Abstract
Carotenoid biosynthesis in plants has been described at the molecular level for most of the biochemical steps in the pathway. However, the cis-trans isomerization of carotenoids, which is known to occur in vivo, has remained a mystery since its discovery five decades ago. To elucidate the molecular mechanism of carotenoid isomerization, we have taken a genetic map-based approach to clone the tangerine locus from tomato. Fruit of tangerine are orange and accumulate prolycopene (7Z,9Z,7′Z,9′Z-tetra-cis-lycopene) instead of the all-trans-lycopene, which normally is synthesized in the wild type. Our data indicate that the tangerine gene, designated CRTISO, encodes an authentic carotenoid isomerase that is required during carotenoid desaturation. CRTISO is a redox-type enzyme structurally related to the bacterial-type phytoene desaturase CRTI. Two alleles of tangerine have been investigated. In tangerinemic, loss of function is attributable to a deletion mutation in CRTISO, and in tangerine3183, expression of this gene is impaired. CRTISO from tomato is expressed in all green tissues but is upregulated during fruit ripening and in flowers. The function of carotene isomerase in plants presumably is to enable carotenoid biosynthesis to occur in the dark and in nonphotosynthetic tissues.
INTRODUCTION
Carotenoid pigments are essential components in all photosynthetic organisms. They assist in harvesting light energy and protect the photosynthetic apparatus against harmful reactive oxygen species that are produced by overexcitation of chlorophyll. They also furnish distinctive yellow, orange, and red colors to fruit and flowers to attract animals. Carotenoids are mainly 40-carbon isoprenoids, which consist of eight isoprene units. The polyene chain in carotenoids contains up to 15 conjugated double bonds, a feature that is responsible for their characteristic absorption spectra and specific photochemical properties (Goodwin, 1980; Britton et al., 1998). These double bonds enable the formation of cis-trans geometric isomers in various positions along the molecule. Indeed, although the bulk of carotenoids in higher plants occur in the all-trans configuration, different cis-isomers exist as well, but in small proportions.
In plants, carotenoids are synthesized within the plastids from the central isoprenoid pathway (reviewed by Hirschberg, 2001) (Figure 1). The first carotenoid in the committed pathway is phytoene, which is produced by the enzyme phytoene synthase through a condensation of two molecules of geranylgeranyl diphosphate. Four double bonds are introduced subsequently into phytoene by two enzymes, phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS), each catalyzing two symmetric dehydrogenation steps to yield ζ-carotene and lycopene, respectively. It is recognized that cis-trans isomerizations take place in vivo because phytoene is synthesized in the 15-cis configuration, whereas most of the other carotenoids are found in the all-trans form (Britton, 1988). Furthermore, a small proportion of cis-isomers exist in many carotenoid species, for example, 9-cis- and 13-cis-isomers of β-carotene, zeaxanthin, and violaxanthin. However, the process of carotenoid isomerization remains unexplained. The existence of a potential carotene isomerase enzyme could be expected from the phenotype of recessive mutations in tomato (Tomes et al., 1953) and Scenedesmus (Ernst and Sandmann, 1988), which accumulate prolycopene (7Z,9Z,7′Z,9′Z tetra-cis-lycopene) as well as poly-cis-isomers of phytofluene, ζ-carotene, and neurosporene. Coexpression of PDS and ZDS from Arabidopsis thaliana in Escherichia coli cells that synthesized phytoene produced mainly prolycopene, whereas all-trans-lycopene was produced in these cells by the bacterial PDS CRTI (Bartley et al., 1999). The authors concluded that an active isomerization function is required in conjunction with the plant-type carotene desaturation reactions to produce all-trans-lycopene.
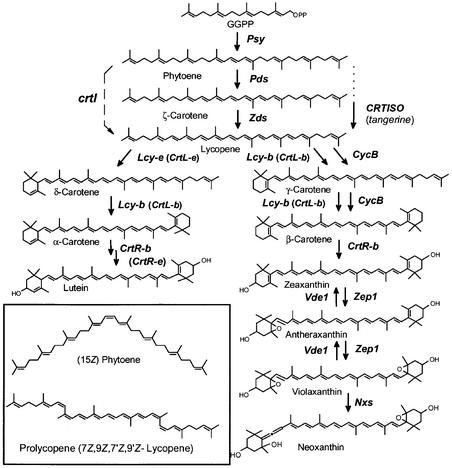
Figure 1.
The Carotenoid Biosynthesis Pathway in Plants.
Enzymes are named according to the designation of their genes. crtI, phytoene desaturase (bacterial type); Lcy-b (CrtL-b), lycopene β-cyclase; CrtL-e, lycopene ɛ-cyclase; CrtR-b, β-ring hydroxylase, CrtR-e, ɛ-ring hydroxylase; CycB, chromoplast-specific lycopene; cyclase; GGPP, geranylgeranyl diphosphate; Nxs, neoxanthin synthase; Pds, phytoene desaturase; Psy, phytoene synthase; Vde, violaxanthin deepoxidase; Zds, ζ-carotene desaturase; Zep, zeaxanthin epoxidase.
To elucidate the mechanism of carotene isomerization, we have studied the tomato gene (Lycopersicon esculentum) that encodes the recessive mutation tangerine (Tomes, 1952). Fruit of tangerine are orange and accumulate prolycopene instead of all-trans-lycopene, which normally is synthesized in wild-type fruit (Figure 2). The structure of prolycopene isolated from the tangerine variety of tomato fruit has been elucidated by NMR (Clough and Pattenden, 1979). The phenotype of tangerinemic is manifested also in yellowish young leaves and sometimes light green foliage and in pale flowers. Using map-based cloning, we identified the gene that encodes tangerine. Our data indicate that this gene, CRTISO, encodes a redox-type enzyme that is related structurally to the bacterial-type phytoene desaturase, CRTI. An ortholog of CRTISO also is found in Arabidopsis and cyanobacteria, where it plays a similar function in carotenoid isomerization.
Figure 2.
Fruits and Seedlings of Tangerine Mutants.
(A) Ripe fruit of the wild type (top) and mutant tangerinemic (bottom).
(B) Immature green fruit of tangerine3183 (top) and mutant tangerinemic (bottom).
