Abstract
The neck region of kinesin constitutes a key component in the enzyme’s walking mechanism. Here we applied cryoelectron microscopy and image reconstruction to investigate the location of the kinesin neck in dimeric and monomeric constructs complexed to microtubules. To this end we enhanced the visibility of this region by engineering an SH3 domain into the transition between neck linker and neck coiled coil. The resulting chimeric kinesin constructs remained functional as verified by physiology assays. In the presence of AMP–PNP the SH3 domains allowed us to identify the position of the neck in a well defined conformation and revealed its high flexibility in the absence of nucleotide. We show here the double-headed binding of dimeric kinesin along the same protofilament, which is characterized by the opposite directionality of neck linkers. In this configuration the neck coiled coil appears fully zipped. The position of the neck region in dimeric constructs is not affected by the presence of the tubulin C-termini as confirmed by subtilisin treatment of microtubules prior to motor decoration.
Keywords: cryoelectron microscopy/kinesin/kinesin neck/microtubules/SH3 domain
Introduction
Active intracellular transport processes are mediated by the interactions of either myosins with filamentous actin or kinesins and dyneins with microtubules. Since the first observation of kinesin-based motility (Brady, 1985; Vale et al., 1985), kinesins have been found to be involved in a large variety of cellular processes such as organelle movement, spindle formation during cell division and fast axonal transport (Hirokawa, 1998). The molecular details underlying kinesin motion and processivity have been the subject of numerous investigations (reviewed in Hirose and Amos, 1999; Mandelkow and Hoenger, 1999; Vale and Milligan, 2000). One of the key elements for kinesin’s ‘walk’ is the neck-linker which connects the catalytic motor domain to the neck helices (Case et al., 1997; Henningsen and Schliwa, 1997; Endow and Waligora, 1998). Structural investigations into the neck linker of monomeric kinesin constructs indicated conformational variability during the ATP hydrolysis cycle (Rice et al., 1999). It became clear from these studies that structural variations in this region are direct determinants of speed and directionality among different kinesins (Endow, 1999). Similarly important, but unclear, is the role of the coiled coil formed by the neck helices following the neck linker. In earlier work (Hoenger et al., 1998) we proposed that the neck coiled coil might zip and unzip during the walking cycle, but it has been suggested that unwinding is not essential for movement (Tomishige and Vale, 2000). Currently, a correlation between the stability at the beginning of the neck coiled coil and the speed of kinesin is being discussed (Hoenger et al., 2000a; Song et al., 2001).
Our earlier work on cryoelectron microscopy of kinesin–tubulin complexes (Hoenger et al., 1998, 2000a) made it clear that it is not possible to visualize the neck region of dimeric kinesins without density labels. Visualization of the neck linker in monomeric kinesin constructs has been achieved using undeca-gold labels linked to cysteines into the linker of monomeric kinesin constructs (Rice et al., 1999). Compared with that study, we expanded our cryoelectron microscopy investigations to dimeric kinesin constructs and, rather than using gold labels, we used for the first time a clonable density marker inserted into a continuous rat kinesin polypeptide chain. Here we engineered a Spectrin SH3 domain (Musacchio et al., 1992) into the neck, between amino acids (aa) 339 and 340 of monomeric and dimeric kinesin constructs (Figure 1B). Ala339 is the transition point from the neck linker into the α-helical neck at the beginning of the long stalk. SH3 domains are globular beta barrels with a molecular weight of ∼7 kDa. This strategy has several advantages over cysteine-linked gold labels; for example, there is no need for cysteine-light mutants and labelling efficiency is 100% (versus typically 50–60% using gold labels). Ensuring the presence of a tag at each position is particularly important for the image reconstruction technique used here. Furthermore, the N- and C-termini of the SH3 domain are close together, and thus its insertion into a polypeptide chain creates minimal extension to the adjacent regions and keeps linker and neck coiled coil in close proximity. ATP hydrolysis and microtubule gliding assays revealed that our constructs remained functional despite the inserted domain, while dimer formation of the longer constructs was not disrupted. Under certain nucleotide conditions and at physiological concentrations, dimeric kinesin constructs will bind with both heads to the microtubule surface (Hoenger et al., 1998, 2000a). To ensure such physiologically relevant binding conditions we had to work at rather low motor-to-tubulin ratios compared with previous cryoelectron microscopy studies (Hirose et al., 1996, 1999). This avoided overcrowding of the microtubule surface and prevented the enforcement of unnatural configurations (Hoenger et al., 2000b; Vilfan et al., 2001). We could show here how a dimeric kinesin binds on microtubules in the presence of the ATP analogue AMP–PNP. New discoveries were also made regarding the dimerization state of the neck coiled-coil helices and about the interaction of the kinesin neck helices with the tubulin C-termini.
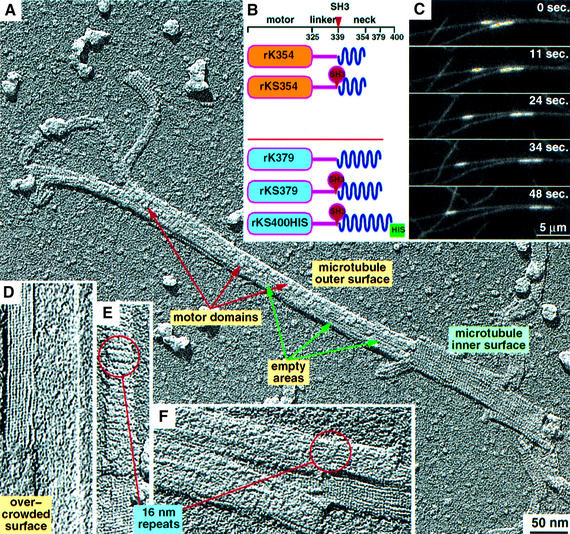
Fig. 1. Design of monomeric and dimeric kinesin constructs, motility assays and unidirectional surface shadowing. (A and D–F) Electron micrographs of unidirectionally shadowed tubulin walls complexed with rKS379 motor constructs in the presence of AMP–PNP. (A) The surface of a partially decorated microtubule leaving open areas on the microtubule outer surface. Note the different appearance of the inner surface, which remains entirely undecorated. (E and F) At slightly higher stoichiometric decoration conditions, generation of 16 nm repeats is sometimes observed, but usually only the typical 8 nm repeat pattern is present. (D) Oversaturation of the microtubule surface with dimeric motor constructs results in an overcrowded surface and destroys the regular pattern. (B) Graphical representations of the kinesin constructs used for cryoelectron microscopy analysis. The red arrowhead indicates the position in which an SH3 domain was inserted. The top two constructs are monomeric while the bottom three are dimeric. (C) TIRF images of microtubules gliding on a glass surface covered with rkS400-HIS. The brighter spots are fluorescent tubulin seeds which were used as nucleation points for tubulin polymerization.
Results
Construction of kinesin–SH3 chimeras
We created a monomeric kinesin–SH3 construct, which was based on rK354 (Sack et al., 1997) (Figure 1B). In this chimera the SH3 domain is inserted after Ala339 (end of neck linker) and continues from the C-terminal end of the SH3 domain to Lys354 of rat kinesin (denoted rKS354). This includes the initial part of the neck helix α7 in the way it was found in the crystal structure (Sack et al., 1997). This helix portion is very short and exhibits a sequence of side chains that are highly unlikely to form a dimeric coiled coil.
