type or U12-type. Cell, 91, 875–879. [DOI] [PubMed] [Google Scholar]
- Smith C.W. and Valcárcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- Sontheimer E.J. and Steitz,J.A. (1993) The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science, 262, 1989–1996. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Tanaka M. and Herr,W. (1990) Differential activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphoryl ation. Cell, 60, 375–386. [DOI] [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz,J.A. (1994) SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev., 8, 2704–2717. [DOI] [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz,J.A. (1997) Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci., 22, 132–137. [DOI] [PubMed] [Google Scholar]
- Thanaraj T.A. and Clark,F. (2001) Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Res., 29, 2581–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg S.R.F., Burns,H.D., Oldridge,M., Heath,J.K. and Wilkie,A.O.M. (1998) Conserved use of a non-canonical 5′ splice site (/GA) in alternative splicing by fibroblast growth factor receptors 1, 2 and 3. Hum. Mol. Genet., 7, 685–691. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I. et al. (1999) Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat. Med., 5, 793–802. [DOI] [PubMed] [Google Scholar]
- Yazaki N., Fujita,H., Ohta,M., Kawasaki,T. and Itoh,N. (1993) The structure and expression of the FGF receptor-1 mRNA isoforms in rat tissues. Biochim. Biophys. Acta, 1172, 37–42. [DOI] [PubMed] [Google Scholar]
- Zhuang Y. and Weiner,A.M. (1986) A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell, 46, 827–835. [DOI] [PubMed] [Google Scholar]