(C) Light-grown seedlings of the wild type (left) and mutant tangerinemic (right).
(D) Light-grown seedlings of mutant tangerinemic (left) and the wild type (right).
RESULTS
Carotenoid Composition in the Wild Type and tangerine
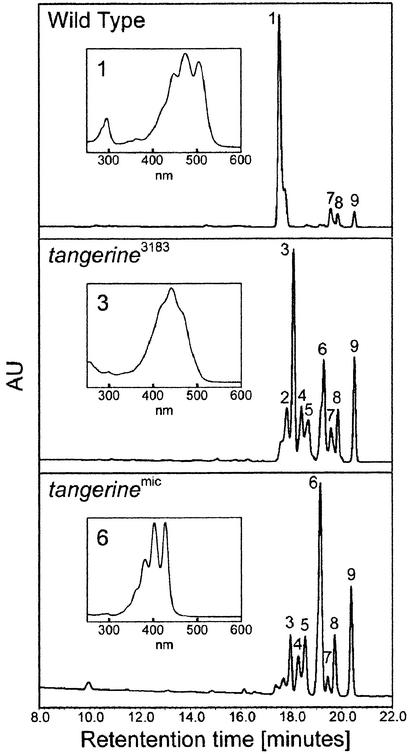
Carotenoids accumulated in fruit and flowers of the wild type and tangerine mutants were extracted and analyzed by HPLC (Figure 3, Table 1). In the wild type, 7 days after the breaker stage, 75% of total carotenoids in ripe fruit consist of all-trans-lycopene and <15% are lycopene precursors (neurosporene, ζ-carotene, phytofluene, and phytoene). In fruit of the mutant tangerine3183, the major carotenoid accumulated is prolycopene, whereas lycopene precursors, mostly in the cis configuration, constitute the rest of the carotenoids. Only a small fraction of <2% is all-trans-lycopene. In tangerinemic, the phenotype is similar but more severe, with the main carotenoids being ζ-carotene (32%) and prolycopene (15%). The identification of geometric isomers of tangerine fruit carotenoids was based on comparison of the HPLC data with previously published analyses that also used NMR (Clough and Pattenden, 1979). Identification of some of the mono-cis- and di-cis-ζ-carotene, neurosporene, and lycopene isomers should be confirmed by the NMR method.
Figure 3.
Analysis by HPLC of Carotenoids in Tomato Fruit of Wild Type and Two Alleles of tangerine Mutants.
Peak 1, all-trans-lycopene; peak 2, di-cis-lycopene; peak 3, prolycopene; peak 4, neurosporene; peak 5, tri-cis-neurosporene; peak 6, ζ-carotene; peak 7, β-carotene; peak 8, phytofluene; peak 9, phytoene. Absorption spectra of peaks are presented in the insets.
Table 1.
Carotenoid Composition in Fruit and Flowers of Wild-Type and tangerine Mutants
| M82 (Wild Type) | tangerine3183 | Micro-Tom | tangerinemic | |
|---|---|---|---|---|
| Fruits | ||||
| Phytoene | 5.2 ± 2.1 | 15.3 ± 2.6 | 6.9 ± 2.1 | 16.0 ± 1.2 |
| Phytofluene | 3.6 ± 0.8 | 8.7 ± 1.53 | 5.9 ± 1.0 | 9.8 ± 1.8 |
| ζ-Carotene | 1.5 ± 2.0 | 23.6 ± 6.3 | 1.1 ± 0.4 | 31.7 ± 8.4 |
| Cis-neurosporene | 8.0 ± 4.1 | 11.4 ± 0.6 | ||
| Neurosporene | 0.2 ± 0.3 | 6.0 ± 3.1 | 0.4 ± 0.4 | 6.2 ± 1.7 |
| Di-cis-lycopene | 6.4 ± 3.2 | 3.6 ± 0.3 | ||
| Prolycopene | 0.4 ± 0.7 | 25.4 ± 7.7 | 15.2 ± 7.8 | |
| Lycopene | 75.2 ± 10.2 | 0.6 ± 1.2 | 78.0 ± 6.0 | |
| β-Carotene | 5.9 ± 3.1 | 2.8 ± 3.3 | 1.2 ± 0.4 | 2.4 ± 1.0 |
| Others/unidentified | 8.0 | 3.2 | 6.5 | 3.7 |
| Total carotenoids (μg g fresh weight) | 77.0 ± 5 | 60.0 ± 17 | 104.0 ± 33 | 53.0 ± 8 |
| Flowers | ||||
| Phytoenea | 11.5 ± 3.4 | 0.3 ± 0.1 | 25.0 ± 7.8 | |
| Phytofluenea | 3.3 ± 0.9 | 7.5 ± 2.8 | ||
| ζ-Carotenea | 4.6 ± 1.9 | 8.8 ± 1.3 | ||
| Cis-neurosporene | 2.6 ± 0.1 | |||
| Neurosporene | 1.6 ± 1.1 | 2.0 ± 0.1 | ||
| Di-cis-lycopene | 4.0 ± 3.8 | |||
| Prolycopene | 1.0 ± 0.9 | 30.2 ± 2.8 | ||
| Lycopene | 1.8 ± 1.8 | |||
| γ-Carotene | 3.2 ± 1.6 | |||
| β-Carotene | 1.1 ± 1.7 | 5.5 ± 1.4 | 0.8 ± 0.7 | 3.5 ± 1.6 |
| Rubixanthin | 11.6 ± 4.5 | 1.8 ± 2.6 | ||
| β-Cryptoxanthin | 2.5 ± 0.7 | 0.9 ± 1.1 | ||
| Violaxanthin | 37.0 ± 7.1 | 11.0 ± 6.1 | 33.2 ± 6.4 | |
| Neoxanthin | 59.4 ± 7.0 | 36.5 ± 9.7 | 57.7 ± 6.3 | 9.7 ± 6.5 |
| Lutein | 2.5 ± 1.0 | 4.8 ± 0.6 | 7.1 ± 2.3 | |
| Others/unidentified | 1.5 | 4.9 | ||
| Total carotenoids (μg g fresh weight) | 770 ± 112 | 490 ± 327 | 1350 ± 521 | 998 |
Numbers correspond to percentage of total carotenoids. Total carotenoid concentration is per fresh weight.