Our dimeric kinesin–SH3 construct was based on rat kinesin construct rK379 (Kozielski et al., 1997). This tagged kinesin included an SH3 domain between linker and neck helix at the same positions as for the monomeric chimera, but it contained the entire neck helix α7 (Figure 1B). The resulting construct (denoted rKS379) indeed forms a dimer, as determined by gel filtration studies, in comparison to dimeric and monomeric kinesins (data not shown). To allow for limited flexibility and efficient coiled-coil formation without steric hindrance, we inserted two additional amino acids (Gly, Ser) at the N-terminus of SH3 and two amino acids (Lys, Leu) at its C-terminus. For motility assays we engineered an identical construct to rKS379 but with the neck region elongated by 21 additional rat kinesin amino acids and the C-terminus flanked by a His6 tag (denoted rKS400-HIS) (Figure 1B).
The SH3 domain insertion reveals a functional kinesin
Since the neck region represents a key element for the regulation of kinesin speed and directionality (Endow, 1999), the effect of the inserted SH3 domain on kinesin function was one of our major concerns. We were able to confirm that functionality was not destroyed by ATP hydrolysis and motility assays (Table I; Figure 1C). The SH3 domain insertion did not abolish the walking properties in our chimeras. Although movement was found to be slower than the wild-type kinesin, the motor was functional.
Table I. ATP hydrolysis rates for the constructs used in this work.
| Construct | Hydrolysis rate (ATP/head/s) |
|---|---|
| rK354 | 200 |
| rKS354 | 78 |
| rK379 | 72 |
| rKS379 | 42 |
| rkS400-HIS | 48 |
ATP hydrolysis assays were based on phosphate quantification by the malachite green method (Lanzetta et al., 1979) and showed that all of our constructs display microtubule stimulated ATPase activity (Table I). The hydrolysis rates of rKS379 and rKS400-HIS were found to be 42 and 48 ATP/head/s respectively, displaying ∼60% the rate of rK379. The similar ATP hydrolysis rates of the tagged dimers suggest that the presence of the additional random coil residues (aa 380–400) does not influence the enzymatic rate. The rates for our monomeric constructs rK354 and rKS354 were 200 and 78 ATP/head/s, respectively. These higher hydrolysis rates of monomers compared with dimers is consistent with previously published results, and indicate multiple hydrolysis cycles prior to detachment (Ma and Taylor, 1997a,b; Moyer et al., 1998).
To test the influence of the added SH3 domain between linker and neck coiled coil on motor activity and motility, we used our dimeric constructs rKS400-HIS. The His tag allowed the construct to be linked to the surface of a glass flow-cell by a monoclonal anti-HIS antibody. The elongated neck was used to ensure proper binding of the motor heads to the glass surface by conserving adequate flexibility. Motility assays showed that rKS400-HIS supported microtubule gliding with a speed >0.1 µm/s (Figure 1C). The movement was uninterrupted and there was no variability in the speeds recorded. Since the enzymatic activity profiles for rKS400-HIS and rKS379 are similar, we assume that rkS739 behaves in a similar way and hence remains a functional dimeric motor.
Direct visualization of the double-binding properties of dimeric kinesins
As a first step we investigated the tubulin-binding properties of rKS379 by electron microscopy using freeze-drying and unidirectional surface shadowing (Figure 1A), and found that its behaviour was comparable with that postulated previously for rK379 (Hoenger et al., 2000a). The strong contrast generated by surface metal shadowing allows a direct interpretation of three dimensional (3D) surface structures at molecular resolution without further image enhancement. When undersaturating conditions (0.25–0.5 heads per tubulin binding site) are used, individual motor heads are visible on the microtubule surface, forming little patches with empty areas between them (Figure 1A). In contrast to the outer surface, the inner side of a microtubule wall exhibits only the typical 4 nm α–β–α–β tubulin repeats (Figure 1A and D–F). Decoration experiments at slightly increased stoichiometric ratios of motors to tubulin binding sites revealed the typical axial 8 nm repeat pattern, indicating the binding of one motor head domain per αβ-tubulin dimer (Figure 1E). Occasionally, 16 nm periodicities appear as well, illustrating the double-binding conformation of dimeric constructs over two tubulin dimers in axial direction (Thormählen et al., 1998; Hoenger et al., 2000a) (Figure 1E and F, rings). Motor-to-tubulin ratios higher than 2:1 reveal an irregular surface pattern due to overcrowding of the surface with dimeric constructs, which can no longer find two free adjacent binding sites (Hoenger et al., 2000b; Vilfan et al., 2001) (Figure 1D). Optimal decoration of dimeric motors was achieved at motor domain-to-tubulin ratios of ∼1.5 for AMP–PNP states and 2.5–3 for nucleotide-free conditions. As monomeric constructs do not produce any overcrowding effects, any motor-to-tubulin ratio above 1:1 usually results in a fully decorated filament.
Localization of SH3 tags by helical 3D reconstructions and difference mapping
The primary goal of this work was the visualization of the kinesin neck region at distinct nucleotide-dependent conformational stages in monomeric and dimeric kinesin constructs (Figures 2–345). The details of the datasets that went into the 3D reconstructions are given in Table II. Here we combined cryoelectron microscopy and 3D image reconstruction with statistical difference mapping (Flicker et al., 1991; Hoenger et al., 2000a). Since the density of the neck-linker region alone is too low to be observed directly with cryoelectron microscopy we tagged it with an SH3 domain as described above. The location of the SH3 domains was determined by comparing 3D maps of SH3-tagged and untagged kinesin–microtubule complexes with the help of a Student’s t-test analysis. Individual datasets, shifted to a common phase origin, were combined to generate variance maps from each group, which were then probed for statistically significant density differences between two groups. All difference maps in Figures 234–5 show mass-related differences with a significance of >99%. The resulting volumes at the illustrations used here do not necessarily reflect the exact shape or the expected mass, but indicate the location of differences at a probability of >99%. Accordingly, flexible parts reveal smaller volumes than stable ones.
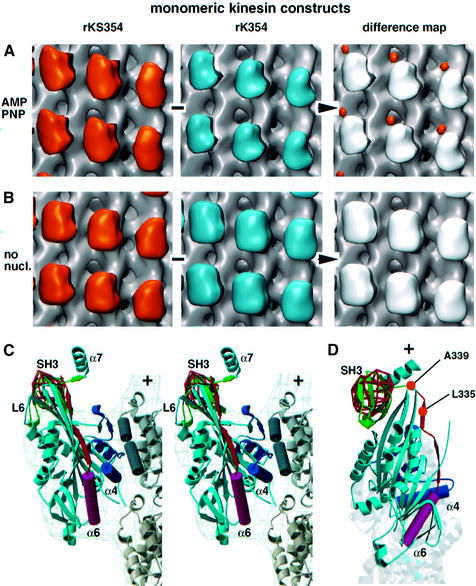
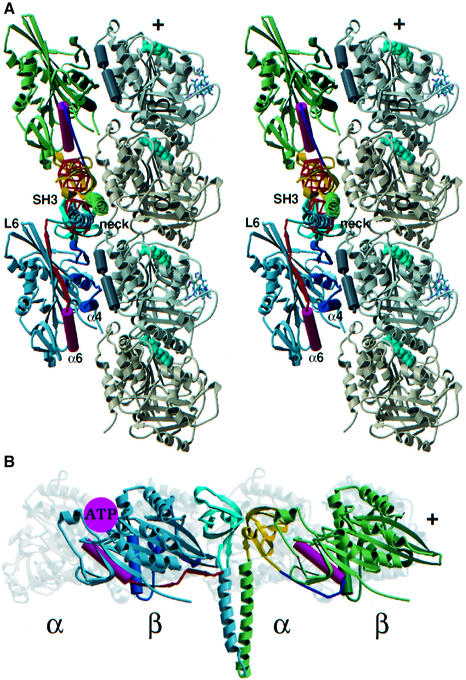
Fig. 2. 3D reconstructions, difference mapping and modelling using monomeric constructs in the presence of AMP–PNP and absence of nucleotide. All comparisons run from left to right, and the volumes representing a statistically significant difference (at 99% probability level) are shown coloured on the right panels. (A) 3D maps of the SH3-tagged rKS354 versus the untagged rK354 construct in the presence of AMP–PNP. The most significant difference is shown as the orange volume in the right panel. (B) No differences were detected using SH3-tagged versus untagged constructs in the absence of nucleotides, suggesting a high flexibility in the neck linker which makes it impossible to localize the SH3 tag (see also Rice et al., 1999). (C) Stereo view of an atomic resolution model of rkS354 complexed to microtubules in the presence of AMP–PNP. The model derives from docking the corresponding X-ray structures in the electron density of rk354 (grey wire frame) and the SH3 difference volume (red wire frame) provided by rkS354 in the presence of AMP–PNP. The motor head (light blue) is interacting with the outer tubulin helices (H11, H12; graphite) mainly via L7/L8 and L12/α5 which follow the ‘switch II’ helix α4 (dark blue). The neck linker (orange) following helix α6 (magenta) is locked on the motor core identically to the way it presented in the X-ray structure of rk354 and slightly modified after residue 338 to meet the N-terminal of the SH3 domain (green). (D) Top view of the model. Rice and coworkers (Rice et al., 1999) labelled position 335 (333 for human kinesin) with a gold cluster, while we inserted the SH3 tag after position 339.
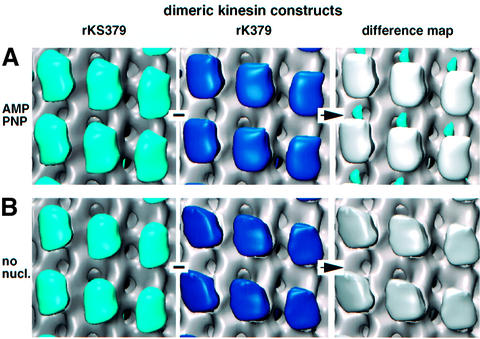
Fig. 3. 3D reconstructions and difference mapping using dimeric constructs in the presence of AMP–PNP and absence of nucleotide. All comparisons run from left to right, and the statistically significant difference is shown as a coloured volume under difference map. (A) In the presence of AMP–PNP the SH3 domains appear as an elongated mass (blue) between two adjacent motor domains along the protofilament. The SH3 tags locate much closer to the microtubule surface than on monomeric constructs. (B) In the nucleotide-free state no difference is detected between tagged and untagged constructs.
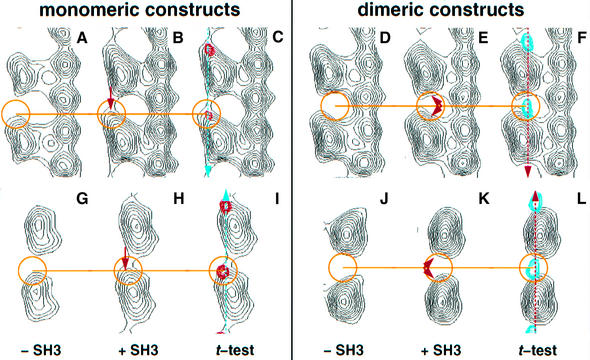
Fig. 4. Vertical (A–F) and horizontal (G–L) cross-sections through motor–tubulin complexes at the height of the corresponding signal attributed to an SH3 domain in the neck at position 339 (monomers, red; dimers, blue). The lines in (C) and (F) mark the plane of the sections in (G), (H) and (I) and in (J), (K) and (L), respectively; similarly, the lines in (I) and (L) mark the planes in (A), (B) and (C) and in (D), (E) and (F), respectively. The monomeric constructs [(A), (B), (C) and (G), (H), (I)] reveal a highly asymmetric signal towards the upper left corner of the kinesin head (towards the plus-end). The additional mass can be seen clearly in (B) and (H). In contrast, the SH3 density in dimeric constructs is located between heads, which show additional mass on both minus- and plus-ends [compare (D) with (E) and (J) with (K)]. This indicates that the difference density is composed from two SH3 domains, one coming from the trailing head and the other coming from the leading head as also shown in Figure 5 and modelled in Figure 6. The resolution of the data combined with the fact that these densities follow a 16 nm repeat does not allow visualization of the domains as directly as for the monomers.
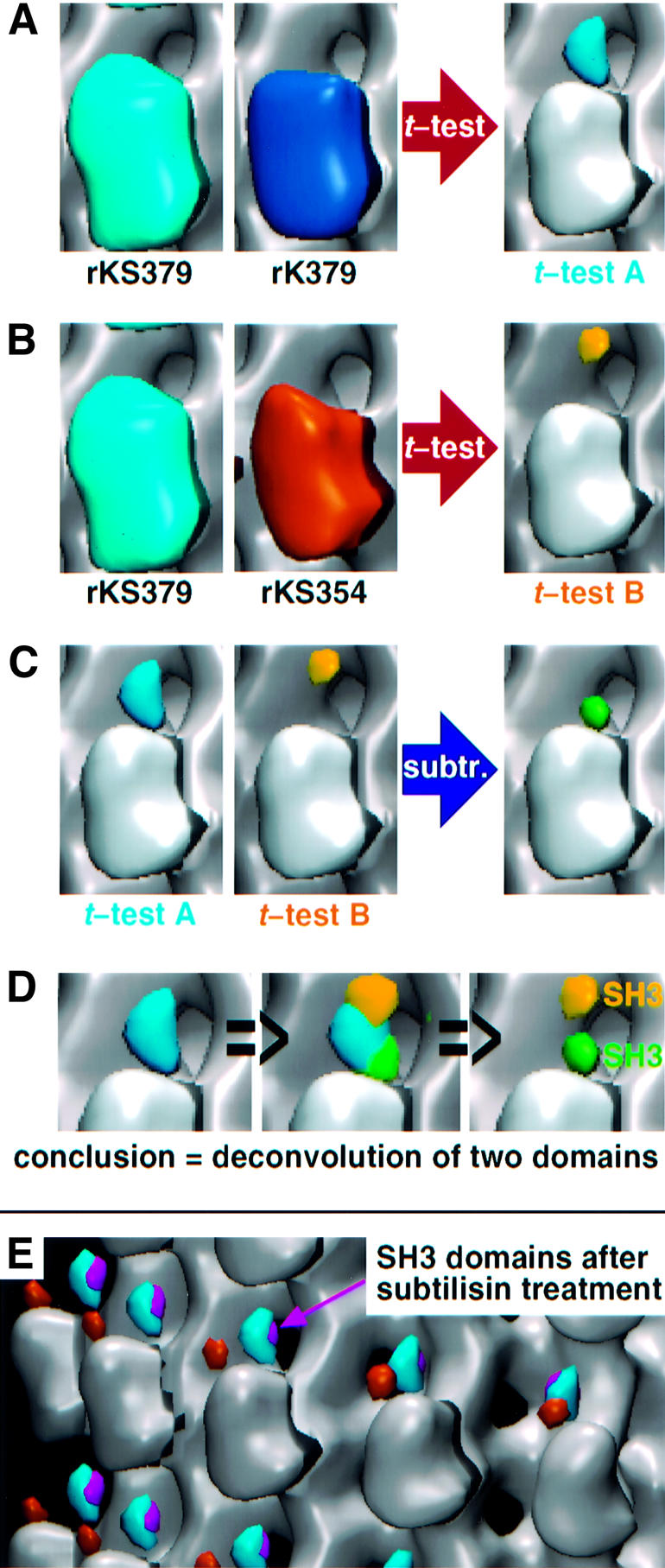
Fig. 5. Comparison and analysis of SH3 signals in the presence of AMP–PNP before and after subtilisin treatment. (A) The t-test comparison between SH3-tagged rkS379 and untagged rk379 in the presence of AMP–PNP reveals an elongated volume (blue; t-test A) at the top of the motor head. For simplicity, all differences shown are superimposed on the map of untagged monomer rk354 with AMP–PNP. (B) The same comparison between the tagged dimer (rkS379; 2 × SH3) and the tagged monomer (rkS354; 1 × SH3) reveals a globular volume (yellow; t-test B). This mass colocalizes with the top part of the overall dimeric SH3 signal (blue) and derives from the additional SH3 of the dimer. It also demonstrates that the C-terminal end of the neck linker in a monomer is rather flexible and at minor populations may occupy a similar position to that of the dimeric SH3 signal. (C) Voxel-by-voxel subtraction of the normalized densities of t-test B from the densities of t-test A results in a second globular volume (green) which corresponds to the lower part of the overall dimeric SH3 signal. This difference represents the second SH3 domain. Hence the elongated mass difference of t-test A can be deconvoluted in two separate domains, implying that each of them correspond to one of the two SH3 domains of rkS379 (D). (E) Subtilisin treatment of microtubules did not change the location of the neck region in complexed dimeric constructs in the presence of AMP–PNP (blue, untreated; magenta, after treatment). The location of the SH3 domain in the monomeric rkS354 construct is shown in orange.