All isomers.
In wild-type flowers, the yellow xanthophylls neoxanthin, violaxanthin, and lutein make up 95% of total carotenoids. In contrast, the fraction of xanthophylls is ∼50% of total carotenoids in flowers of tangerine3183 and <10% in tangerinemic. In both mutants, the flowers accumulate large quantities of prolycopene and its precursors.
The tangerine mutation also affects carotenoid biosynthesis in chloroplasts, as is evident by the yellow color that appears in the newly developed leaves (Figure 2). Leaves of etiolated seedlings of tangerinemic, but not tangerine3183 or the wild type, accumulate prolycopene and its precursors and do not contain any xanthophylls (Table 2). In green leaves of light-grown seedlings of tangerinemic, the proportion of β-xanthophylls increases at the expense of lutein. Green fruit of tangerine contain the normal composition of carotenoids in the outer pericarp, but the inner pericarp and the placental tissue are orange (Figure 2) and contain high levels of prolycopene (data not shown).
Table 2.
Carotenoid Composition in Leaves of Wild-Type (Micro-Tom) and tangerinemic Seedlings Growing on Murashige and Skoog (1962) Medium 7 Days (Dark) or 10 Days (Light) after Germination
| Light
|
Dark
|
|||
|---|---|---|---|---|
| Carotenoid | Micro-Tom | tangerinemic | Micro-Tom | tangerinemic |
| Phytoenea | 2.1 | 17.8 | ||
| Phytofluenea | 7.7 | |||
| ζ-Carotenea | 24.2 | |||
| Cis-neurosporene | 15.1 | |||
| Di-cis-lycopene | 1.2 | |||
| Prolycopene | 33.6 | |||
| β-Carotene | 28.5 | 33.2 | 4.0 | |
| Violaxanthin | 3.3 | 22.5 | 19.8 | |
| Neoxanthin | 7.9 | 10.6 | ||
| Lutein | 59.6 | 29.2 | 76.2 | |
| Others | 0.7 | 0.4 | 0.4 | |
Numbers correspond to percentage of total carotenoids (average of two measurements).
All isomers.
All of the data described above indicate that the locus tangerine is involved in carotenoid isomerization that is essential for the biosynthesis of cyclized carotenes and xanthophylls.
Map-Based Cloning of tangerine
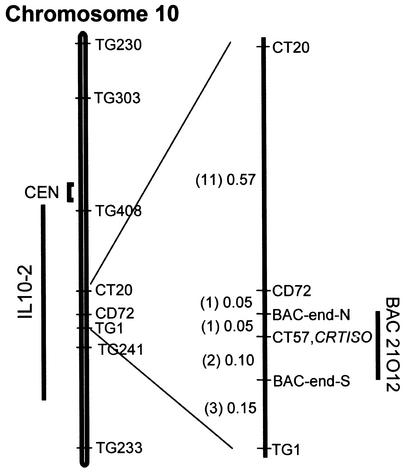
The recessive mutation tangerine was mapped to the long arm of chromosome 10, 4 centimorgans away from the locus l2. This locus is located in a region that overlaps IL10-2. Because none of the known carotenoid biosynthesis genes maps near this locus (data not shown), it has been predicted that tangerine is determined by a new gene. To further map tangerine, we crossed it to IL10-2 and analyzed 1045 F2 plants using the markers TG408 and TG241 that flank tangerine. A total of 218 recombinant plants were obtained, and these individuals were selfed to determine their genotypes with respect to the recessive mutation tangerine. The recombinant plants were probed with additional markers, and CT57 was found to cosegregate with tangerine (Figure 4). A genomic library of tomato in bacterial artificial chromosomes (BACs) (Budiman et al., 2000) was screened with CT57, and BAC 21O12 was identified. Sequences at the ends of the insert of BAC 21O12 were amplified by polymerase chain reaction (PCR) and used as probes in genomic DNA hybridization of the 218 recombinant plants. The results indicated that BAC 21O12 contained the entire region of the tangerine locus because both BAC ends revealed recombinations with the target gene (Figure 4).
Figure 4.
Map-Based Cloning of the tangerine Locus.
Fine mapping of chromosome 10 was performed with the restriction fragment length polymorphism markers indicated in the map. The chromosomal segment in the introgression line IL 10-2 that overlaps tangerine is depicted as a bar. Numbers correspond to centimorgans. Numbers in parentheses describe recombination events. Markers were TG408, CT20, CD72, CT57, TG1, and TG241. CEN, centromere.
The entire insert of BAC 21O12 was sequenced. An open reading frame sequence with similarity to the bacterial gene for PDS, crtI, was found to cosegregate with the tangerine phenotype. The cDNA clone of this gene, designated CRTISO, was obtained by reverse transcriptase–mediated PCR using primers that were designed according to data from the tomato expressed sequence tag database. Comparison between the genomic and cDNA sequences revealed that the gene is composed of 13 exons and 12 introns (Figure 5). DNA gel blot hybridization with total genomic DNA indicated that CRTISO exists as a single copy in the tomato genome (data not shown).
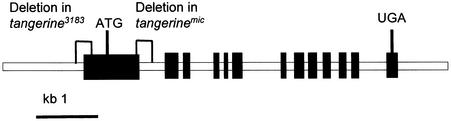
Figure 5.
Organization of the Genomic Sequences of CRTISO.
Closed boxes represent exons. Deletions found in CRTISO of tangerine alleles are indicated. ATG, initiation codon; UGA, stop codon.
Sequence Analysis of CRTISO in Wild-Type and tangerine Alleles
The cDNA of CRTISO contains an open reading frame of 615 codons that encodes a polypeptide with a calculated molecular mass of 67.5 kD. No differences in amino acid sequence were found between CRTISO from the wild type of cv M82, Ailsa Craig, and Micro-Tom and the polypeptide in tangerine3183. In contrast, analysis of both cDNA and genomic sequences of CRTISO from tangerinemic indicated that this allele contains a deletion of 282 bp that encompasses 24 bp of the first exon and 258 bp of the first intron (Figure 5). As a result of this deletion, a splicing site is eliminated and the abnormal mRNA that is produced contains an early stop codon that aborts the synthesis of functional CRTISO. A deletion of 348 bp was discovered in the promoter region of CRTISO of tangerine3183 (Figure 5).