Table II. Numbers of datasets used for 3D reconstructions andstatistical t-test analysis.
| Data sets | Asymmetric units (approx.) | |
|---|---|---|
| rK354 | ||
| AMP–PNP | 15 | 37 500 |
| No nucleotide | 18 | 42 000 |
| rKS354 | ||
| AMP–PNP | 16 | 40 000 |
| No nucleotide | 15 | 34 000 |
| rK379 | ||
| AMP–PNP | 28 | 74 000 |
| No nucleotide | 24 | 63 500 |
| Subtilisin | 22 | 56 500 |
| rKS379 | ||
| AMP–PNP | 30 | 73 000 |
| No nucleotide | 22 | 59 000 |
| Subtilisin | 18 | 46 000 |
Monomeric kinesin constructs
In the first set of experiments we attempted to localize the SH3-tagged neck-linker region in monomeric kinesin constructs (Figure 2). There were two reasons for this: first, it allowed the creation of an initial set of difference maps which were unaffected by the averaging problems expected for dimeric constructs (see explanation below); secondly, this gave us a direct comparison with the results of Rice et al. (1999) where gold labels were used as markers for the linker region. Monomeric constructs were analysed between rK354 and rKS354 in the presence of AMP–PNP and in the absence of nucleotide.
Difference mapping between rK354 and rKS354 in the presence of AMP–PNP revealed a signal attributable to the engineered SH3 domain (Figures 2 and 4, left panel). The difference signal locates in the plus-end direction towards the top left end of the kinesin head (Figure 2A, orange density). The additional mass coming from the SH3 domain can be best seen directly in vertical and horizontal cross-sections along the protofilament axis (Figure 4A–C and G–I). Owing to the limited resolution in our reconstructions the SH3 mass merges into the upper left tip of the motor head domain (in the plus-end direction). This result is in good agreement with previous results (Rice et al., 1999) and indicates a ‘locked’ well-defined configuration for the neck linker. Rice and coworkers (Rice et al., 1999) labelled position 333 of human kinesin (335 in rat kinesin) and detected the signal at the top right of the head. Here we inserted the SH3 tag after position 339 of rat kinesin and detected the signal more towards the top left of the motor head. However, molecular modelling shows that the additional four amino acid residues can easily account for this difference in location (Figure 2C and D). To demonstrate this, we remodelled the neck linker after residue 338, thus maintaining the contacts between β9/β10 and the core motor as found in the X-ray map of rK354–ADP monomer (Sack et al., 1997). The averaged position of the SH3 signal appears to be ∼3.0 nm away from the outer tubulin surface, suggesting little contact between the beginning of the neck helix (α7: aa 340–354; length, 2.25 nm) and the negatively charged C-terminal ends of tubulin (Figure 2C). However, as indicated by our t-tests between monomeric and dimeric kinesin–SH3 constructs, the C-terminal part of the neck linker in a monomer is characterized by limited flexibility and it may also reach positions closer to the microtubule surface (Figure 5B; see below).
In the absence of nucleotides we did not find any significant (>99%) mass-related differences between rK354 and rKS354 (Figure 2B), indicating increased flexibility of the neck under these conditions.
In the presence of excess ADP, monomeric constructs bind weakly and we could only achieve very sparse microtubule–motor decoration at motor-to-tubulin ratios which we consider reasonable (two to three times more motors than binding sites), or even at ratios with many times more motors than available tubulin binding sites (data not shown). Image reconstructions at these conditions were not conclusive. This observation is in good agreement with kinetic (Gilbert et al., 1998) and biophysical (Sosa et al., 2001) data. The latter authors showed convincingly that the ADP state of motor binding is highly flexible and possibly only mediated by loop L11.
Dimeric kinesin constructs
To determine the position of the neck in dimeric constructs we compared 3D maps obtained from construct rK379 (Kozielski et al., 1997) with maps obtained from construct rKS379 (SH3 insertion between aa 339 and 340). Com parisons were made in the presence of AMP–PNP and in the absence of nucleotide. Possible interaction with the highly charged C-terminal ends of α- and β-tubulin were investigated by cleavage of the tubulin C-termini with subtilisin prior to complex formation with motor head domains.
In the presence of AMP–PNP, t-test analysis between rk379 and rkS379 revealed a difference density located between two adjacent motor heads (blue volume in Figures 3A, 4F and L, and 5A). This is compatible with a double-headed binding conformation along the protofilament axis (Hoenger et al., 2000a). The position of the SH3 signal at the 99% confidence level is much closer to the microtubule surface (∼1.5 nm above the outer rim) than in monomeric constructs (e.g. Figures 4C and F, and 5E) and its shape resembles that of a coffee bean aligned along the microtubule axis. The gain in density in the heads carrying an SH3 domain appears more evenly distributed on both ends of the head (Figure 4E and K) than in monomeric constructs (Figure 4B and H). A clear separation of heads from the SH3 densities in such close proximity is not possible owing to the lack of resolution.
Similarly to monomeric constructs, nucleotide-free conditions again did not reveal any statistically significant density differences attributable to the SH3 tag in dimeric constructs (Figure 3B) even after treatment of microtubules with subtilisin (see below).