Functional Expression of CrtISO in E. coli
E. coli cells of the strain XL1-Blue, carrying plasmids pGB-Ipi and pAC-Zeta, accumulate mainly ζ-carotene. The latter plasmid contains the genes CrtE and CrtB, which encode geranylgeranyl pyrophosphate synthase and phytoene synthase, respectively, from Erwinia herbicola, and crtP from Synechococcus PCC7942, which encodes PDS. When cotransformed with plasmid pT-Zds, which encodes ZDS from tomato, the cells also produced prolycopene. A similar result has been reported (Bartley et al., 1999). Expressing both Zds and CRTISO from plasmid pCRTISO-TZds resulted in significant accumulation of lycopene (Table 3). The differences between dark-grown and light-grown bacteria probably are caused by the photoisomerization of 15-cis-ζ-carotene, which releases a barrier in the pathway. These results indicate that the polypeptide encoded by CRTISO is an authentic carotenoid isomerase that is able to convert cis-carotenoids to all-trans-carotenoids in E. coli.
Table 3.
Functional Expression of CRTISO in E. coli
| Genes | Phytoene | Phytofluene | ζ-Carotene | Neurosporene | Di-cis-lycopene | Prolycopene | Lycopene | Others |
|---|---|---|---|---|---|---|---|---|
| Light | ||||||||
| crtE, crtB | 98.4 | 1.6 | ||||||
| crtE, crtB, Zds, CRTISO | 99.4 | 0.6 | ||||||
| crtE, crtB, crtP | 12.2 ± 1.8 | 6.8 ± 0.2 | 79.2 ± 1.5 | 1.8 | ||||
| crtE, crtB, crtP, CRTISO | 15.2 ± 4.6 | 7.0 ± 0.6 | 75.3 ± 4.7 | 2.5 | ||||
| crtE, crtB, crtP, Zds | 17.9 ± 0.1 | 10.4 ± 0.2 | 54.6 ± 0.6 | 3.6 ± 0.2 | 6.9 ± 0.7 | 6.2 ± 0.5 | 0.4 | |
| crtE, crtB, crtP, Zds, CRTISO | 19.7 ± 3.1 | 11.4 ± 1.2 | 38.5 ± 5.6 | 30.1 ± 9.7 | 0.3 | |||
| Dark | ||||||||
| crtE, crtB | 99.0 | 1.0 | ||||||
| crtE, crtB, Zds, CRTISO | 100 | |||||||
| crtE, crtB, crtP | 8.9 ± 1.5 | 6.0 ± 0.8 | 83.2 ± 2.2 | 1.9 | ||||
| crtE, crtB, crtP, CRTISO | 11.7 ± 0.9 | 7.6 ± 0.1 | 78.5 ± 1.3 | 2.2 | ||||
| crtE, crtB, crtP, Zds | 13.7 ± 1.2 | 11.1 ± 1.6 | 66.2 ± 1.9 | 8.3 ± 1.4 | 0.7 | |||
| crtE, crtB, crtP, Zds, CRTISO | 23.2 ± 1.1 | 16.5 ± 0.9 | 50.0 ± 1.9 | 0.6 ± 0.9 | 8.5 ± 2.4 | 1.2 |
Cells of E. coli, all carrying plasmid with the gene Ipi, were transfected with different combinations of carotenoid biosynthesis genes. crtE, geranylgeranyl diphosphate synthase; crtB, phytoene synthase; crtP, phytoene desaturase; Zds, ζ-carotene desaturase; CrtiSO, carotenoid isomerase. Numbers correspond to percentage of total carotenoids.
Expression of CRTISO during Fruit Ripening
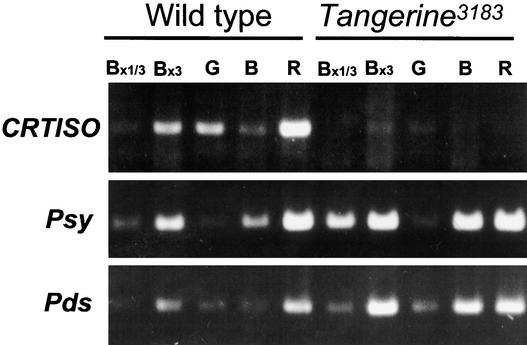
To determine the pattern of expression of CRTISO, we measured its mRNA level in different stages of fruit development (Figure 6). In wild-type fruit, the mRNA levels of CRTISO increased 10-fold during the breaker stage of fruit ripening, similar to the mode of expression of the genes Psy and Pds. In this analysis, no mRNA of CRTISO could be detected in fruit of tangerine3183 during fruit ripening. However, low amounts of mRNA, estimated to be ∼1 to 2% of the wild-type level, were measured after 33 cycles of PCR amplification (data not shown). Expression of CRTISO in fruit of the mutant tangerinemic was similar to that in the wild type (data not shown).
Figure 6.
Expression of CRTISO during Tomato Fruit Development.
Steady state levels of mRNA of CRTISO, Psy, and Pds were measured by reverse transcriptase–mediated PCR from total RNA isolated from different stages of fruit development in wild-type tomato (cv M82) and mutant tangerine3183. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. G, mature green fruit; B, breaker stage; R, ripe stage 7 days after breaker; B×1/3 and B×3, samples that contained one-third and three times the total RNA, respectively, from breaker-stage fruit.
DISCUSSION
CRTISO Carotenoid Isomerase
Several lines of evidence designate CRTISO as a genuine carotenoid isomerase, which serves the indispensable function of carotenoid biosynthesis in oxygenic photosynthetic organisms. In tangerine tomato mutants, CRTISO cosegregates with the phenotype of prolycopene accumulation in fruit; a deletion mutation in CRTISO, which nullifies its function, was discovered in the allele tangerinemic that exhibits a typical tangerine phenotype, and a deletion of 348 bp in the promoter of CRTISO abolishes its expression in fruit of tangerine3183. The definite identification of CRTISO as an isomerase was confirmed by its functional expression in E. coli, in which it demonstrated activity of cis-to-trans isomerization of carotenes, especially prolycopene. The discovery of CRTISO solves a long-standing question that has puzzled investigators of carotenoid biosynthesis since the early 1940s.