To further clarify whether, in the presence of AMP– PNP, the density accounting for SH3 domains in dimeric constructs represent one or two domains we used the t-test to compare tagged monomers rkS354 and tagged dimers rkS379 and detected a single difference that colocalizes with the top part of the SH3 signal in dimers (Figure 5B, yellow volume). In this comparison the remaining densities found for monomers and dimers, respectively, appear only at lower probability levels. This clearly indicates that the degree of flexibility of the SH3 tag is higher in monomeric than in dimeric constructs and shows that the monomeric tag may also occupy locations much closer to the microtubule surface. Molecular modelling made it clear that the yellow volume in Figure 5B cannot be reached by the neck linker from the trailing head for steric reasons. This position can only be reached from the leading head by dissociating the β9/β10 contacts and bending the neck linker backwards to the minus-end. Consequently, the yellow volume cannot represent an SH3 tag from the trailing head, but only one coming from the leading head. Normalization and subsequent subtraction of the yellow volume from the dimeric SH3 signal (blue volume) leaves the green volume shown in Figure 5C. This position coincides nicely with the lower end (towards the trailing head) of the dimeric difference density (blue in Figure 5) and the probable position of a tag coming from the trailing head.
In order to test the interaction of the neck with the C-terminal ends of tubulin (see Thorn et al., 2000) we cleaved these highly charged stretches on intact microtubules with subtilisin (Wolf et al., 1996). The complete cleavage of the C-termini for both α- and β-tubulin was verified by SDS–PAGE (data not shown). In our experiments cleavage of the C-termini from α- and β-tubulin did not change the position of the neck (Figure 5E, compare blue and magenta volumes). Accordingly, the differences betweeen the neck positions in monomeric and dimeric constructs cannot be explained by such an interaction alone.
Discussion
The work presented here revealed new insights into the interaction between dimeric kinesin constructs and microtubules, and into the walking properties of a processive kinesin. Insertion of an SH3 domain as a visibility marker into the neck region of conventional rat kinesin allowed us to monitor the whereabouts of this region during different microtubule binding states. In the meantime we have again successfully applied the strategy of using a clonable density marker for electron microscopy 3D analysis to label the coiled-coil neck of a dimeric ncd construct (Wendt et al., 2002).
Insertion of a clonable density marker into the neck-linker region of kinesin reveals a functional complex
Using motility and ATP hydrolysis assays we could show that the functionality of the kinesin–SH3 constructs remained preserved despite the insertion of an SH3 domain into the neck. With the modifications made, the decrease in speed measured here is not unexpected. The presence of the SH3 domains may produce more drag during head relocalization (Cross, 1995; Hua et al., 2002) or may interfere with a transient conformational state. It seems reasonable to speculate that the limited additional flexibility conveyed by the SH3 insertion somehow affected the fine tuning of the walking mechanism, but walking itself might not depend on any tightly restrained interactions between the two motor head domains or parts of the neck. The conservation of functionality is an interesting result in itself and indicates that the walking mechanism of conventional kinesin may be quite resistant even to such major disturbances in the rather sensitive neck region.
Dimeric kinesin may occupy two adjacent microtubule-binding sites along one protofilament
As already postulated (Hoenger et al., 2000a,b), nucleotide conditions that promote strong microtubule binding states create double-binding configurations. We were able to visualize these conditions directly and to analyse the stoichiometric ratios required to fully decorate a microtubule surface with dimeric motor domains without creating an overload. Since dimeric kinesin constructs are capable of contacting the microtubule surface with both heads at the same time under certain physiological conditions, enforced conformations such as ‘stumbling’ of two dimers on each other are likely to occur at high motor-to-tubulin ratios (Hoenger et al., 2000a). To date, helical 3D reconstruction is probably still the best way to assess the kinds of structures shown here, but this approach requires almost complete decoration of the microtubule surface with motor head domains. Therefore the stoichiometry of motors to potential microtubule binding sites has to be carefully balanced at ratios which achieve a nearly complete decoration but no overcrowding effects. Here we used surface metal shadowing as described above to screen for optimal conditions (see Figure 1A and F for under-decorated surfaces, Figure 1D for an over-decorated surface and Figure 1E for a nearly completely decorated surface). The differences in motor-to-tubulin ratios from nucleotide-free to AMP–PNP-containing conditions reflect the higher tendency to assume a double-binding conformation in the presence of AMP–PNP, while in nucleotide-free conditions the leading head shows a reduced tubulin occupancy (Gilbert et al., 1998).
Helical image reconstructions of dimeric kinesin decorating microtubules in a double-binding conformation have to be interpreted with caution. A straightforward interpretation would require that kinesin dimers pack onto the microtubule surface in a cooperative manner both axially and laterally, which would then generate axial 16 nm repeats (Figure 1E and F, circles; see also Thormählen et al., 1998; Hoenger et al., 2000a). Such repeats would perfectly match the underlying helical symmetry of a 15-protofilament microtubule. However, they do not occur very often and in most cases double-bound kinesin dimers are randomly distributed over the microtubule surface. Therefore most reconstructions of microtubules complexed with dimeric motor in a double-binding configuration average over trailing and leading heads, thereby revealing a protofilament with an 8 nm repeat. Consequently, any detectable signal attributed to the dimerized coiled-coil stalk will appear every 8 nm in the axial direction and not at their actual 16 nm interval. Thus the difference densities from the dimeric constructs in Figures 3 and 4 are in their correct position with respect to the tubulin surface but show up every 8 nm between each head at half their actual weight.
The location of the neck linker in monomeric kinesin constructs
SH3-tagged monomeric kinesin constructs revealed the position of the neck linker in a stable (locked) configuration pointing towards the microtubule plus-end (Figures 2A and 4A–C, G–I). In the absence of nucleotides the neck linker could not be localized, most likely because of intrinsic flexibility (Figure 2B). In essence, these results are in good agreement with electron paramagnetic resonance (EPR) spectroscopy and fluorescence resonance energy transfer (FRET) measurements (Rice et al., 1999) which revealed a flexible neck linker in the absence of nucleotide. The only difference with that work is that their cryoelectron microscopy analysis revealed two distinct locations for the neck linker under nucleotide-free conditions. Our results are a clear indication for an entirely non-localized position of the neck linker in the absence of nucleotide, which is in even better agreement with the EPR spectroscopy and FRET measurements (Rice et al., 1999).
The location of the coiled-coil neck in dimeric kinesin constructs
Similarly to monomeric constructs, we were able to localize the neck region of dimers only under AMP–PNP conditions and not in the absence of nucleotides (Figure 3A and B). The fact that the SH3 tags are not visible in the absence of nucleotides may be related to the following: (i) the neck region in dimers may assume a similarly flexible conformation to that found on monomeric constructs; (ii) in the absence of ATP or AMP–PNP the leading head binds to tubulin in a looser and therefore probably more flexible conformation as shown from kinetic data (Gilbert et al., 1998); and (iii) while there is evidence for a stable coiled coil in the presence of AMP–PNP (Figure 5D), in the absence of nucleotide the neck coiled coil may still be partially melted, which would add even more flexibility to the system (Hoenger et al., 2000a). The last point is still speculative and there is as yet no direct evidence for partial melting of a coiled coil under any conditions. However, the sequence between positions 339 and 355 does not show a strong coiled-coil-forming propensity and does not exhibit any hydrophobic a–d heptad repeat contacts (Kozielski et al., 1997). In addition, this part of the neck forms a stable helix by itself (Sack et al., 1997), which is very atypical for α-helices forming classical homodimeric coiled coils (Tripet et al., 2000).
One of the key questions posed here is whether the difference density detected from dimeric constructs in the presence of AMP–PNP represents one or two SH3 domains (Figure 5). Similarly to the situation with monomeric constructs, the resolution of our maps is not sufficient to resolve directly the SH3-domain-related volumes in such close proximity to the head. Therefore its density will merge with the volumes of the heads. This additional mass in the heads can be best visualized with vertical and horizontal cross-sections through the axis of a protofilament at the position of the difference density. Unlike monomeric constructs (Figure 4, left panel), the additional densities in the SH3-tagged constructs are symmetrically distributed over plus- and minus-ends of the heads (Figure 4, right panel). The situation here is somewhat complicated by the fact that the resulting shape of the heads after helical image reconstruction represents an average over the leading and trailing heads. Nevertheless, the more even gain in density at both ends of the heads compared with monomeric constructs (Figure 4E and K) clearly indicates the presence of two SH3 domains. The additional density in the upper end (towards the plus-end) comes from the SH3 tag on the trailing head, while the additional volume at the lower end is contributed by the tag on the leading head. According to the comparisons between the monomeric and dimeric maps shown in Figure 5A–C, we can demonstrate the deconvolution of the dimeric difference density (blue in Figure 5) into two individual densities as presented in Figure 5D and modelled in Figure 6.
Fig. 6. (A) Stereo view and (B) top view of a model for dimeric rKS379 construct complexed to microtubules. Here we used the monomeric crystal structure of rat kinesin 354 to model the trailing head (blue), leaving β9 and β10 intact (orange). The chain after position 338 connects to the modelled position of the docked trailing head SH3 domain (cyan) according to Figure 5 (red wire frame). The leading head (green) represents the crystal structure up to the end of helix α6 (magenta). From then on the chain is folded backwards (blue) to meet its corresponding SH3 tag (yellow). The configuration shown here suggests that the neck coiled coil remains folded.
The interpretations above are supported by the following considerations. If the dimeric differences represented only one rather than two SH3 domains, this could mean that the dimers would have fallen apart entirely. However, in such a case, we expect a similar image to that found with monomers (i.e. we would not average over leading and trailing head conformations). Alternatively, the neck coiled coil may unfold only partially (e.g. between aa 340 and 354). This scenario would again reveal a gain in mass, mostly towards the plus-end, although at half the weight of the previous one. The SH3 domain coming from the leading head would then be lost due to flexibility. Our data presented here clearly oppose these possibilities and agree well with other previous observations which have directly shown double-binding conformations of dimeric kinesin constructs (e.g. see Thormählen et al., 1998; Figure 1). According to the model presented in Figure 6, the close proximity of the SH3 tags from the trailing and leading heads suggests that the neck coiled coil remains completely folded (Kozielski et al., 1997).
Implications for a kinesin walking mechanism
The data presented here add new insight into the processive walking properties of dimeric kinesin. Several important observations have been made: (i) the insertion of an SH3 domain into the neck region still reveals a functional dimeric motor; (ii) the absence of nucleotide leaves the neck-linker region in an unrestricted conformation in both monomeric (see also Rice et al., 1999) and dimeric complexes; (iii) in the presence of AMP–PNP the neck helices are completely folded in a coiled-coil conformation, although the possibility of coiled-coil unzipping in the absence of nucleotide cannot be entirely ruled out with the current data. Taken together, these observations indicate that the processive walking process of conventional kinesin is not regulated by a series of tight sequential interactions between the two heads or the heads and the tubulin surface. Although cleavage of the tubulin C-terminus had a profound effect on kinesin processivity (Thorn et al., 2000), it does not seem to affect the position of the neck (Figure 5E). This does not exclude a potential interaction between neck and tubulin C-termini, but suggests that additional interactions between heads and tubulin C-termini may play a regulatory role as well.
The opposite directionality of the neck linkers in a double-bound configuration partially explains the alternating head catalysis mechanism in kinesin, which seems to be a key element for processive movement (Cross, 1995; Hua et al., 2002). The forward locking of the neck linker is directly coupled to ATP binding, and in a double-binding configuration this will only be possible in the trailing head. The enforced backward-pointing configuration of the neck linker in the leading head probably prevents ATP uptake. Only ATP hydrolysis in the trailing head and its release from the microtubule surface enables ATP binding in the leading head after relaxation of its linker.
Under physiological conditions (no overcrowding!) we could not find any clear indications of binding states for dimers with only one of the two heads bound to the microtubule surface while the second head is in a stable unbound state (proposed by Arnal et al., 1996; Hirose et al., 1996, 1999), e.g. such as found for ncd (Hirose et al., 1996; Sosa et al., 1997; Wendt et al., 2002). We cannot exclude the possibility that transient states as suggested by Hua et al. (2002) may occur, but in our view hand-over-hand walking (Cross, 1995) appears equally likely. However, a rigid hand-over-hand pathway (e.g. only allowing ‘left steps’ or ‘right steps’) would automatically rotate the stalk and eventually the cargo (discussed by Hoenger et al., 2000a). On the other hand, if the neck becomes flexible at some stage, the forward motion of the heads is achieved by diffusion only. Hence, ‘left steps’ and ‘right steps’ may occur at random. Our results here suggest that in the absence on nucleotides this might be actually the case, and the issue of enforcing torque into the cargo by a hand-over-hand walk may not be relevant. In this respect, the active role of kinesin in a processive walking mechanism, besides maintaining constant contact with the microtubule surface, would simply be to prevent the trailing motor from re-binding backwards into the previous position.
Materials and methods
Cloning, expression and purification of kinesin constructs
Constructs rK379 and rK354 were expressed from a pET-3a derivative vector as described (Hoenger et al., 2000a). For all kinesin–SH3 chimeras, the corresponding fragments were amplified by the polymerase chain reaction (PCR) and cloned in vector pET-22b. Spectrin SH3 in a pBAT4 plasmid (a kind gift from Luis Serrano) was cloned in the desired kinesin position in pET-22b. For all kinesin–SH3 chimeras, apart from rKS400-HIS, a stop codon was introduced after the end of kinesin coding sequence, using the Quikchange method (Stratagene), in order to avoid expression of the His tag. All constructs were expressed in Escherichia coli BL21 (DE3) by induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h at 24°C. Cells were resuspended in 20 mM PIPES pH 6.9, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM MgCl2, 50 mM NaCl and 100 µM MgATP, and lysed with a French press (Aminco). Construct rkS400-HIS was purified on a Talon® column followed by gel filtration. For all the other constructs purification was carried out by two ion exchange columns (SP Sepharose, monoQ) followed by a gel filtration run on G200 Hiload 16/60 (Pharmacia) in 20 mM PIPES pH 7.2, 50 mM NaCl and 5 mM MgCl2.
ATPase measurements
Kinesin constructs were characterized for their ATPase activity by the malachite green method (Lanzetta et al., 1979). Briefly, bovine brain tubulin (Cytoskeleton, Denver, CO) was polymerized for 20 min at 37°C in 80 mM PIPES pH 6.8 and 2 mM MgCl2 at a concentration of 5 mg/ml and in the presence of 7.5% v/v dimethylsulfoxide (DMSO), 2 mM GTP and 20 µM Taxol. Microtubules were used at a final concentration of 1.125 µM. ATP hydrolysis was initiated by adding a mixture of kinesin and MgATP at final concentrations of 0.1–0.225 µM and 1 mM, respectively. Aliquots from the reaction were taken every 30 s for a total of 12 min and were assayed for their phosphate content.