In E. coli, the two desaturases from A. thaliana, PDS and ZDS, produce prolycopene from phytoene (Bartley et al., 1999). Our results confirm this finding in a combination of cyanobacterial PDS and ZDS from tomato. The data clearly indicate that poly-cis-carotenes occur as a default in the general pathway of phytoene desaturation in both chloroplasts and chromoplasts of plants and in cyanobacteria (our unpublished data), as was suggested by Bartley et al. (1999). CRTISO possibly is required as well for isomerization of cis-ζ-carotene and cis-neurosporene, because these species accumulate when the enzyme is deficient (Tables 1 and 3). Either CRTISO must rearrange the ζ-carotene molecule to enable ZDS activity, or ZDS catalysis, which is performed in conjunction with CRTISO, is required in the dark or in nonphotosynthetic tissues to enable the pathway to proceed to all-trans-lycopene and downstream to cyclic carotenoids. Support in favor of the latter hypothesis comes from in vitro experiments with the conversion of ζ-carotene to lycopene with purified ZDS from Capsicum annuum that was expressed in E. coli (Breitenbach et al., 1999). In this system, different lycopene isomers were formed according to the type of isomer that was used as a substrate. All-trans-ζ-carotene gave all-trans-lycopene, whereas cis-ζ-carotene yielded prolycopene via the corresponding cis-neurosporene isomer. This result indicated that although ZDS could use cis-isomers as substrates, it was unable to change their cis-trans molecular configuration, suggesting that the isomer of lycopene formed during desaturation depends strictly on the isomeric state of the substrate. We propose that CRTISO might operate in vivo together with ZDS and hypothesize that the two enzymes form a complex.
A dinucleotide binding motif in the N terminus of the CRTISO polypeptide is characteristic of all carotenoid desaturases identified to date and is present as well in lycopene cyclases. Its existence suggests that the carotene isomerase, possibly a flavoprotein, is engaged in a redox-related reaction in which a temporary abstraction of electrons takes place to allow the rotation of the molecule around the specific C-C bond, which is followed by reconfiguration of the double bond.
Requirement for cis-trans Isomerization for Lycopene Cyclization
The reason for the accumulation of prolycopene in the absence of CRTISO is unclear at present. The most plausible explanation is that prolycopene cannot be cyclized by lycopene cyclases because of steric hindrance. This hypothesis is supported by the correlation observed in flowers of tangerine between the accumulation of di-cis-lycopene and the presence of γ-carotene and rubixanthin (Table 1). We have observed a similar correlation in the Synechococcus PCC6803 mutant, which lacks the isomerase ortholog (data not shown). Formation of the one-ringed carotenoids could have occurred by cyclization at the side of the molecule that is not bent by the cis configuration.
It was determined that the bacterial-type lycopene cyclase, CRTY, converts all-trans-lycopene to β-carotene and does not accept tetra-cis-prolycopene as a substrate (Schnurr et al., 1996). Likewise, it was shown that the plant-type lycopene cyclases efficiently convert all-trans-lycopene to β-carotene or δ-carotene (Cunningham et al., 1993, 1996; Pecker et al., 1996; Ronen et al., 1999). It is inexplicable, therefore, that only tetra-cis-prolycopene was accepted as a substrate for cyclization in isolated membranes of Narcissus pseudonarcissus (Beyer et al., 1994). Because the cyclized carotenes were formed in this system in the all-trans configuration, it was assumed that isomerization occurred during the cyclization of lycopene independent of the cyclization reaction (Beyer et al., 1991). Our data clearly indicate that isomerization takes place independent of cyclization in both plant tissues and E. coli.
Carotenoid Isomerases in Other Species
CRTISO is conserved among photosynthetic organisms in which phytoene conversion to lycopene is performed by two enzymes, PDS and ZDS. In Arabidopsis, a gene designated Pdh encodes a polypeptide that is 75% identical to CRTISO from tomato (86% identical in the predicted mature polypeptide region). Evidence that the Arabidopsis CRTISO ortholog is involved in carotenoid biosynthesis is described by Park et al. (2002). The similarity between the CRTISO genes in tomato and Arabidopsis extends to their genomic structure: both contain 12 introns in the same position of the coding sequence. In the cyanobacterium Synechocystis PCC6803, the polypeptide encoded by sll0033 (http://www.kazusa.or.jp/cyano/) is 60% identical to the presumed mature CRTISO polypeptide. We have generated in this cyanobacterium a null mutation in the gene sll0033 by insertion mutagenesis (Δsll0033; data not shown). Cells of this mutant contain a significant proportion of prolycopene and other cis-carotenoids, similar to the phenotype observed in young or dark-grown green leaves of tangerinemic tomato (data not shown).
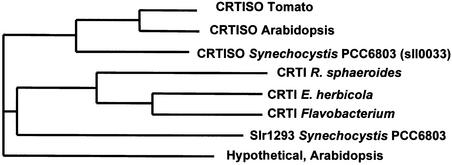
Interestingly, the CRTISO sequences are similar to those of crtI, the bacterial-type PDS. However, CRTISO itself does not have PDS activity (Table 3). A phylogenetic tree based on amino acid sequence similarity demonstrates this relationship (Figure 7). It is possible that CRTISO originated in evolution from crtI. It is important to note that the bacterial phytoene desaturase, CRTI, produces all-trans-lycopene from phytoene through four dehydrogenation reactions, which apparently are mechanistically different in the plant-type enzymes PDS and ZDS.
Figure 7.
Phylogenetic Tree of CRTISO and Related Genes.
CRTI, phytoene desaturase from bacteria. Slr1293 is a hypothetical gene from Synechocystis PCC6803. CRTI R. sphaeroides, CRTI Rhodobacter sphaeroides.
Photoisomerization in Green Tissues
We observed that light could substitute for the lack of isomerase activity in leaves of tangerinemic, in the outer green tissues in immature fruit of tangerine that are exposed to light, and in the Δsll0033 mutant of Synechocystis PCC6803 (data not shown) but not in nonphotosynthetic tissues of flowers (petals), ripe fruit, and the innermost parts of the green fruit of tangerine. A similar light-induced isomerization of prolycopene to all-trans-lycopene was observed in cells of the mutant C-6D of the unicellular green alga Scenedesmus obliquus (Romer et al., 1991; Sandmann, 1991), suggesting that CRTISO is impaired in this mutant.