Motility assays
Rhodamine-labelled microtubules with highly labelled polymerization seeds were prepared from commercial tubulin and rhodamine tubulin (Cytoskeleton). Anti-His antibody (Qiagen, Penta His Ab, BSA-free) was dissolved in BRB80 and incubated briefly with rKS400-His at final concentrations of 0.2 mg/ml and 0.1 mg/ml, respectively. The glass flow chambers were incubated with BRB80 plus 0.1 mg/ml casein and then with the motor-Ab mix for 2 min, and then were rinsed again with BRB80 plus casein. Microtubules at 0.05 mg/ml were given ∼1 min to adsorb and then the chamber was rinsed again with BRB80 plus casein. The success of the absorption process was checked in the microscope before the chamber was finally filled with BRB80 including 2 mM ATP and oxygen scavenging system. We observed gliding using fluorescence excited at 532 nm (LASER2000, Germany), by objective-type total internal reflection (TIRF) through a 100× objective (N.A. 1.4 PlanApo, Olympus, Japan). The images were recorded continuously using an integration time of 486 ms/image on a Cooled-CCD Camera (Hamamatsu Orca ER).
Decoration of tubulin sheets and metal shadowing
Motor-decorated tubulin walls were prepared, metal shadowed and imaged as described (Hoenger et al., 2000b). Briefly, dimeric rKS379 diluted in BRB80 was added to freshly polymerized sheet solution at various stoichiometric ratios, and AMP–PNP was supplemented to a final concentration of 4 mM. The resulting mix was incubated for 2 min and then directly applied to glow-discharged carbon-coated electron microscopy grids. The grids were frozen in liquid nitrogen and transferred to the MIDILAB (Gross et al., 1990) for shadowing and imaging. Images were recorded with a Gatan-794 Multiscan CCD camera (Gatan, Pleasanton, CA) at an electron dose of 500–1000 electrons/nm2.
Specimen preparation for cryoelectron microscopy
Microtubules were polymerized as described above. Decoration of microtubules with kinesin constructs was carried out in solution at a final tubulin concentration of 0.5 mg/ml. For some experiments microtubules were treated with 200 µg/ml subtilisin for 1 h at 30°C in order to remove the C-terminal ends of α- and β-tubulin. Nucleotide-free conditions were generated by treatment of the motor with apyrase, while for ATP-like states we used 4 mM AMP–PNP. For dimeric motors, we used 1.5 times motor per binding site for AMP–PNP decoration, and 2.5–3 times motor per binding site for nucleotide-free conditions. Decoration in the presence of ADP did not reveal sufficient binding even at many times more motor than binding sites. Motors with microtubules were incubated for 2 min, followed by absorption of the sample on perforated carbon grids and fast freezing in liquid ethane (Dubochet et al., 1988). Cryoelectron microscopy was performed with a Philips CM200-FEG microscope using a GATAN-626 cryo-holder. Images were recorded on Kodak SO-163 electron microscopy film, at a magnification of 38 000× and a defocus range between –1.5 and –2.0 µm.
Image processing and 3D reconstruction
For 3D reconstructions we screened for 15-protofilament/2-start helical microtubules (Beuron and Hoenger, 2001). Micrographs were digitized using a Zeiss-SCAI scanner operated at 21 µm on the negative, which corresponds to 0.5526 nm on the sample. Suitable microtubules were helically reconstructed using the program suite PHOELIX (Whittaker et al., 1995). The 3D maps were visualized using SUPRIM (Schroeter and Bretaudiere, 1996) and VolVis (State University of New York, Stonybrook). Significant differences between the 3D maps were calculated by t-test within the PHOELIX suite, comparing all the datasets used for each reconstruction. Only differences with a probability level >99% were accepted.
Molecular docking and modelling
Motor domains from the crystal structure of rK354 (Sack et al., 1997; [2KIN]) were docked in the EM envelopes obtained for rk354 in the presence of AMP–PNP and the absence of nucleotide. This individual docking was based on previous docking models (Hoenger et al., 1998; Kikkawa et al., 2001), which placed the L8 and switch II elements at the tubulin interface. For the double-headed binding, after assigning the AMP–PNP motor head as the trailing head and the nucleotide-free motor head as the leading head, the rest of the modelling concerned the neck linkers and the coiled coil (Kozielski et al., 1997; [3KIN]). The positions of the latter were indirectly provided by the SH3 signals of rKS379-AMP–PNP. The crystal structures of Spectrin SH3 domain (Martinez et al., 1998; [1BK2]) were docked into the corresponding EM envelopes so that their C-termini were close enough to allow coiled-coil formation by the kinesin helices which followed the SH3 domains. A possible position for the coiled coil is one tangential to the protofilament, underlying the possibility of an interaction with tubulin residues. The trailing head neck linker was slightly redesigned from its crystal structure configuration in order to meet the N-terminus of the trailing SH3 domain. The neck linker of the leading head was reversed after helix α6, so that it ends at the N-terminus of the leading SH3 domain.
Acknowledgments
Acknowledgements
We would like to thank Eva-Maria Mandelkow (MPG, DESY-Hamburg), Stephen Fuller (Oxford University), Bettina Boettcher (EMBL-Heidelberg) and Helen Saibil (Birkbeck College) for helpful discussions. We are grateful to Jens Mueller for providing us with kinesin constructs rK354 and rK379. We would like to thank Luis Serrano (EMBL Heidelberg) for the SH3 plasmid pBAT4.
References
- Arnal I., Metoz,F., DeBonis,S. and Wade,R.H. (1996) Three-dimensional structure of functional motor proteins on microtubules. Curr. Biol., 6, 1265–1270. [DOI] [PubMed] [Google Scholar]
- Beuron F. and Hoenger,A. (2001) Structural analysis of the microtubule–kinesin complex by cryo-electron microscopy. In Vernos,I. (ed.) Kinesin Protocols. Humana Press, Totowa, NJ, pp. 235–254. [DOI] [PubMed]
- Brady S.T. (1985) A novel brain ATPase with properties expected for the fast axonal transport motor. Nature, 317, 73–75. [DOI] [PubMed] [Google Scholar]
- Case R.B., Pierce,D.W., Hom-Booher,N., Hart,C.L. and Vale,R.D. (1997) The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell, 90, 959–966. [DOI] [PubMed] [Google Scholar]
- Cross R.A. (1995) On the hand-over-hand footsteps of kinesin heads. J. Muscle Res. Cell Motil., 16, 91–94. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Adrian,M., Chang,J.J., Homo,J.C., Lepault,J., McDowall,A.W. and Schultz,P. (1988) Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys., 21, 129–228. [DOI] [PubMed] [Google Scholar]
- Endow S.A. (1999) Determinants of molecular motor directionality. Nature Cell Biol., 1, E163–E167. [DOI] [PubMed] [Google Scholar]
- Endow S.A. and Waligora,K.W. (1998) Determinants of kinesin motor polarity. Science, 281, 1200–1202. [DOI] [PubMed] [Google Scholar]
- Gilbert S.P., Moyer,M.L. and Johnson,K.A. (1998) Alternating site mechanism of the kinesin ATPase. Biochemistry, 37, 792–799. [DOI] [PubMed] [Google Scholar]
- Gross H., Krusche,K. and Tittmann,P. (1990) Recent progress in high resolution shadowing for biological TEM. XIIth International Congress for Electron Microscopy, Seattle, WA. Springer, New York, NY, pp. 510–511.
- Flicker P.F., Milligan,R.A. and Applegate,D. (1991) Cryo-electron microscopy of S1-decorated actin filaments. Adv. Biophys., 27, 185–196. [DOI] [PubMed] [Google Scholar]
- Henningsen U. and Schliwa,M. (1997) Reversal in the direction of movement of a molecular motor. Nature, 389, 93–96. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science, 279, 519–526. [DOI] [PubMed] [Google Scholar]
- Hirose K. and Amos,L.A. (1999) Three-dimensional structure of motor molecules. Cell. Mol. Life Sci., 56, 184–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K., Lockhart,A., Cross,R.A. and Amos,L.A. (1996) Three-dimensional cryo-electron microscopy of dimeric kinesin and ncd motor domains on microtubules. Proc. Natl Acad. Sci. USA, 93, 9539–9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K., Löwe,J., Alonso,M., Cross,R.A. and Amos,L.A. (1999) Congruent docking of dimeric kinesin and ncd into three-dimensional electron cryomicroscopy maps of microtubule-motor ADP complexes. Mol. Biol. Cell, 10, 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A., Sack,S., Thormählen,M., Marx,A., Muller,J., Gross,H. and Mandelkow,E. (1998) Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: comparison with X-ray structure and implications for motility. J. Cell Biol., 141, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A., Thormählen,M., Diaz-Avalos,R., Doerhoefer,M., Goldie,K.N., Muller,J. and Mandelkow,E. (2000a) A new look at the microtubule binding patterns of dimeric kinesins. J. Mol. Biol., 297, 1087–1103. [DOI] [PubMed] [Google Scholar]
- Hoenger A., Doerhoefer,M., Woehlke,G., Tittmann,P., Gross,H., Song,Y.-H. and Mandelkow,E. (2000b) Surface topography of microtubule walls decorated with monomeric and dimeric kinesin constructs. Biol. Chem. 381, 1001–1011. [DOI] [PubMed] [Google Scholar]
- Hua W., Chung,J. and Gelles,J. (2002) Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science, 295, 844–848. [DOI] [PubMed] [Google Scholar]
- Kikkawa M., Sablin,E.P., Okada,Y., Yajima,H., Fletterick,R.J. and Hirokawa,N. (2001) Switch-based mechanism of kinesin motors. Nature, 411, 439–445. [DOI] [PubMed] [Google Scholar]
- Kozielski F., Sack,S., Marx,A., Thormählen,M., Schönbrunn,E., Biou,V., Thompson,A., Mandelkow,E.-M. and Mandelkow,E. (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell, 91, 985–994. [DOI] [PubMed] [Google Scholar]
- Lanzetta P.A., Alvarez,L.J., Reinach,P.S. and Candia,O.A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem., 100, 95–97. [DOI] [PubMed] [Google Scholar]
- Ma Y.Z. and Taylor,E.W. (1997a) Kinetic mechanism of a monomeric kinesin construct. J. Biol. Chem., 272, 717–723. [DOI] [PubMed] [Google Scholar]
- Ma Y.Z. and Taylor,E.W. (1997b) Interacting head mechanism of microtubule-kinesin ATPase. J. Biol. Chem., 272, 724–730. [DOI] [PubMed] [Google Scholar]
- Mandelkow E. and Hoenger,A. (1999) Structures of kinesin and kinesin–microtubule interactions. Curr. Opin. Cell. Biol., 11, 34–44. [DOI] [PubMed] [Google Scholar]
- Martinez J.C., Pisabarro,M.T. and Serrano,L. (1998) Obligatory steps in protein folding and the conformational diversity of the transition state. Nat. Struct. Biol., 5, 721–729. [DOI] [PubMed] [Google Scholar]
- Moyer M.L., Gilbert,S.P. and Johnson,K.A. (1998) Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry, 37, 800–813. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Noble,M., Pauptit,R., Wierenga,R. and Saraste,M. (1992) Crystal structure of a Src-homology 3 (SH3) domain. Nature, 359, 851–855. [DOI] [PubMed] [Google Scholar]
- Rice S. et al. (1999) A structural change in the kinesin motor protein that drives motility. Nature, 402, 778–784. [DOI] [PubMed] [Google Scholar]
- Sack S., Muller,J., Marx,A., Thormählen,M., Mandelkow,E.-M., Brady,S.T. and Mandelkow,E. (1997) X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry, 36, 16155–16165. [DOI] [PubMed] [Google Scholar]
- Schroeter J.P. and Bretaudiere,J.P. (1996) SUPRIM: easily modified image processing software. J. Struct. Biol., 116, 131–137. [DOI] [PubMed] [Google Scholar]
- Song Y.-H., Marx,A., Muller,J., Woehlke,G., Schliwa,M., Krebs,A., Hoenger,A. and Mandelkow,E. (2001) Structure of a fast kinesin: implications for ATPase mechanism and interactions with microtubules. EMBO J., 20, 6213–6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa H., Dias,D.P., Hoenger,A., Whittaker,M., Wilson-Kubalek,E., Sablin,E., Fletterick,R.J., Vale,R.D. and Milligan,R.A. (1997) A model for the microtubule–Ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell, 90, 217–224. [DOI] [PubMed] [Google Scholar]
- Sosa H., Peterman,E.J., Moerner,W.E. and Goldstein,L.S. (2001) ADP-induced rocking of the kinesin motor domain revealed by single-molecule fluorescence polarization microscopy. Nat. Struct. Biol., 8, 540–544. [DOI] [PubMed] [Google Scholar]
- Thormählen M., Marx,A., Müller,S.A., Song,Y.-H., Mandelkow,E.-M., Aebi,U. and Mandelkow,E. (1998) Interaction of monomeric and dimeric kinesin with microtubules. J. Mol. Biol., 275, 795–809. [DOI] [PubMed] [Google Scholar]
- Thorn K.S., Ubersax,J.A. and Vale,R.D. (2000) Engineering the processive run length of the kinesin motor. J. Cell Biol., 151, 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M. and Vale,R.D. (2000) Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J. Cell Biol., 151, 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Wagschal,K., Lavigne,P., Mant,C.T. and Hodges,R.S. (2000) Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position ‘d’. J. Mol. Biol., 300, 377–402. [DOI] [PubMed] [Google Scholar]
- Vale R.D. and Milligan,R.A. (2000) The way things move: looking under the hood of molecular motor proteins. Science, 288, 88–95. [DOI] [PubMed] [Google Scholar]
- Vale R.D., Schnapp,B.J., Reese,T.S. and Sheetz,M.P. (1985) Movement of organelles along filaments dissociated from the axoplasm of the squid giant axon. Cell, 40, 449–454. [DOI] [PubMed] [Google Scholar]
- Vilfan A., Frey,E., Schwabl,F., Thormählen,M., Song,Y.-H. and Mandelkow,E. (2001) Dynamics and cooperativity of microtubule decoration by the motor protein kinesin. J. Mol. Biol., 312, 1011–1026. [DOI] [PubMed] [Google Scholar]
- Wendt T.G., Volkmann,N., Skiniotis,G., Goldie,K.N., Muller,J., Mandelkow,E. and Hoenger,A. (2002) Microscopic evidence for a minus-end-directed power stroke in the kinesin motor ncd. EMBO J., 21, 5969–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M., Carragher,B.O. and Milligan,R.A. (1995) PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy, 58, 245–259. [DOI] [PubMed] [Google Scholar]
- Wolf S.G., Nogales,E., Kikkawa,M., Gratzinger,D., Hirokawa,N. and Downing,K. (1996) Interpreting a medium-resolution model of tubulin: comparison of zinc-sheet and microtubule structure. J. Mol. Biol., 262, 485–501. [DOI] [PubMed] [Google Scholar]