The chemical mechanism of photoisomerization is unclear. We exclude the possibility that light interacts directly with the cis-isomers because we see no effect of light on prolycopene in chromoplasts of fruit and flowers and in E. coli cells. The differences in carotenoid production between light- and dark-grown E. coli (Table 3) are attributed to photoisomerization of 15-cis-ζ-carotene produced by PDS, which enables the subsequent desaturation by ZDS. This type of photoisomerization had been described previously in Euglena gracilis (Cunningham and Schiff, 1985).
Cis-to-trans isomerization was observed upon incubations of all-trans-isomers of β-carotene, lycopene, and canthaxanthin with HTMD (3-hydroxymethyl-3,4,4-trimethyl-1,2-dioxetane), a thermodissociable source of electronically excited ketones (Sundquist et al., 1993). The ability of chlorophyll compounds to sensitize the photoisomerization of all-trans-β-carotene has been demonstrated (Oneil and Schwartz, 1995). Light-driven isomerization of all-trans violaxanthin to the conformation of 9-cis and 13-cis was found in the light-harvesting pigment-protein complex of photosystem II (Phillip et al., 1999). Up to 9% 13-cis isomer was detected in leaves of barley after illumination, and up to 15% trans-to-cis conversion was achieved in the isolated light-harvesting pigment-protein complex of photosystem II of Secale cereale via a presumed triplet-sensitized photoisomerization reaction with blue light absorbed by chlorophylls and carotenoids as well as by red light absorbed exclusively by chlorophyll pigments (Grudzinski et al., 2001). In Dunaliella salina, the ratio of 9-cis- to all-trans-β-carotene is >2:1 under low light, and this is decreased to <0.45:1 upon exposure to high irradiance (Orset and Young, 2000). However, this isomerization probably occurs after cyclization of lycopene. We conclude that photoisomerization of ζ-carotene, neurosporene, and prolycopene, which occurs in chloroplasts, is associated with the existence of the photosynthetic apparatus. It is unclear at this time whether a component of this system can elicit isomerization using absorbed light energy or if this is an indirect effect of photosynthesis that relates to redox reactions necessary for isomerization.
In view of the possibility of substituting the activity of the carotenoid isomerase for light in green cells, it is hypothesized that the function of carotene isomerase in plants is to enable carotenoid biosynthesis in nonphotosynthetic tissues or in the dark. This is essential in germinating seedlings, as shown by Park et al. (2002) in roots and chromoplasts in the absence of chlorophyll sensitization.
METHODS
Plant Material and Growth Conditions
Tomato (Lycopersicon esculentum cv M82) and the introgression line IL 10-2 (Eshed and Zamir, 1995) served as the wild-type lines. The tangerine mutant LA3183 (tangerine3183), which was kindly provided by Roger Chetelat (Tomato Genetics Resource Center, University of California, Davis), was used to map the tangerine locus and to characterize the phenotype. The mutant tangerinemic was identified among M2 plants of a fast neutron–mutagenized population (David-Schwartz et al., 2001) in the background of the miniature cultivar Micro-Tom (Meissner et al., 1997) and was kindly donated by Dr. Avi Levy (Weizmann Institute, Rehovot, Israel). The two mutations were found to be allelic by genetic crossing (data not shown).
Recombinant plants in the F2 generation of a cross between tangerine3183 and IL10-2 were selfed, and the F3 progeny were screened for homozygous recombination products. Fixed recombinant plants were used to fine map the tangerine locus and served as isogenic lines for carotenoid analysis and measurement of gene expression. Lines 98-802 and 98-818 served as the wild type, and lines 98-823 and 104 served as tangerine3183 (see Results for details).
Seed of the different lines were sterilized by soaking in 70% ethanol for 2 min, in 3.3% NaOCl and 0.1% Tween 20 for 10 min, followed by three washes with sterile water. Seed were sowed on Murashige and Skoog (1962) basal salt mixture with 3% sucrose. The seedlings were grown at 23°C in dark or light for 2 weeks before leaves were analyzed. Plants were grown in the field for crossing and in the greenhouse for fruit analysis.
Carotenoid Analysis
Extraction of carotenoids from tomato fruit followed previously described protocols (Ronen et al., 1999, 2000). Leaf pigments were extracted from ∼70 mg of fresh cotyledons of dark- or light-grown seedlings. Fresh tissue was minced in acetone and filtered. The solvent was dried under a stream of nitrogen and dissolved in acetone. Flower pigments were extracted from petals of fresh single flowers (for cv Micro-Tom, two flowers were extracted for each sample). The tissues were ground in 2 mL of acetone, 2 mL of dichloromethane was added, and the samples were agitated until all pigments were extracted. Saponification of flower carotenoids was performed in 60% (w/v) ethanol/KOH (9:1) for 16 hr at 4°C. The carotenoids were extracted with ether after the addition of NaCl to a final concentration of 1.2%. The samples were dried and dissolved in acetone. Analysis by HPLC using a photodiode array detector has been described previously (Ronen et al., 1999, 2000). Carotenoids were identified by their characteristic absorption spectra, distinctive retention times, and, in some cases, comparison with standards. Quantification was performed by integrating the peak areas of the HPLC results using Millennium chromatography software (Waters, Milford, MA).
Map-Based Cloning
Genomic DNA was prepared from 5 g of leaf tissue as described (Eshed and Zamir, 1995). Restriction fragment length polymorphism in genomic DNA from tomato was performed with markers TG408, CT20, CD72, CT57, TG1, and TG241 (Tanksley et al., 1992). A genomic library in bacterial artificial chromosomes (BACs) of tomato (var Heinz1706; http://www.clemson.edu) was screened with the marker DNA CT57. Sequences at the ends of the insert in BAC 21O12 were amplified by polymerase chain reaction (PCR) using the primers BAC2FA (5′-TGTCATCACCCAATTTTCCA-3′; forward end of BAC2), BAC2FB (5′-TTCCAGGAACTTGGTTTCCTT-3′; forward end of BAC2), BAC2RA, (5′-TGAAAGGGCATACCAAAAGG-3′; reverse end of BAC2), and BAC2RB (5′-GGCTACGCCAAGAAC-TCTGA-3′; reverse end of BAC2). The amplified sequences were used as probes in hybridization with DNA from recombinant plants. DNA fragments of the BAC insert were subcloned in the plasmid vector pBS (Promega) and sequenced using the T3 and T7 primers. Assembly of sequences was accomplished with the Vector NTI Suit software package (InfoMax Inc., Bethesda, MD). cDNA clones were obtained by reverse transcription followed by PCR using total RNA isolated from flowers.
Functional Expression of CRTISO in Escherichia coli
Plasmid pAC-Zeta, which carries the genes crtB and crtE from Erwinia and crtP from Synechococcus PCC7942, has been described (Cunningham et al., 1994). Plasmid pGB-Ipi was constructed by inserting the cDNA of Ipi from Haematococcus pluvialis (Cunningham and Gantt, 2001) (kindly provided by F.X. Cunningham, University of Maryland, College Park) into the HindIII site of plasmid vector pGB2 (Churchward et al., 1984). Plasmid pCRTISO was constructed by subcloning a 1631-bp PCR-amplified fragment from the cDNA of the tomato (cv M82) CRTISO. The primers used for amplification were 5′-GTTCTAGATGTAGACAAAAGAGTGGA-3′ (forward) and 5′-ACA-TCTAGATATCATGCTAGTGTCCTT-3′ (reverse). Both primers contain a single mismatch to create an XbaI restriction site. The PCR fragment was cut with XbaI and subcloned into the XbaI site of vector pBluescript SK− (Stratagene). Plasmid pT-Zds was constructed by subcloning a 1643-bp PCR-amplified sequence from the tomato cDNA of Zds. This DNA fragment was obtained using the primers TZds248 (5′-GCTGATTTGGATATCTATGGTTTC-3′; forward) and TZds1901 (5′-AACTCGAGTTGTATTTGGATGATTTGCA-3′; reverse). Each primer contains a single mismatch to create an EcoRV (TZds248) and a XhoI (TZds1901) restriction site. The PCR fragment was cut with EcoRV and XhoI and subcloned into vector pBluescript SK−, which was cut with SmaI and XhoI. Plasmid pCRTISO-TZds was constructed by subcloning the CRTISO cDNA fragment, which was excised from pCRTISO with the restriction endonucleases Cfr42I and BcuI, into pT-Zds, which was cut with the same enzymes.
E. coli cells of the strain XL1-Blue carrying plasmid pGB-Ipi were cotransformed with plasmids pAC-Zeta, pT-Zds, pCRTISO, and pCRTISO-TZds in various combinations and selected on Luria-Bertani medium containing the appropriate antibiotics: spectinomycin (50 mg/L), ampicillin (100 mg/L), and chloramphenicol (50 mg/L). Cells were incubated either in the dark or under dim light of 10 to 30 μmol·m−2·sec−1 photon flux at room temperature for 48 hr.
Measurement of mRNA by Reverse Transcriptase–Mediated PCR
Protocols for RNA extraction and reverse transcription have been described previously (Ronen et al., 1999, 2000). Total RNA was isolated from 1 g of fruit tissue using the TRI reagent protocol (Molecular Research Center, Cincinnati, OH). After reverse transcription of total mRNA, the cDNAs of Psy, Pds, and CRTISO were amplified by PCR for 24, 26, and 28 cycles, respectively, of 1 min at 95°C, 1 min at 56°C, and 1 min at 72°C. Various initial concentrations of mRNA, ranging over a ninefold difference, were used to demonstrate the linear ratio between the concentration of template mRNA and the final PCR products. The following primers were used for PCR amplification: for Pds, 5′-TTGTGTTTGCCGCTCCAGTGGATAT-3′ (forward) and 5′-GCGCCTTCCATTGAAGCCAAGTAT-3′ (reverse); for Psy, 5′-GGGGAATTTGGGCTTGTTGAGT-3′ (forward) and 5′-CCTTTGATT-CAGGGGCGATACC-3′ (reverse); for CRTISO, 5′-GATCGCCAAATCCTTAGCAA-3′ (forward) and 5′-GCCCTGGGAAGAGTGTTTTT-3′ (reverse). The products of PCR amplification were separated by electrophoresis on 1.5% agarose gels and stained with ethidium bromide.
DNA and Protein Sequence Analysis
The DNA sequence was determined with the ABI Prism 377 DNA Sequencer (Perkin Elmer) and processed with ABI sequence analysis software. Vector NTI Suit software was used for sequence analysis.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are AF416727 (CRTISO), AC011001 (Arabidopsis gene Pdh), and AF195507 (tomato gene Zds).
Acknowledgments
We thank Dr. Avi Levy from the Weizmann Institute for providing the mutant tangerinemic, David Kachanovsky for sharing unpublished data on the inactivation of sll0033 in Synechocystis PCC6803 and for technical assistance, and Dr. V. Mann for valuable comments on the manuscript. This work was supported in part by the European Commission, contract ProVitA (QLK3-CT2000-0809), and by the Israel Ministry of Science. Work in our laboratory is performed under the auspices of the Avron Even-Ari Minerva Center.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010303.
References
- Bartley, G.E., Scolnik, P.A., and Beyer, P. (1999). Two Arabidopsis thaliana carotene desaturases, phytoene desaturase and ζ-carotene desaturase, expressed in Escherichia coli, catalyze a poly-cis pathway to yield pro-lycopene. Eur. J. Biochem. 259, 396–403. [DOI] [PubMed] [Google Scholar]
- Beyer, P., Croncke, U., and Nievelstein, V. (1991). On the mechanism of lycopene isomerase cyclase reaction in Narcissus pseudonarcissus L. chromoplasts. J. Biol. Chem. 266, 17072–17078. [PubMed] [Google Scholar]
- Beyer, P., Nievelstein, V., Albabili, S., Bonk, M., and Kleinig, H. (1994). Biochemical aspects of carotene desaturation and cyclization in chromoplast membranes from Narcissus pseudonarcissus. Pure Appl. Chem. 66, 1047–1056. [Google Scholar]
- Breitenbach, J., Kuntz, M., Takaichi, S., and Sandmann, G. (1999). Catalytic properties of an expressed and purified higher plant type ζ-carotene desaturase from Capsicum annuum. Eur. J. Biochem. 265, 376–383. [DOI] [PubMed] [Google Scholar]
- Britton, G. (1988). Biosynthesis of carotenoids. In Plant Pigments, T.W. Goodwin, ed (London and New York: Academic Press), pp. 133–180.
- Britton, G., Liaaen-Jensen, S., and Pfander, H. (1998). Carotenoids, Vol. 3: Biosynthesis and Metabolism. (Basel, Switzerland: Birkauser Verlag).
- Budiman, M.A., Mao, L., Wood, T.C., and Wing, R.A. (2000). A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res. 10, 129–136. [PMC free article] [PubMed] [Google Scholar]
- Churchward, G., Belin, D., and Nagamine, Y. (1984). A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31, 165–171. [DOI] [PubMed] [Google Scholar]
- Clough, J.M., and Pattenden, G. (1979). Naturally occurring poly-cis carotenoids: Stereochemistry of poly-cis lycopene and in congeners in ‘tangerine’ tomato fruits. J. Chem. Soc. Chem. Commun. 14, 616–619. [Google Scholar]
- Cunningham, F.X., Jr., and Gantt, E. (2001). One ring or two? Determination of ring number in carotenoids by lycopene ɛ-cyclases. Proc. Natl. Acad. Sci. USA 98, 2905–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, F.X., Jr., and Schiff, J.A. (1985). Photoisomerization of ζ-carotene stereoisomers in cells of Euglena gracilis mutant W3BUL and in solution. Photochem. Photobiol. 42, 295–307. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.X., Jr., Chamovitz, D., Misawa, N., Gantt, E., and Hirschberg, J. (1993). Cloning and functional expression in Escherichia coli of a cyanobacterial gene for lycopene cyclase, the enzyme that catalyzes the biosynthesis of β-carotene. FEBS Lett. 328, 130–138. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.X., Jr., Sun, Z.R., Chamovitz, D., Hirschberg, J., and Gantt, E. (1994). Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell 6, 1107–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, F.X., Jr., Pogson, B., Sun, Z.R., Mcdonald, K.A., DellaPenna, D., and Gantt, E. (1996). Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8, 1613–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Schwartz, R., Badani, H., Wininger, S., Levy, A.A., Galili, G., and Kapulnik, Y. (2001). Identification of a novel genetically controlled step in mycorrhizal colonization: Plant resistance to infection by fungal spores but not to extraradical hyphae. Plant J. 27, 561–569. [DOI] [PubMed] [Google Scholar]
- Ernst, S., and Sandmann, G. (1988). Poly-cis carotene pathway in the Scenedesmus mutant C-6D. Arch. Microbiol. 150, 590–594. [Google Scholar]
- Eshed, Y., and Zamir, D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, T.W. (1980). The Biochemistry of the Carotenoids, Vol. 1. (London: Chapman and Hall).
- Grudzinski, W., Matula, M., Sielewiesiuk, J., Kernen, P., Krupa, Z., and Gruszecki, W.I. (2001). Effect of 13-cis violaxanthin on organization of light harvesting complex II in monomolecular layers. Biochim. Biophys. Acta 1503, 291–302. [DOI] [PubMed] [Google Scholar]
- Hirschberg, J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4, 210–218. [DOI] [PubMed] [Google Scholar]
- Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., Elkind, Y., and Levy, A.A. (1997). A new model system for tomato genetics. Plant J. 12, 1465–1472. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revise medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Oneil, C.A., and Schwartz, S.J. (1995). Photoisomerization of β-carotene by photosensitization with chlorophyll derivatives as sensitizers. J. Agric. Food Chem. 43, 631–635. [Google Scholar]
- Orset, S.C., and Young, A.J. (2000). Exposure to low irradiances favors the synthesis of 9-cis β,β-carotene in Dunaliella salina (Teod.). Plant Physiol. 122, 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., Kreunen, S.S., Cuttriss, A.J., DellaPenna, D., and Pogson, B.J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecker, I., Gabbay, R., Cunningham, F.X., Jr., and Hirschberg, J. (1996). Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol. Biol. 30, 807–819. [DOI] [PubMed] [Google Scholar]
- Phillip, D., Molnar, P., Toth, G., and Young, A.J. (1999). Light-induced formation of 13-cis violaxanthin in leaves of Hordeum vulgare. J. Photochem. Photobiol. B Biol. 49, 89–95. [Google Scholar]
- Romer, S., Humbeck, K., and Senger, H. (1991). Dark reactions following photoisomerization of prolycopene. Photochem. Photobiol. 53, 535–538. [Google Scholar]
- Ronen, G., Cohen, M., Zamir, D., and Hirschberg, J. (1999). Regulation of carotenoid biosynthesis during tomato fruit development: Expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant delta. Plant J. 17, 341–351. [DOI] [PubMed] [Google Scholar]
- Ronen, G., Carmel-Goren, L., Zamir, D., and Hirschberg, J. (2000). An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. USA 97, 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann, G. (1991). Light-dependent switch from formation of poly-cis carotenes to all-trans carotenoids in the Scenedesmus mutant C-6D. Arch. Microbiol. 155, 229–233. [Google Scholar]
- Schnurr, G., Misawa, N., and Sandmann, G. (1996). Expression, purification and properties of lycopene cyclase from Erwinia uredovora. Biochem. J. 315, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist, A.R., Hanusch, M., Stahl, W., and Sies, H. (1993). Cis/trans isomerization of carotenoids by the triplet carbonyl source 3-hydroxymethyl-3,4,4-trimethyl-1,2-dioxetane. Photochem. Photobiol. 57, 785–791. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomes, M.L. (1952). Flower color modification associated with the gene t. Rep. Tomato Genet. Coop. 2, 12. [Google Scholar]
- Tomes, M.L., Quackenbush, F.W., Nelson, O.E., and North, B. (1953). The inheritance of carotenoid pigment system in the tomato. Genetics 38, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]